Project
Illumina Body Map 2, young vs old
Navigation
Downloads
Results for PROX1
Z-value: 0.57
Transcription factors associated with PROX1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PROX1
|
ENSG00000117707.11 | prospero homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| PROX1 | hg19_v2_chr1_+_214161272_214161322 | -0.17 | 3.6e-01 | Click! |
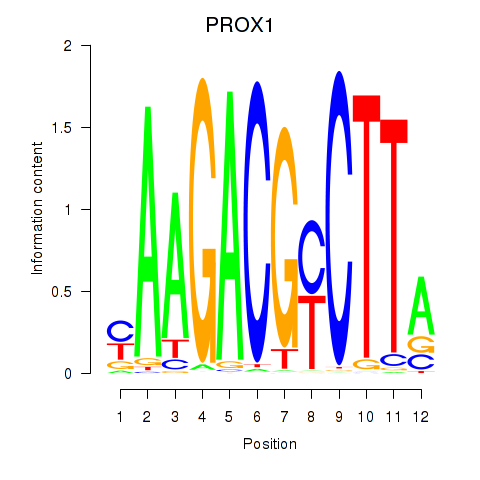
Activity profile of PROX1 motif
Sorted Z-values of PROX1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_+_22180536 | 4.91 |
ENST00000390424.2
|
TRAV2
|
T cell receptor alpha variable 2 |
| chr14_+_22409308 | 4.89 |
ENST00000390441.2
|
TRAV9-2
|
T cell receptor alpha variable 9-2 |
| chr5_+_156569944 | 4.81 |
ENST00000521769.1
|
ITK
|
IL2-inducible T-cell kinase |
| chr14_+_22320634 | 4.21 |
ENST00000390435.1
|
TRAV8-3
|
T cell receptor alpha variable 8-3 |
| chr12_+_113229737 | 3.75 |
ENST00000551052.1
ENST00000415485.3 |
RPH3A
|
rabphilin 3A homolog (mouse) |
| chr12_+_113229543 | 3.23 |
ENST00000447659.2
|
RPH3A
|
rabphilin 3A homolog (mouse) |
| chr14_+_22788560 | 3.18 |
ENST00000390468.1
|
TRAV41
|
T cell receptor alpha variable 41 |
| chr1_-_160832490 | 2.97 |
ENST00000322302.7
ENST00000368033.3 |
CD244
|
CD244 molecule, natural killer cell receptor 2B4 |
| chr1_-_160832642 | 2.92 |
ENST00000368034.4
|
CD244
|
CD244 molecule, natural killer cell receptor 2B4 |
| chr12_+_113229452 | 2.87 |
ENST00000389385.4
|
RPH3A
|
rabphilin 3A homolog (mouse) |
| chr5_-_156569850 | 2.83 |
ENST00000524219.1
|
HAVCR2
|
hepatitis A virus cellular receptor 2 |
| chr1_-_160832670 | 2.62 |
ENST00000368032.2
|
CD244
|
CD244 molecule, natural killer cell receptor 2B4 |
| chr9_+_93564191 | 2.47 |
ENST00000375747.1
|
SYK
|
spleen tyrosine kinase |
| chr4_-_170947485 | 2.44 |
ENST00000504999.1
|
MFAP3L
|
microfibrillar-associated protein 3-like |
| chr14_+_22458631 | 2.17 |
ENST00000390444.1
|
TRAV16
|
T cell receptor alpha variable 16 |
| chr1_-_159046617 | 2.15 |
ENST00000368130.4
|
AIM2
|
absent in melanoma 2 |
| chr5_+_156693159 | 2.10 |
ENST00000347377.6
|
CYFIP2
|
cytoplasmic FMR1 interacting protein 2 |
| chr19_+_15752088 | 2.09 |
ENST00000585846.1
|
CYP4F3
|
cytochrome P450, family 4, subfamily F, polypeptide 3 |
| chr3_-_112218205 | 2.03 |
ENST00000383680.4
|
BTLA
|
B and T lymphocyte associated |
| chr5_-_157002775 | 2.02 |
ENST00000257527.4
|
ADAM19
|
ADAM metallopeptidase domain 19 |
| chr10_+_45869652 | 2.01 |
ENST00000542434.1
ENST00000374391.2 |
ALOX5
|
arachidonate 5-lipoxygenase |
| chr3_-_112218378 | 2.00 |
ENST00000334529.5
|
BTLA
|
B and T lymphocyte associated |
| chr4_-_170947522 | 1.88 |
ENST00000361618.3
|
MFAP3L
|
microfibrillar-associated protein 3-like |
| chr6_+_37475109 | 1.86 |
ENST00000570443.2
|
RP1-153P14.8
|
RP1-153P14.8 |
| chr3_-_130745403 | 1.83 |
ENST00000504725.1
ENST00000509060.1 |
ASTE1
|
asteroid homolog 1 (Drosophila) |
| chr1_+_159770292 | 1.81 |
ENST00000536257.1
ENST00000321935.6 |
FCRL6
|
Fc receptor-like 6 |
| chr4_-_170947446 | 1.79 |
ENST00000507601.1
ENST00000512698.1 |
MFAP3L
|
microfibrillar-associated protein 3-like |
| chr1_-_173176452 | 1.77 |
ENST00000281834.3
|
TNFSF4
|
tumor necrosis factor (ligand) superfamily, member 4 |
| chr11_+_73360024 | 1.73 |
ENST00000540431.1
|
PLEKHB1
|
pleckstrin homology domain containing, family B (evectins) member 1 |
| chr17_+_72209653 | 1.71 |
ENST00000269346.4
|
TTYH2
|
tweety family member 2 |
| chr6_-_25042231 | 1.70 |
ENST00000510784.2
|
FAM65B
|
family with sequence similarity 65, member B |
| chr10_+_134258649 | 1.68 |
ENST00000392630.3
ENST00000321248.2 |
C10orf91
|
chromosome 10 open reading frame 91 |
| chr1_+_207669573 | 1.64 |
ENST00000400960.2
ENST00000534202.1 |
CR1
|
complement component (3b/4b) receptor 1 (Knops blood group) |
| chr4_-_170948361 | 1.61 |
ENST00000393702.3
|
MFAP3L
|
microfibrillar-associated protein 3-like |
| chr2_-_239140011 | 1.61 |
ENST00000409376.1
ENST00000409070.1 ENST00000409942.1 |
AC016757.3
|
Protein LOC151174 |
| chrY_+_22918021 | 1.59 |
ENST00000288666.5
|
RPS4Y2
|
ribosomal protein S4, Y-linked 2 |
| chr19_+_52255261 | 1.58 |
ENST00000600258.1
|
FPR2
|
formyl peptide receptor 2 |
| chr5_-_157002749 | 1.56 |
ENST00000517905.1
ENST00000430702.2 ENST00000394020.1 |
ADAM19
|
ADAM metallopeptidase domain 19 |
| chr2_-_152830479 | 1.48 |
ENST00000360283.6
|
CACNB4
|
calcium channel, voltage-dependent, beta 4 subunit |
| chr11_+_73359936 | 1.47 |
ENST00000542389.1
|
PLEKHB1
|
pleckstrin homology domain containing, family B (evectins) member 1 |
| chr10_-_121296045 | 1.43 |
ENST00000392865.1
|
RGS10
|
regulator of G-protein signaling 10 |
| chr2_+_163175394 | 1.42 |
ENST00000446271.1
ENST00000429691.2 |
GCA
|
grancalcin, EF-hand calcium binding protein |
| chr16_+_50300427 | 1.39 |
ENST00000394697.2
ENST00000566433.2 ENST00000538642.1 |
ADCY7
|
adenylate cyclase 7 |
| chr22_+_24990746 | 1.37 |
ENST00000456869.1
ENST00000411974.1 |
GGT1
|
gamma-glutamyltransferase 1 |
| chr9_+_125133315 | 1.31 |
ENST00000223423.4
ENST00000362012.2 |
PTGS1
|
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) |
| chr2_-_233877912 | 1.31 |
ENST00000264051.3
|
NGEF
|
neuronal guanine nucleotide exchange factor |
| chr15_+_81489213 | 1.31 |
ENST00000559383.1
ENST00000394660.2 |
IL16
|
interleukin 16 |
| chr19_+_917287 | 1.30 |
ENST00000592648.1
ENST00000234371.5 |
KISS1R
|
KISS1 receptor |
| chr3_-_130745571 | 1.25 |
ENST00000514044.1
ENST00000264992.3 |
ASTE1
|
asteroid homolog 1 (Drosophila) |
| chr4_-_48683188 | 1.25 |
ENST00000505759.1
|
FRYL
|
FRY-like |
| chr11_-_126870655 | 1.23 |
ENST00000525144.2
|
KIRREL3
|
kin of IRRE like 3 (Drosophila) |
| chr11_-_111749767 | 1.21 |
ENST00000542429.1
|
FDXACB1
|
ferredoxin-fold anticodon binding domain containing 1 |
| chr14_+_23013015 | 1.19 |
ENST00000390535.2
|
TRAJ2
|
T cell receptor alpha joining 2 (non-functional) |
| chr1_-_109203685 | 1.18 |
ENST00000402983.1
ENST00000420055.1 |
HENMT1
|
HEN1 methyltransferase homolog 1 (Arabidopsis) |
| chr7_-_143105941 | 1.17 |
ENST00000275815.3
|
EPHA1
|
EPH receptor A1 |
| chr9_+_125132803 | 1.16 |
ENST00000540753.1
|
PTGS1
|
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) |
| chr19_+_35773242 | 1.15 |
ENST00000222304.3
|
HAMP
|
hepcidin antimicrobial peptide |
| chr14_+_22782867 | 1.15 |
ENST00000390467.3
|
TRAV40
|
T cell receptor alpha variable 40 |
| chr1_+_241695670 | 1.13 |
ENST00000366557.4
|
KMO
|
kynurenine 3-monooxygenase (kynurenine 3-hydroxylase) |
| chr15_-_74726283 | 1.13 |
ENST00000543145.2
|
SEMA7A
|
semaphorin 7A, GPI membrane anchor (John Milton Hagen blood group) |
| chr5_+_173473662 | 1.12 |
ENST00000519717.1
|
NSG2
|
Neuron-specific protein family member 2 |
| chr20_-_61885826 | 1.11 |
ENST00000370316.3
|
NKAIN4
|
Na+/K+ transporting ATPase interacting 4 |
| chr17_+_8869157 | 1.11 |
ENST00000585297.1
|
CTB-41I6.1
|
CTB-41I6.1 |
| chr19_-_19754404 | 1.10 |
ENST00000587205.1
ENST00000445806.2 ENST00000203556.4 |
GMIP
|
GEM interacting protein |
| chr12_+_113229725 | 1.08 |
ENST00000551198.1
|
RPH3A
|
rabphilin 3A homolog (mouse) |
| chr6_-_134495992 | 1.08 |
ENST00000475719.2
ENST00000367857.5 ENST00000237305.7 |
SGK1
|
serum/glucocorticoid regulated kinase 1 |
| chr11_-_131533462 | 1.08 |
ENST00000416725.1
|
AP003039.3
|
AP003039.3 |
| chr12_+_113229708 | 1.07 |
ENST00000550901.1
|
RPH3A
|
rabphilin 3A homolog (mouse) |
| chr17_+_73629500 | 1.07 |
ENST00000375215.3
|
SMIM5
|
small integral membrane protein 5 |
| chr11_+_61722629 | 1.06 |
ENST00000526988.1
|
BEST1
|
bestrophin 1 |
| chr1_+_241695424 | 1.05 |
ENST00000366558.3
ENST00000366559.4 |
KMO
|
kynurenine 3-monooxygenase (kynurenine 3-hydroxylase) |
| chr19_-_19754354 | 1.05 |
ENST00000587238.1
|
GMIP
|
GEM interacting protein |
| chr12_+_121078355 | 1.04 |
ENST00000316803.3
|
CABP1
|
calcium binding protein 1 |
| chr2_+_28718921 | 1.03 |
ENST00000327757.5
ENST00000422425.2 ENST00000404858.1 |
PLB1
|
phospholipase B1 |
| chr12_+_10331605 | 1.02 |
ENST00000298530.3
|
TMEM52B
|
transmembrane protein 52B |
| chr6_-_35992270 | 1.00 |
ENST00000394602.2
ENST00000355574.2 |
SLC26A8
|
solute carrier family 26 (anion exchanger), member 8 |
| chr4_-_170947565 | 0.99 |
ENST00000506764.1
|
MFAP3L
|
microfibrillar-associated protein 3-like |
| chr1_-_109203648 | 0.99 |
ENST00000370031.1
|
HENMT1
|
HEN1 methyltransferase homolog 1 (Arabidopsis) |
| chr3_-_33138286 | 0.99 |
ENST00000416695.2
ENST00000342462.4 ENST00000399402.3 |
TMPPE
GLB1
|
transmembrane protein with metallophosphoesterase domain galactosidase, beta 1 |
| chr11_-_111749878 | 0.92 |
ENST00000260257.4
|
FDXACB1
|
ferredoxin-fold anticodon binding domain containing 1 |
| chr4_-_38666430 | 0.91 |
ENST00000436901.1
|
AC021860.1
|
Uncharacterized protein |
| chr21_-_46961716 | 0.91 |
ENST00000427839.1
|
SLC19A1
|
solute carrier family 19 (folate transporter), member 1 |
| chr2_-_54087066 | 0.91 |
ENST00000394705.2
ENST00000352846.3 ENST00000406625.2 |
GPR75
GPR75-ASB3
ASB3
|
G protein-coupled receptor 75 GPR75-ASB3 readthrough Ankyrin repeat and SOCS box protein 3 |
| chr9_+_132096166 | 0.90 |
ENST00000436710.1
|
RP11-65J3.1
|
RP11-65J3.1 |
| chr10_+_16478942 | 0.90 |
ENST00000535784.2
ENST00000423462.2 ENST00000378000.1 |
PTER
|
phosphotriesterase related |
| chr9_-_130497565 | 0.89 |
ENST00000336067.6
ENST00000373281.5 ENST00000373284.5 ENST00000458505.3 |
TOR2A
|
torsin family 2, member A |
| chr1_-_225616515 | 0.87 |
ENST00000338179.2
ENST00000425080.1 |
LBR
|
lamin B receptor |
| chr11_-_126870683 | 0.87 |
ENST00000525704.2
|
KIRREL3
|
kin of IRRE like 3 (Drosophila) |
| chr22_+_27042387 | 0.86 |
ENST00000421867.1
|
MIAT
|
myocardial infarction associated transcript (non-protein coding) |
| chr14_+_73603126 | 0.86 |
ENST00000557356.1
ENST00000556864.1 ENST00000556533.1 ENST00000556951.1 ENST00000557293.1 ENST00000553719.1 ENST00000553599.1 ENST00000556011.1 ENST00000394157.3 ENST00000357710.4 ENST00000324501.5 ENST00000560005.2 ENST00000555254.1 ENST00000261970.3 ENST00000344094.3 ENST00000554131.1 ENST00000557037.1 |
PSEN1
|
presenilin 1 |
| chr21_-_16125773 | 0.85 |
ENST00000454128.2
|
AF127936.3
|
AF127936.3 |
| chr14_-_50559361 | 0.85 |
ENST00000305273.1
|
C14orf183
|
chromosome 14 open reading frame 183 |
| chr7_+_18126557 | 0.85 |
ENST00000417496.2
|
HDAC9
|
histone deacetylase 9 |
| chr3_+_186915274 | 0.84 |
ENST00000312295.4
|
RTP1
|
receptor (chemosensory) transporter protein 1 |
| chr3_-_33138592 | 0.84 |
ENST00000415454.1
|
GLB1
|
galactosidase, beta 1 |
| chr2_+_131862900 | 0.84 |
ENST00000438882.2
ENST00000538982.1 ENST00000404460.1 |
PLEKHB2
|
pleckstrin homology domain containing, family B (evectins) member 2 |
| chr7_+_48128316 | 0.83 |
ENST00000341253.4
|
UPP1
|
uridine phosphorylase 1 |
| chr12_+_113229337 | 0.83 |
ENST00000552667.1
|
RPH3A
|
rabphilin 3A homolog (mouse) |
| chr3_-_33138624 | 0.81 |
ENST00000445488.2
ENST00000307377.8 ENST00000440656.1 ENST00000436768.1 ENST00000307363.5 |
GLB1
|
galactosidase, beta 1 |
| chr4_+_2819883 | 0.77 |
ENST00000511747.1
ENST00000503393.2 |
SH3BP2
|
SH3-domain binding protein 2 |
| chr11_+_92085262 | 0.76 |
ENST00000298047.6
ENST00000409404.2 ENST00000541502.1 |
FAT3
|
FAT atypical cadherin 3 |
| chr14_+_78266408 | 0.76 |
ENST00000238561.5
|
ADCK1
|
aarF domain containing kinase 1 |
| chr1_+_179262905 | 0.76 |
ENST00000539888.1
ENST00000540564.1 ENST00000535686.1 ENST00000367619.3 |
SOAT1
|
sterol O-acyltransferase 1 |
| chr2_+_131862872 | 0.76 |
ENST00000439822.2
|
PLEKHB2
|
pleckstrin homology domain containing, family B (evectins) member 2 |
| chr7_+_48128194 | 0.75 |
ENST00000416681.1
ENST00000331803.4 ENST00000432131.1 |
UPP1
|
uridine phosphorylase 1 |
| chr8_-_116681686 | 0.74 |
ENST00000519815.1
|
TRPS1
|
trichorhinophalangeal syndrome I |
| chr8_-_19615382 | 0.74 |
ENST00000544602.1
|
CSGALNACT1
|
chondroitin sulfate N-acetylgalactosaminyltransferase 1 |
| chr1_+_95975672 | 0.74 |
ENST00000440116.2
ENST00000456933.1 |
RP11-286B14.1
|
RP11-286B14.1 |
| chr11_+_1874200 | 0.73 |
ENST00000311604.3
|
LSP1
|
lymphocyte-specific protein 1 |
| chr5_-_180688105 | 0.72 |
ENST00000327767.4
|
TRIM52
|
tripartite motif containing 52 |
| chr4_+_26862400 | 0.72 |
ENST00000467011.1
ENST00000412829.2 |
STIM2
|
stromal interaction molecule 2 |
| chr3_-_18487057 | 0.72 |
ENST00000415069.1
|
SATB1
|
SATB homeobox 1 |
| chr14_+_45605127 | 0.71 |
ENST00000556036.1
ENST00000267430.5 |
FANCM
|
Fanconi anemia, complementation group M |
| chr7_+_64254793 | 0.70 |
ENST00000494380.1
ENST00000440155.2 ENST00000440598.1 ENST00000437743.1 |
ZNF138
|
zinc finger protein 138 |
| chr11_-_83436446 | 0.70 |
ENST00000529399.1
|
DLG2
|
discs, large homolog 2 (Drosophila) |
| chr17_+_41994576 | 0.68 |
ENST00000588043.2
|
FAM215A
|
family with sequence similarity 215, member A (non-protein coding) |
| chr12_-_11091862 | 0.68 |
ENST00000537503.1
|
TAS2R14
|
taste receptor, type 2, member 14 |
| chr2_-_239140276 | 0.68 |
ENST00000334973.4
|
AC016757.3
|
Protein LOC151174 |
| chr2_+_241499719 | 0.67 |
ENST00000405954.1
|
DUSP28
|
dual specificity phosphatase 28 |
| chr1_+_179263308 | 0.67 |
ENST00000426956.1
|
SOAT1
|
sterol O-acyltransferase 1 |
| chr11_-_46408107 | 0.67 |
ENST00000433765.2
|
CHRM4
|
cholinergic receptor, muscarinic 4 |
| chr11_+_61522844 | 0.67 |
ENST00000265460.5
|
MYRF
|
myelin regulatory factor |
| chr1_-_211307404 | 0.67 |
ENST00000367007.4
|
KCNH1
|
potassium voltage-gated channel, subfamily H (eag-related), member 1 |
| chr7_+_66093851 | 0.67 |
ENST00000275532.3
|
KCTD7
|
potassium channel tetramerization domain containing 7 |
| chr10_-_50970382 | 0.63 |
ENST00000419399.1
ENST00000432695.1 |
OGDHL
|
oxoglutarate dehydrogenase-like |
| chr5_+_162930114 | 0.63 |
ENST00000280969.5
|
MAT2B
|
methionine adenosyltransferase II, beta |
| chr8_-_19615538 | 0.62 |
ENST00000517494.1
|
CSGALNACT1
|
chondroitin sulfate N-acetylgalactosaminyltransferase 1 |
| chr19_+_53073526 | 0.62 |
ENST00000596514.1
ENST00000391785.3 ENST00000301093.2 |
ZNF701
|
zinc finger protein 701 |
| chr5_+_175490540 | 0.61 |
ENST00000515817.1
|
FAM153B
|
family with sequence similarity 153, member B |
| chr16_+_12070567 | 0.61 |
ENST00000566228.1
|
SNX29
|
sorting nexin 29 |
| chrX_+_118602363 | 0.60 |
ENST00000317881.8
|
SLC25A5
|
solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 5 |
| chr7_+_66093908 | 0.60 |
ENST00000443322.1
ENST00000449064.1 |
KCTD7
|
potassium channel tetramerization domain containing 7 |
| chr22_-_50050986 | 0.60 |
ENST00000405854.1
|
C22orf34
|
chromosome 22 open reading frame 34 |
| chr17_+_16593539 | 0.60 |
ENST00000340621.5
ENST00000399273.1 ENST00000443444.2 ENST00000360524.8 ENST00000456009.1 |
CCDC144A
|
coiled-coil domain containing 144A |
| chr10_-_98945264 | 0.60 |
ENST00000314867.5
|
SLIT1
|
slit homolog 1 (Drosophila) |
| chr10_-_50970322 | 0.58 |
ENST00000374103.4
|
OGDHL
|
oxoglutarate dehydrogenase-like |
| chr9_+_131267052 | 0.57 |
ENST00000539582.1
|
GLE1
|
GLE1 RNA export mediator |
| chr7_+_64254766 | 0.57 |
ENST00000307355.7
ENST00000359735.3 |
ZNF138
|
zinc finger protein 138 |
| chr3_+_190105909 | 0.57 |
ENST00000456423.1
|
CLDN16
|
claudin 16 |
| chr3_+_9839335 | 0.57 |
ENST00000453882.1
|
ARPC4-TTLL3
|
ARPC4-TTLL3 readthrough |
| chr19_-_9546227 | 0.56 |
ENST00000361451.2
ENST00000361151.1 |
ZNF266
|
zinc finger protein 266 |
| chr2_-_89310012 | 0.56 |
ENST00000493819.1
|
IGKV1-9
|
immunoglobulin kappa variable 1-9 |
| chr7_-_102257139 | 0.55 |
ENST00000521076.1
ENST00000462172.1 ENST00000522801.1 ENST00000449970.2 ENST00000262940.7 |
RASA4
|
RAS p21 protein activator 4 |
| chr14_+_45605157 | 0.54 |
ENST00000542564.2
|
FANCM
|
Fanconi anemia, complementation group M |
| chr12_-_122240792 | 0.54 |
ENST00000545885.1
ENST00000542933.1 ENST00000428029.2 ENST00000541694.1 ENST00000536662.1 ENST00000535643.1 ENST00000541657.1 |
AC084018.1
RHOF
|
AC084018.1 ras homolog family member F (in filopodia) |
| chr6_+_132455526 | 0.53 |
ENST00000443303.1
|
LINC01013
|
long intergenic non-protein coding RNA 1013 |
| chr19_+_23945768 | 0.53 |
ENST00000486528.1
ENST00000496398.1 |
RPSAP58
|
ribosomal protein SA pseudogene 58 |
| chr1_-_179457805 | 0.53 |
ENST00000600581.1
|
AL160286.1
|
Uncharacterized protein |
| chr6_-_8102279 | 0.53 |
ENST00000488226.2
|
EEF1E1
|
eukaryotic translation elongation factor 1 epsilon 1 |
| chr5_-_22853429 | 0.52 |
ENST00000504376.2
|
CDH12
|
cadherin 12, type 2 (N-cadherin 2) |
| chr1_+_10459111 | 0.52 |
ENST00000541529.1
ENST00000270776.8 ENST00000483936.1 ENST00000538557.1 |
PGD
|
phosphogluconate dehydrogenase |
| chr10_+_71075516 | 0.52 |
ENST00000436817.1
|
HK1
|
hexokinase 1 |
| chr4_-_128887069 | 0.51 |
ENST00000541133.1
ENST00000296468.3 ENST00000513559.1 |
MFSD8
|
major facilitator superfamily domain containing 8 |
| chr12_-_58240470 | 0.50 |
ENST00000548823.1
ENST00000398073.2 |
CTDSP2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 2 |
| chr11_-_83435965 | 0.50 |
ENST00000434967.1
ENST00000530800.1 |
DLG2
|
discs, large homolog 2 (Drosophila) |
| chr8_-_19615435 | 0.50 |
ENST00000523262.1
|
CSGALNACT1
|
chondroitin sulfate N-acetylgalactosaminyltransferase 1 |
| chr7_-_91509986 | 0.50 |
ENST00000456229.1
ENST00000442961.1 ENST00000406735.2 ENST00000419292.1 ENST00000351870.3 |
MTERF
|
mitochondrial transcription termination factor |
| chr4_+_26862431 | 0.50 |
ENST00000465503.1
|
STIM2
|
stromal interaction molecule 2 |
| chr6_-_107235287 | 0.50 |
ENST00000436659.1
ENST00000428750.1 ENST00000427903.1 |
RP1-60O19.1
|
RP1-60O19.1 |
| chr19_-_9546177 | 0.49 |
ENST00000592292.1
ENST00000588221.1 |
ZNF266
|
zinc finger protein 266 |
| chr19_-_57335682 | 0.49 |
ENST00000593931.1
|
ZIM2
|
zinc finger, imprinted 2 |
| chr1_-_151431674 | 0.49 |
ENST00000531094.1
|
POGZ
|
pogo transposable element with ZNF domain |
| chr2_+_139259324 | 0.49 |
ENST00000280098.4
|
SPOPL
|
speckle-type POZ protein-like |
| chr21_-_16126181 | 0.48 |
ENST00000455253.2
|
AF127936.3
|
AF127936.3 |
| chr7_-_102158157 | 0.48 |
ENST00000541662.1
ENST00000306682.6 ENST00000465829.1 |
RASA4B
|
RAS p21 protein activator 4B |
| chr4_+_128886532 | 0.48 |
ENST00000444616.1
ENST00000388795.5 |
C4orf29
|
chromosome 4 open reading frame 29 |
| chr4_+_26862313 | 0.48 |
ENST00000467087.1
ENST00000382009.3 ENST00000237364.5 |
STIM2
|
stromal interaction molecule 2 |
| chrX_+_150565038 | 0.48 |
ENST00000370361.1
|
VMA21
|
VMA21 vacuolar H+-ATPase homolog (S. cerevisiae) |
| chr5_-_72861175 | 0.48 |
ENST00000504641.1
|
ANKRA2
|
ankyrin repeat, family A (RFXANK-like), 2 |
| chr20_+_56964253 | 0.47 |
ENST00000395802.3
|
VAPB
|
VAMP (vesicle-associated membrane protein)-associated protein B and C |
| chr2_-_225434538 | 0.47 |
ENST00000409096.1
|
CUL3
|
cullin 3 |
| chr5_-_156569754 | 0.47 |
ENST00000420343.1
|
MED7
|
mediator complex subunit 7 |
| chr3_-_11888246 | 0.47 |
ENST00000455809.1
ENST00000444133.2 ENST00000273037.5 |
TAMM41
|
TAM41, mitochondrial translocator assembly and maintenance protein, homolog (S. cerevisiae) |
| chrX_+_57618269 | 0.47 |
ENST00000374888.1
|
ZXDB
|
zinc finger, X-linked, duplicated B |
| chr8_-_135652051 | 0.46 |
ENST00000522257.1
|
ZFAT
|
zinc finger and AT hook domain containing |
| chr18_-_2571210 | 0.46 |
ENST00000577166.1
|
METTL4
|
methyltransferase like 4 |
| chr9_+_131266963 | 0.45 |
ENST00000309971.4
ENST00000372770.4 |
GLE1
|
GLE1 RNA export mediator |
| chr10_+_71075552 | 0.45 |
ENST00000298649.3
|
HK1
|
hexokinase 1 |
| chr18_+_60190226 | 0.45 |
ENST00000269499.5
|
ZCCHC2
|
zinc finger, CCHC domain containing 2 |
| chr11_+_28131821 | 0.44 |
ENST00000379199.2
ENST00000303459.6 |
METTL15
|
methyltransferase like 15 |
| chr16_+_29817399 | 0.44 |
ENST00000545521.1
|
MAZ
|
MYC-associated zinc finger protein (purine-binding transcription factor) |
| chr5_-_177210399 | 0.44 |
ENST00000510276.1
|
FAM153A
|
family with sequence similarity 153, member A |
| chr18_-_21166841 | 0.44 |
ENST00000269228.5
|
NPC1
|
Niemann-Pick disease, type C1 |
| chr3_-_47555167 | 0.44 |
ENST00000296149.4
|
ELP6
|
elongator acetyltransferase complex subunit 6 |
| chr19_+_4007644 | 0.43 |
ENST00000262971.2
|
PIAS4
|
protein inhibitor of activated STAT, 4 |
| chr6_-_28226984 | 0.43 |
ENST00000423974.2
|
ZKSCAN4
|
zinc finger with KRAB and SCAN domains 4 |
| chr5_+_145583156 | 0.43 |
ENST00000265271.5
|
RBM27
|
RNA binding motif protein 27 |
| chr11_+_46740730 | 0.42 |
ENST00000311907.5
ENST00000530231.1 ENST00000442468.1 |
F2
|
coagulation factor II (thrombin) |
| chr1_+_161719552 | 0.42 |
ENST00000367943.4
|
DUSP12
|
dual specificity phosphatase 12 |
| chr1_+_179050512 | 0.42 |
ENST00000367627.3
|
TOR3A
|
torsin family 3, member A |
| chr3_-_197024965 | 0.42 |
ENST00000392382.2
|
DLG1
|
discs, large homolog 1 (Drosophila) |
| chr2_-_98280383 | 0.42 |
ENST00000289228.5
|
ACTR1B
|
ARP1 actin-related protein 1 homolog B, centractin beta (yeast) |
| chr14_-_36645674 | 0.41 |
ENST00000556013.2
|
PTCSC3
|
papillary thyroid carcinoma susceptibility candidate 3 (non-protein coding) |
| chr5_+_145583107 | 0.41 |
ENST00000506502.1
|
RBM27
|
RNA binding motif protein 27 |
| chr20_-_7921090 | 0.40 |
ENST00000378789.3
|
HAO1
|
hydroxyacid oxidase (glycolate oxidase) 1 |
| chr6_+_132455118 | 0.40 |
ENST00000458028.1
|
LINC01013
|
long intergenic non-protein coding RNA 1013 |
| chr15_+_43809797 | 0.40 |
ENST00000399453.1
ENST00000300231.5 |
MAP1A
|
microtubule-associated protein 1A |
| chr2_+_232826394 | 0.40 |
ENST00000409401.3
ENST00000441279.1 |
DIS3L2
|
DIS3 mitotic control homolog (S. cerevisiae)-like 2 |
| chr7_+_92158083 | 0.39 |
ENST00000265732.5
ENST00000481551.1 ENST00000496410.1 |
RBM48
|
RNA binding motif protein 48 |
| chr16_-_23607598 | 0.38 |
ENST00000562133.1
ENST00000570319.1 ENST00000007516.3 |
NDUFAB1
|
NADH dehydrogenase (ubiquinone) 1, alpha/beta subcomplex, 1, 8kDa |
Network of associatons between targets according to the STRING database.
First level regulatory network of PROX1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 8.5 | GO:0071613 | granzyme B production(GO:0071613) regulation of granzyme B production(GO:0071661) positive regulation of granzyme B production(GO:0071663) |
| 0.9 | 2.8 | GO:0002519 | natural killer cell tolerance induction(GO:0002519) regulation of tolerance induction dependent upon immune response(GO:0002652) negative regulation of response to tumor cell(GO:0002835) negative regulation of immune response to tumor cell(GO:0002838) negative regulation of natural killer cell mediated immune response to tumor cell(GO:0002856) negative regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002859) |
| 0.7 | 2.1 | GO:0002121 | inter-male aggressive behavior(GO:0002121) |
| 0.7 | 2.6 | GO:0051413 | response to cortisone(GO:0051413) |
| 0.6 | 2.5 | GO:0045401 | response to molecule of fungal origin(GO:0002238) regulation of interleukin-3 production(GO:0032672) positive regulation of interleukin-3 production(GO:0032752) interleukin-3 biosynthetic process(GO:0042223) regulation of interleukin-3 biosynthetic process(GO:0045399) positive regulation of interleukin-3 biosynthetic process(GO:0045401) cellular response to molecule of fungal origin(GO:0071226) |
| 0.6 | 1.8 | GO:2000523 | regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 0.5 | 1.6 | GO:0030451 | regulation of complement activation, alternative pathway(GO:0030451) negative regulation of complement activation, alternative pathway(GO:0045957) |
| 0.4 | 1.6 | GO:0006218 | uridine catabolic process(GO:0006218) |
| 0.4 | 1.2 | GO:1903413 | cellular response to bile acid(GO:1903413) response to iron ion starvation(GO:1990641) |
| 0.3 | 2.2 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.3 | 2.1 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.3 | 2.1 | GO:0036101 | leukotriene catabolic process(GO:0036100) leukotriene B4 catabolic process(GO:0036101) leukotriene B4 metabolic process(GO:0036102) icosanoid catabolic process(GO:1901523) fatty acid derivative catabolic process(GO:1901569) |
| 0.3 | 4.8 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 0.2 | 1.4 | GO:0042986 | positive regulation of amyloid precursor protein biosynthetic process(GO:0042986) |
| 0.2 | 1.9 | GO:0050653 | chondroitin sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0050653) |
| 0.2 | 0.9 | GO:0098838 | reduced folate transmembrane transport(GO:0098838) |
| 0.2 | 2.0 | GO:2001300 | lipoxin metabolic process(GO:2001300) |
| 0.2 | 2.2 | GO:0070269 | pyroptosis(GO:0070269) |
| 0.2 | 2.1 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.2 | 0.6 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.2 | 1.7 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.1 | 0.9 | GO:0051563 | smooth endoplasmic reticulum calcium ion homeostasis(GO:0051563) |
| 0.1 | 12.8 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.1 | 0.5 | GO:0019521 | aldonic acid metabolic process(GO:0019520) D-gluconate metabolic process(GO:0019521) |
| 0.1 | 0.4 | GO:0045643 | regulation of eosinophil differentiation(GO:0045643) positive regulation of eosinophil differentiation(GO:0045645) |
| 0.1 | 0.6 | GO:0015853 | adenine transport(GO:0015853) |
| 0.1 | 2.2 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.1 | 0.6 | GO:0033563 | dorsal/ventral axon guidance(GO:0033563) |
| 0.1 | 0.1 | GO:0000022 | mitotic spindle elongation(GO:0000022) |
| 0.1 | 1.6 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.1 | 0.3 | GO:0071139 | resolution of recombination intermediates(GO:0071139) resolution of mitotic recombination intermediates(GO:0071140) |
| 0.1 | 0.5 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 0.1 | 1.3 | GO:0061002 | negative regulation of dendritic spine morphogenesis(GO:0061002) |
| 0.1 | 2.4 | GO:0007213 | G-protein coupled acetylcholine receptor signaling pathway(GO:0007213) |
| 0.1 | 1.0 | GO:0072656 | maintenance of protein location in mitochondrion(GO:0072656) |
| 0.1 | 0.3 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.1 | 1.0 | GO:2000344 | positive regulation of acrosome reaction(GO:2000344) |
| 0.1 | 0.4 | GO:1900738 | cytolysis in other organism involved in symbiotic interaction(GO:0051801) positive regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900738) |
| 0.1 | 1.1 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.1 | 0.2 | GO:1901143 | insulin catabolic process(GO:1901143) |
| 0.1 | 2.2 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.1 | 0.3 | GO:2000434 | regulation of protein neddylation(GO:2000434) negative regulation of protein neddylation(GO:2000435) |
| 0.1 | 1.4 | GO:1901750 | leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.1 | 0.1 | GO:1901979 | regulation of inward rectifier potassium channel activity(GO:1901979) |
| 0.1 | 0.3 | GO:0034476 | U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) |
| 0.1 | 0.7 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.1 | 1.1 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.1 | 0.2 | GO:0035602 | fibroblast growth factor receptor signaling pathway involved in negative regulation of apoptotic process in bone marrow(GO:0035602) fibroblast growth factor receptor signaling pathway involved in hemopoiesis(GO:0035603) fibroblast growth factor receptor signaling pathway involved in positive regulation of cell proliferation in bone marrow(GO:0035604) |
| 0.1 | 1.0 | GO:0006449 | regulation of translational termination(GO:0006449) |
| 0.1 | 1.2 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
| 0.1 | 0.4 | GO:1902231 | positive regulation of keratinocyte apoptotic process(GO:1902174) positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.1 | 0.7 | GO:0032286 | central nervous system myelin maintenance(GO:0032286) |
| 0.1 | 0.4 | GO:0033133 | positive regulation of glucokinase activity(GO:0033133) positive regulation of hexokinase activity(GO:1903301) |
| 0.1 | 0.6 | GO:1903764 | regulation of potassium ion export across plasma membrane(GO:1903764) |
| 0.1 | 0.2 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
| 0.1 | 0.2 | GO:0036292 | DNA rewinding(GO:0036292) |
| 0.1 | 1.2 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.1 | 12.2 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.1 | 1.4 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 0.1 | 3.6 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) |
| 0.1 | 0.4 | GO:0034427 | nuclear-transcribed mRNA catabolic process, exonucleolytic, 3'-5'(GO:0034427) |
| 0.1 | 0.4 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.1 | 0.3 | GO:0034184 | positive regulation of maintenance of sister chromatid cohesion(GO:0034093) positive regulation of maintenance of mitotic sister chromatid cohesion(GO:0034184) |
| 0.1 | 1.0 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.1 | 1.3 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.1 | 0.6 | GO:0018094 | protein polyglycylation(GO:0018094) |
| 0.1 | 0.4 | GO:0009441 | glycolate metabolic process(GO:0009441) |
| 0.0 | 0.8 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.0 | 0.4 | GO:0060501 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 0.0 | 0.4 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.0 | 1.1 | GO:0060907 | positive regulation of macrophage cytokine production(GO:0060907) |
| 0.0 | 0.2 | GO:0015692 | lead ion transport(GO:0015692) |
| 0.0 | 0.9 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.0 | 0.5 | GO:0006987 | activation of signaling protein activity involved in unfolded protein response(GO:0006987) |
| 0.0 | 0.6 | GO:0000050 | urea cycle(GO:0000050) |
| 0.0 | 3.2 | GO:0007602 | phototransduction(GO:0007602) |
| 0.0 | 1.5 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 4.9 | GO:0031295 | T cell costimulation(GO:0031295) |
| 0.0 | 2.6 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.0 | 0.7 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 0.5 | GO:0070072 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.0 | 0.7 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.0 | 0.5 | GO:0032049 | cardiolipin biosynthetic process(GO:0032049) |
| 0.0 | 0.5 | GO:2000774 | positive regulation of cellular senescence(GO:2000774) |
| 0.0 | 0.1 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.0 | 0.1 | GO:2000969 | positive regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000969) |
| 0.0 | 0.4 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.0 | 1.2 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.0 | 0.3 | GO:0006701 | progesterone biosynthetic process(GO:0006701) |
| 0.0 | 0.1 | GO:0048200 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.0 | 1.1 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.1 | GO:0043983 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) histone H4-K12 acetylation(GO:0043983) |
| 0.0 | 1.2 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
| 0.0 | 0.6 | GO:1903830 | magnesium ion transmembrane transport(GO:1903830) |
| 0.0 | 1.5 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.0 | 0.3 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 0.7 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 0.4 | GO:0032025 | response to cobalt ion(GO:0032025) |
| 0.0 | 0.3 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 0.9 | GO:0030855 | epithelial cell differentiation(GO:0030855) |
| 0.0 | 0.5 | GO:1902236 | negative regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway(GO:1902236) |
| 0.0 | 0.1 | GO:1903336 | negative regulation of vacuolar transport(GO:1903336) |
| 0.0 | 0.5 | GO:0060712 | spongiotrophoblast layer development(GO:0060712) |
| 0.0 | 0.3 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.0 | 0.2 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.0 | 0.3 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.2 | GO:0036123 | histone H3-K9 dimethylation(GO:0036123) |
| 0.0 | 0.5 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 0.0 | 0.1 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.4 | GO:1903206 | negative regulation of hydrogen peroxide-induced cell death(GO:1903206) |
| 0.0 | 1.7 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 0.1 | GO:0043504 | mitochondrial DNA repair(GO:0043504) |
| 0.0 | 0.1 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.1 | GO:0009328 | phenylalanine-tRNA ligase complex(GO:0009328) |
| 0.4 | 1.2 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 0.4 | 2.5 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.2 | 2.2 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 0.2 | 0.6 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.1 | 1.0 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 0.1 | 2.0 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.1 | 1.2 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.1 | 0.6 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.1 | 0.4 | GO:0043293 | apoptosome(GO:0043293) |
| 0.1 | 0.9 | GO:0035253 | ciliary rootlet(GO:0035253) gamma-secretase complex(GO:0070765) |
| 0.1 | 2.2 | GO:0043186 | P granule(GO:0043186) pole plasm(GO:0045495) germ plasm(GO:0060293) |
| 0.1 | 10.9 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.1 | 1.2 | GO:0045179 | apical cortex(GO:0045179) |
| 0.1 | 0.5 | GO:1990393 | 3M complex(GO:1990393) |
| 0.1 | 0.8 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 0.5 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.0 | 0.5 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.0 | 0.3 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.0 | 1.4 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.4 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.2 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.0 | 0.3 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.0 | 0.6 | GO:0097025 | lateral loop(GO:0043219) MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 1.1 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.0 | 1.0 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.1 | GO:0097361 | CIA complex(GO:0097361) |
| 0.0 | 0.3 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 2.0 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.1 | GO:0000126 | transcription factor TFIIIB complex(GO:0000126) |
| 0.0 | 0.9 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.0 | 1.2 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 3.8 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.0 | 3.2 | GO:0101003 | ficolin-1-rich granule membrane(GO:0101003) |
| 0.0 | 0.4 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 2.8 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.8 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 0.1 | GO:0031592 | centrosomal corona(GO:0031592) |
| 0.0 | 0.1 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.0 | 0.7 | GO:0000178 | exosome (RNase complex)(GO:0000178) |
| 0.0 | 0.6 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 1.0 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.0 | 10.1 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 2.1 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 0.5 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.0 | 0.1 | GO:0034991 | nuclear meiotic cohesin complex(GO:0034991) |
| 0.0 | 2.5 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 1.5 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 1.6 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 3.8 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 1.5 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 0.3 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 1.9 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.2 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 0.1 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.0 | 0.2 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 0.1 | GO:0000243 | commitment complex(GO:0000243) |
| 0.0 | 0.2 | GO:0031597 | cytosolic proteasome complex(GO:0031597) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.1 | GO:0052870 | tocopherol omega-hydroxylase activity(GO:0052870) alpha-tocopherol omega-hydroxylase activity(GO:0052871) 20-hydroxy-leukotriene B4 omega oxidase activity(GO:0097258) 20-aldehyde-leukotriene B4 20-monooxygenase activity(GO:0097259) |
| 0.6 | 12.8 | GO:0008430 | selenium binding(GO:0008430) |
| 0.5 | 1.6 | GO:0001861 | complement component C4b receptor activity(GO:0001861) complement component C3b receptor activity(GO:0004877) |
| 0.5 | 2.4 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.5 | 1.4 | GO:0034736 | sterol O-acyltransferase activity(GO:0004772) cholesterol O-acyltransferase activity(GO:0034736) |
| 0.4 | 2.6 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.3 | 1.6 | GO:0004850 | uridine phosphorylase activity(GO:0004850) |
| 0.3 | 2.1 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.3 | 0.9 | GO:0050613 | delta14-sterol reductase activity(GO:0050613) |
| 0.3 | 1.9 | GO:0008955 | peptidoglycan glycosyltransferase activity(GO:0008955) |
| 0.3 | 1.0 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.2 | 0.9 | GO:0008518 | reduced folate carrier activity(GO:0008518) |
| 0.2 | 1.0 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 0.2 | 8.5 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.2 | 1.6 | GO:0005124 | N-formyl peptide receptor activity(GO:0004982) scavenger receptor binding(GO:0005124) |
| 0.2 | 1.2 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.2 | 0.5 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.2 | 0.6 | GO:0005471 | ATP:ADP antiporter activity(GO:0005471) adenine transmembrane transporter activity(GO:0015207) |
| 0.1 | 0.4 | GO:0030626 | U12 snRNA binding(GO:0030626) |
| 0.1 | 0.9 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.1 | 2.2 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.1 | 0.4 | GO:0052853 | (S)-2-hydroxy-acid oxidase activity(GO:0003973) very-long-chain-(S)-2-hydroxy-acid oxidase activity(GO:0052852) long-chain-(S)-2-hydroxy-long-chain-acid oxidase activity(GO:0052853) medium-chain-(S)-2-hydroxy-acid oxidase activity(GO:0052854) |
| 0.1 | 0.4 | GO:0031177 | phosphopantetheine binding(GO:0031177) |
| 0.1 | 0.4 | GO:0005137 | interleukin-5 receptor binding(GO:0005137) |
| 0.1 | 2.5 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.1 | 0.8 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.1 | 0.5 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.1 | 1.0 | GO:0004340 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.1 | 0.5 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.1 | 1.2 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.1 | 0.5 | GO:0031208 | POZ domain binding(GO:0031208) |
| 0.1 | 1.7 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.1 | 1.1 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.1 | 0.7 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.1 | 0.2 | GO:0031626 | beta-endorphin binding(GO:0031626) |
| 0.1 | 0.5 | GO:0004609 | phosphatidylserine decarboxylase activity(GO:0004609) |
| 0.1 | 1.4 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.1 | 0.7 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.1 | 0.4 | GO:0016434 | rRNA (cytosine) methyltransferase activity(GO:0016434) |
| 0.1 | 0.6 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.1 | 2.2 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.1 | 1.5 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.1 | 0.3 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.1 | 1.0 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.1 | 0.6 | GO:0070735 | protein-glycine ligase activity(GO:0070735) |
| 0.1 | 2.0 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 0.4 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 0.5 | GO:0009008 | DNA-methyltransferase activity(GO:0009008) |
| 0.0 | 0.3 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.0 | 0.6 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 1.3 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.0 | 4.8 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.2 | GO:0004906 | interferon-gamma receptor activity(GO:0004906) |
| 0.0 | 1.4 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.3 | GO:0042954 | lipoprotein transporter activity(GO:0042954) |
| 0.0 | 1.4 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.9 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 0.5 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.2 | GO:0015639 | cadmium ion transmembrane transporter activity(GO:0015086) cobalt ion transmembrane transporter activity(GO:0015087) lead ion transmembrane transporter activity(GO:0015094) ferrous iron uptake transmembrane transporter activity(GO:0015639) |
| 0.0 | 0.1 | GO:0030627 | pre-mRNA 5'-splice site binding(GO:0030627) |
| 0.0 | 0.3 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.1 | GO:0036033 | mediator complex binding(GO:0036033) |
| 0.0 | 1.8 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.3 | GO:1990948 | ligase inhibitor activity(GO:0055104) ubiquitin ligase inhibitor activity(GO:1990948) |
| 0.0 | 0.7 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 1.0 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.0 | 0.2 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.0 | 1.4 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 1.1 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.1 | GO:0001026 | TFIIIB-type transcription factor activity(GO:0001026) |
| 0.0 | 0.4 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.8 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 0.3 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.0 | 1.0 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.6 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.4 | GO:0003691 | double-stranded telomeric DNA binding(GO:0003691) |
| 0.0 | 1.7 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 0.3 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.0 | 0.1 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 0.0 | 0.2 | GO:1904047 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) S-adenosyl-L-methionine binding(GO:1904047) |
| 0.0 | 0.9 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.1 | GO:0004611 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 0.0 | 0.1 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.0 | 1.6 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 0.8 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.8 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 4.4 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 0.2 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.2 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.0 | 1.7 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.0 | 0.3 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.0 | 5.1 | GO:0004518 | nuclease activity(GO:0004518) |
| 0.0 | 0.1 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.0 | 0.5 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.0 | GO:0008670 | 2,4-dienoyl-CoA reductase (NADPH) activity(GO:0008670) |
| 0.0 | 0.1 | GO:1902282 | voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1902282) |
| 0.0 | 0.5 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.3 | GO:0017127 | cholesterol transporter activity(GO:0017127) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.4 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.1 | 2.5 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.1 | 8.4 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 1.4 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.8 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 2.7 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 0.9 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.9 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.2 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.0 | 0.9 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.4 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 1.5 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 1.6 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 0.7 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.4 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 1.3 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.5 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 0.1 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 0.8 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.6 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.1 | 2.2 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.1 | 1.6 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.1 | 2.1 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.1 | 4.8 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.1 | 1.4 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.1 | 2.2 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.1 | 4.9 | REACTOME COSTIMULATION BY THE CD28 FAMILY | Genes involved in Costimulation by the CD28 family |
| 0.1 | 2.5 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.1 | 0.6 | REACTOME INTERACTIONS OF VPR WITH HOST CELLULAR PROTEINS | Genes involved in Interactions of Vpr with host cellular proteins |
| 0.0 | 1.6 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.9 | REACTOME REGULATED PROTEOLYSIS OF P75NTR | Genes involved in Regulated proteolysis of p75NTR |
| 0.0 | 1.6 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 1.9 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 4.5 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.0 | 1.1 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 1.4 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.3 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.4 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 0.4 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 0.3 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.7 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.6 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 1.5 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 1.3 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.0 | 1.3 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.5 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.6 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.4 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.2 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |