Project
Illumina Body Map 2, young vs old
Navigation
Downloads
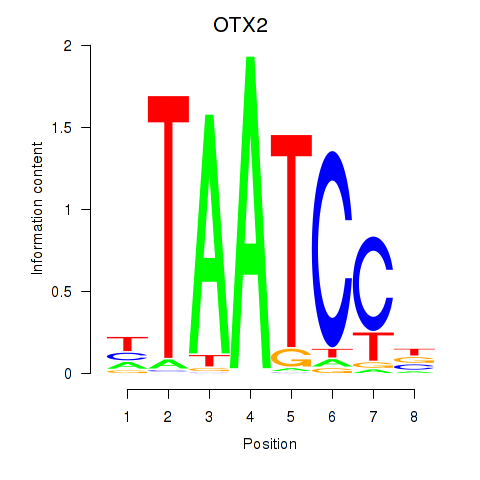
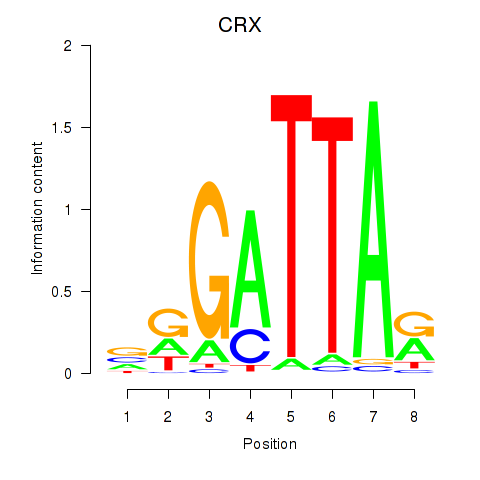
Results for OTX2_CRX
Z-value: 0.63
Transcription factors associated with OTX2_CRX
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
OTX2
|
ENSG00000165588.12 | orthodenticle homeobox 2 |
|
CRX
|
ENSG00000105392.11 | cone-rod homeobox |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| OTX2 | hg19_v2_chr14_-_57277163_57277168 | 0.54 | 1.6e-03 | Click! |
| CRX | hg19_v2_chr19_+_48325097_48325211 | 0.32 | 7.1e-02 | Click! |
Activity profile of OTX2_CRX motif
Sorted Z-values of OTX2_CRX motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr18_+_32290218 | 5.02 |
ENST00000348997.5
ENST00000588949.1 ENST00000597599.1 |
DTNA
|
dystrobrevin, alpha |
| chr12_+_6949964 | 3.22 |
ENST00000541978.1
ENST00000435982.2 |
GNB3
|
guanine nucleotide binding protein (G protein), beta polypeptide 3 |
| chr3_-_179691866 | 2.72 |
ENST00000464614.1
ENST00000476138.1 ENST00000463761.1 |
PEX5L
|
peroxisomal biogenesis factor 5-like |
| chr18_-_40857493 | 2.66 |
ENST00000255224.3
|
SYT4
|
synaptotagmin IV |
| chr15_-_27184664 | 2.18 |
ENST00000541819.2
|
GABRB3
|
gamma-aminobutyric acid (GABA) A receptor, beta 3 |
| chr1_+_169079823 | 2.16 |
ENST00000367813.3
|
ATP1B1
|
ATPase, Na+/K+ transporting, beta 1 polypeptide |
| chrX_-_15619076 | 2.15 |
ENST00000252519.3
|
ACE2
|
angiotensin I converting enzyme 2 |
| chr22_-_38380543 | 2.06 |
ENST00000396884.2
|
SOX10
|
SRY (sex determining region Y)-box 10 |
| chr18_-_64271316 | 1.94 |
ENST00000540086.1
ENST00000580157.1 |
CDH19
|
cadherin 19, type 2 |
| chr17_+_68101117 | 1.94 |
ENST00000587698.1
ENST00000587892.1 |
KCNJ16
|
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr18_-_70532906 | 1.80 |
ENST00000299430.2
ENST00000397929.1 |
NETO1
|
neuropilin (NRP) and tolloid (TLL)-like 1 |
| chr3_-_179692042 | 1.76 |
ENST00000468741.1
|
PEX5L
|
peroxisomal biogenesis factor 5-like |
| chr2_-_175711133 | 1.74 |
ENST00000409597.1
ENST00000413882.1 |
CHN1
|
chimerin 1 |
| chr18_-_64271363 | 1.73 |
ENST00000262150.2
|
CDH19
|
cadherin 19, type 2 |
| chr14_-_22005343 | 1.72 |
ENST00000327430.3
|
SALL2
|
spalt-like transcription factor 2 |
| chr6_+_54173227 | 1.72 |
ENST00000259782.4
ENST00000370864.3 |
TINAG
|
tubulointerstitial nephritis antigen |
| chr6_+_34433844 | 1.64 |
ENST00000244458.2
ENST00000374043.2 |
PACSIN1
|
protein kinase C and casein kinase substrate in neurons 1 |
| chr5_-_149669192 | 1.59 |
ENST00000398376.3
|
CAMK2A
|
calcium/calmodulin-dependent protein kinase II alpha |
| chr1_-_165414414 | 1.47 |
ENST00000359842.5
|
RXRG
|
retinoid X receptor, gamma |
| chr5_+_161494770 | 1.43 |
ENST00000414552.2
ENST00000361925.4 |
GABRG2
|
gamma-aminobutyric acid (GABA) A receptor, gamma 2 |
| chr8_+_104892639 | 1.42 |
ENST00000436393.2
|
RIMS2
|
regulating synaptic membrane exocytosis 2 |
| chr5_+_161494521 | 1.41 |
ENST00000356592.3
|
GABRG2
|
gamma-aminobutyric acid (GABA) A receptor, gamma 2 |
| chr6_-_42690312 | 1.41 |
ENST00000230381.5
|
PRPH2
|
peripherin 2 (retinal degeneration, slow) |
| chr14_-_22005018 | 1.41 |
ENST00000546363.1
|
SALL2
|
spalt-like transcription factor 2 |
| chr17_+_48911942 | 1.40 |
ENST00000426127.1
|
WFIKKN2
|
WAP, follistatin/kazal, immunoglobulin, kunitz and netrin domain containing 2 |
| chr14_-_22005197 | 1.36 |
ENST00000541965.1
|
SALL2
|
spalt-like transcription factor 2 |
| chr5_-_24645078 | 1.35 |
ENST00000264463.4
|
CDH10
|
cadherin 10, type 2 (T2-cadherin) |
| chr17_-_38859996 | 1.34 |
ENST00000264651.2
|
KRT24
|
keratin 24 |
| chr10_+_18240834 | 1.31 |
ENST00000377371.3
ENST00000539911.1 |
SLC39A12
|
solute carrier family 39 (zinc transporter), member 12 |
| chr8_+_70378852 | 1.31 |
ENST00000525061.1
ENST00000458141.2 ENST00000260128.4 |
SULF1
|
sulfatase 1 |
| chr5_-_95768973 | 1.31 |
ENST00000311106.3
|
PCSK1
|
proprotein convertase subtilisin/kexin type 1 |
| chr14_-_22005062 | 1.31 |
ENST00000317492.5
|
SALL2
|
spalt-like transcription factor 2 |
| chr10_+_18240753 | 1.29 |
ENST00000377369.2
|
SLC39A12
|
solute carrier family 39 (zinc transporter), member 12 |
| chr12_+_40787194 | 1.27 |
ENST00000425730.2
ENST00000454784.4 |
MUC19
|
mucin 19, oligomeric |
| chr11_-_62752429 | 1.18 |
ENST00000377871.3
|
SLC22A6
|
solute carrier family 22 (organic anion transporter), member 6 |
| chr11_-_62752455 | 1.18 |
ENST00000360421.4
|
SLC22A6
|
solute carrier family 22 (organic anion transporter), member 6 |
| chr11_-_62752162 | 1.18 |
ENST00000458333.2
ENST00000421062.2 |
SLC22A6
|
solute carrier family 22 (organic anion transporter), member 6 |
| chr5_-_41794313 | 1.18 |
ENST00000512084.1
|
OXCT1
|
3-oxoacid CoA transferase 1 |
| chr5_-_41510725 | 1.17 |
ENST00000328457.3
|
PLCXD3
|
phosphatidylinositol-specific phospholipase C, X domain containing 3 |
| chr15_+_43885252 | 1.17 |
ENST00000453782.1
ENST00000300283.6 ENST00000437924.1 ENST00000450086.2 |
CKMT1B
|
creatine kinase, mitochondrial 1B |
| chr17_+_68100989 | 1.16 |
ENST00000585558.1
ENST00000392670.1 |
KCNJ16
|
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr13_+_36050881 | 1.16 |
ENST00000537702.1
|
NBEA
|
neurobeachin |
| chr15_+_33603147 | 1.14 |
ENST00000415757.3
ENST00000389232.4 |
RYR3
|
ryanodine receptor 3 |
| chr8_-_91095099 | 1.13 |
ENST00000265431.3
|
CALB1
|
calbindin 1, 28kDa |
| chr1_+_159141397 | 1.13 |
ENST00000368124.4
ENST00000368125.4 ENST00000416746.1 |
CADM3
|
cell adhesion molecule 3 |
| chr15_+_43985084 | 1.10 |
ENST00000434505.1
ENST00000411750.1 |
CKMT1A
|
creatine kinase, mitochondrial 1A |
| chr10_+_18240814 | 1.08 |
ENST00000377374.4
|
SLC39A12
|
solute carrier family 39 (zinc transporter), member 12 |
| chr10_-_21186144 | 1.05 |
ENST00000377119.1
|
NEBL
|
nebulette |
| chr11_-_111794446 | 1.04 |
ENST00000527950.1
|
CRYAB
|
crystallin, alpha B |
| chr11_-_111783595 | 1.04 |
ENST00000528628.1
|
CRYAB
|
crystallin, alpha B |
| chr18_-_21891460 | 1.03 |
ENST00000357041.4
|
OSBPL1A
|
oxysterol binding protein-like 1A |
| chr3_+_173302222 | 1.02 |
ENST00000361589.4
|
NLGN1
|
neuroligin 1 |
| chr10_+_95848824 | 1.00 |
ENST00000371385.3
ENST00000371375.1 |
PLCE1
|
phospholipase C, epsilon 1 |
| chr18_-_40857447 | 0.99 |
ENST00000590752.1
ENST00000596867.1 |
SYT4
|
synaptotagmin IV |
| chr17_+_74536115 | 0.96 |
ENST00000592014.1
|
PRCD
|
progressive rod-cone degeneration |
| chr14_-_101034407 | 0.96 |
ENST00000443071.2
ENST00000557378.1 |
BEGAIN
|
brain-enriched guanylate kinase-associated |
| chr9_+_103204553 | 0.96 |
ENST00000502978.1
ENST00000334943.6 |
MSANTD3-TMEFF1
TMEFF1
|
MSANTD3-TMEFF1 readthrough transmembrane protein with EGF-like and two follistatin-like domains 1 |
| chr12_-_16758059 | 0.94 |
ENST00000261169.6
|
LMO3
|
LIM domain only 3 (rhombotin-like 2) |
| chr13_-_36050819 | 0.93 |
ENST00000379919.4
|
MAB21L1
|
mab-21-like 1 (C. elegans) |
| chrX_-_21676442 | 0.92 |
ENST00000379499.2
|
KLHL34
|
kelch-like family member 34 |
| chr8_+_70379072 | 0.92 |
ENST00000529134.1
|
SULF1
|
sulfatase 1 |
| chr16_+_58010339 | 0.91 |
ENST00000290871.5
ENST00000441824.2 |
TEPP
|
testis, prostate and placenta expressed |
| chr1_+_209757051 | 0.91 |
ENST00000009105.1
ENST00000423146.1 ENST00000361322.2 |
CAMK1G
|
calcium/calmodulin-dependent protein kinase IG |
| chr12_-_16758304 | 0.91 |
ENST00000320122.6
|
LMO3
|
LIM domain only 3 (rhombotin-like 2) |
| chr9_+_12693336 | 0.90 |
ENST00000381137.2
ENST00000388918.5 |
TYRP1
|
tyrosinase-related protein 1 |
| chr12_+_78224667 | 0.89 |
ENST00000549464.1
|
NAV3
|
neuron navigator 3 |
| chr7_-_83824169 | 0.89 |
ENST00000265362.4
|
SEMA3A
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chrX_-_6146876 | 0.89 |
ENST00000381095.3
|
NLGN4X
|
neuroligin 4, X-linked |
| chr8_+_134029937 | 0.89 |
ENST00000518108.1
|
TG
|
thyroglobulin |
| chr5_-_41510656 | 0.89 |
ENST00000377801.3
|
PLCXD3
|
phosphatidylinositol-specific phospholipase C, X domain containing 3 |
| chr5_+_50678921 | 0.88 |
ENST00000230658.7
|
ISL1
|
ISL LIM homeobox 1 |
| chr15_+_54901540 | 0.87 |
ENST00000539562.2
|
UNC13C
|
unc-13 homolog C (C. elegans) |
| chrX_+_17653413 | 0.85 |
ENST00000398097.3
|
NHS
|
Nance-Horan syndrome (congenital cataracts and dental anomalies) |
| chr10_-_116418053 | 0.84 |
ENST00000277895.5
|
ABLIM1
|
actin binding LIM protein 1 |
| chr13_-_62001982 | 0.84 |
ENST00000409186.1
|
PCDH20
|
protocadherin 20 |
| chr16_-_58004992 | 0.81 |
ENST00000564448.1
ENST00000251102.8 ENST00000311183.4 |
CNGB1
|
cyclic nucleotide gated channel beta 1 |
| chr7_+_73868220 | 0.80 |
ENST00000455841.2
|
GTF2IRD1
|
GTF2I repeat domain containing 1 |
| chr8_+_54764346 | 0.78 |
ENST00000297313.3
ENST00000344277.6 |
RGS20
|
regulator of G-protein signaling 20 |
| chr6_-_154568815 | 0.76 |
ENST00000519344.1
|
IPCEF1
|
interaction protein for cytohesin exchange factors 1 |
| chr2_+_219472488 | 0.75 |
ENST00000450993.2
|
PLCD4
|
phospholipase C, delta 4 |
| chrX_+_28605516 | 0.75 |
ENST00000378993.1
|
IL1RAPL1
|
interleukin 1 receptor accessory protein-like 1 |
| chr10_-_86001210 | 0.75 |
ENST00000372105.3
|
LRIT1
|
leucine-rich repeat, immunoglobulin-like and transmembrane domains 1 |
| chr11_-_36619771 | 0.75 |
ENST00000311485.3
ENST00000527033.1 ENST00000532616.1 |
RAG2
|
recombination activating gene 2 |
| chr14_-_101034811 | 0.74 |
ENST00000553553.1
|
BEGAIN
|
brain-enriched guanylate kinase-associated |
| chr18_+_616672 | 0.74 |
ENST00000338387.7
|
CLUL1
|
clusterin-like 1 (retinal) |
| chr10_-_104178857 | 0.74 |
ENST00000020673.5
|
PSD
|
pleckstrin and Sec7 domain containing |
| chr16_+_81272287 | 0.74 |
ENST00000425577.2
ENST00000564552.1 |
BCMO1
|
beta-carotene 15,15'-monooxygenase 1 |
| chr3_-_149095652 | 0.73 |
ENST00000305366.3
|
TM4SF1
|
transmembrane 4 L six family member 1 |
| chr5_-_36301984 | 0.73 |
ENST00000502994.1
ENST00000515759.1 ENST00000296604.3 |
RANBP3L
|
RAN binding protein 3-like |
| chr1_-_102312517 | 0.72 |
ENST00000338858.5
|
OLFM3
|
olfactomedin 3 |
| chr9_-_28670283 | 0.72 |
ENST00000379992.2
|
LINGO2
|
leucine rich repeat and Ig domain containing 2 |
| chr10_-_61122934 | 0.71 |
ENST00000512919.1
|
FAM13C
|
family with sequence similarity 13, member C |
| chr1_+_20396649 | 0.70 |
ENST00000375108.3
|
PLA2G5
|
phospholipase A2, group V |
| chr18_+_616711 | 0.68 |
ENST00000579494.1
|
CLUL1
|
clusterin-like 1 (retinal) |
| chr20_-_18774614 | 0.68 |
ENST00000412553.1
|
LINC00652
|
long intergenic non-protein coding RNA 652 |
| chr22_-_38480100 | 0.67 |
ENST00000427592.1
|
SLC16A8
|
solute carrier family 16 (monocarboxylate transporter), member 8 |
| chr1_+_202431859 | 0.67 |
ENST00000391959.3
ENST00000367270.4 |
PPP1R12B
|
protein phosphatase 1, regulatory subunit 12B |
| chr3_+_4535025 | 0.65 |
ENST00000302640.8
ENST00000354582.6 ENST00000423119.2 ENST00000357086.4 ENST00000456211.2 |
ITPR1
|
inositol 1,4,5-trisphosphate receptor, type 1 |
| chr2_+_219472637 | 0.65 |
ENST00000417849.1
|
PLCD4
|
phospholipase C, delta 4 |
| chr2_+_115219171 | 0.64 |
ENST00000409163.1
|
DPP10
|
dipeptidyl-peptidase 10 (non-functional) |
| chr6_+_155537771 | 0.64 |
ENST00000275246.7
|
TIAM2
|
T-cell lymphoma invasion and metastasis 2 |
| chr11_-_71752571 | 0.63 |
ENST00000544238.1
|
NUMA1
|
nuclear mitotic apparatus protein 1 |
| chr19_-_55865908 | 0.62 |
ENST00000590900.1
|
COX6B2
|
cytochrome c oxidase subunit VIb polypeptide 2 (testis) |
| chr1_-_39395165 | 0.61 |
ENST00000372985.3
|
RHBDL2
|
rhomboid, veinlet-like 2 (Drosophila) |
| chr7_-_17598506 | 0.61 |
ENST00000451792.1
|
AC017060.1
|
AC017060.1 |
| chr1_+_66999268 | 0.60 |
ENST00000371039.1
ENST00000424320.1 |
SGIP1
|
SH3-domain GRB2-like (endophilin) interacting protein 1 |
| chr18_-_40857329 | 0.60 |
ENST00000593720.1
|
SYT4
|
synaptotagmin IV |
| chr3_-_9291063 | 0.58 |
ENST00000383836.3
|
SRGAP3
|
SLIT-ROBO Rho GTPase activating protein 3 |
| chr9_+_34992846 | 0.58 |
ENST00000443266.1
|
DNAJB5
|
DnaJ (Hsp40) homolog, subfamily B, member 5 |
| chr2_-_30144432 | 0.58 |
ENST00000389048.3
|
ALK
|
anaplastic lymphoma receptor tyrosine kinase |
| chr7_+_123295861 | 0.57 |
ENST00000458573.2
ENST00000456238.2 |
LMOD2
|
leiomodin 2 (cardiac) |
| chr11_-_71753188 | 0.57 |
ENST00000543009.1
|
NUMA1
|
nuclear mitotic apparatus protein 1 |
| chr1_-_117664317 | 0.57 |
ENST00000256649.4
ENST00000369464.3 ENST00000485032.1 |
TRIM45
|
tripartite motif containing 45 |
| chr1_+_180875711 | 0.57 |
ENST00000434447.1
|
RP11-46A10.2
|
RP11-46A10.2 |
| chr2_+_170366203 | 0.56 |
ENST00000284669.1
|
KLHL41
|
kelch-like family member 41 |
| chr3_-_48229846 | 0.56 |
ENST00000302506.3
ENST00000351231.3 ENST00000437972.1 |
CDC25A
|
cell division cycle 25A |
| chr1_+_171939240 | 0.56 |
ENST00000523513.1
|
DNM3
|
dynamin 3 |
| chr1_+_144989309 | 0.56 |
ENST00000596396.1
|
AL590452.1
|
Uncharacterized protein |
| chrX_-_33357558 | 0.56 |
ENST00000288447.4
|
DMD
|
dystrophin |
| chr9_-_39288092 | 0.55 |
ENST00000323947.7
ENST00000297668.6 ENST00000377656.2 ENST00000377659.1 |
CNTNAP3
|
contactin associated protein-like 3 |
| chr6_-_154568551 | 0.54 |
ENST00000519190.1
|
IPCEF1
|
interaction protein for cytohesin exchange factors 1 |
| chr18_-_35145689 | 0.54 |
ENST00000591287.1
ENST00000601019.1 ENST00000601392.1 |
CELF4
|
CUGBP, Elav-like family member 4 |
| chr8_-_114449112 | 0.54 |
ENST00000455883.2
ENST00000352409.3 ENST00000297405.5 |
CSMD3
|
CUB and Sushi multiple domains 3 |
| chr4_+_154125565 | 0.53 |
ENST00000338700.5
|
TRIM2
|
tripartite motif containing 2 |
| chr12_-_86650045 | 0.52 |
ENST00000604798.1
|
MGAT4C
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme C (putative) |
| chr10_-_69455873 | 0.51 |
ENST00000433211.2
|
CTNNA3
|
catenin (cadherin-associated protein), alpha 3 |
| chr17_-_43209862 | 0.51 |
ENST00000322765.5
|
PLCD3
|
phospholipase C, delta 3 |
| chr12_-_66275350 | 0.50 |
ENST00000536648.1
|
RP11-366L20.2
|
Uncharacterized protein |
| chr7_-_83824449 | 0.50 |
ENST00000420047.1
|
SEMA3A
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chr1_+_164528866 | 0.50 |
ENST00000420696.2
|
PBX1
|
pre-B-cell leukemia homeobox 1 |
| chr13_-_49975632 | 0.50 |
ENST00000457041.1
ENST00000355854.4 |
CAB39L
|
calcium binding protein 39-like |
| chr11_-_71752838 | 0.50 |
ENST00000537930.1
|
NUMA1
|
nuclear mitotic apparatus protein 1 |
| chr6_+_148593425 | 0.50 |
ENST00000367469.1
|
SASH1
|
SAM and SH3 domain containing 1 |
| chr3_+_111393501 | 0.49 |
ENST00000393934.3
|
PLCXD2
|
phosphatidylinositol-specific phospholipase C, X domain containing 2 |
| chr11_-_128894053 | 0.49 |
ENST00000392657.3
|
ARHGAP32
|
Rho GTPase activating protein 32 |
| chr1_-_102312600 | 0.49 |
ENST00000359814.3
|
OLFM3
|
olfactomedin 3 |
| chr12_-_13248598 | 0.49 |
ENST00000337630.6
ENST00000545699.1 |
GSG1
|
germ cell associated 1 |
| chr12_-_13248562 | 0.48 |
ENST00000457134.2
ENST00000537302.1 |
GSG1
|
germ cell associated 1 |
| chr5_-_11589131 | 0.48 |
ENST00000511377.1
|
CTNND2
|
catenin (cadherin-associated protein), delta 2 |
| chr7_+_123241908 | 0.48 |
ENST00000434204.1
ENST00000437535.1 ENST00000451215.1 |
ASB15
|
ankyrin repeat and SOCS box containing 15 |
| chr9_-_8857776 | 0.48 |
ENST00000481079.1
|
PTPRD
|
protein tyrosine phosphatase, receptor type, D |
| chr5_-_11588907 | 0.48 |
ENST00000513598.1
ENST00000503622.1 |
CTNND2
|
catenin (cadherin-associated protein), delta 2 |
| chr17_+_12626199 | 0.48 |
ENST00000609971.1
|
AC005358.1
|
AC005358.1 |
| chr5_-_88178964 | 0.48 |
ENST00000513252.1
ENST00000508569.1 ENST00000510942.1 ENST00000506554.1 |
MEF2C
|
myocyte enhancer factor 2C |
| chr4_+_96012614 | 0.48 |
ENST00000264568.4
|
BMPR1B
|
bone morphogenetic protein receptor, type IB |
| chr1_+_164528616 | 0.48 |
ENST00000340699.3
|
PBX1
|
pre-B-cell leukemia homeobox 1 |
| chr17_-_43210580 | 0.48 |
ENST00000538093.1
ENST00000590644.1 |
PLCD3
|
phospholipase C, delta 3 |
| chr17_-_39661849 | 0.48 |
ENST00000246635.3
ENST00000336861.3 ENST00000587544.1 ENST00000587435.1 |
KRT13
|
keratin 13 |
| chr4_+_96012585 | 0.47 |
ENST00000502683.1
|
BMPR1B
|
bone morphogenetic protein receptor, type IB |
| chr5_-_58882219 | 0.47 |
ENST00000505453.1
ENST00000360047.5 |
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chr17_-_9808887 | 0.47 |
ENST00000226193.5
|
RCVRN
|
recoverin |
| chr21_-_19775973 | 0.46 |
ENST00000284885.3
|
TMPRSS15
|
transmembrane protease, serine 15 |
| chr10_+_52750930 | 0.46 |
ENST00000401604.2
|
PRKG1
|
protein kinase, cGMP-dependent, type I |
| chr3_-_50605150 | 0.45 |
ENST00000357203.3
|
C3orf18
|
chromosome 3 open reading frame 18 |
| chr18_-_35145593 | 0.45 |
ENST00000334919.5
ENST00000591282.1 ENST00000588597.1 |
CELF4
|
CUGBP, Elav-like family member 4 |
| chr15_-_74495188 | 0.43 |
ENST00000563965.1
ENST00000395105.4 |
STRA6
|
stimulated by retinoic acid 6 |
| chr5_+_63461642 | 0.43 |
ENST00000296615.6
ENST00000381081.2 ENST00000389100.4 |
RNF180
|
ring finger protein 180 |
| chr11_-_128712362 | 0.43 |
ENST00000392664.2
|
KCNJ1
|
potassium inwardly-rectifying channel, subfamily J, member 1 |
| chr6_-_35480640 | 0.43 |
ENST00000428978.1
ENST00000322263.4 |
TULP1
|
tubby like protein 1 |
| chr11_-_30608413 | 0.42 |
ENST00000528686.1
|
MPPED2
|
metallophosphoesterase domain containing 2 |
| chr6_-_139613269 | 0.42 |
ENST00000358430.3
|
TXLNB
|
taxilin beta |
| chr17_-_36499693 | 0.42 |
ENST00000342292.4
|
GPR179
|
G protein-coupled receptor 179 |
| chr5_+_50679506 | 0.42 |
ENST00000511384.1
|
ISL1
|
ISL LIM homeobox 1 |
| chr3_-_50605077 | 0.42 |
ENST00000426034.1
ENST00000441239.1 |
C3orf18
|
chromosome 3 open reading frame 18 |
| chr14_+_23846328 | 0.42 |
ENST00000382809.2
|
CMTM5
|
CKLF-like MARVEL transmembrane domain containing 5 |
| chr9_+_75229616 | 0.41 |
ENST00000340019.3
|
TMC1
|
transmembrane channel-like 1 |
| chr12_-_86650154 | 0.41 |
ENST00000552435.2
|
MGAT4C
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme C (putative) |
| chr15_+_63050785 | 0.41 |
ENST00000472902.1
|
TLN2
|
talin 2 |
| chr3_+_69915385 | 0.40 |
ENST00000314589.5
|
MITF
|
microphthalmia-associated transcription factor |
| chrX_+_135279179 | 0.40 |
ENST00000370676.3
|
FHL1
|
four and a half LIM domains 1 |
| chr12_-_55375622 | 0.40 |
ENST00000316577.8
|
TESPA1
|
thymocyte expressed, positive selection associated 1 |
| chr9_-_13279589 | 0.40 |
ENST00000319217.7
|
MPDZ
|
multiple PDZ domain protein |
| chr7_+_73868439 | 0.39 |
ENST00000424337.2
|
GTF2IRD1
|
GTF2I repeat domain containing 1 |
| chr12_-_67197760 | 0.39 |
ENST00000539540.1
ENST00000540433.1 ENST00000541947.1 ENST00000538373.1 |
GRIP1
|
glutamate receptor interacting protein 1 |
| chr9_+_82188077 | 0.39 |
ENST00000425506.1
|
TLE4
|
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) |
| chr15_-_35047166 | 0.39 |
ENST00000290374.4
|
GJD2
|
gap junction protein, delta 2, 36kDa |
| chr19_+_39989580 | 0.39 |
ENST00000596614.1
ENST00000205143.4 |
DLL3
|
delta-like 3 (Drosophila) |
| chr19_+_50183032 | 0.38 |
ENST00000527412.1
|
PRMT1
|
protein arginine methyltransferase 1 |
| chr7_+_73868120 | 0.38 |
ENST00000265755.3
|
GTF2IRD1
|
GTF2I repeat domain containing 1 |
| chr5_+_150040403 | 0.37 |
ENST00000517768.1
ENST00000297130.4 |
MYOZ3
|
myozenin 3 |
| chr3_-_47950745 | 0.37 |
ENST00000429422.1
|
MAP4
|
microtubule-associated protein 4 |
| chr5_-_33984741 | 0.37 |
ENST00000382102.3
ENST00000509381.1 ENST00000342059.3 ENST00000345083.5 |
SLC45A2
|
solute carrier family 45, member 2 |
| chr2_-_214016314 | 0.36 |
ENST00000434687.1
ENST00000374319.4 |
IKZF2
|
IKAROS family zinc finger 2 (Helios) |
| chr15_-_34610962 | 0.36 |
ENST00000290209.5
|
SLC12A6
|
solute carrier family 12 (potassium/chloride transporter), member 6 |
| chr12_-_21556577 | 0.36 |
ENST00000450590.1
ENST00000453443.1 |
SLCO1A2
|
solute carrier organic anion transporter family, member 1A2 |
| chr13_-_95131923 | 0.36 |
ENST00000377028.5
ENST00000446125.1 |
DCT
|
dopachrome tautomerase |
| chr19_-_44123734 | 0.36 |
ENST00000598676.1
|
ZNF428
|
zinc finger protein 428 |
| chr10_-_49482907 | 0.36 |
ENST00000374201.3
ENST00000407470.4 |
FRMPD2
|
FERM and PDZ domain containing 2 |
| chr19_-_44124019 | 0.36 |
ENST00000300811.3
|
ZNF428
|
zinc finger protein 428 |
| chr7_+_136553824 | 0.36 |
ENST00000320658.5
ENST00000453373.1 ENST00000397608.3 ENST00000402486.3 ENST00000401861.1 |
CHRM2
|
cholinergic receptor, muscarinic 2 |
| chr16_-_325910 | 0.35 |
ENST00000359740.5
ENST00000316163.5 ENST00000431291.2 ENST00000397770.3 ENST00000397768.3 |
RGS11
|
regulator of G-protein signaling 11 |
| chr3_-_98241358 | 0.35 |
ENST00000503004.1
ENST00000506575.1 ENST00000513452.1 ENST00000515620.1 |
CLDND1
|
claudin domain containing 1 |
| chr2_+_55746722 | 0.35 |
ENST00000339012.3
|
CCDC104
|
coiled-coil domain containing 104 |
| chr18_-_35145728 | 0.35 |
ENST00000361795.5
ENST00000603232.1 |
CELF4
|
CUGBP, Elav-like family member 4 |
| chr10_+_102505468 | 0.35 |
ENST00000361791.3
ENST00000355243.3 ENST00000428433.1 ENST00000370296.2 |
PAX2
|
paired box 2 |
| chrX_-_100129128 | 0.35 |
ENST00000372960.4
ENST00000372964.1 ENST00000217885.5 |
NOX1
|
NADPH oxidase 1 |
| chr5_-_524445 | 0.34 |
ENST00000514375.1
ENST00000264938.3 |
SLC9A3
|
solute carrier family 9, subfamily A (NHE3, cation proton antiporter 3), member 3 |
| chr8_-_87755878 | 0.34 |
ENST00000320005.5
|
CNGB3
|
cyclic nucleotide gated channel beta 3 |
| chr11_-_40315640 | 0.34 |
ENST00000278198.2
|
LRRC4C
|
leucine rich repeat containing 4C |
| chr5_-_88179017 | 0.34 |
ENST00000514028.1
ENST00000514015.1 ENST00000503075.1 ENST00000437473.2 |
MEF2C
|
myocyte enhancer factor 2C |
Network of associatons between targets according to the STRING database.
First level regulatory network of OTX2_CRX
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 4.2 | GO:0048174 | negative regulation of short-term neuronal synaptic plasticity(GO:0048174) |
| 1.2 | 3.5 | GO:0097254 | renal tubular secretion(GO:0097254) |
| 0.5 | 2.1 | GO:0071393 | cellular response to progesterone stimulus(GO:0071393) |
| 0.4 | 2.1 | GO:0003051 | angiotensin-mediated drinking behavior(GO:0003051) |
| 0.4 | 4.5 | GO:0016560 | protein import into peroxisome matrix, docking(GO:0016560) |
| 0.4 | 1.8 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.3 | 1.4 | GO:1903045 | neural crest cell migration involved in sympathetic nervous system development(GO:1903045) |
| 0.3 | 1.3 | GO:0048936 | visceral motor neuron differentiation(GO:0021524) peripheral nervous system neuron axonogenesis(GO:0048936) cardiac cell fate determination(GO:0060913) positive regulation of granulocyte colony-stimulating factor production(GO:0071657) positive regulation of macrophage colony-stimulating factor production(GO:1901258) |
| 0.3 | 2.2 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.3 | 1.6 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.3 | 2.2 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) positive regulation of calcium:sodium antiporter activity(GO:1903281) |
| 0.3 | 0.9 | GO:0043438 | acetoacetic acid metabolic process(GO:0043438) |
| 0.3 | 1.0 | GO:0098942 | cytoskeletal matrix organization at active zone(GO:0048789) neurexin clustering involved in presynaptic membrane assembly(GO:0097115) retrograde trans-synaptic signaling by trans-synaptic protein complex(GO:0098942) |
| 0.2 | 5.0 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.2 | 2.1 | GO:0032888 | regulation of spindle elongation(GO:0032887) regulation of mitotic spindle elongation(GO:0032888) anastral spindle assembly(GO:0055048) protein localization to spindle pole body(GO:0071988) regulation of protein localization to spindle pole body(GO:1902363) positive regulation of protein localization to spindle pole body(GO:1902365) positive regulation of mitotic spindle elongation(GO:1902846) |
| 0.2 | 0.6 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.2 | 0.7 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.2 | 1.2 | GO:0003185 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.2 | 1.0 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.2 | 1.8 | GO:0097105 | presynaptic membrane assembly(GO:0097105) |
| 0.2 | 3.7 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.1 | 1.2 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.1 | 0.6 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.1 | 0.9 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.1 | 1.5 | GO:0072025 | distal convoluted tubule development(GO:0072025) metanephric distal convoluted tubule development(GO:0072221) |
| 0.1 | 1.8 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.1 | 0.5 | GO:0015770 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.1 | 1.3 | GO:1902866 | regulation of retina development in camera-type eye(GO:1902866) |
| 0.1 | 1.4 | GO:0061669 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.1 | 0.7 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.1 | 0.6 | GO:0002331 | pre-B cell allelic exclusion(GO:0002331) |
| 0.1 | 1.6 | GO:0038166 | angiotensin-activated signaling pathway(GO:0038166) |
| 0.1 | 0.7 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.1 | 1.1 | GO:0071286 | cellular response to magnesium ion(GO:0071286) |
| 0.1 | 0.9 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.1 | 0.3 | GO:0045726 | positive regulation of integrin biosynthetic process(GO:0045726) |
| 0.1 | 5.0 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.1 | 0.6 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 0.1 | 0.3 | GO:1905205 | positive regulation of connective tissue replacement(GO:1905205) |
| 0.1 | 1.8 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.1 | 0.4 | GO:0042539 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.1 | 2.3 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.1 | 0.4 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.1 | 0.5 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.1 | 1.1 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.1 | 0.9 | GO:0015705 | iodide transport(GO:0015705) |
| 0.1 | 0.5 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.1 | 0.4 | GO:1901631 | positive regulation of presynaptic membrane organization(GO:1901631) positive regulation of synaptic vesicle clustering(GO:2000809) |
| 0.1 | 1.5 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.1 | 0.2 | GO:2000824 | negative regulation of androgen receptor activity(GO:2000824) |
| 0.1 | 0.4 | GO:0010387 | COP9 signalosome assembly(GO:0010387) |
| 0.1 | 3.5 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.1 | 0.5 | GO:0086073 | bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 0.1 | 0.4 | GO:0046985 | positive regulation of hemoglobin biosynthetic process(GO:0046985) |
| 0.1 | 0.4 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.1 | 0.6 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.1 | 0.8 | GO:0060087 | relaxation of vascular smooth muscle(GO:0060087) |
| 0.1 | 0.4 | GO:0021847 | ventricular zone neuroblast division(GO:0021847) |
| 0.1 | 0.8 | GO:0031284 | positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.0 | 0.7 | GO:0015727 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) |
| 0.0 | 0.5 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.0 | 0.6 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.0 | 3.2 | GO:0071377 | cellular response to glucagon stimulus(GO:0071377) |
| 0.0 | 0.4 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.0 | 0.2 | GO:0008616 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.0 | 0.4 | GO:0042415 | norepinephrine metabolic process(GO:0042415) |
| 0.0 | 0.4 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) |
| 0.0 | 0.2 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 0.0 | 0.1 | GO:1903980 | negative regulation of macrophage colony-stimulating factor signaling pathway(GO:1902227) negative regulation of response to macrophage colony-stimulating factor(GO:1903970) negative regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903973) positive regulation of microglial cell activation(GO:1903980) |
| 0.0 | 0.3 | GO:0048859 | formation of anatomical boundary(GO:0048859) |
| 0.0 | 1.5 | GO:0035162 | embryonic hemopoiesis(GO:0035162) |
| 0.0 | 1.7 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 0.6 | GO:0031272 | regulation of pseudopodium assembly(GO:0031272) |
| 0.0 | 0.6 | GO:0043587 | tongue morphogenesis(GO:0043587) |
| 0.0 | 0.5 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.0 | 1.0 | GO:0060716 | labyrinthine layer blood vessel development(GO:0060716) |
| 0.0 | 1.3 | GO:1901522 | positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus(GO:1901522) |
| 0.0 | 0.4 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.0 | 1.1 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.0 | 0.4 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.0 | 6.1 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.4 | GO:0015824 | proline transport(GO:0015824) |
| 0.0 | 0.1 | GO:0033122 | negative regulation of cyclic nucleotide catabolic process(GO:0030806) negative regulation of cAMP catabolic process(GO:0030821) negative regulation of purine nucleotide catabolic process(GO:0033122) |
| 0.0 | 0.4 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.0 | 1.4 | GO:0007340 | acrosome reaction(GO:0007340) |
| 0.0 | 0.2 | GO:0019918 | peptidyl-arginine methylation, to symmetrical-dimethyl arginine(GO:0019918) |
| 0.0 | 0.2 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.0 | 0.3 | GO:0021860 | pyramidal neuron development(GO:0021860) |
| 0.0 | 0.7 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.1 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 0.0 | 0.2 | GO:0045905 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.0 | 0.1 | GO:1903625 | negative regulation of DNA catabolic process(GO:1903625) |
| 0.0 | 0.2 | GO:0032252 | secretory granule localization(GO:0032252) |
| 0.0 | 0.3 | GO:1990118 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.3 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.0 | 0.4 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 | 0.4 | GO:0060044 | negative regulation of cardiac muscle cell proliferation(GO:0060044) |
| 0.0 | 1.5 | GO:0048747 | muscle fiber development(GO:0048747) |
| 0.0 | 0.3 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.0 | 0.3 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.0 | 1.0 | GO:0006699 | bile acid biosynthetic process(GO:0006699) |
| 0.0 | 0.7 | GO:0030318 | melanocyte differentiation(GO:0030318) |
| 0.0 | 0.3 | GO:0051256 | mitotic spindle midzone assembly(GO:0051256) |
| 0.0 | 0.3 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.0 | 0.2 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.0 | 0.2 | GO:0032494 | response to peptidoglycan(GO:0032494) |
| 0.0 | 0.6 | GO:1902751 | positive regulation of cell cycle G2/M phase transition(GO:1902751) |
| 0.0 | 0.3 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.3 | GO:0006878 | cellular copper ion homeostasis(GO:0006878) |
| 0.0 | 7.1 | GO:0001654 | eye development(GO:0001654) |
| 0.0 | 0.7 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.0 | 0.3 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.0 | 0.5 | GO:0008037 | cell recognition(GO:0008037) |
| 0.0 | 0.2 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.0 | 0.2 | GO:0007097 | nuclear migration(GO:0007097) |
| 0.0 | 1.0 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.0 | 1.3 | GO:1901379 | regulation of potassium ion transmembrane transport(GO:1901379) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 4.2 | GO:0032127 | dense core granule membrane(GO:0032127) |
| 0.2 | 2.1 | GO:0055028 | cortical microtubule(GO:0055028) |
| 0.2 | 0.6 | GO:0098844 | postsynaptic endocytic zone membrane(GO:0098844) |
| 0.1 | 5.8 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.1 | 5.0 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.1 | 1.8 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.1 | 2.1 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.1 | 0.6 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.1 | 0.9 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.1 | 0.9 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.1 | 2.2 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.1 | 1.1 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.1 | 1.7 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.1 | 0.9 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 1.8 | GO:0030137 | COPI-coated vesicle(GO:0030137) |
| 0.0 | 0.6 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.0 | 2.1 | GO:0031312 | extrinsic component of organelle membrane(GO:0031312) |
| 0.0 | 0.3 | GO:0097441 | basilar dendrite(GO:0097441) |
| 0.0 | 1.0 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 4.5 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 0.3 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.0 | 0.2 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 0.2 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 0.0 | 0.4 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.0 | 0.4 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 0.6 | GO:0034709 | methylosome(GO:0034709) |
| 0.0 | 0.4 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.0 | 1.2 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.1 | GO:0035841 | new growing cell tip(GO:0035841) |
| 0.0 | 0.2 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.0 | 1.4 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.5 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 0.3 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.0 | 1.7 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 5.0 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.0 | 2.8 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 0.8 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 3.5 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 0.5 | GO:0043220 | Schmidt-Lanterman incisure(GO:0043220) |
| 0.0 | 0.2 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.0 | 0.6 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.0 | 1.6 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 1.7 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 1.2 | GO:0031672 | A band(GO:0031672) |
| 0.0 | 0.7 | GO:0030660 | Golgi-associated vesicle membrane(GO:0030660) |
| 0.0 | 0.2 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.4 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 1.9 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.4 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.4 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 0.3 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 0.2 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.0 | 1.1 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 7.9 | GO:0030425 | dendrite(GO:0030425) |
| 0.0 | 2.2 | GO:0005795 | Golgi stack(GO:0005795) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 4.5 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.4 | 1.5 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.3 | 3.7 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.3 | 1.2 | GO:0008260 | 3-oxoacid CoA-transferase activity(GO:0008260) |
| 0.3 | 1.1 | GO:0048763 | calcium-induced calcium release activity(GO:0048763) |
| 0.3 | 2.1 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.2 | 0.9 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.2 | 2.2 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.2 | 4.4 | GO:0031404 | chloride ion binding(GO:0031404) |
| 0.2 | 0.6 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.2 | 2.8 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.2 | 0.6 | GO:0031798 | type 1 metabotropic glutamate receptor binding(GO:0031798) |
| 0.2 | 2.2 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 0.2 | 0.7 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.2 | 1.1 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.2 | 0.8 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.2 | 0.8 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.1 | 1.3 | GO:0042835 | BRE binding(GO:0042835) |
| 0.1 | 1.3 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.1 | 3.1 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.1 | 0.5 | GO:0015154 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.1 | 1.1 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.1 | 0.8 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.1 | 0.2 | GO:0004947 | bradykinin receptor activity(GO:0004947) |
| 0.1 | 2.1 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.1 | 2.2 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.1 | 0.9 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.1 | 3.4 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.1 | 3.7 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 1.5 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.1 | 1.7 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.1 | 0.4 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.1 | 0.4 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.1 | 0.6 | GO:0044020 | histone methyltransferase activity (H4-R3 specific)(GO:0044020) |
| 0.1 | 1.4 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 0.4 | GO:0004167 | dopachrome isomerase activity(GO:0004167) |
| 0.1 | 0.7 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.1 | 1.8 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 0.3 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.1 | 0.6 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.1 | 1.7 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.1 | 0.3 | GO:0005223 | intracellular cGMP activated cation channel activity(GO:0005223) |
| 0.1 | 1.2 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.1 | 0.3 | GO:0008665 | 2'-phosphotransferase activity(GO:0008665) |
| 0.1 | 0.4 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.1 | 0.7 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 2.9 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 3.2 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 1.6 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.5 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.0 | 0.5 | GO:0045029 | UDP-activated nucleotide receptor activity(GO:0045029) |
| 0.0 | 0.4 | GO:0030267 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.0 | 1.4 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.2 | GO:0031208 | POZ domain binding(GO:0031208) |
| 0.0 | 0.1 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.0 | 1.1 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.7 | GO:0019966 | interleukin-1 binding(GO:0019966) |
| 0.0 | 0.3 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.0 | 5.9 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.2 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.0 | 0.8 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 0.6 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.0 | 0.4 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.0 | 1.7 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 1.8 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.0 | 0.4 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 0.7 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.4 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.3 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 1.0 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 0.7 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.4 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 0.3 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.5 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.3 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.6 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.9 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.3 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.0 | 0.1 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.2 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 1.2 | GO:0070063 | RNA polymerase binding(GO:0070063) |
| 0.0 | 0.1 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.0 | 0.4 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.4 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.0 | 0.5 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.4 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.0 | 1.3 | GO:0008081 | phosphoric diester hydrolase activity(GO:0008081) |
| 0.0 | 0.2 | GO:0016918 | retinal binding(GO:0016918) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.8 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 1.9 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 0.6 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 1.5 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 1.1 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 1.3 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 2.4 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 1.8 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 0.8 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.0 | 2.1 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.8 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 0.9 | PID BMP PATHWAY | BMP receptor signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 5.8 | REACTOME INCRETIN SYNTHESIS SECRETION AND INACTIVATION | Genes involved in Incretin Synthesis, Secretion, and Inactivation |
| 0.1 | 5.0 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.1 | 3.5 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.1 | 2.1 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.1 | 1.4 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 1.6 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.0 | 0.6 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.0 | 2.8 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 2.1 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.7 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 1.3 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.4 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.0 | 1.3 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.8 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 1.2 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 0.9 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.5 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.3 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 0.8 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 1.0 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.5 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.0 | 0.4 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.8 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.8 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.4 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.7 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.0 | 0.6 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |