Project
Illumina Body Map 2, young vs old
Navigation
Downloads
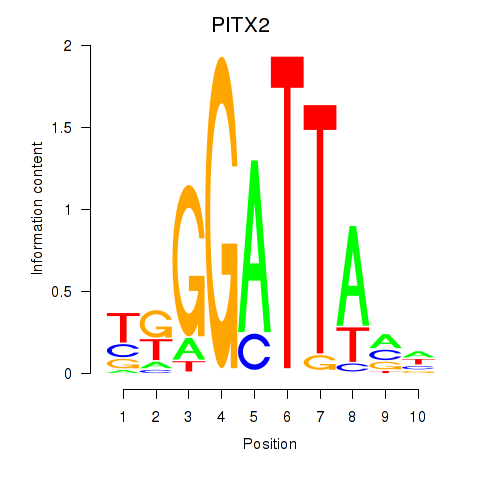
Results for PITX2
Z-value: 0.21
Transcription factors associated with PITX2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PITX2
|
ENSG00000164093.11 | paired like homeodomain 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| PITX2 | hg19_v2_chr4_-_111544207_111544240 | 0.12 | 5.1e-01 | Click! |
Activity profile of PITX2 motif
Sorted Z-values of PITX2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_+_34458771 | 1.55 |
ENST00000437363.1
ENST00000242317.4 |
DNAI1
|
dynein, axonemal, intermediate chain 1 |
| chr10_-_49482907 | 1.31 |
ENST00000374201.3
ENST00000407470.4 |
FRMPD2
|
FERM and PDZ domain containing 2 |
| chr15_+_54901540 | 1.26 |
ENST00000539562.2
|
UNC13C
|
unc-13 homolog C (C. elegans) |
| chr9_+_34458851 | 1.04 |
ENST00000545019.1
|
DNAI1
|
dynein, axonemal, intermediate chain 1 |
| chr11_+_61522844 | 0.94 |
ENST00000265460.5
|
MYRF
|
myelin regulatory factor |
| chr10_+_18240834 | 0.89 |
ENST00000377371.3
ENST00000539911.1 |
SLC39A12
|
solute carrier family 39 (zinc transporter), member 12 |
| chr10_+_18240814 | 0.88 |
ENST00000377374.4
|
SLC39A12
|
solute carrier family 39 (zinc transporter), member 12 |
| chr9_-_8857776 | 0.87 |
ENST00000481079.1
|
PTPRD
|
protein tyrosine phosphatase, receptor type, D |
| chr10_+_18240753 | 0.86 |
ENST00000377369.2
|
SLC39A12
|
solute carrier family 39 (zinc transporter), member 12 |
| chr9_+_12693336 | 0.85 |
ENST00000381137.2
ENST00000388918.5 |
TYRP1
|
tyrosinase-related protein 1 |
| chr4_-_36245561 | 0.73 |
ENST00000506189.1
|
ARAP2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
| chr11_-_36619771 | 0.68 |
ENST00000311485.3
ENST00000527033.1 ENST00000532616.1 |
RAG2
|
recombination activating gene 2 |
| chr13_-_36429763 | 0.66 |
ENST00000379893.1
|
DCLK1
|
doublecortin-like kinase 1 |
| chr7_-_122339162 | 0.66 |
ENST00000340112.2
|
RNF133
|
ring finger protein 133 |
| chr15_-_89764929 | 0.62 |
ENST00000268125.5
|
RLBP1
|
retinaldehyde binding protein 1 |
| chr16_-_5116025 | 0.58 |
ENST00000472572.3
ENST00000315997.5 ENST00000422873.1 ENST00000350219.4 |
C16orf89
|
chromosome 16 open reading frame 89 |
| chr16_-_68014732 | 0.56 |
ENST00000268793.4
|
DPEP3
|
dipeptidase 3 |
| chr18_-_3880051 | 0.55 |
ENST00000584874.1
|
DLGAP1
|
discs, large (Drosophila) homolog-associated protein 1 |
| chr1_+_92417716 | 0.51 |
ENST00000402388.1
|
BRDT
|
bromodomain, testis-specific |
| chr3_+_119421849 | 0.49 |
ENST00000273390.5
ENST00000463700.1 |
MAATS1
|
MYCBP-associated, testis expressed 1 |
| chr19_-_51893827 | 0.49 |
ENST00000574814.1
|
CTD-2616J11.4
|
chromosome 19 open reading frame 84 |
| chr19_-_51893782 | 0.47 |
ENST00000570516.1
|
CTD-2616J11.4
|
chromosome 19 open reading frame 84 |
| chr20_+_44350968 | 0.46 |
ENST00000279058.3
|
SPINT4
|
serine peptidase inhibitor, Kunitz type 4 |
| chr12_-_90049828 | 0.46 |
ENST00000261173.2
ENST00000348959.3 |
ATP2B1
|
ATPase, Ca++ transporting, plasma membrane 1 |
| chr1_+_205538165 | 0.46 |
ENST00000536357.1
|
MFSD4
|
major facilitator superfamily domain containing 4 |
| chr1_+_2407754 | 0.44 |
ENST00000419816.2
ENST00000378486.3 ENST00000378488.3 ENST00000288766.5 |
PLCH2
|
phospholipase C, eta 2 |
| chr12_-_90049878 | 0.43 |
ENST00000359142.3
|
ATP2B1
|
ATPase, Ca++ transporting, plasma membrane 1 |
| chr6_+_43215727 | 0.43 |
ENST00000304139.5
|
TTBK1
|
tau tubulin kinase 1 |
| chr6_+_46661575 | 0.38 |
ENST00000450697.1
|
TDRD6
|
tudor domain containing 6 |
| chr13_-_95131923 | 0.36 |
ENST00000377028.5
ENST00000446125.1 |
DCT
|
dopachrome tautomerase |
| chr11_-_5248294 | 0.36 |
ENST00000335295.4
|
HBB
|
hemoglobin, beta |
| chr17_+_4643337 | 0.32 |
ENST00000592813.1
|
ZMYND15
|
zinc finger, MYND-type containing 15 |
| chr3_-_49837254 | 0.31 |
ENST00000412678.2
ENST00000343366.4 ENST00000487256.1 |
CDHR4
|
cadherin-related family member 4 |
| chr8_-_42360015 | 0.31 |
ENST00000522707.1
|
SLC20A2
|
solute carrier family 20 (phosphate transporter), member 2 |
| chr1_-_165414414 | 0.31 |
ENST00000359842.5
|
RXRG
|
retinoid X receptor, gamma |
| chr5_-_64920115 | 0.30 |
ENST00000381018.3
ENST00000274327.7 |
TRIM23
|
tripartite motif containing 23 |
| chr3_+_158787041 | 0.29 |
ENST00000471575.1
ENST00000476809.1 ENST00000485419.1 |
IQCJ-SCHIP1
|
IQCJ-SCHIP1 readthrough |
| chr8_-_10512569 | 0.26 |
ENST00000382483.3
|
RP1L1
|
retinitis pigmentosa 1-like 1 |
| chr17_+_4643300 | 0.26 |
ENST00000433935.1
|
ZMYND15
|
zinc finger, MYND-type containing 15 |
| chr17_+_79495397 | 0.25 |
ENST00000417245.2
ENST00000334850.7 |
FSCN2
|
fascin homolog 2, actin-bundling protein, retinal (Strongylocentrotus purpuratus) |
| chr17_-_40021656 | 0.25 |
ENST00000319121.3
|
KLHL11
|
kelch-like family member 11 |
| chr3_-_69249863 | 0.23 |
ENST00000478263.1
ENST00000462512.1 |
FRMD4B
|
FERM domain containing 4B |
| chr10_-_61900762 | 0.21 |
ENST00000355288.2
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G) |
| chr17_-_4643161 | 0.21 |
ENST00000574412.1
|
CXCL16
|
chemokine (C-X-C motif) ligand 16 |
| chr16_+_58010339 | 0.20 |
ENST00000290871.5
ENST00000441824.2 |
TEPP
|
testis, prostate and placenta expressed |
| chr1_-_98515395 | 0.17 |
ENST00000424528.2
|
MIR137HG
|
MIR137 host gene (non-protein coding) |
| chr4_-_68620053 | 0.17 |
ENST00000420975.2
ENST00000226413.4 |
GNRHR
|
gonadotropin-releasing hormone receptor |
| chr12_+_40787194 | 0.16 |
ENST00000425730.2
ENST00000454784.4 |
MUC19
|
mucin 19, oligomeric |
| chr5_-_92023923 | 0.16 |
ENST00000503489.1
|
CTC-529L17.2
|
CTC-529L17.2 |
| chr19_+_2476116 | 0.15 |
ENST00000215631.4
ENST00000587345.1 |
GADD45B
|
growth arrest and DNA-damage-inducible, beta |
| chr2_+_120770581 | 0.15 |
ENST00000263713.5
|
EPB41L5
|
erythrocyte membrane protein band 4.1 like 5 |
| chr2_+_120770645 | 0.14 |
ENST00000443902.2
|
EPB41L5
|
erythrocyte membrane protein band 4.1 like 5 |
| chr9_-_98268883 | 0.13 |
ENST00000551630.1
ENST00000548420.1 |
PTCH1
|
patched 1 |
| chr5_+_64920543 | 0.13 |
ENST00000399438.3
ENST00000510585.2 |
TRAPPC13
CTC-534A2.2
|
trafficking protein particle complex 13 CDNA FLJ26957 fis, clone SLV00486; Uncharacterized protein |
| chr19_-_46142362 | 0.11 |
ENST00000586770.1
ENST00000591721.1 |
EML2
|
echinoderm microtubule associated protein like 2 |
| chr1_+_22328144 | 0.10 |
ENST00000290122.3
ENST00000374663.1 |
CELA3A
|
chymotrypsin-like elastase family, member 3A |
| chr1_+_180875711 | 0.10 |
ENST00000434447.1
|
RP11-46A10.2
|
RP11-46A10.2 |
| chr7_+_73868220 | 0.09 |
ENST00000455841.2
|
GTF2IRD1
|
GTF2I repeat domain containing 1 |
| chrX_-_48931648 | 0.08 |
ENST00000376386.3
ENST00000376390.4 |
PRAF2
|
PRA1 domain family, member 2 |
| chr10_+_103986085 | 0.08 |
ENST00000370005.3
|
ELOVL3
|
ELOVL fatty acid elongase 3 |
| chr7_+_73868439 | 0.08 |
ENST00000424337.2
|
GTF2IRD1
|
GTF2I repeat domain containing 1 |
| chrX_-_21676442 | 0.08 |
ENST00000379499.2
|
KLHL34
|
kelch-like family member 34 |
| chr21_-_19775973 | 0.08 |
ENST00000284885.3
|
TMPRSS15
|
transmembrane protease, serine 15 |
| chr8_+_123875624 | 0.07 |
ENST00000534247.1
|
ZHX2
|
zinc fingers and homeoboxes 2 |
| chr9_-_127358087 | 0.06 |
ENST00000475178.1
|
NR6A1
|
nuclear receptor subfamily 6, group A, member 1 |
| chrX_-_6146876 | 0.06 |
ENST00000381095.3
|
NLGN4X
|
neuroligin 4, X-linked |
| chr2_+_120770686 | 0.05 |
ENST00000331393.4
ENST00000443124.1 |
EPB41L5
|
erythrocyte membrane protein band 4.1 like 5 |
| chr16_+_28722684 | 0.05 |
ENST00000331666.6
ENST00000395587.1 ENST00000569690.1 ENST00000564243.1 |
EIF3C
|
eukaryotic translation initiation factor 3, subunit C |
| chr8_-_27462822 | 0.05 |
ENST00000522098.1
|
CLU
|
clusterin |
| chr3_+_158787098 | 0.04 |
ENST00000397832.2
ENST00000451172.1 ENST00000482126.1 |
IQCJ
|
IQ motif containing J |
| chr16_-_12070468 | 0.03 |
ENST00000538896.1
|
RP11-166B2.1
|
Putative NPIP-like protein LOC729978 |
| chr3_+_139062838 | 0.03 |
ENST00000310776.4
ENST00000465056.1 ENST00000465373.1 |
MRPS22
|
mitochondrial ribosomal protein S22 |
| chr2_-_121223697 | 0.02 |
ENST00000593290.1
|
FLJ14816
|
long intergenic non-protein coding RNA 1101 |
| chr4_+_70916119 | 0.01 |
ENST00000246896.3
ENST00000511674.1 |
HTN1
|
histatin 1 |
| chr1_+_22303503 | 0.00 |
ENST00000337107.6
|
CELA3B
|
chymotrypsin-like elastase family, member 3B |
| chr4_-_76912070 | 0.00 |
ENST00000395711.4
ENST00000356260.5 |
SDAD1
|
SDA1 domain containing 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of PITX2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0043438 | acetoacetic acid metabolic process(GO:0043438) |
| 0.2 | 0.5 | GO:0051039 | histone displacement(GO:0001207) positive regulation of transcription involved in meiotic cell cycle(GO:0051039) |
| 0.1 | 1.3 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.1 | 2.6 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.1 | 0.7 | GO:0002331 | pre-B cell allelic exclusion(GO:0002331) |
| 0.1 | 0.9 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.1 | 2.6 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.1 | 0.9 | GO:1990034 | cellular response to corticosterone stimulus(GO:0071386) calcium ion export from cell(GO:1990034) |
| 0.1 | 0.4 | GO:0030185 | nitric oxide transport(GO:0030185) |
| 0.1 | 1.0 | GO:0032286 | central nervous system myelin maintenance(GO:0032286) |
| 0.1 | 0.3 | GO:0007509 | mesoderm migration involved in gastrulation(GO:0007509) |
| 0.1 | 0.4 | GO:0021847 | ventricular zone neuroblast division(GO:0021847) |
| 0.0 | 0.2 | GO:1900827 | maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.0 | 0.2 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.0 | 0.5 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.0 | 0.1 | GO:0010157 | response to chlorate(GO:0010157) |
| 0.0 | 0.2 | GO:0097210 | response to gonadotropin-releasing hormone(GO:0097210) cellular response to gonadotropin-releasing hormone(GO:0097211) |
| 0.0 | 0.3 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.0 | 0.7 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.0 | 1.3 | GO:0070830 | bicellular tight junction assembly(GO:0070830) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.6 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.2 | 0.5 | GO:0001534 | radial spoke(GO:0001534) |
| 0.1 | 1.3 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.1 | 0.4 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 1.2 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 0.9 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.4 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.0 | 0.1 | GO:0044294 | dendritic growth cone(GO:0044294) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.1 | 2.6 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.1 | 0.6 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.1 | 0.3 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.1 | 0.4 | GO:0004167 | dopachrome isomerase activity(GO:0004167) |
| 0.1 | 0.4 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.1 | 0.2 | GO:0004968 | gonadotropin-releasing hormone receptor activity(GO:0004968) |
| 0.1 | 2.6 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 1.3 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.3 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.0 | 1.3 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.9 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.9 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.7 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.1 | GO:0005119 | smoothened binding(GO:0005119) |
| 0.0 | 0.6 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.7 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 0.5 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.4 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |