Project
Illumina Body Map 2, young vs old
Navigation
Downloads
Results for PTF1A
Z-value: 0.26
Transcription factors associated with PTF1A
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PTF1A
|
ENSG00000168267.5 | pancreas associated transcription factor 1a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| PTF1A | hg19_v2_chr10_+_23481233_23481256 | 0.43 | 1.3e-02 | Click! |
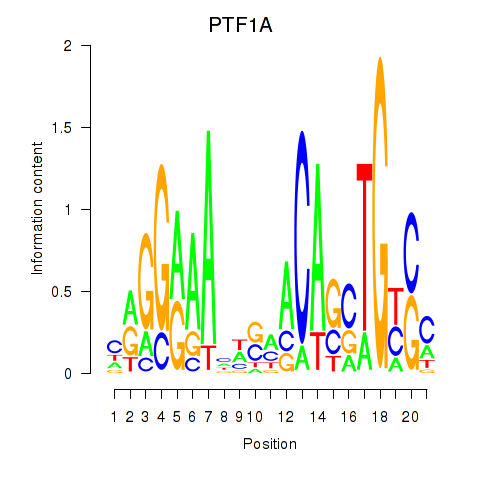
Activity profile of PTF1A motif
Sorted Z-values of PTF1A motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_35721106 | 2.70 |
ENST00000474696.1
ENST00000412048.1 ENST00000396482.2 ENST00000432682.1 |
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa |
| chr8_-_22089533 | 2.39 |
ENST00000321613.3
|
PHYHIP
|
phytanoyl-CoA 2-hydroxylase interacting protein |
| chr8_-_22089845 | 2.32 |
ENST00000454243.2
|
PHYHIP
|
phytanoyl-CoA 2-hydroxylase interacting protein |
| chr8_+_11351494 | 2.27 |
ENST00000259089.4
|
BLK
|
B lymphoid tyrosine kinase |
| chr3_+_35721182 | 2.20 |
ENST00000413378.1
ENST00000417925.1 |
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa |
| chr1_-_9129598 | 2.16 |
ENST00000535586.1
|
SLC2A5
|
solute carrier family 2 (facilitated glucose/fructose transporter), member 5 |
| chr1_-_9129631 | 2.15 |
ENST00000377414.3
|
SLC2A5
|
solute carrier family 2 (facilitated glucose/fructose transporter), member 5 |
| chr22_-_50523807 | 2.09 |
ENST00000442311.1
ENST00000538737.1 |
MLC1
|
megalencephalic leukoencephalopathy with subcortical cysts 1 |
| chr3_+_35681081 | 2.09 |
ENST00000428373.1
|
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa |
| chr22_-_50523688 | 2.07 |
ENST00000450140.2
|
MLC1
|
megalencephalic leukoencephalopathy with subcortical cysts 1 |
| chr22_-_50523760 | 1.98 |
ENST00000395876.2
|
MLC1
|
megalencephalic leukoencephalopathy with subcortical cysts 1 |
| chr8_+_11351876 | 1.84 |
ENST00000529894.1
|
BLK
|
B lymphoid tyrosine kinase |
| chr6_-_159466042 | 1.82 |
ENST00000338313.5
|
TAGAP
|
T-cell activation RhoGTPase activating protein |
| chrX_-_13956497 | 1.78 |
ENST00000398361.3
|
GPM6B
|
glycoprotein M6B |
| chr1_-_9129895 | 1.78 |
ENST00000473209.1
|
SLC2A5
|
solute carrier family 2 (facilitated glucose/fructose transporter), member 5 |
| chr6_-_159466136 | 1.61 |
ENST00000367066.3
ENST00000326965.6 |
TAGAP
|
T-cell activation RhoGTPase activating protein |
| chrX_-_13956737 | 1.58 |
ENST00000454189.2
|
GPM6B
|
glycoprotein M6B |
| chr3_+_35721130 | 1.57 |
ENST00000432450.1
|
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa |
| chr14_-_77737543 | 1.53 |
ENST00000298352.4
|
NGB
|
neuroglobin |
| chr12_-_75603482 | 1.53 |
ENST00000341669.3
ENST00000298972.1 ENST00000350228.2 |
KCNC2
|
potassium voltage-gated channel, Shaw-related subfamily, member 2 |
| chr2_-_239140011 | 1.50 |
ENST00000409376.1
ENST00000409070.1 ENST00000409942.1 |
AC016757.3
|
Protein LOC151174 |
| chr4_-_83719983 | 1.47 |
ENST00000319540.4
|
SCD5
|
stearoyl-CoA desaturase 5 |
| chr12_-_75603643 | 1.46 |
ENST00000549446.1
|
KCNC2
|
potassium voltage-gated channel, Shaw-related subfamily, member 2 |
| chr7_-_142169013 | 1.46 |
ENST00000454561.2
|
TRBV5-4
|
T cell receptor beta variable 5-4 |
| chr3_-_193272874 | 1.39 |
ENST00000342695.4
|
ATP13A4
|
ATPase type 13A4 |
| chr3_+_35680339 | 1.36 |
ENST00000450234.1
|
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa |
| chr15_-_89764929 | 1.34 |
ENST00000268125.5
|
RLBP1
|
retinaldehyde binding protein 1 |
| chr1_-_40782347 | 1.34 |
ENST00000417105.1
|
COL9A2
|
collagen, type IX, alpha 2 |
| chr2_+_89196746 | 1.32 |
ENST00000390244.2
|
IGKV5-2
|
immunoglobulin kappa variable 5-2 |
| chr4_-_87278857 | 1.31 |
ENST00000509464.1
ENST00000511167.1 |
MAPK10
|
mitogen-activated protein kinase 10 |
| chr3_+_167453493 | 1.29 |
ENST00000295777.5
ENST00000472747.2 |
SERPINI1
|
serpin peptidase inhibitor, clade I (neuroserpin), member 1 |
| chr7_-_142207004 | 1.28 |
ENST00000426318.2
|
TRBV10-2
|
T cell receptor beta variable 10-2 |
| chr13_+_107029084 | 1.28 |
ENST00000444865.1
|
LINC00460
|
long intergenic non-protein coding RNA 460 |
| chr1_-_9129735 | 1.26 |
ENST00000377424.4
|
SLC2A5
|
solute carrier family 2 (facilitated glucose/fructose transporter), member 5 |
| chr4_+_40198527 | 1.25 |
ENST00000381799.5
|
RHOH
|
ras homolog family member H |
| chr12_+_50451331 | 1.24 |
ENST00000228468.4
|
ASIC1
|
acid-sensing (proton-gated) ion channel 1 |
| chr11_-_83878041 | 1.23 |
ENST00000398299.1
|
DLG2
|
discs, large homolog 2 (Drosophila) |
| chr16_+_71392616 | 1.21 |
ENST00000349553.5
ENST00000302628.4 ENST00000562305.1 |
CALB2
|
calbindin 2 |
| chr14_-_24042184 | 1.20 |
ENST00000544177.1
|
JPH4
|
junctophilin 4 |
| chr3_+_167453026 | 1.20 |
ENST00000472941.1
|
SERPINI1
|
serpin peptidase inhibitor, clade I (neuroserpin), member 1 |
| chr11_+_73357614 | 1.15 |
ENST00000536527.1
|
PLEKHB1
|
pleckstrin homology domain containing, family B (evectins) member 1 |
| chr6_-_62996066 | 1.15 |
ENST00000281156.4
|
KHDRBS2
|
KH domain containing, RNA binding, signal transduction associated 2 |
| chr10_-_10504285 | 1.10 |
ENST00000602311.1
|
RP11-271F18.4
|
RP11-271F18.4 |
| chr2_-_237412331 | 1.02 |
ENST00000418802.1
|
IQCA1
|
IQ motif containing with AAA domain 1 |
| chr1_+_23037323 | 0.99 |
ENST00000544305.1
ENST00000374630.3 ENST00000400191.3 ENST00000374632.3 |
EPHB2
|
EPH receptor B2 |
| chr6_-_127780510 | 0.97 |
ENST00000487331.2
ENST00000483725.3 |
KIAA0408
|
KIAA0408 |
| chr6_+_27107053 | 0.95 |
ENST00000354348.2
|
HIST1H4I
|
histone cluster 1, H4i |
| chr17_-_61778528 | 0.94 |
ENST00000584645.1
|
LIMD2
|
LIM domain containing 2 |
| chrX_-_48216101 | 0.93 |
ENST00000298396.2
ENST00000376893.3 |
SSX3
|
synovial sarcoma, X breakpoint 3 |
| chr12_+_1929644 | 0.92 |
ENST00000299194.1
|
LRTM2
|
leucine-rich repeats and transmembrane domains 2 |
| chr10_-_1779663 | 0.91 |
ENST00000381312.1
|
ADARB2
|
adenosine deaminase, RNA-specific, B2 (non-functional) |
| chr22_+_23101182 | 0.91 |
ENST00000390312.2
|
IGLV2-14
|
immunoglobulin lambda variable 2-14 |
| chr5_+_132149017 | 0.90 |
ENST00000378693.2
|
SOWAHA
|
sosondowah ankyrin repeat domain family member A |
| chr9_+_33795533 | 0.88 |
ENST00000379405.3
|
PRSS3
|
protease, serine, 3 |
| chr2_-_237416071 | 0.88 |
ENST00000309507.5
ENST00000431676.2 |
IQCA1
|
IQ motif containing with AAA domain 1 |
| chr7_-_142099977 | 0.88 |
ENST00000390359.3
|
TRBV7-8
|
T cell receptor beta variable 7-8 |
| chr7_-_142198049 | 0.84 |
ENST00000471935.1
|
TRBV11-2
|
T cell receptor beta variable 11-2 |
| chr11_+_56949221 | 0.82 |
ENST00000497933.1
|
LRRC55
|
leucine rich repeat containing 55 |
| chr17_+_40996590 | 0.80 |
ENST00000253799.3
ENST00000452774.2 |
AOC2
|
amine oxidase, copper containing 2 (retina-specific) |
| chr11_-_45687128 | 0.78 |
ENST00000308064.2
|
CHST1
|
carbohydrate (keratan sulfate Gal-6) sulfotransferase 1 |
| chr19_+_29493456 | 0.76 |
ENST00000591143.1
ENST00000592653.1 |
CTD-2081K17.2
|
CTD-2081K17.2 |
| chr12_+_1929421 | 0.75 |
ENST00000543818.1
|
LRTM2
|
leucine-rich repeats and transmembrane domains 2 |
| chr1_+_26856236 | 0.75 |
ENST00000374168.2
ENST00000374166.4 |
RPS6KA1
|
ribosomal protein S6 kinase, 90kDa, polypeptide 1 |
| chr1_+_2005126 | 0.75 |
ENST00000495347.1
|
PRKCZ
|
protein kinase C, zeta |
| chr16_+_89696692 | 0.74 |
ENST00000261615.4
|
DPEP1
|
dipeptidase 1 (renal) |
| chr2_-_237416181 | 0.74 |
ENST00000409907.3
|
IQCA1
|
IQ motif containing with AAA domain 1 |
| chr12_-_10095997 | 0.73 |
ENST00000412084.1
|
AC091814.2
|
AC091814.2 |
| chr13_+_107028897 | 0.70 |
ENST00000439790.1
ENST00000435024.1 |
LINC00460
|
long intergenic non-protein coding RNA 460 |
| chr15_+_81225699 | 0.70 |
ENST00000560027.1
|
KIAA1199
|
KIAA1199 |
| chr2_-_89417335 | 0.69 |
ENST00000490686.1
|
IGKV1-17
|
immunoglobulin kappa variable 1-17 |
| chr10_+_124739911 | 0.67 |
ENST00000405485.1
|
PSTK
|
phosphoseryl-tRNA kinase |
| chr1_+_228337553 | 0.67 |
ENST00000366714.2
|
GJC2
|
gap junction protein, gamma 2, 47kDa |
| chr2_+_173600514 | 0.67 |
ENST00000264111.6
|
RAPGEF4
|
Rap guanine nucleotide exchange factor (GEF) 4 |
| chr12_-_90102594 | 0.67 |
ENST00000428670.3
|
ATP2B1
|
ATPase, Ca++ transporting, plasma membrane 1 |
| chr12_-_57644952 | 0.66 |
ENST00000554578.1
ENST00000546246.2 ENST00000553489.1 ENST00000332782.2 |
STAC3
|
SH3 and cysteine rich domain 3 |
| chr11_-_1629693 | 0.65 |
ENST00000399685.1
|
KRTAP5-3
|
keratin associated protein 5-3 |
| chr2_+_173600565 | 0.64 |
ENST00000397081.3
|
RAPGEF4
|
Rap guanine nucleotide exchange factor (GEF) 4 |
| chr8_-_9762871 | 0.64 |
ENST00000521242.1
|
LINC00599
|
long intergenic non-protein coding RNA 599 |
| chr4_-_5990166 | 0.62 |
ENST00000324058.5
|
C4orf50
|
chromosome 4 open reading frame 50 |
| chr2_+_85922491 | 0.61 |
ENST00000526018.1
|
GNLY
|
granulysin |
| chr10_+_124739964 | 0.60 |
ENST00000406217.2
|
PSTK
|
phosphoseryl-tRNA kinase |
| chr6_-_24877490 | 0.60 |
ENST00000540914.1
ENST00000378023.4 |
FAM65B
|
family with sequence similarity 65, member B |
| chr15_+_68924327 | 0.60 |
ENST00000543950.1
|
CORO2B
|
coronin, actin binding protein, 2B |
| chr11_+_71276609 | 0.58 |
ENST00000398531.1
ENST00000376536.4 |
KRTAP5-10
|
keratin associated protein 5-10 |
| chr15_+_51633826 | 0.56 |
ENST00000335449.6
|
GLDN
|
gliomedin |
| chr11_+_64107663 | 0.55 |
ENST00000356786.5
|
CCDC88B
|
coiled-coil domain containing 88B |
| chr2_-_89278535 | 0.55 |
ENST00000390247.2
|
IGKV3-7
|
immunoglobulin kappa variable 3-7 (non-functional) |
| chr4_-_175750364 | 0.55 |
ENST00000340217.5
ENST00000274093.3 |
GLRA3
|
glycine receptor, alpha 3 |
| chr4_-_83719884 | 0.54 |
ENST00000282709.4
ENST00000273908.4 |
SCD5
|
stearoyl-CoA desaturase 5 |
| chr17_-_3461092 | 0.54 |
ENST00000301365.4
ENST00000572519.1 |
TRPV3
|
transient receptor potential cation channel, subfamily V, member 3 |
| chr17_+_67498538 | 0.54 |
ENST00000589647.1
|
MAP2K6
|
mitogen-activated protein kinase kinase 6 |
| chr17_+_67498396 | 0.52 |
ENST00000588110.1
|
MAP2K6
|
mitogen-activated protein kinase kinase 6 |
| chr1_-_114447683 | 0.51 |
ENST00000256658.4
ENST00000369564.1 |
AP4B1
|
adaptor-related protein complex 4, beta 1 subunit |
| chr1_-_45956822 | 0.51 |
ENST00000372086.3
ENST00000341771.6 |
TESK2
|
testis-specific kinase 2 |
| chr1_-_45956800 | 0.51 |
ENST00000538496.1
|
TESK2
|
testis-specific kinase 2 |
| chr1_-_231114542 | 0.51 |
ENST00000522821.1
ENST00000366661.4 ENST00000366662.4 ENST00000414259.1 ENST00000522399.1 |
TTC13
|
tetratricopeptide repeat domain 13 |
| chr20_+_42544782 | 0.50 |
ENST00000423191.2
ENST00000372999.1 |
TOX2
|
TOX high mobility group box family member 2 |
| chr1_+_114447763 | 0.50 |
ENST00000369563.3
|
DCLRE1B
|
DNA cross-link repair 1B |
| chr10_+_49892904 | 0.50 |
ENST00000360890.2
|
WDFY4
|
WDFY family member 4 |
| chr8_-_110660975 | 0.50 |
ENST00000528045.1
|
SYBU
|
syntabulin (syntaxin-interacting) |
| chr17_-_36499693 | 0.50 |
ENST00000342292.4
|
GPR179
|
G protein-coupled receptor 179 |
| chr17_-_73892992 | 0.49 |
ENST00000540128.1
ENST00000269383.3 |
TRIM65
|
tripartite motif containing 65 |
| chr19_-_7764960 | 0.47 |
ENST00000593418.1
|
FCER2
|
Fc fragment of IgE, low affinity II, receptor for (CD23) |
| chr1_-_114447620 | 0.47 |
ENST00000369569.1
ENST00000432415.1 ENST00000369571.2 |
AP4B1
|
adaptor-related protein complex 4, beta 1 subunit |
| chr7_-_142251148 | 0.47 |
ENST00000390360.3
|
TRBV6-4
|
T cell receptor beta variable 6-4 |
| chr11_-_40315640 | 0.46 |
ENST00000278198.2
|
LRRC4C
|
leucine rich repeat containing 4C |
| chr19_+_19779686 | 0.46 |
ENST00000415784.2
|
ZNF101
|
zinc finger protein 101 |
| chr1_-_45956868 | 0.46 |
ENST00000451835.2
|
TESK2
|
testis-specific kinase 2 |
| chr11_+_64058758 | 0.44 |
ENST00000538767.1
|
KCNK4
|
potassium channel, subfamily K, member 4 |
| chr9_+_107266455 | 0.44 |
ENST00000334726.2
|
OR13F1
|
olfactory receptor, family 13, subfamily F, member 1 |
| chr1_-_205649580 | 0.43 |
ENST00000367145.3
|
SLC45A3
|
solute carrier family 45, member 3 |
| chr5_-_55902050 | 0.42 |
ENST00000438651.1
|
AC022431.2
|
Homo sapiens uncharacterized LOC101928448 (LOC101928448), mRNA. |
| chr5_-_75013193 | 0.42 |
ENST00000514838.2
ENST00000506164.1 ENST00000502826.1 ENST00000503835.1 ENST00000428202.2 ENST00000380475.2 |
POC5
|
POC5 centriolar protein |
| chr3_+_49027308 | 0.41 |
ENST00000383729.4
ENST00000343546.4 |
P4HTM
|
prolyl 4-hydroxylase, transmembrane (endoplasmic reticulum) |
| chr13_-_31736132 | 0.41 |
ENST00000429785.2
|
HSPH1
|
heat shock 105kDa/110kDa protein 1 |
| chr13_-_111522162 | 0.40 |
ENST00000538077.1
|
LINC00346
|
long intergenic non-protein coding RNA 346 |
| chr13_-_31736027 | 0.39 |
ENST00000380406.5
ENST00000320027.5 ENST00000380405.4 |
HSPH1
|
heat shock 105kDa/110kDa protein 1 |
| chr3_-_69249863 | 0.39 |
ENST00000478263.1
ENST00000462512.1 |
FRMD4B
|
FERM domain containing 4B |
| chr16_-_75569068 | 0.39 |
ENST00000336257.3
ENST00000565039.1 |
CHST5
|
carbohydrate (N-acetylglucosamine 6-O) sulfotransferase 5 |
| chr19_+_54495542 | 0.39 |
ENST00000252729.2
ENST00000352529.1 |
CACNG6
|
calcium channel, voltage-dependent, gamma subunit 6 |
| chr11_+_71249071 | 0.38 |
ENST00000398534.3
|
KRTAP5-8
|
keratin associated protein 5-8 |
| chr22_-_50051151 | 0.37 |
ENST00000400023.1
ENST00000444628.1 |
C22orf34
|
chromosome 22 open reading frame 34 |
| chr12_+_112451120 | 0.36 |
ENST00000261735.3
ENST00000455836.1 |
ERP29
|
endoplasmic reticulum protein 29 |
| chr3_+_2140565 | 0.34 |
ENST00000455083.1
ENST00000418658.1 |
CNTN4
|
contactin 4 |
| chr19_+_11670245 | 0.34 |
ENST00000585493.1
|
ZNF627
|
zinc finger protein 627 |
| chr1_-_33647267 | 0.33 |
ENST00000291416.5
|
TRIM62
|
tripartite motif containing 62 |
| chr2_-_228497888 | 0.33 |
ENST00000264387.4
ENST00000409066.1 |
C2orf83
|
chromosome 2 open reading frame 83 |
| chr11_-_96123022 | 0.32 |
ENST00000542662.1
|
CCDC82
|
coiled-coil domain containing 82 |
| chr17_+_67498295 | 0.31 |
ENST00000589295.1
|
MAP2K6
|
mitogen-activated protein kinase kinase 6 |
| chr20_-_44937124 | 0.31 |
ENST00000537909.1
|
CDH22
|
cadherin 22, type 2 |
| chr11_+_7597639 | 0.30 |
ENST00000533792.1
|
PPFIBP2
|
PTPRF interacting protein, binding protein 2 (liprin beta 2) |
| chrX_-_134305322 | 0.30 |
ENST00000276241.6
ENST00000344129.2 |
CXorf48
|
cancer/testis antigen 55 |
| chr12_+_31079362 | 0.29 |
ENST00000545802.1
|
TSPAN11
|
tetraspanin 11 |
| chr5_-_173217916 | 0.29 |
ENST00000523617.1
|
CTB-43E15.4
|
CTB-43E15.4 |
| chr17_+_12692774 | 0.28 |
ENST00000379672.5
ENST00000340825.3 |
ARHGAP44
|
Rho GTPase activating protein 44 |
| chr19_+_29493486 | 0.27 |
ENST00000589821.1
|
CTD-2081K17.2
|
CTD-2081K17.2 |
| chr17_-_33415837 | 0.27 |
ENST00000414419.2
|
RFFL
|
ring finger and FYVE-like domain containing E3 ubiquitin protein ligase |
| chr20_+_44637526 | 0.27 |
ENST00000372330.3
|
MMP9
|
matrix metallopeptidase 9 (gelatinase B, 92kDa gelatinase, 92kDa type IV collagenase) |
| chr13_-_79979952 | 0.27 |
ENST00000438724.1
|
RBM26
|
RNA binding motif protein 26 |
| chr11_-_1606513 | 0.26 |
ENST00000382171.2
|
KRTAP5-1
|
keratin associated protein 5-1 |
| chr10_+_43572475 | 0.26 |
ENST00000355710.3
ENST00000498820.1 ENST00000340058.5 |
RET
|
ret proto-oncogene |
| chr15_+_91446961 | 0.26 |
ENST00000559965.1
|
MAN2A2
|
mannosidase, alpha, class 2A, member 2 |
| chr5_-_162887071 | 0.25 |
ENST00000302764.4
|
NUDCD2
|
NudC domain containing 2 |
| chr11_-_102576537 | 0.25 |
ENST00000260229.4
|
MMP27
|
matrix metallopeptidase 27 |
| chr3_+_2140476 | 0.25 |
ENST00000422330.1
|
CNTN4
|
contactin 4 |
| chr17_+_79859985 | 0.24 |
ENST00000333383.7
|
NPB
|
neuropeptide B |
| chrX_+_15808569 | 0.23 |
ENST00000380308.3
ENST00000307771.7 |
ZRSR2
|
zinc finger (CCCH type), RNA-binding motif and serine/arginine rich 2 |
| chr11_+_6806248 | 0.22 |
ENST00000307401.4
|
OR2AG1
|
olfactory receptor, family 2, subfamily AG, member 1 |
| chr13_+_113633620 | 0.22 |
ENST00000421756.1
ENST00000375601.3 |
MCF2L
|
MCF.2 cell line derived transforming sequence-like |
| chr1_-_20446020 | 0.22 |
ENST00000375105.3
|
PLA2G2D
|
phospholipase A2, group IID |
| chr2_-_136288113 | 0.21 |
ENST00000401392.1
|
ZRANB3
|
zinc finger, RAN-binding domain containing 3 |
| chr11_+_1651033 | 0.21 |
ENST00000399676.2
|
KRTAP5-5
|
keratin associated protein 5-5 |
| chr9_-_35042824 | 0.21 |
ENST00000595331.1
|
FLJ00273
|
FLJ00273 |
| chr19_+_17326521 | 0.20 |
ENST00000593597.1
|
USE1
|
unconventional SNARE in the ER 1 homolog (S. cerevisiae) |
| chr2_-_89513402 | 0.20 |
ENST00000498435.1
|
IGKV1-27
|
immunoglobulin kappa variable 1-27 |
| chr22_-_50050986 | 0.20 |
ENST00000405854.1
|
C22orf34
|
chromosome 22 open reading frame 34 |
| chr9_-_139219640 | 0.20 |
ENST00000601276.1
|
DKFZP434A062
|
HCG2022364; Putative uncharacterized protein DKFZp434A062; Uncharacterized protein |
| chr11_+_64052692 | 0.19 |
ENST00000377702.4
|
GPR137
|
G protein-coupled receptor 137 |
| chr20_+_44441304 | 0.18 |
ENST00000352551.5
|
UBE2C
|
ubiquitin-conjugating enzyme E2C |
| chr5_+_140557371 | 0.18 |
ENST00000239444.2
|
PCDHB8
|
protocadherin beta 8 |
| chr8_-_145642203 | 0.18 |
ENST00000526658.1
|
SLC39A4
|
solute carrier family 39 (zinc transporter), member 4 |
| chr1_-_114447412 | 0.18 |
ENST00000369567.1
ENST00000369566.3 |
AP4B1
|
adaptor-related protein complex 4, beta 1 subunit |
| chr14_+_70346125 | 0.18 |
ENST00000361956.3
ENST00000381280.4 |
SMOC1
|
SPARC related modular calcium binding 1 |
| chr13_-_79979919 | 0.18 |
ENST00000267229.7
|
RBM26
|
RNA binding motif protein 26 |
| chr5_+_125800836 | 0.18 |
ENST00000511134.1
|
GRAMD3
|
GRAM domain containing 3 |
| chr1_-_204654826 | 0.18 |
ENST00000367177.3
|
LRRN2
|
leucine rich repeat neuronal 2 |
| chr14_+_22981834 | 0.17 |
ENST00000390507.1
|
TRAJ30
|
T cell receptor alpha joining 30 |
| chr18_+_5238055 | 0.16 |
ENST00000582363.1
ENST00000582008.1 ENST00000580082.1 |
LINC00667
|
long intergenic non-protein coding RNA 667 |
| chr16_-_46723066 | 0.16 |
ENST00000299138.7
|
VPS35
|
vacuolar protein sorting 35 homolog (S. cerevisiae) |
| chr19_-_44258733 | 0.16 |
ENST00000597586.1
ENST00000596714.1 |
SMG9
|
SMG9 nonsense mediated mRNA decay factor |
| chr2_+_202122826 | 0.15 |
ENST00000413726.1
|
CASP8
|
caspase 8, apoptosis-related cysteine peptidase |
| chr11_+_64052266 | 0.15 |
ENST00000539851.1
|
GPR137
|
G protein-coupled receptor 137 |
| chr9_+_99691286 | 0.14 |
ENST00000372322.3
|
NUTM2G
|
NUT family member 2G |
| chr20_+_18125727 | 0.14 |
ENST00000489634.2
|
CSRP2BP
|
CSRP2 binding protein |
| chr9_+_138235095 | 0.14 |
ENST00000320778.2
|
C9orf62
|
chromosome 9 open reading frame 62 |
| chr17_-_33416231 | 0.14 |
ENST00000584655.1
ENST00000447669.2 ENST00000315249.7 |
RFFL
|
ring finger and FYVE-like domain containing E3 ubiquitin protein ligase |
| chr1_+_100503643 | 0.13 |
ENST00000370152.3
|
HIAT1
|
hippocampus abundant transcript 1 |
| chr11_+_96123158 | 0.13 |
ENST00000332349.4
ENST00000458427.1 |
JRKL
|
jerky homolog-like (mouse) |
| chr5_-_162887054 | 0.13 |
ENST00000517501.1
|
NUDCD2
|
NudC domain containing 2 |
| chr3_-_169587621 | 0.13 |
ENST00000523069.1
ENST00000316428.5 ENST00000264676.5 |
LRRC31
|
leucine rich repeat containing 31 |
| chr4_+_183370146 | 0.13 |
ENST00000510504.1
|
TENM3
|
teneurin transmembrane protein 3 |
| chr3_+_49027771 | 0.13 |
ENST00000475629.1
ENST00000444213.1 |
P4HTM
|
prolyl 4-hydroxylase, transmembrane (endoplasmic reticulum) |
| chr2_+_202122703 | 0.12 |
ENST00000447616.1
ENST00000358485.4 |
CASP8
|
caspase 8, apoptosis-related cysteine peptidase |
| chr18_-_53177984 | 0.11 |
ENST00000543082.1
|
TCF4
|
transcription factor 4 |
| chr1_-_76076759 | 0.11 |
ENST00000370855.5
|
SLC44A5
|
solute carrier family 44, member 5 |
| chr5_+_109218883 | 0.11 |
ENST00000523446.1
|
AC011366.3
|
Uncharacterized protein |
| chr2_-_196933536 | 0.10 |
ENST00000312428.6
ENST00000410072.1 |
DNAH7
|
dynein, axonemal, heavy chain 7 |
| chr20_+_54823788 | 0.10 |
ENST00000243911.2
|
MC3R
|
melanocortin 3 receptor |
| chr9_-_104145795 | 0.10 |
ENST00000259407.2
|
BAAT
|
bile acid CoA: amino acid N-acyltransferase (glycine N-choloyltransferase) |
| chr1_+_40204538 | 0.10 |
ENST00000324379.5
ENST00000356511.2 ENST00000497370.1 ENST00000470213.1 ENST00000372835.5 ENST00000372830.1 |
PPIE
|
peptidylprolyl isomerase E (cyclophilin E) |
| chr11_-_130298888 | 0.09 |
ENST00000257359.6
|
ADAMTS8
|
ADAM metallopeptidase with thrombospondin type 1 motif, 8 |
| chr17_-_3375033 | 0.09 |
ENST00000268981.5
ENST00000397168.3 ENST00000572969.1 ENST00000355380.4 ENST00000571553.1 ENST00000574797.1 ENST00000575375.1 |
SPATA22
|
spermatogenesis associated 22 |
| chr14_-_58893832 | 0.09 |
ENST00000556007.2
|
TIMM9
|
translocase of inner mitochondrial membrane 9 homolog (yeast) |
| chr7_-_100844193 | 0.09 |
ENST00000440203.2
ENST00000379423.3 ENST00000223114.4 |
MOGAT3
|
monoacylglycerol O-acyltransferase 3 |
| chr11_+_64052454 | 0.09 |
ENST00000539833.1
|
GPR137
|
G protein-coupled receptor 137 |
| chr7_-_152373216 | 0.08 |
ENST00000359321.1
|
XRCC2
|
X-ray repair complementing defective repair in Chinese hamster cells 2 |
| chr7_+_141811539 | 0.08 |
ENST00000550469.2
ENST00000477922.3 |
RP11-1220K2.2
|
Putative inactive maltase-glucoamylase-like protein LOC93432 |
| chr14_+_67656110 | 0.07 |
ENST00000524532.1
ENST00000530728.1 |
FAM71D
|
family with sequence similarity 71, member D |
| chr9_-_97090926 | 0.06 |
ENST00000335456.7
ENST00000253262.4 ENST00000341207.4 |
NUTM2F
|
NUT family member 2F |
| chr8_-_145641864 | 0.06 |
ENST00000276833.5
|
SLC39A4
|
solute carrier family 39 (zinc transporter), member 4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of PTF1A
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 7.4 | GO:0032445 | fructose import(GO:0032445) carbohydrate import into cell(GO:0097319) carbohydrate import across plasma membrane(GO:0098704) fructose import across plasma membrane(GO:1990539) |
| 0.7 | 2.0 | GO:1903964 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.4 | 3.0 | GO:0038060 | nitric oxide-cGMP-mediated signaling pathway(GO:0038060) |
| 0.3 | 1.0 | GO:0051389 | inactivation of MAPKK activity(GO:0051389) |
| 0.3 | 3.4 | GO:0051612 | negative regulation of neurotransmitter uptake(GO:0051581) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.3 | 1.4 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.3 | 5.9 | GO:0047484 | regulation of response to osmotic stress(GO:0047484) |
| 0.2 | 0.7 | GO:0016999 | antibiotic metabolic process(GO:0016999) |
| 0.2 | 0.6 | GO:0002818 | intracellular defense response(GO:0002818) |
| 0.2 | 1.3 | GO:0097056 | selenocysteinyl-tRNA(Sec) biosynthetic process(GO:0097056) |
| 0.2 | 1.2 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.2 | 0.2 | GO:1903181 | regulation of dopamine biosynthetic process(GO:1903179) positive regulation of dopamine biosynthetic process(GO:1903181) |
| 0.1 | 0.4 | GO:0071469 | detection of mechanical stimulus involved in sensory perception of touch(GO:0050976) cellular response to alkaline pH(GO:0071469) |
| 0.1 | 0.4 | GO:0015772 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.1 | 0.8 | GO:1903750 | regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903750) negative regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903751) |
| 0.1 | 1.3 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.1 | 0.3 | GO:0007497 | posterior midgut development(GO:0007497) |
| 0.1 | 1.2 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.1 | 0.5 | GO:0031627 | telomeric loop formation(GO:0031627) telomeric 3' overhang formation(GO:0031860) |
| 0.1 | 0.4 | GO:0043335 | protein unfolding(GO:0043335) |
| 0.1 | 0.5 | GO:0002925 | positive regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002925) |
| 0.1 | 1.3 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.1 | 4.1 | GO:0050855 | regulation of B cell receptor signaling pathway(GO:0050855) |
| 0.1 | 0.7 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.1 | 0.7 | GO:1990034 | cellular response to corticosterone stimulus(GO:0071386) calcium ion export from cell(GO:1990034) |
| 0.1 | 0.4 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.1 | 0.3 | GO:0071934 | thiamine transmembrane transport(GO:0071934) |
| 0.1 | 0.7 | GO:2001181 | positive regulation of interleukin-10 secretion(GO:2001181) |
| 0.1 | 0.5 | GO:0042636 | negative regulation of hair cycle(GO:0042636) |
| 0.1 | 1.5 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.1 | 0.8 | GO:1903818 | positive regulation of voltage-gated potassium channel activity(GO:1903818) |
| 0.1 | 0.5 | GO:0060012 | synaptic transmission, glycinergic(GO:0060012) |
| 0.1 | 0.3 | GO:0039650 | modulation by virus of host molecular function(GO:0039506) suppression by virus of host molecular function(GO:0039507) suppression by virus of host catalytic activity(GO:0039513) modulation by virus of host catalytic activity(GO:0039516) suppression by virus of host cysteine-type endopeptidase activity involved in apoptotic process(GO:0039650) negative regulation by symbiont of host catalytic activity(GO:0052053) negative regulation by symbiont of host molecular function(GO:0052056) modulation by symbiont of host catalytic activity(GO:0052148) |
| 0.1 | 0.2 | GO:0002361 | CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0002361) |
| 0.1 | 0.2 | GO:0036292 | DNA rewinding(GO:0036292) |
| 0.0 | 0.2 | GO:0034224 | cellular response to zinc ion starvation(GO:0034224) |
| 0.0 | 0.3 | GO:0098886 | modification of dendritic spine(GO:0098886) |
| 0.0 | 8.9 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.0 | 0.4 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.0 | 0.8 | GO:0006012 | galactose metabolic process(GO:0006012) |
| 0.0 | 0.3 | GO:0051549 | positive regulation of keratinocyte migration(GO:0051549) |
| 0.0 | 0.6 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.4 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.0 | 0.8 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.0 | 0.2 | GO:0031536 | positive regulation of exit from mitosis(GO:0031536) |
| 0.0 | 0.3 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.0 | 0.1 | GO:0000711 | meiotic DNA repair synthesis(GO:0000711) |
| 0.0 | 1.2 | GO:0046710 | GDP metabolic process(GO:0046710) receptor localization to synapse(GO:0097120) |
| 0.0 | 1.7 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 2.4 | GO:0007422 | peripheral nervous system development(GO:0007422) |
| 0.0 | 0.6 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.0 | 2.9 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.0 | 1.3 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.0 | 1.2 | GO:0007602 | phototransduction(GO:0007602) |
| 0.0 | 1.3 | GO:0019933 | cAMP-mediated signaling(GO:0019933) |
| 0.0 | 0.7 | GO:0010644 | cell communication by electrical coupling(GO:0010644) |
| 0.0 | 0.7 | GO:0048741 | skeletal muscle fiber development(GO:0048741) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.3 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.2 | 0.5 | GO:0016935 | glycine-gated chloride channel complex(GO:0016935) |
| 0.1 | 0.4 | GO:0002947 | tumor necrosis factor receptor superfamily complex(GO:0002947) |
| 0.1 | 1.2 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.1 | 0.6 | GO:0097013 | phagocytic vesicle lumen(GO:0097013) |
| 0.1 | 3.7 | GO:0044298 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 1.9 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 0.7 | GO:0045179 | apical cortex(GO:0045179) |
| 0.0 | 5.6 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 7.1 | GO:0035579 | specific granule membrane(GO:0035579) |
| 0.0 | 1.2 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 0.2 | GO:0089701 | U2AF(GO:0089701) |
| 0.0 | 0.2 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
| 0.0 | 0.3 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.0 | 0.8 | GO:0071682 | endocytic vesicle lumen(GO:0071682) |
| 0.0 | 0.5 | GO:0097433 | dense body(GO:0097433) |
| 0.0 | 0.7 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.0 | 0.1 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.0 | 1.2 | GO:0031904 | endosome lumen(GO:0031904) |
| 0.0 | 1.3 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 3.1 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 3.8 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 1.1 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.3 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) ripoptosome(GO:0097342) |
| 0.0 | 0.1 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 7.4 | GO:0005353 | fructose transmembrane transporter activity(GO:0005353) |
| 0.2 | 2.0 | GO:0016215 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) |
| 0.2 | 1.2 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.2 | 0.8 | GO:0052594 | tryptamine:oxygen oxidoreductase (deaminating) activity(GO:0052593) aminoacetone:oxygen oxidoreductase(deaminating) activity(GO:0052594) aliphatic-amine oxidase activity(GO:0052595) phenethylamine:oxygen oxidoreductase (deaminating) activity(GO:0052596) |
| 0.2 | 1.3 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.1 | 0.4 | GO:0098782 | mechanically-gated potassium channel activity(GO:0098782) |
| 0.1 | 1.2 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.1 | 1.3 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.1 | 1.3 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.1 | 0.4 | GO:0015157 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.1 | 0.5 | GO:0016933 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.1 | 3.0 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.1 | 0.3 | GO:0015403 | thiamine uptake transmembrane transporter activity(GO:0015403) |
| 0.1 | 1.0 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.1 | 1.5 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.1 | 0.7 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.1 | 0.7 | GO:1903763 | gap junction channel activity involved in cell communication by electrical coupling(GO:1903763) |
| 0.1 | 0.8 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.1 | 0.3 | GO:0004572 | mannosyl-oligosaccharide 1,3-1,6-alpha-mannosidase activity(GO:0004572) |
| 0.0 | 0.9 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.0 | 0.5 | GO:0019863 | IgE binding(GO:0019863) |
| 0.0 | 1.7 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 1.4 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.3 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.0 | 0.5 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.0 | 1.3 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 6.6 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 0.4 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 2.0 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 3.6 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 1.2 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.7 | GO:0043560 | insulin receptor substrate binding(GO:0043560) |
| 0.0 | 0.4 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.2 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.0 | 0.1 | GO:0004980 | melanocyte-stimulating hormone receptor activity(GO:0004980) |
| 0.0 | 1.2 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 8.3 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 0.6 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.0 | 1.3 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 1.3 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.0 | 0.2 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.0 | 0.4 | GO:0003756 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.0 | 0.1 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.0 | 0.6 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.0 | 0.1 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.0 | 0.5 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 5.5 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 2.1 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.0 | 0.7 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 1.6 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 2.5 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 1.4 | PID TNF PATHWAY | TNF receptor signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 7.4 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.1 | 4.1 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 1.3 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 4.3 | REACTOME REGULATION OF INSULIN SECRETION BY GLUCAGON LIKE PEPTIDE1 | Genes involved in Regulation of Insulin Secretion by Glucagon-like Peptide-1 |
| 0.0 | 1.4 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.0 | 1.2 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.0 | 0.7 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.7 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.4 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.7 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 1.3 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 1.0 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 4.5 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.3 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.0 | 0.5 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.0 | 0.2 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |