Project
Illumina Body Map 2, young vs old
Navigation
Downloads
Results for NR2E1
Z-value: 0.94
Transcription factors associated with NR2E1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NR2E1
|
ENSG00000112333.7 | nuclear receptor subfamily 2 group E member 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NR2E1 | hg19_v2_chr6_+_108487245_108487262 | -0.13 | 4.8e-01 | Click! |
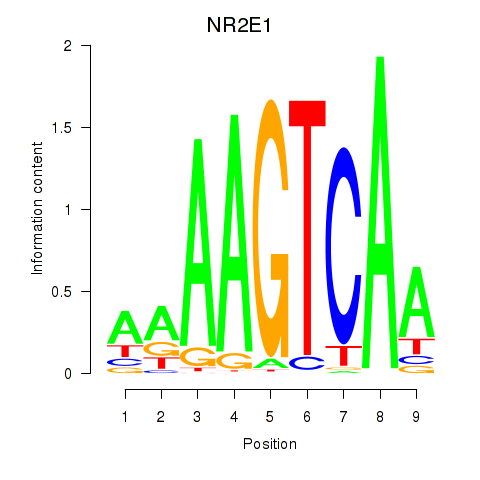
Activity profile of NR2E1 motif
Sorted Z-values of NR2E1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chrX_+_114874727 | 3.64 |
ENST00000543070.1
|
PLS3
|
plastin 3 |
| chr6_-_46922659 | 3.61 |
ENST00000265417.7
|
GPR116
|
G protein-coupled receptor 116 |
| chr1_+_185703513 | 3.36 |
ENST00000271588.4
ENST00000367492.2 |
HMCN1
|
hemicentin 1 |
| chr9_-_16728161 | 3.26 |
ENST00000603713.1
ENST00000603313.1 |
BNC2
|
basonuclin 2 |
| chr12_-_91539918 | 3.26 |
ENST00000548218.1
|
DCN
|
decorin |
| chr3_-_64673668 | 3.23 |
ENST00000498707.1
|
ADAMTS9
|
ADAM metallopeptidase with thrombospondin type 1 motif, 9 |
| chr7_+_116166331 | 3.16 |
ENST00000393468.1
ENST00000393467.1 |
CAV1
|
caveolin 1, caveolae protein, 22kDa |
| chr12_-_91572278 | 3.12 |
ENST00000425043.1
ENST00000420120.2 ENST00000441303.2 ENST00000456569.2 |
DCN
|
decorin |
| chr3_-_64673289 | 3.11 |
ENST00000295903.4
|
ADAMTS9
|
ADAM metallopeptidase with thrombospondin type 1 motif, 9 |
| chr8_-_86253888 | 3.00 |
ENST00000522389.1
ENST00000432364.2 ENST00000517618.1 |
CA1
|
carbonic anhydrase I |
| chr17_-_66951474 | 3.00 |
ENST00000269080.2
|
ABCA8
|
ATP-binding cassette, sub-family A (ABC1), member 8 |
| chrX_-_73061339 | 2.84 |
ENST00000602863.1
|
XIST
|
X inactive specific transcript (non-protein coding) |
| chr6_+_31916733 | 2.77 |
ENST00000483004.1
|
CFB
|
complement factor B |
| chr9_-_95166976 | 2.71 |
ENST00000447356.1
|
OGN
|
osteoglycin |
| chr17_+_12569306 | 2.57 |
ENST00000425538.1
|
MYOCD
|
myocardin |
| chr9_-_95166884 | 2.56 |
ENST00000375561.5
|
OGN
|
osteoglycin |
| chr17_-_66951382 | 2.56 |
ENST00000586539.1
|
ABCA8
|
ATP-binding cassette, sub-family A (ABC1), member 8 |
| chr3_-_64673656 | 2.56 |
ENST00000459780.1
|
ADAMTS9
|
ADAM metallopeptidase with thrombospondin type 1 motif, 9 |
| chr3_-_112356944 | 2.55 |
ENST00000461431.1
|
CCDC80
|
coiled-coil domain containing 80 |
| chr7_-_107443652 | 2.54 |
ENST00000340010.5
ENST00000422236.2 ENST00000453332.1 |
SLC26A3
|
solute carrier family 26 (anion exchanger), member 3 |
| chr12_+_54384370 | 2.54 |
ENST00000504315.1
|
HOXC6
|
homeobox C6 |
| chr9_-_95166841 | 2.53 |
ENST00000262551.4
|
OGN
|
osteoglycin |
| chr12_+_7169887 | 2.53 |
ENST00000542978.1
|
C1S
|
complement component 1, s subcomponent |
| chr5_-_33892204 | 2.50 |
ENST00000504830.1
|
ADAMTS12
|
ADAM metallopeptidase with thrombospondin type 1 motif, 12 |
| chr17_-_46667594 | 2.49 |
ENST00000476342.1
ENST00000460160.1 ENST00000472863.1 |
HOXB3
|
homeobox B3 |
| chr2_+_192141611 | 2.46 |
ENST00000392316.1
|
MYO1B
|
myosin IB |
| chr3_-_93747425 | 2.45 |
ENST00000315099.2
|
STX19
|
syntaxin 19 |
| chr13_+_110958124 | 2.39 |
ENST00000400163.2
|
COL4A2
|
collagen, type IV, alpha 2 |
| chr3_-_148939598 | 2.35 |
ENST00000455472.3
|
CP
|
ceruloplasmin (ferroxidase) |
| chr1_-_116311402 | 2.27 |
ENST00000261448.5
|
CASQ2
|
calsequestrin 2 (cardiac muscle) |
| chr11_-_111794446 | 2.19 |
ENST00000527950.1
|
CRYAB
|
crystallin, alpha B |
| chr3_-_116164306 | 2.17 |
ENST00000490035.2
|
LSAMP
|
limbic system-associated membrane protein |
| chr17_-_46667628 | 2.13 |
ENST00000498678.1
|
HOXB3
|
homeobox B3 |
| chr4_+_71063641 | 2.08 |
ENST00000514097.1
|
ODAM
|
odontogenic, ameloblast asssociated |
| chr3_+_12392971 | 2.07 |
ENST00000287820.6
|
PPARG
|
peroxisome proliferator-activated receptor gamma |
| chr5_-_42811986 | 2.01 |
ENST00000511224.1
ENST00000507920.1 ENST00000510965.1 |
SEPP1
|
selenoprotein P, plasma, 1 |
| chr16_-_10652993 | 1.99 |
ENST00000536829.1
|
EMP2
|
epithelial membrane protein 2 |
| chr1_+_32042105 | 1.98 |
ENST00000457433.2
ENST00000441210.2 |
TINAGL1
|
tubulointerstitial nephritis antigen-like 1 |
| chr7_+_142829162 | 1.94 |
ENST00000291009.3
|
PIP
|
prolactin-induced protein |
| chr1_-_116311323 | 1.93 |
ENST00000456138.2
|
CASQ2
|
calsequestrin 2 (cardiac muscle) |
| chr5_+_32711829 | 1.90 |
ENST00000415167.2
|
NPR3
|
natriuretic peptide receptor C/guanylate cyclase C (atrionatriuretic peptide receptor C) |
| chr4_-_76008706 | 1.89 |
ENST00000562355.1
ENST00000563602.1 |
RP11-44F21.5
|
RP11-44F21.5 |
| chr5_-_33892118 | 1.87 |
ENST00000515401.1
|
ADAMTS12
|
ADAM metallopeptidase with thrombospondin type 1 motif, 12 |
| chr12_-_53343633 | 1.83 |
ENST00000546826.1
|
KRT8
|
keratin 8 |
| chr17_+_12626199 | 1.76 |
ENST00000609971.1
|
AC005358.1
|
AC005358.1 |
| chr9_-_119162885 | 1.75 |
ENST00000445861.2
|
PAPPA-AS1
|
PAPPA antisense RNA 1 |
| chr12_-_71031185 | 1.68 |
ENST00000548122.1
ENST00000551525.1 ENST00000550358.1 |
PTPRB
|
protein tyrosine phosphatase, receptor type, B |
| chrX_-_128657457 | 1.67 |
ENST00000371121.3
ENST00000371123.1 ENST00000371122.4 |
SMARCA1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 1 |
| chrX_-_33146477 | 1.65 |
ENST00000378677.2
|
DMD
|
dystrophin |
| chr5_-_149516966 | 1.63 |
ENST00000517957.1
|
PDGFRB
|
platelet-derived growth factor receptor, beta polypeptide |
| chr5_+_9546306 | 1.62 |
ENST00000508179.1
|
SNHG18
|
small nucleolar RNA host gene 18 (non-protein coding) |
| chr1_+_170633047 | 1.62 |
ENST00000239461.6
ENST00000497230.2 |
PRRX1
|
paired related homeobox 1 |
| chr3_-_168864315 | 1.60 |
ENST00000475754.1
ENST00000484519.1 |
MECOM
|
MDS1 and EVI1 complex locus |
| chr5_+_32711419 | 1.56 |
ENST00000265074.8
|
NPR3
|
natriuretic peptide receptor C/guanylate cyclase C (atrionatriuretic peptide receptor C) |
| chr5_-_33892046 | 1.56 |
ENST00000352040.3
|
ADAMTS12
|
ADAM metallopeptidase with thrombospondin type 1 motif, 12 |
| chr9_+_87285539 | 1.56 |
ENST00000359847.3
|
NTRK2
|
neurotrophic tyrosine kinase, receptor, type 2 |
| chr9_-_95298927 | 1.55 |
ENST00000395534.2
|
ECM2
|
extracellular matrix protein 2, female organ and adipocyte specific |
| chr8_+_24241969 | 1.54 |
ENST00000522298.1
|
ADAMDEC1
|
ADAM-like, decysin 1 |
| chr2_-_183291741 | 1.52 |
ENST00000351439.5
ENST00000409365.1 |
PDE1A
|
phosphodiesterase 1A, calmodulin-dependent |
| chr8_-_122653630 | 1.51 |
ENST00000303924.4
|
HAS2
|
hyaluronan synthase 2 |
| chr1_+_159141397 | 1.51 |
ENST00000368124.4
ENST00000368125.4 ENST00000416746.1 |
CADM3
|
cell adhesion molecule 3 |
| chr12_-_71031220 | 1.49 |
ENST00000334414.6
|
PTPRB
|
protein tyrosine phosphatase, receptor type, B |
| chr13_+_28527647 | 1.47 |
ENST00000567234.1
|
LINC00543
|
long intergenic non-protein coding RNA 543 |
| chr12_-_50643664 | 1.46 |
ENST00000550592.1
|
LIMA1
|
LIM domain and actin binding 1 |
| chr8_+_24241789 | 1.44 |
ENST00000256412.4
ENST00000538205.1 |
ADAMDEC1
|
ADAM-like, decysin 1 |
| chr10_+_85933494 | 1.44 |
ENST00000372126.3
|
C10orf99
|
chromosome 10 open reading frame 99 |
| chr9_-_16727978 | 1.43 |
ENST00000418777.1
ENST00000468187.2 |
BNC2
|
basonuclin 2 |
| chr14_+_67656110 | 1.43 |
ENST00000524532.1
ENST00000530728.1 |
FAM71D
|
family with sequence similarity 71, member D |
| chr11_+_31833939 | 1.41 |
ENST00000530348.1
|
RCN1
|
reticulocalbin 1, EF-hand calcium binding domain |
| chr3_-_116163830 | 1.39 |
ENST00000333617.4
|
LSAMP
|
limbic system-associated membrane protein |
| chr2_-_183387064 | 1.38 |
ENST00000536095.1
ENST00000331935.6 ENST00000358139.2 ENST00000456212.1 |
PDE1A
|
phosphodiesterase 1A, calmodulin-dependent |
| chr7_+_70597109 | 1.34 |
ENST00000333538.5
|
WBSCR17
|
Williams-Beuren syndrome chromosome region 17 |
| chr12_+_56624436 | 1.34 |
ENST00000266980.4
ENST00000437277.1 |
SLC39A5
|
solute carrier family 39 (zinc transporter), member 5 |
| chr8_+_11561660 | 1.32 |
ENST00000526716.1
ENST00000335135.4 ENST00000528027.1 |
GATA4
|
GATA binding protein 4 |
| chr3_-_148939835 | 1.31 |
ENST00000264613.6
|
CP
|
ceruloplasmin (ferroxidase) |
| chr6_+_125540951 | 1.30 |
ENST00000524679.1
|
TPD52L1
|
tumor protein D52-like 1 |
| chr5_+_9546376 | 1.30 |
ENST00000509788.1
|
SNHG18
|
small nucleolar RNA host gene 18 (non-protein coding) |
| chr4_-_175041663 | 1.29 |
ENST00000503140.1
|
RP11-148L24.1
|
RP11-148L24.1 |
| chr4_+_77177512 | 1.29 |
ENST00000606246.1
|
FAM47E
|
family with sequence similarity 47, member E |
| chr4_-_47983519 | 1.28 |
ENST00000358519.4
ENST00000544810.1 ENST00000402813.3 |
CNGA1
|
cyclic nucleotide gated channel alpha 1 |
| chr11_-_102714534 | 1.27 |
ENST00000299855.5
|
MMP3
|
matrix metallopeptidase 3 (stromelysin 1, progelatinase) |
| chr3_-_168864427 | 1.26 |
ENST00000468789.1
|
MECOM
|
MDS1 and EVI1 complex locus |
| chr8_+_104384616 | 1.26 |
ENST00000520337.1
|
CTHRC1
|
collagen triple helix repeat containing 1 |
| chr1_+_240408560 | 1.26 |
ENST00000441342.1
ENST00000545751.1 |
FMN2
|
formin 2 |
| chr17_+_12569472 | 1.26 |
ENST00000343344.4
|
MYOCD
|
myocardin |
| chr2_-_106013400 | 1.25 |
ENST00000409807.1
|
FHL2
|
four and a half LIM domains 2 |
| chr12_-_56236711 | 1.24 |
ENST00000409200.3
|
MMP19
|
matrix metallopeptidase 19 |
| chr8_+_76452097 | 1.24 |
ENST00000396423.2
|
HNF4G
|
hepatocyte nuclear factor 4, gamma |
| chr4_+_96012614 | 1.22 |
ENST00000264568.4
|
BMPR1B
|
bone morphogenetic protein receptor, type IB |
| chr12_-_28124903 | 1.22 |
ENST00000395872.1
ENST00000354417.3 ENST00000201015.4 |
PTHLH
|
parathyroid hormone-like hormone |
| chr8_+_80523962 | 1.19 |
ENST00000518491.1
|
STMN2
|
stathmin-like 2 |
| chr6_-_41715128 | 1.19 |
ENST00000356667.4
ENST00000373025.3 ENST00000425343.2 |
PGC
|
progastricsin (pepsinogen C) |
| chr1_+_212738676 | 1.18 |
ENST00000366981.4
ENST00000366987.2 |
ATF3
|
activating transcription factor 3 |
| chr7_-_14026123 | 1.18 |
ENST00000420159.2
ENST00000399357.3 ENST00000403527.1 |
ETV1
|
ets variant 1 |
| chr12_-_52887034 | 1.17 |
ENST00000330722.6
|
KRT6A
|
keratin 6A |
| chr4_-_135122903 | 1.14 |
ENST00000421491.3
ENST00000529122.2 |
PABPC4L
|
poly(A) binding protein, cytoplasmic 4-like |
| chr3_+_113616317 | 1.14 |
ENST00000440446.2
ENST00000488680.1 |
GRAMD1C
|
GRAM domain containing 1C |
| chr9_-_95244781 | 1.14 |
ENST00000375544.3
ENST00000375543.1 ENST00000395538.3 ENST00000450139.2 |
ASPN
|
asporin |
| chr11_+_100784231 | 1.12 |
ENST00000531183.1
|
ARHGAP42
|
Rho GTPase activating protein 42 |
| chr20_-_22559211 | 1.11 |
ENST00000564492.1
|
LINC00261
|
long intergenic non-protein coding RNA 261 |
| chr12_+_64238539 | 1.11 |
ENST00000357825.3
|
SRGAP1
|
SLIT-ROBO Rho GTPase activating protein 1 |
| chr4_-_13546632 | 1.11 |
ENST00000382438.5
|
NKX3-2
|
NK3 homeobox 2 |
| chr11_-_111781554 | 1.10 |
ENST00000526167.1
ENST00000528961.1 |
CRYAB
|
crystallin, alpha B |
| chr7_+_116312411 | 1.10 |
ENST00000456159.1
ENST00000397752.3 ENST00000318493.6 |
MET
|
met proto-oncogene |
| chrX_-_65259914 | 1.09 |
ENST00000374737.4
ENST00000455586.2 |
VSIG4
|
V-set and immunoglobulin domain containing 4 |
| chr5_-_160279207 | 1.07 |
ENST00000327245.5
|
ATP10B
|
ATPase, class V, type 10B |
| chr12_-_24102576 | 1.07 |
ENST00000537393.1
ENST00000309359.1 ENST00000381381.2 ENST00000451604.2 |
SOX5
|
SRY (sex determining region Y)-box 5 |
| chr12_-_56236690 | 1.06 |
ENST00000322569.4
|
MMP19
|
matrix metallopeptidase 19 |
| chr9_-_91793675 | 1.06 |
ENST00000375835.4
ENST00000375830.1 |
SHC3
|
SHC (Src homology 2 domain containing) transforming protein 3 |
| chr7_+_114055052 | 1.05 |
ENST00000462331.1
ENST00000408937.3 ENST00000403559.4 ENST00000350908.4 ENST00000393498.2 ENST00000393495.3 ENST00000378237.3 ENST00000393489.3 |
FOXP2
|
forkhead box P2 |
| chr5_+_125706998 | 1.05 |
ENST00000506445.1
|
GRAMD3
|
GRAM domain containing 3 |
| chr14_+_37131058 | 1.05 |
ENST00000361487.6
|
PAX9
|
paired box 9 |
| chr8_+_77593474 | 1.04 |
ENST00000455469.2
ENST00000050961.6 |
ZFHX4
|
zinc finger homeobox 4 |
| chr14_+_51955831 | 1.04 |
ENST00000356218.4
|
FRMD6
|
FERM domain containing 6 |
| chr6_-_138866823 | 1.04 |
ENST00000342260.5
|
NHSL1
|
NHS-like 1 |
| chr14_+_69865401 | 1.03 |
ENST00000556605.1
ENST00000336643.5 ENST00000031146.4 |
SLC39A9
|
solute carrier family 39, member 9 |
| chr19_-_47157914 | 1.03 |
ENST00000300875.4
|
DACT3
|
dishevelled-binding antagonist of beta-catenin 3 |
| chr20_-_22565101 | 1.02 |
ENST00000419308.2
|
FOXA2
|
forkhead box A2 |
| chr12_-_56236734 | 1.02 |
ENST00000548629.1
|
MMP19
|
matrix metallopeptidase 19 |
| chrX_-_65259900 | 1.01 |
ENST00000412866.2
|
VSIG4
|
V-set and immunoglobulin domain containing 4 |
| chr17_+_1959369 | 1.00 |
ENST00000576444.1
ENST00000322941.3 |
HIC1
|
hypermethylated in cancer 1 |
| chr10_-_21786179 | 0.99 |
ENST00000377113.5
|
CASC10
|
cancer susceptibility candidate 10 |
| chr11_-_111781454 | 0.99 |
ENST00000533280.1
|
CRYAB
|
crystallin, alpha B |
| chr2_+_177015950 | 0.99 |
ENST00000306324.3
|
HOXD4
|
homeobox D4 |
| chr6_+_148663729 | 0.98 |
ENST00000367467.3
|
SASH1
|
SAM and SH3 domain containing 1 |
| chrX_-_65253506 | 0.98 |
ENST00000427538.1
|
VSIG4
|
V-set and immunoglobulin domain containing 4 |
| chr17_-_67057203 | 0.98 |
ENST00000340001.4
|
ABCA9
|
ATP-binding cassette, sub-family A (ABC1), member 9 |
| chr5_+_140743859 | 0.98 |
ENST00000518069.1
|
PCDHGA5
|
protocadherin gamma subfamily A, 5 |
| chr7_-_16505440 | 0.96 |
ENST00000307068.4
|
SOSTDC1
|
sclerostin domain containing 1 |
| chr1_-_227505289 | 0.96 |
ENST00000366765.3
|
CDC42BPA
|
CDC42 binding protein kinase alpha (DMPK-like) |
| chr3_+_108855558 | 0.95 |
ENST00000467240.1
ENST00000477643.1 ENST00000479039.1 ENST00000593799.1 |
RP11-59E19.1
|
RP11-59E19.1 |
| chr6_+_142623758 | 0.95 |
ENST00000541199.1
ENST00000435011.2 |
GPR126
|
G protein-coupled receptor 126 |
| chr17_-_26695013 | 0.94 |
ENST00000555059.2
|
CTB-96E2.2
|
Homeobox protein SEBOX |
| chr11_-_8832182 | 0.94 |
ENST00000527510.1
ENST00000528527.1 ENST00000528523.1 ENST00000313726.6 |
ST5
|
suppression of tumorigenicity 5 |
| chr5_+_39520499 | 0.94 |
ENST00000604954.1
|
CTD-2078B5.2
|
CTD-2078B5.2 |
| chr4_+_52917451 | 0.94 |
ENST00000295213.4
ENST00000419395.2 |
SPATA18
|
spermatogenesis associated 18 |
| chr2_+_101437487 | 0.93 |
ENST00000427413.1
ENST00000542504.1 |
NPAS2
|
neuronal PAS domain protein 2 |
| chr6_+_143447322 | 0.92 |
ENST00000458219.1
|
AIG1
|
androgen-induced 1 |
| chr17_-_67057114 | 0.92 |
ENST00000370732.2
|
ABCA9
|
ATP-binding cassette, sub-family A (ABC1), member 9 |
| chr3_+_136676707 | 0.90 |
ENST00000329582.4
|
IL20RB
|
interleukin 20 receptor beta |
| chr15_-_88799384 | 0.89 |
ENST00000540489.2
ENST00000557856.1 ENST00000558676.1 |
NTRK3
|
neurotrophic tyrosine kinase, receptor, type 3 |
| chr9_+_1050331 | 0.88 |
ENST00000382255.3
ENST00000382251.3 ENST00000412350.2 |
DMRT2
|
doublesex and mab-3 related transcription factor 2 |
| chr11_-_5271122 | 0.88 |
ENST00000330597.3
|
HBG1
|
hemoglobin, gamma A |
| chr6_+_54173227 | 0.87 |
ENST00000259782.4
ENST00000370864.3 |
TINAG
|
tubulointerstitial nephritis antigen |
| chr5_-_135290651 | 0.86 |
ENST00000522943.1
ENST00000514447.2 |
LECT2
|
leukocyte cell-derived chemotaxin 2 |
| chr12_-_91546926 | 0.86 |
ENST00000550758.1
|
DCN
|
decorin |
| chr7_+_80267949 | 0.86 |
ENST00000482059.2
|
CD36
|
CD36 molecule (thrombospondin receptor) |
| chr5_-_9546180 | 0.85 |
ENST00000382496.5
|
SEMA5A
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5A |
| chr11_-_57004658 | 0.84 |
ENST00000606794.1
|
APLNR
|
apelin receptor |
| chr4_-_141348999 | 0.84 |
ENST00000325617.5
|
CLGN
|
calmegin |
| chr6_+_12290586 | 0.84 |
ENST00000379375.5
|
EDN1
|
endothelin 1 |
| chr2_-_118943930 | 0.83 |
ENST00000449075.1
ENST00000414886.1 ENST00000449819.1 |
AC093901.1
|
AC093901.1 |
| chr20_-_7238861 | 0.83 |
ENST00000428954.1
|
RP11-19D2.1
|
RP11-19D2.1 |
| chr4_+_183370146 | 0.83 |
ENST00000510504.1
|
TENM3
|
teneurin transmembrane protein 3 |
| chr2_+_74685527 | 0.82 |
ENST00000393972.3
ENST00000409737.1 ENST00000428943.1 |
WBP1
|
WW domain binding protein 1 |
| chr18_-_22804637 | 0.82 |
ENST00000577775.1
|
ZNF521
|
zinc finger protein 521 |
| chrX_-_114253536 | 0.82 |
ENST00000371936.1
|
IL13RA2
|
interleukin 13 receptor, alpha 2 |
| chr2_-_169887827 | 0.82 |
ENST00000263817.6
|
ABCB11
|
ATP-binding cassette, sub-family B (MDR/TAP), member 11 |
| chrX_+_135614293 | 0.82 |
ENST00000370634.3
|
VGLL1
|
vestigial like 1 (Drosophila) |
| chr12_-_75784669 | 0.81 |
ENST00000552497.1
ENST00000551829.1 ENST00000436898.1 ENST00000442339.2 |
CAPS2
|
calcyphosine 2 |
| chr10_-_21463116 | 0.80 |
ENST00000417816.2
|
NEBL
|
nebulette |
| chr14_-_103434869 | 0.80 |
ENST00000559043.1
|
CDC42BPB
|
CDC42 binding protein kinase beta (DMPK-like) |
| chr20_+_32782375 | 0.80 |
ENST00000568305.1
|
ASIP
|
agouti signaling protein |
| chr3_+_42897512 | 0.79 |
ENST00000493193.1
|
ACKR2
|
atypical chemokine receptor 2 |
| chr3_+_148508845 | 0.79 |
ENST00000491148.1
|
CPB1
|
carboxypeptidase B1 (tissue) |
| chr13_-_49975632 | 0.79 |
ENST00000457041.1
ENST00000355854.4 |
CAB39L
|
calcium binding protein 39-like |
| chr17_-_26694979 | 0.79 |
ENST00000438614.1
|
VTN
|
vitronectin |
| chr12_+_80603233 | 0.79 |
ENST00000547103.1
ENST00000458043.2 |
OTOGL
|
otogelin-like |
| chr15_-_51535208 | 0.78 |
ENST00000405913.3
ENST00000559878.1 |
CYP19A1
|
cytochrome P450, family 19, subfamily A, polypeptide 1 |
| chr1_+_77333117 | 0.77 |
ENST00000477717.1
|
ST6GALNAC5
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 5 |
| chr2_-_231989808 | 0.76 |
ENST00000258400.3
|
HTR2B
|
5-hydroxytryptamine (serotonin) receptor 2B, G protein-coupled |
| chr5_-_82969405 | 0.76 |
ENST00000510978.1
|
HAPLN1
|
hyaluronan and proteoglycan link protein 1 |
| chr7_+_134671234 | 0.75 |
ENST00000436302.2
ENST00000359383.3 ENST00000458078.1 ENST00000435976.2 ENST00000455283.2 |
AGBL3
|
ATP/GTP binding protein-like 3 |
| chr16_-_66952779 | 0.74 |
ENST00000570262.1
ENST00000394055.3 ENST00000299752.4 |
CDH16
|
cadherin 16, KSP-cadherin |
| chr6_+_74405501 | 0.74 |
ENST00000437994.2
ENST00000422508.2 |
CD109
|
CD109 molecule |
| chr3_-_50360192 | 0.73 |
ENST00000442581.1
ENST00000447092.1 ENST00000357750.4 |
HYAL2
|
hyaluronoglucosaminidase 2 |
| chr2_-_183387283 | 0.73 |
ENST00000435564.1
|
PDE1A
|
phosphodiesterase 1A, calmodulin-dependent |
| chr6_-_138820624 | 0.72 |
ENST00000343505.5
|
NHSL1
|
NHS-like 1 |
| chr12_+_26348582 | 0.72 |
ENST00000535504.1
|
SSPN
|
sarcospan |
| chr1_+_152850787 | 0.72 |
ENST00000368765.3
|
SMCP
|
sperm mitochondria-associated cysteine-rich protein |
| chr9_-_75567962 | 0.72 |
ENST00000297785.3
ENST00000376939.1 |
ALDH1A1
|
aldehyde dehydrogenase 1 family, member A1 |
| chr18_+_6774000 | 0.71 |
ENST00000532723.1
|
ARHGAP28
|
Rho GTPase activating protein 28 |
| chr3_-_24536453 | 0.70 |
ENST00000453729.2
ENST00000413780.1 |
THRB
|
thyroid hormone receptor, beta |
| chr2_+_201173667 | 0.68 |
ENST00000409755.3
|
SPATS2L
|
spermatogenesis associated, serine-rich 2-like |
| chr3_+_46616017 | 0.68 |
ENST00000542931.1
|
TDGF1
|
teratocarcinoma-derived growth factor 1 |
| chr7_-_8301768 | 0.67 |
ENST00000265577.7
|
ICA1
|
islet cell autoantigen 1, 69kDa |
| chr12_-_82152444 | 0.67 |
ENST00000549325.1
ENST00000550584.2 |
PPFIA2
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2 |
| chr3_+_69788576 | 0.66 |
ENST00000352241.4
ENST00000448226.2 |
MITF
|
microphthalmia-associated transcription factor |
| chr17_-_67057047 | 0.65 |
ENST00000495634.1
ENST00000453985.2 ENST00000585714.1 |
ABCA9
|
ATP-binding cassette, sub-family A (ABC1), member 9 |
| chr4_+_126315091 | 0.65 |
ENST00000335110.5
|
FAT4
|
FAT atypical cadherin 4 |
| chr17_+_19186292 | 0.64 |
ENST00000395626.1
ENST00000571254.1 |
EPN2
|
epsin 2 |
| chr1_-_169703203 | 0.63 |
ENST00000333360.7
ENST00000367781.4 ENST00000367782.4 ENST00000367780.4 ENST00000367779.4 |
SELE
|
selectin E |
| chr2_-_207630033 | 0.62 |
ENST00000449792.1
|
MDH1B
|
malate dehydrogenase 1B, NAD (soluble) |
| chr5_+_71475449 | 0.62 |
ENST00000504492.1
|
MAP1B
|
microtubule-associated protein 1B |
| chr4_-_111119804 | 0.62 |
ENST00000394607.3
ENST00000302274.3 |
ELOVL6
|
ELOVL fatty acid elongase 6 |
| chr16_-_77465450 | 0.62 |
ENST00000562345.1
|
ADAMTS18
|
ADAM metallopeptidase with thrombospondin type 1 motif, 18 |
| chr7_+_80267973 | 0.62 |
ENST00000394788.3
ENST00000447544.2 |
CD36
|
CD36 molecule (thrombospondin receptor) |
| chr17_-_42327236 | 0.62 |
ENST00000399246.2
|
AC003102.1
|
AC003102.1 |
| chrX_+_115301975 | 0.62 |
ENST00000371906.4
|
AGTR2
|
angiotensin II receptor, type 2 |
| chr5_+_127873657 | 0.62 |
ENST00000508645.1
|
SLC27A6
|
solute carrier family 27 (fatty acid transporter), member 6 |
Network of associatons between targets according to the STRING database.
First level regulatory network of NR2E1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 5.9 | GO:2001112 | negative regulation of hepatocyte growth factor receptor signaling pathway(GO:1902203) regulation of cellular response to hepatocyte growth factor stimulus(GO:2001112) negative regulation of cellular response to hepatocyte growth factor stimulus(GO:2001113) |
| 1.1 | 3.2 | GO:1903609 | negative regulation of peptidyl-tyrosine autophosphorylation(GO:1900085) negative regulation of inward rectifier potassium channel activity(GO:1903609) |
| 1.0 | 3.8 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) |
| 0.6 | 4.6 | GO:0021546 | rhombomere development(GO:0021546) |
| 0.5 | 1.6 | GO:0060981 | cell migration involved in coronary angiogenesis(GO:0060981) |
| 0.5 | 2.1 | GO:2000230 | negative regulation of pancreatic stellate cell proliferation(GO:2000230) |
| 0.5 | 7.3 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.5 | 2.8 | GO:0070295 | renal water absorption(GO:0070295) |
| 0.5 | 8.9 | GO:0045636 | positive regulation of melanocyte differentiation(GO:0045636) |
| 0.4 | 0.8 | GO:0030185 | nitric oxide transport(GO:0030185) |
| 0.4 | 1.3 | GO:0051758 | homologous chromosome movement towards spindle pole involved in homologous chromosome segregation(GO:0051758) |
| 0.4 | 2.1 | GO:0060054 | positive regulation of epithelial cell proliferation involved in wound healing(GO:0060054) |
| 0.4 | 1.2 | GO:0061394 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) |
| 0.3 | 5.6 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 0.3 | 7.8 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.3 | 1.3 | GO:0035054 | embryonic heart tube anterior/posterior pattern specification(GO:0035054) |
| 0.3 | 3.3 | GO:0001554 | luteolysis(GO:0001554) |
| 0.3 | 4.2 | GO:0086029 | Purkinje myocyte to ventricular cardiac muscle cell signaling(GO:0086029) Purkinje myocyte to ventricular cardiac muscle cell communication(GO:0086068) |
| 0.3 | 1.2 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
| 0.3 | 1.2 | GO:0051710 | cytolysis by symbiont of host cells(GO:0001897) regulation of cytolysis in other organism(GO:0051710) |
| 0.3 | 1.3 | GO:0034224 | cellular response to zinc ion starvation(GO:0034224) |
| 0.3 | 1.1 | GO:0098582 | innate vocalization behavior(GO:0098582) |
| 0.3 | 1.6 | GO:0099540 | synaptic signaling via neuropeptide(GO:0099538) trans-synaptic signaling by neuropeptide(GO:0099540) trans-synaptic signaling by neuropeptide, modulating synaptic transmission(GO:0099551) |
| 0.3 | 1.0 | GO:0045014 | carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) |
| 0.2 | 5.9 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.2 | 1.0 | GO:0060648 | mammary gland bud morphogenesis(GO:0060648) |
| 0.2 | 1.1 | GO:0070171 | negative regulation of tooth mineralization(GO:0070171) |
| 0.2 | 0.9 | GO:0001808 | negative regulation of type IV hypersensitivity(GO:0001808) |
| 0.2 | 0.9 | GO:0052026 | modulation by virus of host transcription(GO:0019056) positive regulation of sprouting of injured axon(GO:0048687) positive regulation of axon extension involved in regeneration(GO:0048691) modulation by symbiont of host transcription(GO:0052026) |
| 0.2 | 2.0 | GO:0070836 | caveola assembly(GO:0070836) |
| 0.2 | 2.5 | GO:0051454 | pH elevation(GO:0045852) intracellular pH elevation(GO:0051454) |
| 0.2 | 1.4 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.2 | 0.6 | GO:0035566 | negative regulation of icosanoid secretion(GO:0032304) regulation of metanephros size(GO:0035566) |
| 0.2 | 0.8 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.2 | 1.2 | GO:0002760 | positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antimicrobial humoral response(GO:0002760) positive regulation of antibacterial peptide production(GO:0002803) |
| 0.2 | 0.8 | GO:2000866 | positive regulation of estrogen secretion(GO:2000863) positive regulation of estradiol secretion(GO:2000866) |
| 0.2 | 3.6 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.2 | 0.7 | GO:1904425 | negative regulation of GTP binding(GO:1904425) |
| 0.2 | 3.4 | GO:0002158 | osteoclast proliferation(GO:0002158) |
| 0.2 | 4.3 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.2 | 0.8 | GO:0048842 | positive regulation of axon extension involved in axon guidance(GO:0048842) |
| 0.2 | 0.7 | GO:0045875 | negative regulation of sister chromatid cohesion(GO:0045875) |
| 0.2 | 1.0 | GO:0034628 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.2 | 0.5 | GO:0090427 | activation of meiosis(GO:0090427) |
| 0.2 | 2.5 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.2 | 1.3 | GO:0043932 | ossification involved in bone remodeling(GO:0043932) |
| 0.2 | 1.1 | GO:1905098 | negative regulation of guanyl-nucleotide exchange factor activity(GO:1905098) |
| 0.2 | 0.8 | GO:0010513 | positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) |
| 0.1 | 0.7 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.1 | 0.7 | GO:0008050 | courtship behavior(GO:0007619) female courtship behavior(GO:0008050) |
| 0.1 | 1.1 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.1 | 0.4 | GO:0070676 | intralumenal vesicle formation(GO:0070676) |
| 0.1 | 1.2 | GO:0055014 | atrial cardiac muscle cell differentiation(GO:0055011) atrial cardiac muscle cell development(GO:0055014) |
| 0.1 | 1.6 | GO:0048664 | neuron fate determination(GO:0048664) |
| 0.1 | 3.6 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.1 | 0.8 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.1 | 0.6 | GO:0072137 | condensed mesenchymal cell proliferation(GO:0072137) |
| 0.1 | 1.7 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.1 | 4.7 | GO:0003416 | endochondral bone growth(GO:0003416) |
| 0.1 | 0.9 | GO:0061055 | myotome development(GO:0061055) |
| 0.1 | 1.5 | GO:2000332 | blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.1 | 0.7 | GO:0006001 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 0.1 | 0.5 | GO:0001994 | norepinephrine-epinephrine vasoconstriction involved in regulation of systemic arterial blood pressure(GO:0001994) |
| 0.1 | 0.9 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.1 | 1.0 | GO:0003383 | apical constriction(GO:0003383) |
| 0.1 | 0.8 | GO:0019566 | arabinose metabolic process(GO:0019566) L-arabinose metabolic process(GO:0046373) |
| 0.1 | 1.8 | GO:0051599 | response to hydrostatic pressure(GO:0051599) |
| 0.1 | 3.7 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.1 | 1.7 | GO:0000733 | DNA strand renaturation(GO:0000733) |
| 0.1 | 1.8 | GO:0097475 | motor neuron migration(GO:0097475) |
| 0.1 | 0.8 | GO:0048023 | positive regulation of melanin biosynthetic process(GO:0048023) positive regulation of secondary metabolite biosynthetic process(GO:1900378) |
| 0.1 | 0.2 | GO:0034627 | 'de novo' NAD biosynthetic process(GO:0034627) |
| 0.1 | 1.0 | GO:0031666 | positive regulation of lipopolysaccharide-mediated signaling pathway(GO:0031666) |
| 0.1 | 0.5 | GO:0070173 | regulation of enamel mineralization(GO:0070173) |
| 0.1 | 2.4 | GO:0099500 | synaptic vesicle fusion to presynaptic active zone membrane(GO:0031629) vesicle fusion to plasma membrane(GO:0099500) |
| 0.1 | 0.8 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.1 | 0.7 | GO:0061370 | testosterone biosynthetic process(GO:0061370) |
| 0.1 | 2.2 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.1 | 0.7 | GO:0072674 | multinuclear osteoclast differentiation(GO:0072674) osteoclast fusion(GO:0072675) |
| 0.1 | 0.9 | GO:0048021 | regulation of melanin biosynthetic process(GO:0048021) negative regulation of melanin biosynthetic process(GO:0048022) regulation of secondary metabolite biosynthetic process(GO:1900376) negative regulation of secondary metabolite biosynthetic process(GO:1900377) |
| 0.1 | 1.3 | GO:0010727 | negative regulation of hydrogen peroxide metabolic process(GO:0010727) |
| 0.1 | 1.6 | GO:0048935 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.1 | 2.3 | GO:0032331 | negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.1 | 1.1 | GO:0055059 | asymmetric neuroblast division(GO:0055059) |
| 0.1 | 0.8 | GO:0043305 | negative regulation of mast cell degranulation(GO:0043305) |
| 0.1 | 0.2 | GO:0006533 | aspartate catabolic process(GO:0006533) |
| 0.1 | 1.2 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.1 | 0.4 | GO:1901164 | negative regulation of trophoblast cell migration(GO:1901164) |
| 0.1 | 0.7 | GO:0010623 | programmed cell death involved in cell development(GO:0010623) |
| 0.1 | 0.4 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.1 | 0.8 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.1 | 2.4 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
| 0.1 | 1.5 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.1 | 0.3 | GO:0001827 | inner cell mass cell fate commitment(GO:0001827) inner cell mass cellular morphogenesis(GO:0001828) |
| 0.1 | 1.7 | GO:0070233 | negative regulation of T cell apoptotic process(GO:0070233) |
| 0.1 | 0.7 | GO:2000074 | regulation of type B pancreatic cell development(GO:2000074) |
| 0.1 | 1.0 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.1 | 0.4 | GO:0045204 | MAPK export from nucleus(GO:0045204) |
| 0.1 | 0.6 | GO:0034625 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.1 | 0.2 | GO:0018874 | benzoate metabolic process(GO:0018874) |
| 0.0 | 0.9 | GO:0051775 | response to redox state(GO:0051775) |
| 0.0 | 3.0 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.0 | 1.0 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.7 | GO:0008595 | tripartite regional subdivision(GO:0007351) anterior/posterior axis specification, embryo(GO:0008595) |
| 0.0 | 1.0 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.0 | 0.8 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.0 | 0.1 | GO:0061056 | sclerotome development(GO:0061056) |
| 0.0 | 0.1 | GO:1903414 | regulation of transcription from RNA polymerase II promoter in response to iron(GO:0034395) spleen trabecula formation(GO:0060345) iron cation export(GO:1903414) ferrous iron export(GO:1903988) |
| 0.0 | 1.2 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 0.3 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.0 | 0.7 | GO:0042481 | regulation of odontogenesis(GO:0042481) |
| 0.0 | 0.6 | GO:0061339 | establishment of monopolar cell polarity(GO:0061162) establishment or maintenance of monopolar cell polarity(GO:0061339) |
| 0.0 | 3.9 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.0 | 1.2 | GO:0022400 | regulation of rhodopsin mediated signaling pathway(GO:0022400) |
| 0.0 | 0.2 | GO:0072660 | maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.0 | 0.1 | GO:1903251 | multi-ciliated epithelial cell differentiation(GO:1903251) |
| 0.0 | 0.3 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 1.3 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 1.4 | GO:0050832 | defense response to fungus(GO:0050832) |
| 0.0 | 0.8 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.0 | 1.5 | GO:2000144 | positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.0 | 0.4 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.0 | 1.9 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.5 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.0 | 0.6 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.3 | GO:0000212 | meiotic spindle organization(GO:0000212) |
| 0.0 | 0.7 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.0 | 0.6 | GO:0090331 | negative regulation of platelet aggregation(GO:0090331) |
| 0.0 | 1.0 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.0 | 0.1 | GO:0010909 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.0 | 0.8 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.0 | 0.1 | GO:0002385 | mucosal immune response(GO:0002385) |
| 0.0 | 0.1 | GO:0019046 | release from viral latency(GO:0019046) |
| 0.0 | 0.2 | GO:0006933 | negative regulation of cell adhesion involved in substrate-bound cell migration(GO:0006933) |
| 0.0 | 0.7 | GO:0000038 | very long-chain fatty acid metabolic process(GO:0000038) |
| 0.0 | 0.5 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.0 | 1.5 | GO:0031529 | ruffle organization(GO:0031529) |
| 0.0 | 0.4 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.2 | GO:0019695 | choline metabolic process(GO:0019695) |
| 0.0 | 0.7 | GO:0030252 | growth hormone secretion(GO:0030252) |
| 0.0 | 0.2 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.0 | 0.2 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.0 | 2.6 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 2.3 | GO:0006892 | post-Golgi vesicle-mediated transport(GO:0006892) |
| 0.0 | 0.4 | GO:0048873 | homeostasis of number of cells within a tissue(GO:0048873) |
| 0.0 | 0.3 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.3 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.0 | 0.2 | GO:0045003 | DNA recombinase assembly(GO:0000730) double-strand break repair via synthesis-dependent strand annealing(GO:0045003) |
| 0.0 | 1.1 | GO:0007173 | epidermal growth factor receptor signaling pathway(GO:0007173) |
| 0.0 | 0.8 | GO:0048663 | neuron fate commitment(GO:0048663) |
| 0.0 | 0.4 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.0 | 0.9 | GO:0006897 | endocytosis(GO:0006897) |
| 0.0 | 0.3 | GO:0019731 | antibacterial humoral response(GO:0019731) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 7.2 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.4 | 1.6 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.4 | 1.2 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 0.3 | 4.2 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.3 | 2.4 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.2 | 4.4 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.2 | 2.9 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.2 | 1.7 | GO:0016589 | NURF complex(GO:0016589) |
| 0.1 | 1.7 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.1 | 4.1 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 1.8 | GO:0042641 | actomyosin(GO:0042641) |
| 0.1 | 2.5 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.1 | 0.5 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.1 | 1.5 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.1 | 0.3 | GO:0071745 | IgA immunoglobulin complex(GO:0071745) IgA immunoglobulin complex, circulating(GO:0071746) monomeric IgA immunoglobulin complex(GO:0071748) polymeric IgA immunoglobulin complex(GO:0071749) secretory IgA immunoglobulin complex(GO:0071751) |
| 0.1 | 1.4 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.1 | 0.4 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.1 | 2.6 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.1 | 0.8 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.1 | 0.2 | GO:0032798 | Swi5-Sfr1 complex(GO:0032798) |
| 0.1 | 1.3 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.1 | 2.2 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.1 | 7.8 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 0.2 | GO:0097224 | sperm connecting piece(GO:0097224) |
| 0.0 | 4.6 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.4 | GO:0032009 | early phagosome(GO:0032009) |
| 0.0 | 0.3 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.0 | 17.1 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 1.2 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.3 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.0 | 0.9 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.0 | 6.5 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 1.1 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.2 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) |
| 0.0 | 2.2 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 0.6 | GO:0030128 | clathrin coat of endocytic vesicle(GO:0030128) |
| 0.0 | 2.6 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.3 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 1.2 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.0 | 1.0 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 4.5 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 2.0 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 0.3 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.0 | 1.7 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 1.0 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 1.0 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 0.2 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 2.1 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 1.8 | GO:0032432 | actin filament bundle(GO:0032432) |
| 0.0 | 0.7 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 2.4 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 1.1 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 1.9 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.0 | 1.3 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.0 | 0.2 | GO:0033391 | chromatoid body(GO:0033391) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.2 | GO:0070320 | inward rectifier potassium channel inhibitor activity(GO:0070320) |
| 0.8 | 0.8 | GO:0015432 | bile acid-exporting ATPase activity(GO:0015432) |
| 0.5 | 3.6 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.4 | 3.5 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.4 | 5.6 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.4 | 1.6 | GO:0005019 | platelet-derived growth factor beta-receptor activity(GO:0005019) |
| 0.4 | 3.0 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.3 | 1.5 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.3 | 3.7 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.3 | 1.5 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.3 | 1.3 | GO:0033906 | protein tyrosine kinase inhibitor activity(GO:0030292) hyaluronoglucuronidase activity(GO:0033906) |
| 0.3 | 1.6 | GO:0060175 | brain-derived neurotrophic factor-activated receptor activity(GO:0060175) |
| 0.2 | 1.9 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.2 | 0.8 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.2 | 1.0 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.2 | 1.0 | GO:0000309 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) |
| 0.2 | 0.5 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.1 | 0.9 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.1 | 4.3 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 1.5 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.1 | 2.5 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.1 | 1.2 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.1 | 3.9 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.1 | 22.9 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 1.4 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.1 | 1.7 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.1 | 0.9 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.1 | 0.8 | GO:0046556 | alpha-L-arabinofuranosidase activity(GO:0046556) |
| 0.1 | 0.4 | GO:0004140 | dephospho-CoA kinase activity(GO:0004140) |
| 0.1 | 0.7 | GO:0047045 | testosterone 17-beta-dehydrogenase (NADP+) activity(GO:0047045) |
| 0.1 | 1.0 | GO:0005497 | androgen binding(GO:0005497) |
| 0.1 | 2.5 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.1 | 1.9 | GO:0019864 | IgG binding(GO:0019864) |
| 0.1 | 1.3 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.1 | 0.8 | GO:0031779 | melanocortin receptor binding(GO:0031779) type 3 melanocortin receptor binding(GO:0031781) type 4 melanocortin receptor binding(GO:0031782) |
| 0.1 | 0.9 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.1 | 1.0 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.1 | 0.6 | GO:0004945 | angiotensin receptor activity(GO:0001595) angiotensin type II receptor activity(GO:0004945) |
| 0.1 | 3.2 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.1 | 1.3 | GO:0043855 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.1 | 0.8 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.1 | 2.8 | GO:0001848 | complement binding(GO:0001848) |
| 0.1 | 1.7 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.1 | 1.5 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.1 | 3.4 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.1 | 0.8 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.1 | 0.8 | GO:0043176 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.1 | 1.9 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.1 | 1.0 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.1 | 0.3 | GO:0032564 | adenyl deoxyribonucleotide binding(GO:0032558) dATP binding(GO:0032564) |
| 0.1 | 7.1 | GO:0005518 | collagen binding(GO:0005518) |
| 0.1 | 0.6 | GO:0102337 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.7 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.0 | 0.8 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 5.1 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 1.2 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 2.2 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.3 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.0 | 1.3 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.2 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.0 | 1.5 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.6 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.1 | GO:0097689 | iron channel activity(GO:0097689) |
| 0.0 | 1.0 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 1.2 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 2.9 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.0 | 1.3 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.6 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.0 | 2.3 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.0 | 0.8 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.0 | 3.3 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.0 | 0.8 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 1.3 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.9 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 7.0 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.0 | 0.7 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.0 | 0.6 | GO:0030955 | potassium ion binding(GO:0030955) |
| 0.0 | 0.6 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.2 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.0 | 0.1 | GO:0050509 | N-acetylglucosaminyl-proteoglycan 4-beta-glucuronosyltransferase activity(GO:0050509) |
| 0.0 | 0.4 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.2 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 1.5 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.1 | GO:0016530 | metallochaperone activity(GO:0016530) copper chaperone activity(GO:0016531) |
| 0.0 | 0.1 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.0 | 1.1 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 2.0 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.0 | 0.8 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.6 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.2 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.0 | 0.2 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.0 | 0.5 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 2.5 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 1.0 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.0 | 0.5 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.0 | 0.3 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 1.2 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.0 | 1.1 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.0 | 0.6 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.1 | GO:0050115 | myosin-light-chain-phosphatase activity(GO:0050115) |
| 0.0 | 0.2 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 14.2 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 3.2 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.1 | 5.7 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.1 | 1.3 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.1 | 2.5 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.1 | 0.8 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.1 | 23.8 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 3.3 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 1.2 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 1.6 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 1.5 | PID S1P S1P3 PATHWAY | S1P3 pathway |
| 0.0 | 4.1 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.9 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 1.6 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 1.3 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 2.6 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 0.6 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.0 | 0.3 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.0 | 0.8 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.5 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 1.5 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 1.6 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 1.5 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 1.0 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.7 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 1.0 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.7 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.7 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 0.9 | PID LKB1 PATHWAY | LKB1 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.1 | REACTOME SHC MEDIATED SIGNALLING | Genes involved in SHC-mediated signalling |
| 0.4 | 7.8 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.3 | 7.2 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.2 | 5.3 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.2 | 2.5 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.1 | 8.1 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.1 | 5.1 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.1 | 1.8 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.1 | 3.2 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.1 | 3.6 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.1 | 1.3 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.1 | 2.7 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.1 | 1.6 | REACTOME NGF SIGNALLING VIA TRKA FROM THE PLASMA MEMBRANE | Genes involved in NGF signalling via TRKA from the plasma membrane |
| 0.1 | 0.8 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.8 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 0.7 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.0 | 0.5 | REACTOME G ALPHA1213 SIGNALLING EVENTS | Genes involved in G alpha (12/13) signalling events |
| 0.0 | 0.7 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.8 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.0 | 1.5 | REACTOME ENERGY DEPENDENT REGULATION OF MTOR BY LKB1 AMPK | Genes involved in Energy dependent regulation of mTOR by LKB1-AMPK |
| 0.0 | 2.4 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 1.3 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 1.4 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.8 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.8 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 5.1 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.7 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 1.1 | REACTOME SEMA4D IN SEMAPHORIN SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 1.2 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 1.7 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.5 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.3 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 1.3 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 1.3 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.2 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.6 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |