Project
Illumina Body Map 2, young vs old
Navigation
Downloads
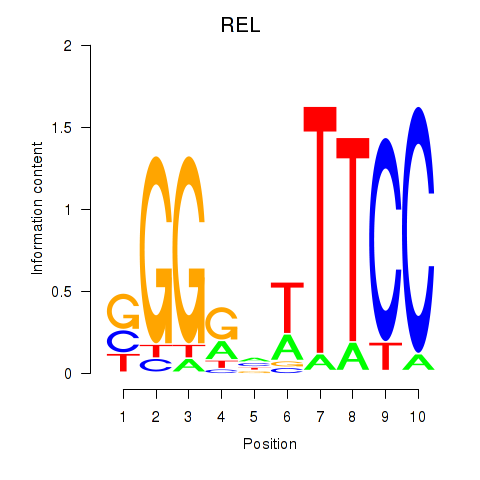
Results for REL
Z-value: 0.36
Transcription factors associated with REL
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
REL
|
ENSG00000162924.9 | REL proto-oncogene, NF-kB subunit |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| REL | hg19_v2_chr2_+_61108771_61108791 | 0.15 | 4.1e-01 | Click! |
Activity profile of REL motif
Sorted Z-values of REL motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_90077680 | 4.86 |
ENST00000390270.2
|
IGKV3D-20
|
immunoglobulin kappa variable 3D-20 |
| chr2_+_90153696 | 3.98 |
ENST00000417279.2
|
IGKV3D-15
|
immunoglobulin kappa variable 3D-15 (gene/pseudogene) |
| chr22_+_22712087 | 3.97 |
ENST00000390294.2
|
IGLV1-47
|
immunoglobulin lambda variable 1-47 |
| chr22_-_19512893 | 3.74 |
ENST00000403084.1
ENST00000413119.2 |
CLDN5
|
claudin 5 |
| chr2_-_89247338 | 3.72 |
ENST00000496168.1
|
IGKV1-5
|
immunoglobulin kappa variable 1-5 |
| chr6_+_138188378 | 3.47 |
ENST00000420009.1
|
TNFAIP3
|
tumor necrosis factor, alpha-induced protein 3 |
| chr14_-_106610852 | 3.47 |
ENST00000390603.2
|
IGHV3-15
|
immunoglobulin heavy variable 3-15 |
| chr2_-_89310012 | 3.44 |
ENST00000493819.1
|
IGKV1-9
|
immunoglobulin kappa variable 1-9 |
| chr8_-_23712312 | 3.36 |
ENST00000290271.2
|
STC1
|
stanniocalcin 1 |
| chr6_+_138188351 | 3.16 |
ENST00000421450.1
|
TNFAIP3
|
tumor necrosis factor, alpha-induced protein 3 |
| chr19_-_6591113 | 3.05 |
ENST00000423145.3
ENST00000245903.3 |
CD70
|
CD70 molecule |
| chr6_+_138188551 | 3.00 |
ENST00000237289.4
ENST00000433680.1 |
TNFAIP3
|
tumor necrosis factor, alpha-induced protein 3 |
| chr7_-_16844611 | 2.67 |
ENST00000401412.1
ENST00000419304.2 |
AGR2
|
anterior gradient 2 |
| chr10_-_97050777 | 2.56 |
ENST00000329399.6
|
PDLIM1
|
PDZ and LIM domain 1 |
| chr10_+_89419370 | 2.45 |
ENST00000361175.4
ENST00000456849.1 |
PAPSS2
|
3'-phosphoadenosine 5'-phosphosulfate synthase 2 |
| chr2_+_228678550 | 2.44 |
ENST00000409189.3
ENST00000358813.4 |
CCL20
|
chemokine (C-C motif) ligand 20 |
| chr12_-_9913489 | 2.42 |
ENST00000228434.3
ENST00000536709.1 |
CD69
|
CD69 molecule |
| chr10_+_30723045 | 2.42 |
ENST00000542547.1
ENST00000415139.1 |
MAP3K8
|
mitogen-activated protein kinase kinase kinase 8 |
| chr22_+_22676808 | 2.36 |
ENST00000390290.2
|
IGLV1-51
|
immunoglobulin lambda variable 1-51 |
| chr2_+_87808725 | 2.30 |
ENST00000413202.1
|
LINC00152
|
long intergenic non-protein coding RNA 152 |
| chr19_-_46272106 | 2.26 |
ENST00000560168.1
|
SIX5
|
SIX homeobox 5 |
| chr17_-_73511504 | 2.22 |
ENST00000581870.1
|
CASKIN2
|
CASK interacting protein 2 |
| chr9_+_101705893 | 2.21 |
ENST00000375001.3
|
COL15A1
|
collagen, type XV, alpha 1 |
| chr6_-_29527702 | 2.16 |
ENST00000377050.4
|
UBD
|
ubiquitin D |
| chr9_-_117880477 | 2.10 |
ENST00000534839.1
ENST00000340094.3 ENST00000535648.1 ENST00000346706.3 ENST00000345230.3 ENST00000350763.4 |
TNC
|
tenascin C |
| chr10_+_30722866 | 2.07 |
ENST00000263056.1
|
MAP3K8
|
mitogen-activated protein kinase kinase kinase 8 |
| chr20_+_43803517 | 2.03 |
ENST00000243924.3
|
PI3
|
peptidase inhibitor 3, skin-derived |
| chr9_+_72658490 | 1.96 |
ENST00000377182.4
|
MAMDC2
|
MAM domain containing 2 |
| chr18_+_21529811 | 1.92 |
ENST00000588004.1
|
LAMA3
|
laminin, alpha 3 |
| chr15_-_20193370 | 1.88 |
ENST00000558565.2
|
IGHV3OR15-7
|
immunoglobulin heavy variable 3/OR15-7 (pseudogene) |
| chr11_-_5271122 | 1.84 |
ENST00000330597.3
|
HBG1
|
hemoglobin, gamma A |
| chr5_-_138842286 | 1.83 |
ENST00000515823.1
|
ECSCR
|
endothelial cell surface expressed chemotaxis and apoptosis regulator |
| chr3_-_112360116 | 1.83 |
ENST00000206423.3
ENST00000439685.2 |
CCDC80
|
coiled-coil domain containing 80 |
| chr3_-_168864315 | 1.77 |
ENST00000475754.1
ENST00000484519.1 |
MECOM
|
MDS1 and EVI1 complex locus |
| chr17_-_73511584 | 1.73 |
ENST00000321617.3
|
CASKIN2
|
CASK interacting protein 2 |
| chr12_+_102271436 | 1.68 |
ENST00000544152.1
|
DRAM1
|
DNA-damage regulated autophagy modulator 1 |
| chr10_-_6104253 | 1.61 |
ENST00000256876.6
ENST00000379954.1 ENST00000379959.3 |
IL2RA
|
interleukin 2 receptor, alpha |
| chr11_+_102188224 | 1.59 |
ENST00000263464.3
|
BIRC3
|
baculoviral IAP repeat containing 3 |
| chr14_+_103589789 | 1.59 |
ENST00000558056.1
ENST00000560869.1 |
TNFAIP2
|
tumor necrosis factor, alpha-induced protein 2 |
| chr11_+_102188272 | 1.59 |
ENST00000532808.1
|
BIRC3
|
baculoviral IAP repeat containing 3 |
| chr16_-_88717482 | 1.56 |
ENST00000261623.3
|
CYBA
|
cytochrome b-245, alpha polypeptide |
| chr6_-_31550192 | 1.55 |
ENST00000429299.2
ENST00000446745.2 |
LTB
|
lymphotoxin beta (TNF superfamily, member 3) |
| chr18_+_3252265 | 1.55 |
ENST00000580887.1
ENST00000536605.1 |
MYL12A
|
myosin, light chain 12A, regulatory, non-sarcomeric |
| chrX_-_153599578 | 1.54 |
ENST00000360319.4
ENST00000344736.4 |
FLNA
|
filamin A, alpha |
| chr13_-_29069232 | 1.54 |
ENST00000282397.4
ENST00000541932.1 ENST00000539099.1 |
FLT1
|
fms-related tyrosine kinase 1 |
| chr19_-_46272462 | 1.54 |
ENST00000317578.6
|
SIX5
|
SIX homeobox 5 |
| chr1_-_159894319 | 1.54 |
ENST00000320307.4
|
TAGLN2
|
transgelin 2 |
| chr4_+_74735102 | 1.53 |
ENST00000395761.3
|
CXCL1
|
chemokine (C-X-C motif) ligand 1 (melanoma growth stimulating activity, alpha) |
| chr1_-_113249948 | 1.52 |
ENST00000339083.7
ENST00000369642.3 |
RHOC
|
ras homolog family member C |
| chr1_-_29508321 | 1.50 |
ENST00000546138.1
|
SRSF4
|
serine/arginine-rich splicing factor 4 |
| chr22_+_37318082 | 1.49 |
ENST00000406230.1
|
CSF2RB
|
colony stimulating factor 2 receptor, beta, low-affinity (granulocyte-macrophage) |
| chr19_+_41725140 | 1.48 |
ENST00000359092.3
|
AXL
|
AXL receptor tyrosine kinase |
| chr11_-_128392085 | 1.46 |
ENST00000526145.2
ENST00000531611.1 ENST00000319397.6 ENST00000345075.4 ENST00000535549.1 |
ETS1
|
v-ets avian erythroblastosis virus E26 oncogene homolog 1 |
| chr5_+_118604385 | 1.43 |
ENST00000274456.6
|
TNFAIP8
|
tumor necrosis factor, alpha-induced protein 8 |
| chr20_+_44746939 | 1.43 |
ENST00000372276.3
|
CD40
|
CD40 molecule, TNF receptor superfamily member 5 |
| chr1_-_204436344 | 1.43 |
ENST00000367184.2
|
PIK3C2B
|
phosphatidylinositol-4-phosphate 3-kinase, catalytic subunit type 2 beta |
| chr4_-_76957214 | 1.41 |
ENST00000306621.3
|
CXCL11
|
chemokine (C-X-C motif) ligand 11 |
| chr12_+_53491220 | 1.39 |
ENST00000548547.1
ENST00000301464.3 |
IGFBP6
|
insulin-like growth factor binding protein 6 |
| chr12_-_7281469 | 1.38 |
ENST00000542370.1
ENST00000266560.3 |
RBP5
|
retinol binding protein 5, cellular |
| chr22_-_24181174 | 1.38 |
ENST00000318109.7
ENST00000406855.3 ENST00000404056.1 ENST00000476077.1 |
DERL3
|
derlin 3 |
| chr22_+_46546406 | 1.37 |
ENST00000440343.1
ENST00000415785.1 |
PPARA
|
peroxisome proliferator-activated receptor alpha |
| chr1_+_201979743 | 1.35 |
ENST00000446188.1
|
ELF3
|
E74-like factor 3 (ets domain transcription factor, epithelial-specific ) |
| chr12_+_50497784 | 1.35 |
ENST00000548814.1
|
GPD1
|
glycerol-3-phosphate dehydrogenase 1 (soluble) |
| chr19_+_41725088 | 1.34 |
ENST00000301178.4
|
AXL
|
AXL receptor tyrosine kinase |
| chr12_-_77272765 | 1.32 |
ENST00000547435.1
ENST00000552330.1 ENST00000546966.1 ENST00000311083.5 |
CSRP2
|
cysteine and glycine-rich protein 2 |
| chr20_+_44746885 | 1.31 |
ENST00000372285.3
|
CD40
|
CD40 molecule, TNF receptor superfamily member 5 |
| chr8_-_119964434 | 1.30 |
ENST00000297350.4
|
TNFRSF11B
|
tumor necrosis factor receptor superfamily, member 11b |
| chr8_-_49834299 | 1.29 |
ENST00000396822.1
|
SNAI2
|
snail family zinc finger 2 |
| chr18_+_21452964 | 1.29 |
ENST00000587184.1
|
LAMA3
|
laminin, alpha 3 |
| chr12_+_102271129 | 1.29 |
ENST00000258534.8
|
DRAM1
|
DNA-damage regulated autophagy modulator 1 |
| chr11_-_65359947 | 1.28 |
ENST00000597463.1
|
AP001362.1
|
Uncharacterized protein |
| chrX_-_17878827 | 1.27 |
ENST00000360011.1
|
RAI2
|
retinoic acid induced 2 |
| chr7_+_143771275 | 1.26 |
ENST00000408898.2
|
OR2A25
|
olfactory receptor, family 2, subfamily A, member 25 |
| chr19_-_47734448 | 1.25 |
ENST00000439096.2
|
BBC3
|
BCL2 binding component 3 |
| chr15_+_33010175 | 1.23 |
ENST00000300177.4
ENST00000560677.1 ENST00000560830.1 |
GREM1
|
gremlin 1, DAN family BMP antagonist |
| chr10_+_104154229 | 1.23 |
ENST00000428099.1
ENST00000369966.3 |
NFKB2
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 (p49/p100) |
| chr5_-_141703713 | 1.22 |
ENST00000511815.1
|
SPRY4
|
sprouty homolog 4 (Drosophila) |
| chr5_-_150467221 | 1.21 |
ENST00000522226.1
|
TNIP1
|
TNFAIP3 interacting protein 1 |
| chr4_-_74904398 | 1.20 |
ENST00000296026.4
|
CXCL3
|
chemokine (C-X-C motif) ligand 3 |
| chr19_-_17958771 | 1.20 |
ENST00000534444.1
|
JAK3
|
Janus kinase 3 |
| chr16_+_31044812 | 1.20 |
ENST00000313843.3
|
STX4
|
syntaxin 4 |
| chr16_-_88717423 | 1.18 |
ENST00000568278.1
ENST00000569359.1 ENST00000567174.1 |
CYBA
|
cytochrome b-245, alpha polypeptide |
| chr3_+_89156674 | 1.18 |
ENST00000336596.2
|
EPHA3
|
EPH receptor A3 |
| chr10_+_30723105 | 1.17 |
ENST00000375322.2
|
MAP3K8
|
mitogen-activated protein kinase kinase kinase 8 |
| chr1_+_201979645 | 1.17 |
ENST00000367284.5
ENST00000367283.3 |
ELF3
|
E74-like factor 3 (ets domain transcription factor, epithelial-specific ) |
| chr17_-_76356148 | 1.15 |
ENST00000587578.1
ENST00000330871.2 |
SOCS3
|
suppressor of cytokine signaling 3 |
| chr18_+_21452804 | 1.15 |
ENST00000269217.6
|
LAMA3
|
laminin, alpha 3 |
| chr10_+_90750378 | 1.14 |
ENST00000355740.2
ENST00000352159.4 |
FAS
|
Fas cell surface death receptor |
| chr1_-_204380919 | 1.14 |
ENST00000367188.4
|
PPP1R15B
|
protein phosphatase 1, regulatory subunit 15B |
| chr10_+_104155450 | 1.12 |
ENST00000471698.1
ENST00000189444.6 |
NFKB2
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 (p49/p100) |
| chr5_-_115890554 | 1.11 |
ENST00000509665.1
|
SEMA6A
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A |
| chr3_+_89156799 | 1.10 |
ENST00000452448.2
ENST00000494014.1 |
EPHA3
|
EPH receptor A3 |
| chr4_+_169418195 | 1.10 |
ENST00000261509.6
ENST00000335742.7 |
PALLD
|
palladin, cytoskeletal associated protein |
| chr4_-_74864386 | 1.08 |
ENST00000296027.4
|
CXCL5
|
chemokine (C-X-C motif) ligand 5 |
| chr3_+_101546827 | 1.08 |
ENST00000461724.1
ENST00000483180.1 ENST00000394054.2 |
NFKBIZ
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, zeta |
| chr8_-_49833978 | 1.07 |
ENST00000020945.1
|
SNAI2
|
snail family zinc finger 2 |
| chr19_+_14142535 | 1.07 |
ENST00000263379.2
|
IL27RA
|
interleukin 27 receptor, alpha |
| chr2_+_201987200 | 1.06 |
ENST00000425030.1
|
CFLAR
|
CASP8 and FADD-like apoptosis regulator |
| chr2_-_43453734 | 1.05 |
ENST00000282388.3
|
ZFP36L2
|
ZFP36 ring finger protein-like 2 |
| chr1_-_207143802 | 1.05 |
ENST00000324852.4
ENST00000400962.3 |
FCAMR
|
Fc receptor, IgA, IgM, high affinity |
| chr12_+_51632638 | 1.04 |
ENST00000549732.2
|
DAZAP2
|
DAZ associated protein 2 |
| chr6_+_152126790 | 1.04 |
ENST00000456483.2
|
ESR1
|
estrogen receptor 1 |
| chr4_+_169418255 | 1.03 |
ENST00000505667.1
ENST00000511948.1 |
PALLD
|
palladin, cytoskeletal associated protein |
| chr15_-_51610687 | 1.03 |
ENST00000559646.1
|
CYP19A1
|
cytochrome P450, family 19, subfamily A, polypeptide 1 |
| chr12_+_51632666 | 1.02 |
ENST00000604900.1
|
DAZAP2
|
DAZ associated protein 2 |
| chr6_+_135502408 | 1.01 |
ENST00000341911.5
ENST00000442647.2 ENST00000316528.8 |
MYB
|
v-myb avian myeloblastosis viral oncogene homolog |
| chr5_-_150466692 | 1.00 |
ENST00000315050.7
ENST00000523338.1 ENST00000522100.1 |
TNIP1
|
TNFAIP3 interacting protein 1 |
| chr1_+_16767195 | 0.97 |
ENST00000504551.2
ENST00000457722.2 ENST00000406746.1 ENST00000443980.2 |
NECAP2
|
NECAP endocytosis associated 2 |
| chr19_+_41222998 | 0.96 |
ENST00000263370.2
|
ITPKC
|
inositol-trisphosphate 3-kinase C |
| chr19_-_4338838 | 0.92 |
ENST00000594605.1
|
STAP2
|
signal transducing adaptor family member 2 |
| chr2_+_202047843 | 0.91 |
ENST00000272879.5
ENST00000374650.3 ENST00000346817.5 ENST00000313728.7 ENST00000448480.1 |
CASP10
|
caspase 10, apoptosis-related cysteine peptidase |
| chr1_-_209824643 | 0.91 |
ENST00000391911.1
ENST00000415782.1 |
LAMB3
|
laminin, beta 3 |
| chr12_-_57504069 | 0.90 |
ENST00000543873.2
ENST00000554663.1 ENST00000557635.1 |
STAT6
|
signal transducer and activator of transcription 6, interleukin-4 induced |
| chr11_-_104893863 | 0.90 |
ENST00000260315.3
ENST00000526056.1 ENST00000531367.1 ENST00000456094.1 ENST00000444749.2 ENST00000393141.2 ENST00000418434.1 ENST00000393139.2 |
CASP5
|
caspase 5, apoptosis-related cysteine peptidase |
| chr17_+_40440481 | 0.87 |
ENST00000590726.2
ENST00000452307.2 ENST00000444283.1 ENST00000588868.1 |
STAT5A
|
signal transducer and activator of transcription 5A |
| chr1_+_110453203 | 0.85 |
ENST00000357302.4
ENST00000344188.5 ENST00000329608.6 |
CSF1
|
colony stimulating factor 1 (macrophage) |
| chr1_+_45274154 | 0.85 |
ENST00000450269.1
ENST00000453418.1 ENST00000409335.2 |
BTBD19
|
BTB (POZ) domain containing 19 |
| chr3_-_156878540 | 0.84 |
ENST00000461804.1
|
CCNL1
|
cyclin L1 |
| chr3_-_156878482 | 0.84 |
ENST00000295925.4
|
CCNL1
|
cyclin L1 |
| chr14_-_53417732 | 0.82 |
ENST00000399304.3
ENST00000395631.2 ENST00000341590.3 ENST00000343279.4 |
FERMT2
|
fermitin family member 2 |
| chr20_-_45530365 | 0.82 |
ENST00000414085.1
|
RP11-323C15.2
|
RP11-323C15.2 |
| chr16_-_2059748 | 0.82 |
ENST00000562103.1
ENST00000431526.1 |
ZNF598
|
zinc finger protein 598 |
| chr14_+_75988768 | 0.81 |
ENST00000286639.6
|
BATF
|
basic leucine zipper transcription factor, ATF-like |
| chr9_+_130547958 | 0.81 |
ENST00000421939.1
ENST00000373265.2 |
CDK9
|
cyclin-dependent kinase 9 |
| chr16_+_27325202 | 0.81 |
ENST00000395762.2
ENST00000562142.1 ENST00000561742.1 ENST00000543915.2 ENST00000449195.1 ENST00000380922.3 ENST00000563002.1 |
IL4R
|
interleukin 4 receptor |
| chr9_+_130911723 | 0.80 |
ENST00000277480.2
ENST00000373013.2 ENST00000540948.1 |
LCN2
|
lipocalin 2 |
| chr16_-_122619 | 0.80 |
ENST00000262316.6
|
RHBDF1
|
rhomboid 5 homolog 1 (Drosophila) |
| chr11_+_46299199 | 0.80 |
ENST00000529193.1
ENST00000288400.3 |
CREB3L1
|
cAMP responsive element binding protein 3-like 1 |
| chr19_-_17958832 | 0.80 |
ENST00000458235.1
|
JAK3
|
Janus kinase 3 |
| chr17_+_40440094 | 0.80 |
ENST00000546010.2
|
STAT5A
|
signal transducer and activator of transcription 5A |
| chr13_-_38443860 | 0.80 |
ENST00000426868.2
ENST00000379681.3 ENST00000338947.5 ENST00000355779.2 ENST00000358477.2 ENST00000379673.2 |
TRPC4
|
transient receptor potential cation channel, subfamily C, member 4 |
| chr21_+_34775698 | 0.78 |
ENST00000381995.1
|
IFNGR2
|
interferon gamma receptor 2 (interferon gamma transducer 1) |
| chr10_+_17270214 | 0.77 |
ENST00000544301.1
|
VIM
|
vimentin |
| chr9_+_130911770 | 0.77 |
ENST00000372998.1
|
LCN2
|
lipocalin 2 |
| chr12_+_57388230 | 0.75 |
ENST00000300098.1
|
GPR182
|
G protein-coupled receptor 182 |
| chr16_+_27324983 | 0.75 |
ENST00000566117.1
|
IL4R
|
interleukin 4 receptor |
| chr8_+_70404996 | 0.75 |
ENST00000402687.4
ENST00000419716.3 |
SULF1
|
sulfatase 1 |
| chr21_+_34775772 | 0.74 |
ENST00000405436.1
|
IFNGR2
|
interferon gamma receptor 2 (interferon gamma transducer 1) |
| chrX_+_115567767 | 0.74 |
ENST00000371900.4
|
SLC6A14
|
solute carrier family 6 (amino acid transporter), member 14 |
| chr6_+_29910301 | 0.74 |
ENST00000376809.5
ENST00000376802.2 |
HLA-A
|
major histocompatibility complex, class I, A |
| chr21_+_46875424 | 0.73 |
ENST00000359759.4
|
COL18A1
|
collagen, type XVIII, alpha 1 |
| chr2_+_220094479 | 0.73 |
ENST00000323348.5
ENST00000453432.1 ENST00000409849.1 ENST00000416565.1 ENST00000410034.3 ENST00000447157.1 |
ANKZF1
|
ankyrin repeat and zinc finger domain containing 1 |
| chr6_-_131291572 | 0.72 |
ENST00000529208.1
|
EPB41L2
|
erythrocyte membrane protein band 4.1-like 2 |
| chr2_+_162016916 | 0.72 |
ENST00000405852.1
|
TANK
|
TRAF family member-associated NFKB activator |
| chr15_+_57891609 | 0.71 |
ENST00000569089.1
|
MYZAP
|
myocardial zonula adherens protein |
| chr3_-_183967296 | 0.71 |
ENST00000455059.1
ENST00000445626.2 |
ALG3
|
ALG3, alpha-1,3- mannosyltransferase |
| chr11_+_120255997 | 0.70 |
ENST00000532993.1
|
ARHGEF12
|
Rho guanine nucleotide exchange factor (GEF) 12 |
| chr1_+_145477060 | 0.70 |
ENST00000369308.3
|
LIX1L
|
Lix1 homolog (mouse)-like |
| chr10_+_90750493 | 0.69 |
ENST00000357339.2
ENST00000355279.2 |
FAS
|
Fas cell surface death receptor |
| chr17_+_34639793 | 0.68 |
ENST00000394465.2
ENST00000394463.2 ENST00000378342.4 |
CCL4L2
|
chemokine (C-C motif) ligand 4-like 2 |
| chr17_+_38474489 | 0.68 |
ENST00000394089.2
ENST00000425707.3 |
RARA
|
retinoic acid receptor, alpha |
| chr5_-_149792295 | 0.68 |
ENST00000518797.1
ENST00000524315.1 ENST00000009530.7 ENST00000377795.3 |
CD74
|
CD74 molecule, major histocompatibility complex, class II invariant chain |
| chr1_-_154474589 | 0.68 |
ENST00000304760.2
|
SHE
|
Src homology 2 domain containing E |
| chr1_-_89738528 | 0.67 |
ENST00000343435.5
|
GBP5
|
guanylate binding protein 5 |
| chr1_+_165796753 | 0.66 |
ENST00000367879.4
|
UCK2
|
uridine-cytidine kinase 2 |
| chr1_-_9189144 | 0.66 |
ENST00000414642.2
|
GPR157
|
G protein-coupled receptor 157 |
| chr6_+_151042224 | 0.66 |
ENST00000358517.2
|
PLEKHG1
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 1 |
| chr19_+_41256764 | 0.66 |
ENST00000243563.3
ENST00000601253.1 ENST00000597353.1 ENST00000599362.1 |
SNRPA
|
small nuclear ribonucleoprotein polypeptide A |
| chr9_-_115095123 | 0.65 |
ENST00000458258.1
|
PTBP3
|
polypyrimidine tract binding protein 3 |
| chr3_+_93698974 | 0.64 |
ENST00000535334.1
ENST00000478400.1 ENST00000303097.7 ENST00000394222.3 ENST00000471138.1 ENST00000539730.1 |
ARL13B
|
ADP-ribosylation factor-like 13B |
| chr19_+_35940486 | 0.63 |
ENST00000246549.2
|
FFAR2
|
free fatty acid receptor 2 |
| chr17_+_26662730 | 0.63 |
ENST00000226225.2
|
TNFAIP1
|
tumor necrosis factor, alpha-induced protein 1 (endothelial) |
| chr6_+_32121908 | 0.63 |
ENST00000375143.2
ENST00000424499.1 |
PPT2
|
palmitoyl-protein thioesterase 2 |
| chr12_-_76425368 | 0.62 |
ENST00000602540.1
|
PHLDA1
|
pleckstrin homology-like domain, family A, member 1 |
| chr11_+_46316677 | 0.62 |
ENST00000534787.1
|
CREB3L1
|
cAMP responsive element binding protein 3-like 1 |
| chr12_-_49504655 | 0.62 |
ENST00000551782.1
ENST00000267102.8 |
LMBR1L
|
limb development membrane protein 1-like |
| chr21_+_34638656 | 0.61 |
ENST00000290200.2
|
IL10RB
|
interleukin 10 receptor, beta |
| chr14_-_69446034 | 0.61 |
ENST00000193403.6
|
ACTN1
|
actinin, alpha 1 |
| chr2_+_173940668 | 0.61 |
ENST00000375213.3
|
MLTK
|
Mitogen-activated protein kinase kinase kinase MLT |
| chr11_-_75062730 | 0.61 |
ENST00000420843.2
ENST00000360025.3 |
ARRB1
|
arrestin, beta 1 |
| chr2_+_131975882 | 0.61 |
ENST00000356920.5
ENST00000358087.5 |
POTEE
|
POTE ankyrin domain family, member E |
| chr19_+_4229495 | 0.60 |
ENST00000221847.5
|
EBI3
|
Epstein-Barr virus induced 3 |
| chr14_-_69262947 | 0.60 |
ENST00000557086.1
|
ZFP36L1
|
ZFP36 ring finger protein-like 1 |
| chr1_+_110453514 | 0.60 |
ENST00000369802.3
ENST00000420111.2 |
CSF1
|
colony stimulating factor 1 (macrophage) |
| chr15_+_85923797 | 0.59 |
ENST00000559362.1
|
AKAP13
|
A kinase (PRKA) anchor protein 13 |
| chr1_+_144339738 | 0.59 |
ENST00000538264.1
|
AL592284.1
|
Protein LOC642441 |
| chr2_-_114647327 | 0.58 |
ENST00000602760.1
|
RP11-141B14.1
|
RP11-141B14.1 |
| chr5_+_82767284 | 0.58 |
ENST00000265077.3
|
VCAN
|
versican |
| chr9_-_13279406 | 0.58 |
ENST00000546205.1
|
MPDZ
|
multiple PDZ domain protein |
| chr16_+_3096638 | 0.57 |
ENST00000336577.4
|
MMP25
|
matrix metallopeptidase 25 |
| chr1_+_110453462 | 0.57 |
ENST00000488198.1
|
CSF1
|
colony stimulating factor 1 (macrophage) |
| chr4_-_185395191 | 0.57 |
ENST00000510814.1
ENST00000507523.1 ENST00000506230.1 |
IRF2
|
interferon regulatory factor 2 |
| chr11_+_66406149 | 0.57 |
ENST00000578778.1
ENST00000483858.1 ENST00000398692.4 ENST00000510173.2 ENST00000506523.2 ENST00000530235.1 ENST00000532968.1 |
RBM4
|
RNA binding motif protein 4 |
| chr17_-_4643114 | 0.55 |
ENST00000293778.6
|
CXCL16
|
chemokine (C-X-C motif) ligand 16 |
| chr12_-_40013553 | 0.55 |
ENST00000308666.3
|
ABCD2
|
ATP-binding cassette, sub-family D (ALD), member 2 |
| chr1_-_8086343 | 0.55 |
ENST00000474874.1
ENST00000469499.1 ENST00000377482.5 |
ERRFI1
|
ERBB receptor feedback inhibitor 1 |
| chr6_+_32121789 | 0.55 |
ENST00000437001.2
ENST00000375137.2 |
PPT2
|
palmitoyl-protein thioesterase 2 |
| chr2_-_232571621 | 0.55 |
ENST00000595658.1
|
MGC4771
|
MGC4771 |
| chr14_-_69262916 | 0.54 |
ENST00000553375.1
|
ZFP36L1
|
ZFP36 ring finger protein-like 1 |
| chr10_-_29923893 | 0.54 |
ENST00000355867.4
|
SVIL
|
supervillin |
| chr13_+_97928395 | 0.53 |
ENST00000445661.2
|
MBNL2
|
muscleblind-like splicing regulator 2 |
| chr22_-_39268308 | 0.53 |
ENST00000407418.3
|
CBX6
|
chromobox homolog 6 |
| chr12_+_56211703 | 0.53 |
ENST00000243045.5
ENST00000552672.1 ENST00000550836.1 |
ORMDL2
|
ORM1-like 2 (S. cerevisiae) |
| chr10_-_100027943 | 0.52 |
ENST00000260702.3
|
LOXL4
|
lysyl oxidase-like 4 |
| chr12_+_51632600 | 0.51 |
ENST00000549555.1
ENST00000439799.2 ENST00000425012.2 |
DAZAP2
|
DAZ associated protein 2 |
| chr6_-_143832820 | 0.51 |
ENST00000002165.6
|
FUCA2
|
fucosidase, alpha-L- 2, plasma |
| chr11_-_58345569 | 0.51 |
ENST00000528954.1
ENST00000528489.1 |
LPXN
|
leupaxin |
| chr17_-_8286484 | 0.51 |
ENST00000582556.1
ENST00000584164.1 ENST00000293842.5 ENST00000584343.1 ENST00000578812.1 ENST00000583011.1 |
RPL26
|
ribosomal protein L26 |
| chr17_-_4852332 | 0.51 |
ENST00000572383.1
|
PFN1
|
profilin 1 |
| chr12_-_49318715 | 0.51 |
ENST00000444214.2
|
FKBP11
|
FK506 binding protein 11, 19 kDa |
Network of associatons between targets according to the STRING database.
First level regulatory network of REL
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.2 | 9.6 | GO:0072573 | regulation of toll-like receptor 5 signaling pathway(GO:0034147) negative regulation of toll-like receptor 5 signaling pathway(GO:0034148) negative regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070429) tolerance induction to lipopolysaccharide(GO:0072573) |
| 1.0 | 3.8 | GO:0014859 | negative regulation of skeletal muscle cell proliferation(GO:0014859) negative regulation of skeletal muscle satellite cell proliferation(GO:1902723) |
| 0.9 | 2.6 | GO:0045210 | FasL biosynthetic process(GO:0045210) |
| 0.8 | 2.4 | GO:0045362 | regulation of interleukin-1 biosynthetic process(GO:0045360) positive regulation of interleukin-1 biosynthetic process(GO:0045362) |
| 0.8 | 2.4 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 0.6 | 1.2 | GO:0090291 | negative regulation of osteoclast proliferation(GO:0090291) |
| 0.6 | 2.5 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.6 | 2.3 | GO:0002268 | follicular dendritic cell differentiation(GO:0002268) |
| 0.6 | 2.3 | GO:0097156 | fasciculation of motor neuron axon(GO:0097156) |
| 0.5 | 2.7 | GO:1904844 | response to L-glutamine(GO:1904844) cellular response to L-glutamine(GO:1904845) |
| 0.5 | 1.5 | GO:1905000 | regulation of membrane repolarization during atrial cardiac muscle cell action potential(GO:1905000) |
| 0.5 | 2.5 | GO:1903971 | mammary gland fat development(GO:0060611) positive regulation of macrophage colony-stimulating factor signaling pathway(GO:1902228) positive regulation of response to macrophage colony-stimulating factor(GO:1903971) positive regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903974) positive regulation of microglial cell migration(GO:1904141) |
| 0.5 | 2.4 | GO:0052027 | modulation of signal transduction in other organism(GO:0044501) modulation by symbiont of host signal transduction pathway(GO:0052027) modulation of signal transduction in other organism involved in symbiotic interaction(GO:0052250) modulation by symbiont of host I-kappaB kinase/NF-kappaB cascade(GO:0085032) |
| 0.5 | 1.4 | GO:0046168 | glycerol-3-phosphate catabolic process(GO:0046168) |
| 0.4 | 1.3 | GO:0042489 | negative regulation of odontogenesis of dentin-containing tooth(GO:0042489) |
| 0.4 | 1.3 | GO:0072361 | regulation of glycolytic process by regulation of transcription from RNA polymerase II promoter(GO:0072361) |
| 0.4 | 2.1 | GO:0060447 | bud outgrowth involved in lung branching(GO:0060447) |
| 0.4 | 2.8 | GO:2000669 | negative regulation of dendritic cell apoptotic process(GO:2000669) |
| 0.4 | 1.6 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.4 | 1.2 | GO:1902568 | positive regulation of eosinophil degranulation(GO:0043311) positive regulation of eosinophil activation(GO:1902568) |
| 0.4 | 1.6 | GO:0045626 | negative regulation of T-helper 1 cell differentiation(GO:0045626) |
| 0.4 | 1.6 | GO:0045084 | positive regulation of interleukin-12 biosynthetic process(GO:0045084) |
| 0.4 | 2.7 | GO:1903899 | lung goblet cell differentiation(GO:0060480) positive regulation of PERK-mediated unfolded protein response(GO:1903899) |
| 0.3 | 2.7 | GO:0033590 | response to cobalamin(GO:0033590) cellular response to erythropoietin(GO:0036018) |
| 0.3 | 4.1 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.3 | 1.0 | GO:1904899 | regulation of hepatic stellate cell proliferation(GO:1904897) positive regulation of hepatic stellate cell proliferation(GO:1904899) hepatic stellate cell proliferation(GO:1990922) |
| 0.3 | 1.6 | GO:0015891 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 0.3 | 3.7 | GO:1903142 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.3 | 1.5 | GO:0038043 | interleukin-5-mediated signaling pathway(GO:0038043) interleukin-3-mediated signaling pathway(GO:0038156) |
| 0.3 | 3.2 | GO:0070424 | regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070424) |
| 0.3 | 1.8 | GO:0097527 | necroptotic signaling pathway(GO:0097527) |
| 0.3 | 1.0 | GO:2000863 | positive regulation of estrogen secretion(GO:2000863) positive regulation of estradiol secretion(GO:2000866) |
| 0.3 | 1.5 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 0.2 | 1.7 | GO:0000255 | allantoin metabolic process(GO:0000255) |
| 0.2 | 0.5 | GO:1902523 | positive regulation of protein K63-linked ubiquitination(GO:1902523) |
| 0.2 | 1.2 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.2 | 1.1 | GO:0097403 | cellular response to raffinose(GO:0097403) response to raffinose(GO:1901545) |
| 0.2 | 0.7 | GO:0072616 | interleukin-18 secretion(GO:0072616) |
| 0.2 | 0.7 | GO:0006425 | glutaminyl-tRNA aminoacylation(GO:0006425) |
| 0.2 | 0.6 | GO:0002752 | cell surface pattern recognition receptor signaling pathway(GO:0002752) |
| 0.2 | 0.8 | GO:0072344 | rescue of stalled ribosome(GO:0072344) |
| 0.2 | 2.2 | GO:0070842 | aggresome assembly(GO:0070842) |
| 0.2 | 0.9 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.2 | 1.5 | GO:0030578 | PML body organization(GO:0030578) |
| 0.2 | 0.9 | GO:0002296 | T-helper 1 cell lineage commitment(GO:0002296) |
| 0.2 | 1.0 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.2 | 1.0 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.2 | 0.7 | GO:0006238 | CMP salvage(GO:0006238) CMP biosynthetic process(GO:0009224) CMP metabolic process(GO:0046035) |
| 0.1 | 0.7 | GO:1902534 | single-organism membrane invagination(GO:1902534) |
| 0.1 | 5.3 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 1.4 | GO:0070278 | extracellular matrix constituent secretion(GO:0070278) |
| 0.1 | 4.4 | GO:0090023 | positive regulation of neutrophil chemotaxis(GO:0090023) |
| 0.1 | 2.5 | GO:0060056 | mammary gland involution(GO:0060056) |
| 0.1 | 0.4 | GO:0097252 | oligodendrocyte apoptotic process(GO:0097252) |
| 0.1 | 0.7 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.1 | 1.7 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.1 | 1.1 | GO:1903898 | negative regulation of PERK-mediated unfolded protein response(GO:1903898) negative regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation(GO:1903912) |
| 0.1 | 1.8 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.1 | 20.4 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.1 | 1.4 | GO:0043950 | positive regulation of cAMP-mediated signaling(GO:0043950) |
| 0.1 | 1.4 | GO:0070862 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.1 | 0.8 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.1 | 0.5 | GO:0090156 | cellular sphingolipid homeostasis(GO:0090156) |
| 0.1 | 1.1 | GO:1904628 | response to phorbol 13-acetate 12-myristate(GO:1904627) cellular response to phorbol 13-acetate 12-myristate(GO:1904628) |
| 0.1 | 1.0 | GO:0060523 | prostate epithelial cord elongation(GO:0060523) |
| 0.1 | 0.3 | GO:0002522 | leukocyte migration involved in immune response(GO:0002522) |
| 0.1 | 1.1 | GO:0097167 | circadian regulation of translation(GO:0097167) |
| 0.1 | 1.0 | GO:0042699 | follicle-stimulating hormone signaling pathway(GO:0042699) |
| 0.1 | 0.6 | GO:0035234 | ectopic germ cell programmed cell death(GO:0035234) |
| 0.1 | 1.8 | GO:2000353 | positive regulation of endothelial cell apoptotic process(GO:2000353) |
| 0.1 | 0.4 | GO:0046967 | cytosol to ER transport(GO:0046967) |
| 0.1 | 1.1 | GO:1903943 | negative regulation of myoblast fusion(GO:1901740) regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.1 | 0.5 | GO:2001153 | regulation of renal water transport(GO:2001151) positive regulation of renal water transport(GO:2001153) |
| 0.1 | 0.3 | GO:0070427 | nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070427) |
| 0.1 | 1.2 | GO:0098734 | macromolecule depalmitoylation(GO:0098734) |
| 0.1 | 1.0 | GO:1903243 | negative regulation of cardiac muscle adaptation(GO:0010616) negative regulation of cardiac muscle hypertrophy in response to stress(GO:1903243) |
| 0.1 | 0.8 | GO:2001168 | regulation of histone H2B ubiquitination(GO:2001166) positive regulation of histone H2B ubiquitination(GO:2001168) |
| 0.1 | 1.6 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.1 | 0.8 | GO:0014051 | gamma-aminobutyric acid secretion(GO:0014051) |
| 0.1 | 2.1 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.1 | 0.6 | GO:0016344 | meiotic chromosome movement towards spindle pole(GO:0016344) |
| 0.1 | 1.1 | GO:0048302 | regulation of isotype switching to IgG isotypes(GO:0048302) |
| 0.1 | 0.7 | GO:0002480 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-independent(GO:0002480) |
| 0.1 | 1.8 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.1 | 0.5 | GO:2000535 | regulation of entry of bacterium into host cell(GO:2000535) |
| 0.1 | 0.4 | GO:0038110 | interleukin-2-mediated signaling pathway(GO:0038110) |
| 0.1 | 0.5 | GO:1902231 | regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902162) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.1 | 0.5 | GO:0071630 | nucleus-associated proteasomal ubiquitin-dependent protein catabolic process(GO:0071630) |
| 0.1 | 0.6 | GO:0042760 | very long-chain fatty acid catabolic process(GO:0042760) |
| 0.1 | 2.0 | GO:0007620 | copulation(GO:0007620) |
| 0.1 | 1.5 | GO:0030949 | positive regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030949) |
| 0.1 | 0.7 | GO:0050859 | negative regulation of B cell receptor signaling pathway(GO:0050859) |
| 0.1 | 0.8 | GO:0060020 | Bergmann glial cell differentiation(GO:0060020) |
| 0.1 | 0.6 | GO:0021943 | formation of radial glial scaffolds(GO:0021943) |
| 0.1 | 1.1 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.1 | 0.7 | GO:0060019 | radial glial cell differentiation(GO:0060019) |
| 0.1 | 5.6 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.1 | 0.4 | GO:1902527 | positive regulation of protein monoubiquitination(GO:1902527) |
| 0.1 | 0.5 | GO:1903288 | positive regulation of potassium ion import(GO:1903288) |
| 0.1 | 0.3 | GO:0098502 | DNA dephosphorylation(GO:0098502) |
| 0.1 | 1.2 | GO:0072540 | T-helper 17 cell lineage commitment(GO:0072540) |
| 0.1 | 0.6 | GO:0045078 | positive regulation of interferon-gamma biosynthetic process(GO:0045078) |
| 0.1 | 1.9 | GO:0060334 | regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
| 0.1 | 0.4 | GO:0034670 | chemotaxis to arachidonic acid(GO:0034670) response to arachidonic acid(GO:1904550) |
| 0.0 | 0.2 | GO:0048861 | leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.0 | 0.2 | GO:1903553 | positive regulation of extracellular exosome assembly(GO:1903553) |
| 0.0 | 0.7 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.0 | 1.1 | GO:0010818 | T cell chemotaxis(GO:0010818) |
| 0.0 | 0.6 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.0 | 0.4 | GO:0051599 | response to hydrostatic pressure(GO:0051599) |
| 0.0 | 1.2 | GO:1904776 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.0 | 0.4 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.0 | 0.6 | GO:0060022 | hard palate development(GO:0060022) |
| 0.0 | 0.0 | GO:0042940 | D-amino acid transport(GO:0042940) |
| 0.0 | 0.2 | GO:0060355 | positive regulation of cell adhesion molecule production(GO:0060355) |
| 0.0 | 1.8 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
| 0.0 | 7.4 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.0 | 1.5 | GO:0048025 | negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.0 | 1.4 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.0 | 0.5 | GO:1904936 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.0 | 0.4 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.0 | 0.4 | GO:0045714 | low-density lipoprotein particle receptor biosynthetic process(GO:0045713) regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045714) |
| 0.0 | 0.2 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.0 | 0.7 | GO:1900363 | regulation of mRNA polyadenylation(GO:1900363) |
| 0.0 | 0.2 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.0 | 0.4 | GO:0033235 | positive regulation of protein sumoylation(GO:0033235) |
| 0.0 | 0.6 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 0.4 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) |
| 0.0 | 0.1 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.0 | 0.8 | GO:0033622 | integrin activation(GO:0033622) |
| 0.0 | 0.1 | GO:0061083 | regulation of protein refolding(GO:0061083) negative regulation of protein refolding(GO:0061084) |
| 0.0 | 0.8 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.0 | 0.6 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.0 | 0.3 | GO:0042487 | regulation of odontogenesis of dentin-containing tooth(GO:0042487) |
| 0.0 | 1.2 | GO:0042531 | positive regulation of tyrosine phosphorylation of STAT protein(GO:0042531) |
| 0.0 | 0.4 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.0 | 2.2 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.0 | 0.4 | GO:0060628 | regulation of ER to Golgi vesicle-mediated transport(GO:0060628) |
| 0.0 | 0.4 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.0 | 0.6 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.0 | 0.2 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.0 | 1.4 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 0.9 | GO:0090200 | positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.0 | 0.0 | GO:1902598 | creatine transport(GO:0015881) creatine transmembrane transport(GO:1902598) |
| 0.0 | 1.2 | GO:0070373 | negative regulation of ERK1 and ERK2 cascade(GO:0070373) |
| 0.0 | 0.1 | GO:0006880 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.0 | 1.0 | GO:0000380 | alternative mRNA splicing, via spliceosome(GO:0000380) |
| 0.0 | 0.3 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.0 | 0.3 | GO:0036148 | phosphatidylglycerol acyl-chain remodeling(GO:0036148) |
| 0.0 | 1.2 | GO:0035690 | cellular response to drug(GO:0035690) |
| 0.0 | 0.7 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.0 | 0.2 | GO:0010628 | positive regulation of gene expression(GO:0010628) |
| 0.0 | 0.5 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 1.6 | GO:0043154 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0043154) |
| 0.0 | 0.1 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.3 | GO:0033257 | Bcl3/NF-kappaB2 complex(GO:0033257) |
| 0.5 | 1.5 | GO:0030526 | granulocyte macrophage colony-stimulating factor receptor complex(GO:0030526) |
| 0.5 | 5.3 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.4 | 1.5 | GO:0031523 | Myb complex(GO:0031523) |
| 0.4 | 2.5 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 0.3 | 2.7 | GO:0043196 | varicosity(GO:0043196) |
| 0.2 | 0.6 | GO:0032002 | interleukin-28 receptor complex(GO:0032002) |
| 0.2 | 2.2 | GO:0098651 | basement membrane collagen trimer(GO:0098651) |
| 0.2 | 1.4 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.2 | 4.1 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.1 | 0.4 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.1 | 0.4 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) |
| 0.1 | 0.9 | GO:0072558 | NLRP1 inflammasome complex(GO:0072558) |
| 0.1 | 1.4 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.1 | 2.7 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.1 | 2.8 | GO:0033643 | host cell part(GO:0033643) |
| 0.1 | 3.8 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 1.4 | GO:0016942 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) |
| 0.1 | 1.8 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.1 | 4.7 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.1 | 5.3 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.1 | 0.5 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.1 | 1.2 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.1 | 0.7 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.1 | 0.2 | GO:0005900 | oncostatin-M receptor complex(GO:0005900) |
| 0.1 | 1.0 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.1 | 0.8 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.1 | 0.7 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.1 | 0.5 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.1 | 1.2 | GO:0008091 | spectrin(GO:0008091) |
| 0.1 | 0.4 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.1 | 1.1 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.1 | 0.8 | GO:0098799 | outer mitochondrial membrane protein complex(GO:0098799) |
| 0.1 | 0.4 | GO:0042825 | TAP complex(GO:0042825) |
| 0.0 | 1.6 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.2 | GO:0031251 | PAN complex(GO:0031251) |
| 0.0 | 3.5 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.4 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.0 | 8.8 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 1.4 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.4 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 1.6 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.2 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.0 | 0.8 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 3.4 | GO:0035580 | specific granule lumen(GO:0035580) |
| 0.0 | 0.1 | GO:0070288 | intracellular ferritin complex(GO:0008043) ferritin complex(GO:0070288) |
| 0.0 | 0.7 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.0 | 0.2 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.0 | 2.2 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 0.8 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.0 | 2.0 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 1.7 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.9 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 1.5 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 0.6 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.8 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 1.0 | GO:0030125 | clathrin vesicle coat(GO:0030125) |
| 0.0 | 0.1 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) |
| 0.0 | 1.9 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 0.7 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.0 | 0.9 | GO:0030863 | cortical cytoskeleton(GO:0030863) |
| 0.0 | 0.4 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 2.6 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 0.5 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.0 | 0.3 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 1.6 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 8.7 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 0.2 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 15.1 | GO:0000323 | lytic vacuole(GO:0000323) lysosome(GO:0005764) |
| 0.0 | 0.4 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.0 | 3.0 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 0.2 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 4.4 | GO:0045121 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 1.4 | GO:0016363 | nuclear matrix(GO:0016363) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.5 | GO:0004779 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.8 | 2.4 | GO:0031731 | CCR6 chemokine receptor binding(GO:0031731) |
| 0.5 | 1.6 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) interleukin-2 binding(GO:0019976) |
| 0.5 | 9.0 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.5 | 1.4 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.4 | 1.6 | GO:0004913 | interleukin-4 receptor activity(GO:0004913) |
| 0.4 | 1.1 | GO:0097158 | pre-mRNA intronic pyrimidine-rich binding(GO:0097158) |
| 0.3 | 5.1 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.3 | 1.0 | GO:0031896 | V2 vasopressin receptor binding(GO:0031896) |
| 0.3 | 1.9 | GO:0004906 | interferon-gamma receptor activity(GO:0004906) |
| 0.3 | 2.5 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.3 | 1.2 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
| 0.3 | 1.5 | GO:0004914 | interleukin-3 receptor activity(GO:0004912) interleukin-5 receptor activity(GO:0004914) |
| 0.3 | 1.8 | GO:0043120 | tumor necrosis factor binding(GO:0043120) |
| 0.2 | 0.7 | GO:0004819 | glutamine-tRNA ligase activity(GO:0004819) |
| 0.2 | 1.5 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.2 | 2.1 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.2 | 0.7 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.2 | 1.2 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.2 | 0.5 | GO:0004560 | alpha-L-fucosidase activity(GO:0004560) fucosidase activity(GO:0015928) |
| 0.1 | 2.3 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.1 | 2.8 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.1 | 0.6 | GO:0004641 | phosphoribosylamine-glycine ligase activity(GO:0004637) phosphoribosylformylglycinamidine cyclo-ligase activity(GO:0004641) phosphoribosylglycinamide formyltransferase activity(GO:0004644) |
| 0.1 | 2.0 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.1 | 1.3 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.1 | 1.4 | GO:0035005 | lipid kinase activity(GO:0001727) 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.1 | 1.0 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.1 | 1.5 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.1 | 32.2 | GO:0003823 | antigen binding(GO:0003823) |
| 0.1 | 5.2 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.1 | 1.0 | GO:0038052 | RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.1 | 2.7 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.1 | 0.8 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.1 | 1.2 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.1 | 1.2 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.1 | 0.6 | GO:0004920 | interleukin-10 receptor activity(GO:0004920) |
| 0.1 | 1.5 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.1 | 1.4 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.1 | 1.0 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.1 | 0.7 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.1 | 1.5 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.1 | 4.4 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.1 | 4.8 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.1 | 1.2 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.1 | 0.9 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.1 | 1.8 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.1 | 2.7 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.1 | 0.6 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.1 | 2.2 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.1 | 0.8 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.1 | 0.3 | GO:0004727 | prenylated protein tyrosine phosphatase activity(GO:0004727) |
| 0.1 | 0.3 | GO:0051733 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 0.1 | 2.2 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.1 | 0.7 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.1 | 0.8 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.1 | 1.0 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.1 | 0.5 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.1 | 0.4 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.1 | 2.0 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.1 | 0.9 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.1 | 0.7 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.1 | 2.6 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.1 | 2.1 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.1 | 2.1 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.1 | 1.8 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 3.3 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 2.4 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.0 | 0.8 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 1.0 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.0 | 0.7 | GO:0048185 | activin binding(GO:0048185) |
| 0.0 | 1.7 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.4 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.0 | 0.5 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.1 | GO:0032558 | adenyl deoxyribonucleotide binding(GO:0032558) dATP binding(GO:0032564) |
| 0.0 | 0.3 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.0 | 0.4 | GO:0000990 | transcription factor activity, core RNA polymerase binding(GO:0000990) |
| 0.0 | 1.1 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.0 | 0.4 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.0 | 0.3 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.0 | 0.3 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.5 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.0 | 0.3 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.7 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.0 | 0.5 | GO:0016641 | oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor(GO:0016641) |
| 0.0 | 0.0 | GO:0005308 | creatine transmembrane transporter activity(GO:0005308) |
| 0.0 | 0.5 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.0 | 0.2 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 3.4 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 1.3 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 1.9 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.0 | 0.8 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 2.8 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.4 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) intronic transcription regulatory region DNA binding(GO:0044213) |
| 0.0 | 0.6 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.8 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.5 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 0.5 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.4 | GO:0001163 | RNA polymerase I regulatory region DNA binding(GO:0001013) RNA polymerase I regulatory region sequence-specific DNA binding(GO:0001163) RNA polymerase I CORE element sequence-specific DNA binding(GO:0001164) |
| 0.0 | 2.0 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.1 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 0.8 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.4 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.0 | 0.3 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.4 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.0 | 0.2 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.0 | 1.8 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.0 | 0.8 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 2.7 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 5.9 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) |
| 0.0 | 0.0 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.0 | 0.2 | GO:0001055 | RNA polymerase I activity(GO:0001054) RNA polymerase II activity(GO:0001055) |
| 0.0 | 0.8 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 10.6 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.0 | 1.8 | GO:0002020 | protease binding(GO:0002020) |
| 0.0 | 0.3 | GO:0019841 | retinol binding(GO:0019841) |
| 0.0 | 0.1 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 20.0 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.2 | 5.9 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.2 | 2.3 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.2 | 5.3 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.1 | 2.7 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.1 | 2.9 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.1 | 1.5 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.1 | 2.6 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.1 | 1.5 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.1 | 2.2 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.1 | 2.9 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.1 | 2.2 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.1 | 2.0 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.1 | 3.9 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.1 | 1.8 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 2.9 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 3.0 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 2.3 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 0.6 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 2.2 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 2.4 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.0 | 1.5 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 2.6 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 1.2 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 3.9 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 2.8 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 2.3 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.8 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 1.0 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.0 | 0.4 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 1.3 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.4 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.8 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 0.5 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 1.3 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 4.6 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 1.3 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.8 | PID AURORA B PATHWAY | Aurora B signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 12.6 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.1 | 4.1 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.1 | 3.6 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.1 | 5.4 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.1 | 3.7 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.1 | 8.6 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.1 | 2.4 | REACTOME TAK1 ACTIVATES NFKB BY PHOSPHORYLATION AND ACTIVATION OF IKKS COMPLEX | Genes involved in TAK1 activates NFkB by phosphorylation and activation of IKKs complex |
| 0.1 | 3.7 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.1 | 3.1 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.1 | 2.5 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.1 | 0.7 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.1 | 1.5 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.1 | 2.7 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.0 | 1.4 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.8 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.0 | 1.4 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 2.2 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 1.7 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 1.2 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 1.0 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.0 | 0.9 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.0 | 1.4 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 1.5 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 1.4 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.7 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 1.5 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.6 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.7 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 2.2 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 1.5 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.0 | 2.7 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 1.3 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 0.8 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.2 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 1.0 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 0.6 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 2.2 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.4 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.0 | 1.3 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.0 | 0.6 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.6 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 1.5 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.2 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.7 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.6 | REACTOME EXTRACELLULAR MATRIX ORGANIZATION | Genes involved in Extracellular matrix organization |
| 0.0 | 0.7 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 0.4 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.2 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 1.2 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.7 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |