Project
Illumina Body Map 2, young vs old
Navigation
Downloads
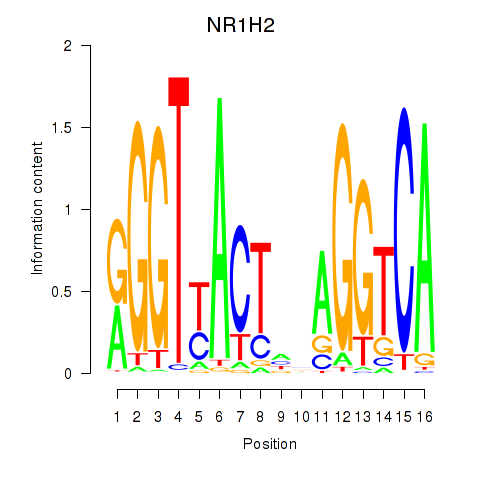
Results for NR1H2
Z-value: 0.07
Transcription factors associated with NR1H2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NR1H2
|
ENSG00000131408.9 | nuclear receptor subfamily 1 group H member 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NR1H2 | hg19_v2_chr19_+_50832895_50832928 | -0.36 | 4.2e-02 | Click! |
Activity profile of NR1H2 motif
Sorted Z-values of NR1H2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_+_77774897 | 6.40 |
ENST00000281030.2
|
THRSP
|
thyroid hormone responsive |
| chr7_+_80253387 | 2.74 |
ENST00000438020.1
|
CD36
|
CD36 molecule (thrombospondin receptor) |
| chr9_-_107690420 | 2.57 |
ENST00000423487.2
ENST00000374733.1 ENST00000374736.3 |
ABCA1
|
ATP-binding cassette, sub-family A (ABC1), member 1 |
| chr12_-_47226152 | 2.30 |
ENST00000546940.1
|
SLC38A4
|
solute carrier family 38, member 4 |
| chr16_+_56995762 | 2.28 |
ENST00000200676.3
ENST00000379780.2 |
CETP
|
cholesteryl ester transfer protein, plasma |
| chr11_-_118083600 | 2.27 |
ENST00000524477.1
|
AMICA1
|
adhesion molecule, interacts with CXADR antigen 1 |
| chr2_-_216946500 | 2.25 |
ENST00000265322.7
|
PECR
|
peroxisomal trans-2-enoyl-CoA reductase |
| chr20_+_5987890 | 2.22 |
ENST00000378868.4
|
CRLS1
|
cardiolipin synthase 1 |
| chr1_-_237167718 | 2.20 |
ENST00000464121.2
|
MT1HL1
|
metallothionein 1H-like 1 |
| chr12_+_32115400 | 2.18 |
ENST00000381054.3
|
KIAA1551
|
KIAA1551 |
| chr2_+_234104079 | 2.15 |
ENST00000417661.1
|
INPP5D
|
inositol polyphosphate-5-phosphatase, 145kDa |
| chr19_-_2042065 | 2.07 |
ENST00000591588.1
ENST00000591142.1 |
MKNK2
|
MAP kinase interacting serine/threonine kinase 2 |
| chr11_+_10326612 | 2.03 |
ENST00000534464.1
ENST00000530439.1 ENST00000524948.1 ENST00000528655.1 ENST00000526492.1 ENST00000525063.1 |
ADM
|
adrenomedullin |
| chr4_-_71532601 | 2.01 |
ENST00000510614.1
|
IGJ
|
immunoglobulin J polypeptide, linker protein for immunoglobulin alpha and mu polypeptides |
| chr4_-_71532668 | 1.97 |
ENST00000510437.1
|
IGJ
|
immunoglobulin J polypeptide, linker protein for immunoglobulin alpha and mu polypeptides |
| chr16_-_68034452 | 1.91 |
ENST00000575510.1
|
DPEP2
|
dipeptidase 2 |
| chr15_-_90358564 | 1.88 |
ENST00000559874.1
|
ANPEP
|
alanyl (membrane) aminopeptidase |
| chr22_-_24096562 | 1.88 |
ENST00000398465.3
|
VPREB3
|
pre-B lymphocyte 3 |
| chr10_-_70287172 | 1.81 |
ENST00000539557.1
|
SLC25A16
|
solute carrier family 25 (mitochondrial carrier; Graves disease autoantigen), member 16 |
| chr12_+_56511943 | 1.79 |
ENST00000257940.2
ENST00000552345.1 ENST00000551880.1 ENST00000546903.1 ENST00000551790.1 |
ZC3H10
ESYT1
|
zinc finger CCCH-type containing 10 extended synaptotagmin-like protein 1 |
| chr16_+_72042487 | 1.74 |
ENST00000572887.1
ENST00000219240.4 ENST00000574309.1 ENST00000576145.1 |
DHODH
|
dihydroorotate dehydrogenase (quinone) |
| chr10_-_72648541 | 1.73 |
ENST00000299299.3
|
PCBD1
|
pterin-4 alpha-carbinolamine dehydratase/dimerization cofactor of hepatocyte nuclear factor 1 alpha |
| chr14_+_105046094 | 1.70 |
ENST00000331952.2
|
C14orf180
|
chromosome 14 open reading frame 180 |
| chr10_-_70287231 | 1.70 |
ENST00000609923.1
|
SLC25A16
|
solute carrier family 25 (mitochondrial carrier; Graves disease autoantigen), member 16 |
| chr16_+_56995854 | 1.64 |
ENST00000566128.1
|
CETP
|
cholesteryl ester transfer protein, plasma |
| chr17_+_7184986 | 1.62 |
ENST00000317370.8
ENST00000571308.1 |
SLC2A4
|
solute carrier family 2 (facilitated glucose transporter), member 4 |
| chr8_-_48651648 | 1.59 |
ENST00000408965.3
|
CEBPD
|
CCAAT/enhancer binding protein (C/EBP), delta |
| chr5_+_35856951 | 1.55 |
ENST00000303115.3
ENST00000343305.4 ENST00000506850.1 ENST00000511982.1 |
IL7R
|
interleukin 7 receptor |
| chr7_+_16700806 | 1.55 |
ENST00000446596.1
ENST00000438834.1 |
BZW2
|
basic leucine zipper and W2 domains 2 |
| chr17_+_32683456 | 1.52 |
ENST00000225844.2
|
CCL13
|
chemokine (C-C motif) ligand 13 |
| chr17_-_26694979 | 1.52 |
ENST00000438614.1
|
VTN
|
vitronectin |
| chr17_+_77018896 | 1.52 |
ENST00000578229.1
|
C1QTNF1
|
C1q and tumor necrosis factor related protein 1 |
| chr21_+_43619796 | 1.51 |
ENST00000398457.2
|
ABCG1
|
ATP-binding cassette, sub-family G (WHITE), member 1 |
| chr17_-_26695013 | 1.50 |
ENST00000555059.2
|
CTB-96E2.2
|
Homeobox protein SEBOX |
| chr11_+_32851487 | 1.48 |
ENST00000257836.3
|
PRRG4
|
proline rich Gla (G-carboxyglutamic acid) 4 (transmembrane) |
| chr19_+_41092680 | 1.48 |
ENST00000594298.1
ENST00000597396.1 |
SHKBP1
|
SH3KBP1 binding protein 1 |
| chr16_-_68034470 | 1.47 |
ENST00000412757.2
|
DPEP2
|
dipeptidase 2 |
| chr12_-_7125770 | 1.45 |
ENST00000261407.4
|
LPCAT3
|
lysophosphatidylcholine acyltransferase 3 |
| chr22_-_24096630 | 1.39 |
ENST00000248948.3
|
VPREB3
|
pre-B lymphocyte 3 |
| chr20_+_43343886 | 1.37 |
ENST00000190983.4
|
WISP2
|
WNT1 inducible signaling pathway protein 2 |
| chr5_+_96424779 | 1.36 |
ENST00000509481.1
|
CTD-2215E18.1
|
Uncharacterized protein |
| chr17_+_8213539 | 1.35 |
ENST00000583529.1
|
ARHGEF15
|
Rho guanine nucleotide exchange factor (GEF) 15 |
| chr6_+_30687978 | 1.34 |
ENST00000327892.8
ENST00000435534.1 |
TUBB
|
tubulin, beta class I |
| chr16_+_21623392 | 1.32 |
ENST00000562961.1
|
METTL9
|
methyltransferase like 9 |
| chr4_+_108910870 | 1.31 |
ENST00000403312.1
ENST00000603302.1 ENST00000309522.3 |
HADH
|
hydroxyacyl-CoA dehydrogenase |
| chr16_-_11723066 | 1.31 |
ENST00000576036.1
|
LITAF
|
lipopolysaccharide-induced TNF factor |
| chr14_-_102976135 | 1.28 |
ENST00000560748.1
|
ANKRD9
|
ankyrin repeat domain 9 |
| chr4_-_84205905 | 1.28 |
ENST00000311461.7
ENST00000311469.4 ENST00000439031.2 |
COQ2
|
coenzyme Q2 4-hydroxybenzoate polyprenyltransferase |
| chr20_+_43343517 | 1.28 |
ENST00000372865.4
|
WISP2
|
WNT1 inducible signaling pathway protein 2 |
| chr2_+_238877424 | 1.27 |
ENST00000434655.1
|
UBE2F
|
ubiquitin-conjugating enzyme E2F (putative) |
| chr19_-_49658387 | 1.26 |
ENST00000595625.1
|
HRC
|
histidine rich calcium binding protein |
| chr5_-_133968529 | 1.26 |
ENST00000402673.2
|
SAR1B
|
SAR1 homolog B (S. cerevisiae) |
| chr19_-_38806540 | 1.26 |
ENST00000592694.1
|
YIF1B
|
Yip1 interacting factor homolog B (S. cerevisiae) |
| chr17_-_3595042 | 1.21 |
ENST00000552723.1
|
P2RX5
|
purinergic receptor P2X, ligand-gated ion channel, 5 |
| chr1_+_87595433 | 1.20 |
ENST00000469312.2
ENST00000490006.2 |
RP5-1052I5.1
|
long intergenic non-protein coding RNA 1140 |
| chr1_-_153950098 | 1.20 |
ENST00000356648.1
|
JTB
|
jumping translocation breakpoint |
| chr2_+_27255806 | 1.19 |
ENST00000238788.9
ENST00000404032.3 |
TMEM214
|
transmembrane protein 214 |
| chr3_+_39371255 | 1.19 |
ENST00000414803.1
ENST00000545843.1 |
CCR8
|
chemokine (C-C motif) receptor 8 |
| chr18_+_21693306 | 1.18 |
ENST00000540918.2
|
TTC39C
|
tetratricopeptide repeat domain 39C |
| chr11_+_86748957 | 1.18 |
ENST00000526733.1
ENST00000532959.1 |
TMEM135
|
transmembrane protein 135 |
| chr10_-_76818272 | 1.17 |
ENST00000338487.5
|
DUPD1
|
dual specificity phosphatase and pro isomerase domain containing 1 |
| chr4_-_71705027 | 1.17 |
ENST00000545193.1
|
GRSF1
|
G-rich RNA sequence binding factor 1 |
| chr4_-_71705060 | 1.16 |
ENST00000514161.1
|
GRSF1
|
G-rich RNA sequence binding factor 1 |
| chr7_+_45067265 | 1.15 |
ENST00000474617.1
|
CCM2
|
cerebral cavernous malformation 2 |
| chr20_-_46041046 | 1.14 |
ENST00000437920.1
|
RP1-148H17.1
|
RP1-148H17.1 |
| chr22_-_33968239 | 1.14 |
ENST00000452586.2
ENST00000421768.1 |
LARGE
|
like-glycosyltransferase |
| chr22_+_23247030 | 1.14 |
ENST00000390324.2
|
IGLJ3
|
immunoglobulin lambda joining 3 |
| chr11_+_844406 | 1.13 |
ENST00000397404.1
|
TSPAN4
|
tetraspanin 4 |
| chr3_-_127309550 | 1.13 |
ENST00000296210.7
ENST00000355552.3 |
TPRA1
|
transmembrane protein, adipocyte asscociated 1 |
| chr12_-_113841678 | 1.12 |
ENST00000552280.1
ENST00000257549.4 |
SDS
|
serine dehydratase |
| chr15_-_89010607 | 1.11 |
ENST00000312475.4
|
MRPL46
|
mitochondrial ribosomal protein L46 |
| chr2_+_168043793 | 1.11 |
ENST00000409273.1
ENST00000409605.1 |
XIRP2
|
xin actin-binding repeat containing 2 |
| chr17_+_27047570 | 1.10 |
ENST00000472628.1
ENST00000578181.1 |
RPL23A
|
ribosomal protein L23a |
| chr19_+_52264104 | 1.09 |
ENST00000340023.6
|
FPR2
|
formyl peptide receptor 2 |
| chr15_+_81589254 | 1.09 |
ENST00000394652.2
|
IL16
|
interleukin 16 |
| chr11_+_827248 | 1.09 |
ENST00000527089.1
ENST00000530183.1 |
EFCAB4A
|
EF-hand calcium binding domain 4A |
| chr19_-_38806390 | 1.07 |
ENST00000589247.1
ENST00000329420.8 ENST00000591784.1 |
YIF1B
|
Yip1 interacting factor homolog B (S. cerevisiae) |
| chr19_+_15752088 | 1.07 |
ENST00000585846.1
|
CYP4F3
|
cytochrome P450, family 4, subfamily F, polypeptide 3 |
| chr17_-_17480779 | 1.07 |
ENST00000395782.1
|
PEMT
|
phosphatidylethanolamine N-methyltransferase |
| chr11_-_82611448 | 1.06 |
ENST00000393399.2
ENST00000313010.3 |
PRCP
|
prolylcarboxypeptidase (angiotensinase C) |
| chr5_+_76145826 | 1.06 |
ENST00000513010.1
|
S100Z
|
S100 calcium binding protein Z |
| chr17_+_77019030 | 1.05 |
ENST00000580454.1
|
C1QTNF1
|
C1q and tumor necrosis factor related protein 1 |
| chr16_+_30484021 | 1.03 |
ENST00000358164.5
|
ITGAL
|
integrin, alpha L (antigen CD11A (p180), lymphocyte function-associated antigen 1; alpha polypeptide) |
| chr2_-_74699770 | 1.03 |
ENST00000409710.1
|
MRPL53
|
mitochondrial ribosomal protein L53 |
| chr1_-_153949751 | 1.02 |
ENST00000428469.1
|
JTB
|
jumping translocation breakpoint |
| chr19_-_38806560 | 1.01 |
ENST00000591755.1
ENST00000337679.8 ENST00000339413.6 |
YIF1B
|
Yip1 interacting factor homolog B (S. cerevisiae) |
| chr1_-_161207986 | 1.00 |
ENST00000506209.1
ENST00000367980.2 |
NR1I3
|
nuclear receptor subfamily 1, group I, member 3 |
| chr14_-_73925225 | 0.99 |
ENST00000356296.4
ENST00000355058.3 ENST00000359560.3 ENST00000557597.1 ENST00000554394.1 ENST00000555238.1 ENST00000535282.1 ENST00000555987.1 ENST00000555394.1 ENST00000554546.1 |
NUMB
|
numb homolog (Drosophila) |
| chr11_+_86749035 | 0.99 |
ENST00000305494.5
ENST00000535167.1 |
TMEM135
|
transmembrane protein 135 |
| chr11_-_118789613 | 0.99 |
ENST00000532899.1
|
BCL9L
|
B-cell CLL/lymphoma 9-like |
| chr12_-_6677422 | 0.98 |
ENST00000382421.3
ENST00000545200.1 ENST00000399466.2 ENST00000536124.1 ENST00000540228.1 ENST00000542867.1 ENST00000545492.1 ENST00000322166.5 ENST00000545915.1 |
NOP2
|
NOP2 nucleolar protein |
| chr1_-_161207953 | 0.96 |
ENST00000367982.4
|
NR1I3
|
nuclear receptor subfamily 1, group I, member 3 |
| chr12_-_56694142 | 0.96 |
ENST00000550655.1
ENST00000548567.1 ENST00000551430.2 ENST00000351328.3 |
CS
|
citrate synthase |
| chr1_-_9714644 | 0.95 |
ENST00000377320.3
|
C1orf200
|
chromosome 1 open reading frame 200 |
| chr5_-_180671172 | 0.95 |
ENST00000512805.1
|
GNB2L1
|
guanine nucleotide binding protein (G protein), beta polypeptide 2-like 1 |
| chr2_-_219146839 | 0.94 |
ENST00000425694.1
|
TMBIM1
|
transmembrane BAX inhibitor motif containing 1 |
| chr16_+_53133070 | 0.92 |
ENST00000565832.1
|
CHD9
|
chromodomain helicase DNA binding protein 9 |
| chrX_-_2847366 | 0.92 |
ENST00000381154.1
|
ARSD
|
arylsulfatase D |
| chr14_-_74462922 | 0.92 |
ENST00000553284.1
|
ENTPD5
|
ectonucleoside triphosphate diphosphohydrolase 5 |
| chr15_+_89010923 | 0.90 |
ENST00000353598.6
|
MRPS11
|
mitochondrial ribosomal protein S11 |
| chr13_+_49684445 | 0.89 |
ENST00000398316.3
|
FNDC3A
|
fibronectin type III domain containing 3A |
| chr5_+_179135246 | 0.89 |
ENST00000508787.1
|
CANX
|
calnexin |
| chr17_+_4675175 | 0.88 |
ENST00000270560.3
|
TM4SF5
|
transmembrane 4 L six family member 5 |
| chr14_-_94970559 | 0.87 |
ENST00000556881.1
|
SERPINA12
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 12 |
| chr12_-_9913489 | 0.86 |
ENST00000228434.3
ENST00000536709.1 |
CD69
|
CD69 molecule |
| chr4_-_140216948 | 0.85 |
ENST00000265500.4
|
NDUFC1
|
NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 1, 6kDa |
| chr20_+_3052264 | 0.85 |
ENST00000217386.2
|
OXT
|
oxytocin/neurophysin I prepropeptide |
| chr11_+_119039414 | 0.84 |
ENST00000409991.1
ENST00000292199.2 ENST00000409265.4 ENST00000409109.1 |
NLRX1
|
NLR family member X1 |
| chr10_+_101491968 | 0.84 |
ENST00000370476.5
ENST00000370472.4 |
CUTC
|
cutC copper transporter |
| chr9_-_130829588 | 0.83 |
ENST00000373078.4
|
NAIF1
|
nuclear apoptosis inducing factor 1 |
| chr3_-_127309879 | 0.83 |
ENST00000489960.1
ENST00000490290.1 |
TPRA1
|
transmembrane protein, adipocyte asscociated 1 |
| chr2_-_165697920 | 0.82 |
ENST00000342193.4
ENST00000375458.2 |
COBLL1
|
cordon-bleu WH2 repeat protein-like 1 |
| chr22_+_36113919 | 0.81 |
ENST00000249044.2
|
APOL5
|
apolipoprotein L, 5 |
| chr17_-_56350797 | 0.81 |
ENST00000577220.1
|
MPO
|
myeloperoxidase |
| chr19_+_35899569 | 0.80 |
ENST00000600405.1
|
AC002511.1
|
AC002511.1 |
| chr19_+_15751689 | 0.80 |
ENST00000586182.2
ENST00000591058.1 ENST00000221307.8 |
CYP4F3
|
cytochrome P450, family 4, subfamily F, polypeptide 3 |
| chr17_-_8151353 | 0.80 |
ENST00000315684.8
|
CTC1
|
CTS telomere maintenance complex component 1 |
| chr6_+_37475109 | 0.80 |
ENST00000570443.2
|
RP1-153P14.8
|
RP1-153P14.8 |
| chr12_+_120972606 | 0.79 |
ENST00000413266.2
|
RNF10
|
ring finger protein 10 |
| chr8_-_144700212 | 0.79 |
ENST00000526290.1
|
TSTA3
|
tissue specific transplantation antigen P35B |
| chr4_-_2965052 | 0.78 |
ENST00000398071.4
ENST00000502735.1 ENST00000314262.6 ENST00000416614.2 |
NOP14
|
NOP14 nucleolar protein |
| chr11_+_86748998 | 0.77 |
ENST00000525018.1
ENST00000355734.4 |
TMEM135
|
transmembrane protein 135 |
| chr9_+_95736758 | 0.75 |
ENST00000337352.6
|
FGD3
|
FYVE, RhoGEF and PH domain containing 3 |
| chr21_-_35281399 | 0.74 |
ENST00000418933.1
|
ATP5O
|
ATP synthase, H+ transporting, mitochondrial F1 complex, O subunit |
| chr16_+_577697 | 0.74 |
ENST00000562370.1
ENST00000568988.1 ENST00000219611.2 |
CAPN15
|
calpain 15 |
| chrX_+_48542168 | 0.73 |
ENST00000376701.4
|
WAS
|
Wiskott-Aldrich syndrome |
| chr21_-_46340807 | 0.73 |
ENST00000397846.3
ENST00000524251.1 ENST00000522688.1 |
ITGB2
|
integrin, beta 2 (complement component 3 receptor 3 and 4 subunit) |
| chr6_-_34639733 | 0.72 |
ENST00000374021.1
|
C6orf106
|
chromosome 6 open reading frame 106 |
| chr20_+_19738792 | 0.72 |
ENST00000412571.1
|
RP1-122P22.2
|
RP1-122P22.2 |
| chr19_-_49121054 | 0.71 |
ENST00000546623.1
ENST00000084795.5 |
RPL18
|
ribosomal protein L18 |
| chr8_+_38065104 | 0.70 |
ENST00000521311.1
|
BAG4
|
BCL2-associated athanogene 4 |
| chr19_+_1026298 | 0.70 |
ENST00000263097.4
|
CNN2
|
calponin 2 |
| chr1_+_22962948 | 0.70 |
ENST00000374642.3
|
C1QA
|
complement component 1, q subcomponent, A chain |
| chr16_-_19533404 | 0.69 |
ENST00000353258.3
|
GDE1
|
glycerophosphodiester phosphodiesterase 1 |
| chr5_+_76145920 | 0.69 |
ENST00000317593.4
|
S100Z
|
S100 calcium binding protein Z |
| chr16_+_4674814 | 0.69 |
ENST00000415496.1
ENST00000587747.1 ENST00000399577.5 ENST00000588994.1 ENST00000586183.1 |
MGRN1
|
mahogunin ring finger 1, E3 ubiquitin protein ligase |
| chr3_+_39371191 | 0.69 |
ENST00000326306.4
|
CCR8
|
chemokine (C-C motif) receptor 8 |
| chr14_-_73924954 | 0.68 |
ENST00000555307.1
ENST00000554818.1 |
NUMB
|
numb homolog (Drosophila) |
| chr2_-_3521518 | 0.67 |
ENST00000382093.5
|
ADI1
|
acireductone dioxygenase 1 |
| chr11_-_62420757 | 0.67 |
ENST00000330574.2
|
INTS5
|
integrator complex subunit 5 |
| chr20_-_48747662 | 0.67 |
ENST00000371656.2
|
TMEM189
|
transmembrane protein 189 |
| chr17_-_5026397 | 0.67 |
ENST00000250076.3
|
ZNF232
|
zinc finger protein 232 |
| chr12_+_52281744 | 0.66 |
ENST00000301190.6
ENST00000538991.1 |
ANKRD33
|
ankyrin repeat domain 33 |
| chr16_+_29467780 | 0.66 |
ENST00000395400.3
|
SULT1A4
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 4 |
| chr17_+_76183398 | 0.65 |
ENST00000409257.5
|
AFMID
|
arylformamidase |
| chr19_-_6670128 | 0.65 |
ENST00000245912.3
|
TNFSF14
|
tumor necrosis factor (ligand) superfamily, member 14 |
| chr14_-_106114739 | 0.65 |
ENST00000460164.1
|
RP11-731F5.2
|
RP11-731F5.2 |
| chr8_+_132916318 | 0.65 |
ENST00000254624.5
ENST00000522709.1 |
EFR3A
|
EFR3 homolog A (S. cerevisiae) |
| chr8_-_13134246 | 0.64 |
ENST00000503161.2
|
DLC1
|
deleted in liver cancer 1 |
| chr17_-_79604075 | 0.64 |
ENST00000374747.5
ENST00000539314.1 ENST00000331134.6 |
NPLOC4
|
nuclear protein localization 4 homolog (S. cerevisiae) |
| chr2_+_207630081 | 0.63 |
ENST00000236980.6
ENST00000418289.1 ENST00000402774.3 ENST00000403094.3 |
FASTKD2
|
FAST kinase domains 2 |
| chr19_+_16254488 | 0.63 |
ENST00000588246.1
ENST00000593031.1 |
HSH2D
|
hematopoietic SH2 domain containing |
| chr11_+_119039069 | 0.63 |
ENST00000422249.1
|
NLRX1
|
NLR family member X1 |
| chr11_+_119038897 | 0.63 |
ENST00000454811.1
ENST00000449394.1 |
NLRX1
|
NLR family member X1 |
| chr15_+_96876340 | 0.62 |
ENST00000453270.2
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr12_-_56123444 | 0.62 |
ENST00000546457.1
ENST00000549117.1 |
CD63
|
CD63 molecule |
| chr11_+_65190245 | 0.61 |
ENST00000499732.1
ENST00000501122.2 ENST00000601801.1 |
NEAT1
|
nuclear paraspeckle assembly transcript 1 (non-protein coding) |
| chr1_+_10290822 | 0.61 |
ENST00000377083.1
|
KIF1B
|
kinesin family member 1B |
| chr16_+_56969284 | 0.61 |
ENST00000568358.1
|
HERPUD1
|
homocysteine-inducible, endoplasmic reticulum stress-inducible, ubiquitin-like domain member 1 |
| chr1_+_36335351 | 0.61 |
ENST00000373206.1
|
AGO1
|
argonaute RISC catalytic component 1 |
| chr5_-_138862326 | 0.60 |
ENST00000330794.4
|
TMEM173
|
transmembrane protein 173 |
| chr19_+_50094866 | 0.60 |
ENST00000418929.2
|
PRR12
|
proline rich 12 |
| chr19_-_50420918 | 0.59 |
ENST00000595761.1
|
NUP62
|
nucleoporin 62kDa |
| chr11_+_86748863 | 0.58 |
ENST00000340353.7
|
TMEM135
|
transmembrane protein 135 |
| chr19_+_17622415 | 0.58 |
ENST00000252603.2
ENST00000600923.1 |
PGLS
|
6-phosphogluconolactonase |
| chr2_+_223725723 | 0.57 |
ENST00000535678.1
|
ACSL3
|
acyl-CoA synthetase long-chain family member 3 |
| chr1_+_161035655 | 0.56 |
ENST00000600454.1
|
AL591806.1
|
Uncharacterized protein |
| chr1_-_161208013 | 0.56 |
ENST00000515452.1
ENST00000367983.4 |
NR1I3
|
nuclear receptor subfamily 1, group I, member 3 |
| chr21_-_46340770 | 0.56 |
ENST00000397854.3
|
ITGB2
|
integrin, beta 2 (complement component 3 receptor 3 and 4 subunit) |
| chr14_-_102976091 | 0.56 |
ENST00000286918.4
|
ANKRD9
|
ankyrin repeat domain 9 |
| chr11_+_62652649 | 0.55 |
ENST00000539507.1
|
SLC3A2
|
solute carrier family 3 (amino acid transporter heavy chain), member 2 |
| chrX_+_1455478 | 0.55 |
ENST00000331035.4
|
IL3RA
|
interleukin 3 receptor, alpha (low affinity) |
| chr17_-_76183111 | 0.54 |
ENST00000405273.1
ENST00000590862.1 ENST00000590430.1 ENST00000586613.1 |
TK1
|
thymidine kinase 1, soluble |
| chr4_-_39640700 | 0.53 |
ENST00000295958.5
|
SMIM14
|
small integral membrane protein 14 |
| chr8_-_27941359 | 0.53 |
ENST00000418860.1
|
NUGGC
|
nuclear GTPase, germinal center associated |
| chr17_-_17726907 | 0.53 |
ENST00000423161.3
|
SREBF1
|
sterol regulatory element binding transcription factor 1 |
| chr8_+_142402089 | 0.53 |
ENST00000521578.1
ENST00000520105.1 ENST00000523147.1 |
PTP4A3
|
protein tyrosine phosphatase type IVA, member 3 |
| chr14_-_57197224 | 0.52 |
ENST00000554597.1
ENST00000556696.1 |
RP11-1085N6.3
|
Uncharacterized protein |
| chr12_-_125399573 | 0.52 |
ENST00000339647.5
|
UBC
|
ubiquitin C |
| chr8_-_121457332 | 0.52 |
ENST00000518918.1
|
MRPL13
|
mitochondrial ribosomal protein L13 |
| chr19_-_36391434 | 0.52 |
ENST00000396901.1
ENST00000585925.1 |
NFKBID
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, delta |
| chr19_-_45873642 | 0.51 |
ENST00000485403.2
ENST00000586856.1 ENST00000586131.1 ENST00000391940.4 ENST00000221481.6 ENST00000391944.3 ENST00000391945.4 |
ERCC2
|
excision repair cross-complementing rodent repair deficiency, complementation group 2 |
| chr12_+_52282001 | 0.51 |
ENST00000340970.4
|
ANKRD33
|
ankyrin repeat domain 33 |
| chr8_+_74903580 | 0.50 |
ENST00000284818.2
ENST00000518893.1 |
LY96
|
lymphocyte antigen 96 |
| chr11_+_67171391 | 0.49 |
ENST00000312390.5
|
TBC1D10C
|
TBC1 domain family, member 10C |
| chr12_+_123942188 | 0.49 |
ENST00000526639.2
|
SNRNP35
|
small nuclear ribonucleoprotein 35kDa (U11/U12) |
| chr14_-_70826438 | 0.49 |
ENST00000389912.6
|
COX16
|
COX16 cytochrome c oxidase assembly homolog (S. cerevisiae) |
| chr11_-_62521614 | 0.49 |
ENST00000527994.1
ENST00000394807.3 |
ZBTB3
|
zinc finger and BTB domain containing 3 |
| chr1_+_10459111 | 0.47 |
ENST00000541529.1
ENST00000270776.8 ENST00000483936.1 ENST00000538557.1 |
PGD
|
phosphogluconate dehydrogenase |
| chr1_+_210001309 | 0.47 |
ENST00000491415.2
|
DIEXF
|
digestive organ expansion factor homolog (zebrafish) |
| chr17_-_62097904 | 0.46 |
ENST00000583366.1
|
ICAM2
|
intercellular adhesion molecule 2 |
| chr9_-_36400213 | 0.46 |
ENST00000259605.6
ENST00000353739.4 |
RNF38
|
ring finger protein 38 |
| chr12_-_56122761 | 0.46 |
ENST00000552164.1
ENST00000420846.3 ENST00000257857.4 |
CD63
|
CD63 molecule |
| chr16_+_21312170 | 0.46 |
ENST00000338573.5
ENST00000561968.1 |
CRYM-AS1
|
CRYM antisense RNA 1 |
| chr7_+_2687173 | 0.46 |
ENST00000403167.1
|
TTYH3
|
tweety family member 3 |
| chr1_+_24117627 | 0.46 |
ENST00000400061.1
|
LYPLA2
|
lysophospholipase II |
| chr16_+_30194118 | 0.45 |
ENST00000563778.1
|
CORO1A
|
coronin, actin binding protein, 1A |
| chr7_+_56131917 | 0.45 |
ENST00000434526.2
ENST00000275607.9 ENST00000395435.2 ENST00000413952.2 ENST00000342190.6 ENST00000437307.2 ENST00000413756.1 ENST00000451338.1 |
SUMF2
|
sulfatase modifying factor 2 |
| chrX_+_107334983 | 0.45 |
ENST00000457035.1
ENST00000545696.1 |
ATG4A
|
autophagy related 4A, cysteine peptidase |
| chr6_-_31763408 | 0.45 |
ENST00000444930.2
|
VARS
|
valyl-tRNA synthetase |
Network of associatons between targets according to the STRING database.
First level regulatory network of NR1H2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.0 | GO:0055099 | response to high density lipoprotein particle(GO:0055099) |
| 0.6 | 1.7 | GO:0044205 | 'de novo' UMP biosynthetic process(GO:0044205) |
| 0.5 | 6.5 | GO:0055091 | phospholipid homeostasis(GO:0055091) |
| 0.4 | 2.2 | GO:0045659 | regulation of neutrophil differentiation(GO:0045658) negative regulation of neutrophil differentiation(GO:0045659) |
| 0.4 | 1.3 | GO:1901899 | positive regulation of relaxation of cardiac muscle(GO:1901899) regulation of calcium ion import into sarcoplasmic reticulum(GO:1902080) negative regulation of calcium ion import into sarcoplasmic reticulum(GO:1902081) |
| 0.4 | 1.7 | GO:0021849 | neuroblast division in subventricular zone(GO:0021849) |
| 0.3 | 2.6 | GO:2000857 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.3 | 2.0 | GO:0036100 | leukotriene catabolic process(GO:0036100) leukotriene B4 catabolic process(GO:0036101) leukotriene B4 metabolic process(GO:0036102) icosanoid catabolic process(GO:1901523) fatty acid derivative catabolic process(GO:1901569) |
| 0.3 | 0.8 | GO:0002125 | maternal aggressive behavior(GO:0002125) positive regulation of female receptivity(GO:0045925) |
| 0.3 | 1.3 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 0.3 | 0.8 | GO:0034471 | endonucleolytic cleavage to generate mature 5'-end of SSU-rRNA from (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000472) rRNA 5'-end processing(GO:0000967) ncRNA 5'-end processing(GO:0034471) |
| 0.3 | 6.4 | GO:0006853 | carnitine shuttle(GO:0006853) |
| 0.2 | 2.2 | GO:0097068 | response to thyroxine(GO:0097068) response to L-phenylalanine derivative(GO:1904386) |
| 0.2 | 2.7 | GO:2000332 | blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.2 | 0.7 | GO:2001247 | positive regulation of phosphatidylcholine biosynthetic process(GO:2001247) |
| 0.2 | 1.1 | GO:0002155 | regulation of thyroid hormone mediated signaling pathway(GO:0002155) |
| 0.2 | 2.0 | GO:0097646 | calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.2 | 0.8 | GO:0042351 | 'de novo' GDP-L-fucose biosynthetic process(GO:0042351) |
| 0.2 | 0.7 | GO:0002625 | regulation of T cell antigen processing and presentation(GO:0002625) |
| 0.2 | 2.7 | GO:0039536 | negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.2 | 1.0 | GO:0009051 | pentose-phosphate shunt, oxidative branch(GO:0009051) |
| 0.2 | 2.2 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.2 | 0.5 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.2 | 1.7 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.2 | 0.9 | GO:1902045 | negative regulation of Fas signaling pathway(GO:1902045) |
| 0.2 | 0.6 | GO:1904647 | response to rotenone(GO:1904647) |
| 0.1 | 0.4 | GO:0032918 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.1 | 0.7 | GO:1903215 | negative regulation of protein targeting to mitochondrion(GO:1903215) |
| 0.1 | 0.8 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.1 | 2.0 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.1 | 0.7 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.1 | 0.6 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.1 | 0.9 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.1 | 1.1 | GO:0006565 | L-serine catabolic process(GO:0006565) |
| 0.1 | 1.0 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.1 | 0.7 | GO:0006438 | valyl-tRNA aminoacylation(GO:0006438) |
| 0.1 | 0.4 | GO:0048075 | positive regulation of eye pigmentation(GO:0048075) |
| 0.1 | 0.6 | GO:0038156 | interleukin-3-mediated signaling pathway(GO:0038156) |
| 0.1 | 0.4 | GO:2000342 | negative regulation of chemokine (C-X-C motif) ligand 2 production(GO:2000342) |
| 0.1 | 1.1 | GO:0050747 | positive regulation of lipoprotein metabolic process(GO:0050747) |
| 0.1 | 1.5 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.1 | 0.6 | GO:1903438 | regulation of cytokinetic process(GO:0032954) regulation of mitotic cytokinetic process(GO:1903436) positive regulation of mitotic cytokinetic process(GO:1903438) positive regulation of mitotic cytokinesis(GO:1903490) |
| 0.1 | 0.9 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.1 | 0.5 | GO:0032796 | uropod organization(GO:0032796) |
| 0.1 | 0.8 | GO:0001878 | response to yeast(GO:0001878) |
| 0.1 | 0.4 | GO:0000966 | RNA 5'-end processing(GO:0000966) |
| 0.1 | 0.4 | GO:0009956 | radial pattern formation(GO:0009956) |
| 0.1 | 0.6 | GO:0090625 | mRNA cleavage involved in gene silencing by siRNA(GO:0090625) |
| 0.1 | 0.9 | GO:0002084 | protein depalmitoylation(GO:0002084) |
| 0.1 | 1.1 | GO:2000680 | rubidium ion transport(GO:0035826) regulation of rubidium ion transport(GO:2000680) |
| 0.1 | 1.1 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.1 | 0.2 | GO:2000397 | regulation of ubiquitin-dependent endocytosis(GO:2000395) positive regulation of ubiquitin-dependent endocytosis(GO:2000397) |
| 0.1 | 0.6 | GO:0060356 | leucine import(GO:0060356) |
| 0.1 | 0.5 | GO:0046104 | thymidine metabolic process(GO:0046104) pyrimidine deoxyribonucleoside metabolic process(GO:0046125) |
| 0.1 | 0.2 | GO:0042245 | RNA repair(GO:0042245) |
| 0.1 | 4.2 | GO:0032094 | response to food(GO:0032094) |
| 0.1 | 0.8 | GO:0010626 | regulation of Schwann cell proliferation(GO:0010624) negative regulation of Schwann cell proliferation(GO:0010626) |
| 0.1 | 0.5 | GO:0032497 | detection of lipopolysaccharide(GO:0032497) |
| 0.1 | 1.3 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.1 | 1.0 | GO:0002291 | T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:0002291) |
| 0.1 | 0.7 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.1 | 0.5 | GO:0070245 | positive regulation of thymocyte apoptotic process(GO:0070245) |
| 0.1 | 1.3 | GO:0006071 | glycerol metabolic process(GO:0006071) |
| 0.1 | 0.2 | GO:0034402 | recruitment of 3'-end processing factors to RNA polymerase II holoenzyme complex(GO:0034402) |
| 0.1 | 1.8 | GO:0071243 | cellular response to arsenic-containing substance(GO:0071243) |
| 0.1 | 1.1 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.1 | 0.6 | GO:0051902 | negative regulation of mitochondrial depolarization(GO:0051902) |
| 0.1 | 1.1 | GO:0048845 | venous blood vessel morphogenesis(GO:0048845) |
| 0.1 | 1.3 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.1 | 1.3 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 0.1 | 0.3 | GO:0042986 | positive regulation of amyloid precursor protein biosynthetic process(GO:0042986) |
| 0.1 | 0.9 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.0 | 1.1 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.0 | 0.4 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.1 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.0 | 0.7 | GO:0019441 | tryptophan catabolic process to kynurenine(GO:0019441) |
| 0.0 | 0.4 | GO:0042983 | amyloid precursor protein biosynthetic process(GO:0042983) regulation of amyloid precursor protein biosynthetic process(GO:0042984) |
| 0.0 | 0.5 | GO:0043117 | positive regulation of vascular permeability(GO:0043117) |
| 0.0 | 1.1 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.9 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.9 | GO:0034656 | nucleobase-containing small molecule catabolic process(GO:0034656) |
| 0.0 | 0.9 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.9 | GO:0060009 | Sertoli cell development(GO:0060009) |
| 0.0 | 1.1 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.5 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.0 | 1.2 | GO:2000678 | negative regulation of transcription regulatory region DNA binding(GO:2000678) |
| 0.0 | 1.6 | GO:0044381 | glucose import in response to insulin stimulus(GO:0044381) |
| 0.0 | 2.3 | GO:0045742 | positive regulation of epidermal growth factor receptor signaling pathway(GO:0045742) |
| 0.0 | 0.3 | GO:0097398 | response to interleukin-17(GO:0097396) cellular response to interleukin-17(GO:0097398) |
| 0.0 | 0.8 | GO:0055070 | copper ion homeostasis(GO:0055070) |
| 0.0 | 2.6 | GO:0006378 | mRNA polyadenylation(GO:0006378) |
| 0.0 | 1.9 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.0 | 0.6 | GO:0035458 | positive regulation of protein import into nucleus, translocation(GO:0033160) cellular response to interferon-beta(GO:0035458) |
| 0.0 | 1.2 | GO:0035590 | purinergic nucleotide receptor signaling pathway(GO:0035590) |
| 0.0 | 0.6 | GO:1903071 | positive regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903071) |
| 0.0 | 0.2 | GO:0072434 | signal transduction involved in G2 DNA damage checkpoint(GO:0072425) signal transduction involved in mitotic G2 DNA damage checkpoint(GO:0072434) |
| 0.0 | 1.3 | GO:0046676 | negative regulation of insulin secretion(GO:0046676) |
| 0.0 | 0.7 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.0 | 0.2 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 2.6 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.0 | 0.4 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 0.2 | GO:0035897 | proteolysis in other organism(GO:0035897) |
| 0.0 | 0.7 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.0 | 0.7 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.0 | 2.7 | GO:0006687 | glycosphingolipid metabolic process(GO:0006687) |
| 0.0 | 1.0 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
| 0.0 | 0.4 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.0 | 1.0 | GO:1904837 | beta-catenin-TCF complex assembly(GO:1904837) |
| 0.0 | 0.3 | GO:0060732 | positive regulation of inositol phosphate biosynthetic process(GO:0060732) |
| 0.0 | 0.1 | GO:1903644 | regulation of chaperone-mediated protein folding(GO:1903644) |
| 0.0 | 2.0 | GO:0045669 | positive regulation of osteoblast differentiation(GO:0045669) |
| 0.0 | 0.1 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.0 | 3.3 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.0 | 2.1 | GO:0030521 | androgen receptor signaling pathway(GO:0030521) |
| 0.0 | 0.8 | GO:0010039 | response to iron ion(GO:0010039) |
| 0.0 | 3.5 | GO:0009108 | coenzyme biosynthetic process(GO:0009108) |
| 0.0 | 0.5 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 1.6 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.0 | 4.6 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.0 | 0.4 | GO:0060330 | regulation of response to interferon-gamma(GO:0060330) regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
| 0.0 | 0.3 | GO:0000460 | maturation of 5.8S rRNA(GO:0000460) |
| 0.0 | 0.7 | GO:0034035 | purine ribonucleoside bisphosphate metabolic process(GO:0034035) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) |
| 0.0 | 0.4 | GO:0036150 | phosphatidylserine acyl-chain remodeling(GO:0036150) |
| 0.0 | 1.1 | GO:0042267 | natural killer cell mediated cytotoxicity(GO:0042267) |
| 0.0 | 0.9 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 1.1 | GO:0003281 | ventricular septum development(GO:0003281) |
| 0.0 | 0.3 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 0.2 | GO:0061088 | sequestering of zinc ion(GO:0032119) regulation of sequestering of zinc ion(GO:0061088) |
| 0.0 | 0.4 | GO:0061049 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 0.6 | GO:0005980 | glycogen catabolic process(GO:0005980) |
| 0.0 | 0.2 | GO:0070987 | error-free translesion synthesis(GO:0070987) |
| 0.0 | 0.4 | GO:0042347 | negative regulation of NF-kappaB import into nucleus(GO:0042347) |
| 0.0 | 0.3 | GO:0007035 | vacuolar acidification(GO:0007035) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.5 | GO:0044299 | C-fiber(GO:0044299) |
| 0.4 | 1.5 | GO:0071062 | rough endoplasmic reticulum lumen(GO:0048237) alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.3 | 0.8 | GO:0030689 | Noc complex(GO:0030689) |
| 0.2 | 2.3 | GO:0034687 | integrin alphaL-beta2 complex(GO:0034687) |
| 0.2 | 1.6 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.1 | 0.7 | GO:0005602 | complement component C1 complex(GO:0005602) |
| 0.1 | 6.5 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.1 | 1.3 | GO:0098559 | cytoplasmic side of early endosome membrane(GO:0098559) cytoplasmic side of late endosome membrane(GO:0098560) |
| 0.1 | 0.6 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.1 | 0.4 | GO:0031085 | BLOC-3 complex(GO:0031085) |
| 0.1 | 2.7 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.1 | 0.5 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.1 | 1.3 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.1 | 1.3 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.1 | 1.1 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.1 | 0.9 | GO:0000243 | commitment complex(GO:0000243) |
| 0.1 | 0.6 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.1 | 0.5 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.1 | 3.6 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.1 | 6.4 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.1 | 2.2 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.6 | GO:0035068 | micro-ribonucleoprotein complex(GO:0035068) |
| 0.0 | 0.7 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.6 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.0 | 5.1 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.0 | 1.1 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.2 | GO:0070939 | Dsl1p complex(GO:0070939) |
| 0.0 | 1.2 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.0 | 0.6 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.0 | 1.3 | GO:0031304 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.3 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 2.7 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.8 | GO:0000782 | telomere cap complex(GO:0000782) nuclear telomere cap complex(GO:0000783) |
| 0.0 | 3.5 | GO:0101003 | ficolin-1-rich granule membrane(GO:0101003) |
| 0.0 | 0.5 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.4 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 0.2 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.0 | 0.3 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.0 | 0.9 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.1 | GO:0032021 | NELF complex(GO:0032021) |
| 0.0 | 0.7 | GO:0005753 | mitochondrial proton-transporting ATP synthase complex(GO:0005753) |
| 0.0 | 0.9 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 0.5 | GO:0001931 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 0.8 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.0 | 0.1 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.0 | 0.5 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.5 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 1.1 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 5.6 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 2.4 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 1.2 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.9 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.2 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 0.1 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.0 | 9.1 | GO:0031966 | mitochondrial membrane(GO:0031966) |
| 0.0 | 0.4 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.2 | GO:0043194 | axon initial segment(GO:0043194) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 3.9 | GO:0017129 | triglyceride binding(GO:0017129) |
| 0.7 | 2.2 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.7 | 2.2 | GO:0030572 | cardiolipin synthase activity(GO:0008808) phosphatidyltransferase activity(GO:0030572) CDP-diacylglycerol-phosphatidylglycerol phosphatidyltransferase activity(GO:0043337) |
| 0.6 | 2.6 | GO:0090556 | apolipoprotein A-I receptor activity(GO:0034188) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.6 | 1.9 | GO:0097259 | tocopherol omega-hydroxylase activity(GO:0052870) alpha-tocopherol omega-hydroxylase activity(GO:0052871) 20-hydroxy-leukotriene B4 omega oxidase activity(GO:0097258) 20-aldehyde-leukotriene B4 20-monooxygenase activity(GO:0097259) |
| 0.6 | 1.7 | GO:0008124 | 4-alpha-hydroxytetrahydrobiopterin dehydratase activity(GO:0008124) |
| 0.5 | 1.5 | GO:0034041 | sterol-transporting ATPase activity(GO:0034041) |
| 0.4 | 1.7 | GO:0016635 | oxidoreductase activity, acting on the CH-CH group of donors, quinone or related compound as acceptor(GO:0016635) |
| 0.4 | 2.5 | GO:0004882 | androgen receptor activity(GO:0004882) |
| 0.4 | 1.5 | GO:0004917 | interleukin-7 receptor activity(GO:0004917) |
| 0.4 | 1.1 | GO:0004794 | L-threonine ammonia-lyase activity(GO:0004794) |
| 0.4 | 1.1 | GO:0000773 | phosphatidyl-N-methylethanolamine N-methyltransferase activity(GO:0000773) phosphatidylethanolamine N-methyltransferase activity(GO:0004608) phosphatidyl-N-dimethylethanolamine N-methyltransferase activity(GO:0080101) |
| 0.3 | 1.3 | GO:0002094 | polyprenyltransferase activity(GO:0002094) |
| 0.3 | 1.0 | GO:0036440 | citrate (Si)-synthase activity(GO:0004108) citrate synthase activity(GO:0036440) |
| 0.3 | 1.5 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.3 | 1.5 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.3 | 2.2 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.3 | 0.8 | GO:0050577 | GDP-4-dehydro-D-rhamnose reductase activity(GO:0042356) GDP-L-fucose synthase activity(GO:0050577) |
| 0.2 | 2.7 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.2 | 1.0 | GO:0009383 | rRNA (cytosine-C5-)-methyltransferase activity(GO:0009383) |
| 0.2 | 0.7 | GO:0010309 | acireductone dioxygenase [iron(II)-requiring] activity(GO:0010309) |
| 0.2 | 2.3 | GO:0030369 | ICAM-3 receptor activity(GO:0030369) |
| 0.2 | 0.6 | GO:0017057 | 6-phosphogluconolactonase activity(GO:0017057) |
| 0.2 | 0.9 | GO:0004382 | guanosine-diphosphatase activity(GO:0004382) |
| 0.1 | 0.4 | GO:0004145 | diamine N-acetyltransferase activity(GO:0004145) polyamine binding(GO:0019808) |
| 0.1 | 1.1 | GO:0005124 | N-formyl peptide receptor activity(GO:0004982) scavenger receptor binding(GO:0005124) |
| 0.1 | 0.5 | GO:0004727 | prenylated protein tyrosine phosphatase activity(GO:0004727) |
| 0.1 | 1.3 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.1 | 0.7 | GO:0004832 | valine-tRNA ligase activity(GO:0004832) |
| 0.1 | 0.6 | GO:0004912 | interleukin-3 receptor activity(GO:0004912) |
| 0.1 | 0.3 | GO:0034736 | sterol O-acyltransferase activity(GO:0004772) cholesterol O-acyltransferase activity(GO:0034736) |
| 0.1 | 1.2 | GO:0004931 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.1 | 0.7 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.1 | 0.5 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.1 | 0.5 | GO:0004797 | thymidine kinase activity(GO:0004797) |
| 0.1 | 0.4 | GO:0016426 | tRNA (adenine) methyltransferase activity(GO:0016426) tRNA (adenine-N1-)-methyltransferase activity(GO:0016429) |
| 0.1 | 1.9 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.1 | 3.4 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.1 | 2.1 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.1 | 0.8 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.1 | 1.6 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.1 | 0.4 | GO:0004906 | interferon-gamma receptor activity(GO:0004906) |
| 0.1 | 0.6 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.1 | 0.5 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.1 | 0.5 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.1 | 0.9 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.1 | 0.2 | GO:0036361 | racemase and epimerase activity, acting on amino acids and derivatives(GO:0016855) racemase activity, acting on amino acids and derivatives(GO:0036361) amino-acid racemase activity(GO:0047661) |
| 0.1 | 0.3 | GO:0004991 | parathyroid hormone receptor activity(GO:0004991) |
| 0.1 | 1.7 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.1 | 1.3 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) |
| 0.1 | 1.9 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.1 | 2.7 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.1 | 0.5 | GO:0032810 | sterol response element binding(GO:0032810) |
| 0.1 | 0.8 | GO:0008035 | high-density lipoprotein particle binding(GO:0008035) |
| 0.1 | 0.2 | GO:0035514 | DNA demethylase activity(GO:0035514) |
| 0.1 | 0.6 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.1 | 1.5 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.1 | 1.3 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.1 | 0.5 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 0.1 | 0.7 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 1.1 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.8 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.0 | 0.1 | GO:0015130 | mevalonate transmembrane transporter activity(GO:0015130) |
| 0.0 | 0.2 | GO:0018685 | alkane 1-monooxygenase activity(GO:0018685) |
| 0.0 | 0.9 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.0 | 1.1 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.0 | 0.9 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 0.6 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.1 | GO:0010698 | acetyltransferase activator activity(GO:0010698) |
| 0.0 | 0.8 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 0.7 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 1.7 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 0.2 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.7 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.0 | 0.3 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.0 | 0.7 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.0 | 3.5 | GO:0015297 | antiporter activity(GO:0015297) |
| 0.0 | 0.5 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.0 | 0.7 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 0.8 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.9 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.0 | 0.4 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.6 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 1.3 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.4 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.7 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 2.0 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 1.1 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.0 | 0.4 | GO:0034483 | heparan sulfate sulfotransferase activity(GO:0034483) |
| 0.0 | 0.7 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.8 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.7 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 2.0 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.9 | GO:0017069 | snRNA binding(GO:0017069) |
| 0.0 | 0.1 | GO:0033857 | diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.0 | 0.9 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 1.2 | GO:0015171 | amino acid transmembrane transporter activity(GO:0015171) |
| 0.0 | 0.5 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.3 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 0.1 | GO:0015087 | cadmium ion transmembrane transporter activity(GO:0015086) cobalt ion transmembrane transporter activity(GO:0015087) lead ion transmembrane transporter activity(GO:0015094) ferrous iron uptake transmembrane transporter activity(GO:0015639) |
| 0.0 | 2.2 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.0 | 0.9 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.0 | 0.5 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 2.3 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.4 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 3.1 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 3.9 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 2.2 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.0 | 0.3 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.0 | 2.1 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 2.9 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 1.6 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 1.5 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.9 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.0 | 1.5 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 1.5 | ST ADRENERGIC | Adrenergic Pathway |
| 0.0 | 2.0 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 1.3 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 0.8 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 0.4 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 2.5 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.5 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 1.7 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 2.0 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.7 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.0 | 0.4 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.0 | 0.4 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 1.9 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.7 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 0.5 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.4 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 1.1 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.5 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.3 | 8.0 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.2 | 3.5 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.1 | 1.3 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.1 | 2.1 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.1 | 1.1 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.1 | 2.3 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.1 | 0.7 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 1.5 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 6.5 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 1.1 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.5 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.0 | 4.9 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 1.1 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 2.2 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 2.1 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 1.7 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 0.7 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 1.1 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.7 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 2.0 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 1.9 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.2 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 4.5 | REACTOME INFLUENZA VIRAL RNA TRANSCRIPTION AND REPLICATION | Genes involved in Influenza Viral RNA Transcription and Replication |
| 0.0 | 2.5 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.4 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.5 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.5 | REACTOME MRNA CAPPING | Genes involved in mRNA Capping |
| 0.0 | 0.7 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.6 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.7 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.6 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.4 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 2.4 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.6 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.0 | 0.6 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.7 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 1.3 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 1.9 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.4 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.6 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |