Project
Illumina Body Map 2, young vs old
Navigation
Downloads
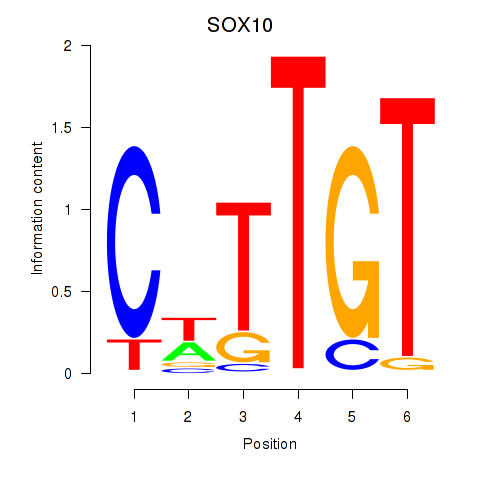
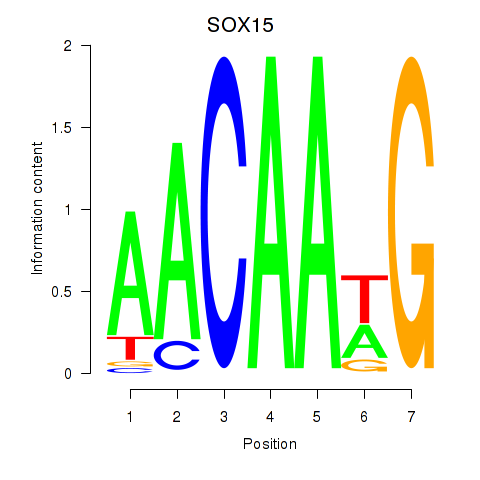
Results for SOX10_SOX15
Z-value: 0.45
Transcription factors associated with SOX10_SOX15
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SOX10
|
ENSG00000100146.12 | SRY-box transcription factor 10 |
|
SOX15
|
ENSG00000129194.3 | SRY-box transcription factor 15 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SOX10 | hg19_v2_chr22_-_38380543_38380569 | -0.39 | 2.9e-02 | Click! |
| SOX15 | hg19_v2_chr17_-_7493390_7493488 | -0.38 | 3.2e-02 | Click! |
Activity profile of SOX10_SOX15 motif
Sorted Z-values of SOX10_SOX15 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_111260856 | 7.56 |
ENST00000352690.4
|
CD96
|
CD96 molecule |
| chr3_+_111260954 | 7.33 |
ENST00000283285.5
|
CD96
|
CD96 molecule |
| chr3_+_111260980 | 6.80 |
ENST00000438817.2
|
CD96
|
CD96 molecule |
| chr18_-_74728998 | 6.15 |
ENST00000359645.3
ENST00000397875.3 ENST00000397869.3 ENST00000578193.1 ENST00000578873.1 ENST00000397866.4 ENST00000528160.1 ENST00000527041.1 ENST00000526111.1 ENST00000397865.5 ENST00000382582.3 |
MBP
|
myelin basic protein |
| chr1_-_200992827 | 5.90 |
ENST00000332129.2
ENST00000422435.2 |
KIF21B
|
kinesin family member 21B |
| chr4_-_109090106 | 4.92 |
ENST00000379951.2
|
LEF1
|
lymphoid enhancer-binding factor 1 |
| chr11_+_128563652 | 4.56 |
ENST00000527786.2
|
FLI1
|
Fli-1 proto-oncogene, ETS transcription factor |
| chr6_-_128222103 | 4.20 |
ENST00000434358.1
ENST00000543064.1 ENST00000368248.2 |
THEMIS
|
thymocyte selection associated |
| chrX_+_12993202 | 4.16 |
ENST00000451311.2
ENST00000380636.1 |
TMSB4X
|
thymosin beta 4, X-linked |
| chr4_+_154387480 | 3.66 |
ENST00000409663.3
ENST00000440693.1 ENST00000409959.3 |
KIAA0922
|
KIAA0922 |
| chr13_-_41593425 | 3.58 |
ENST00000239882.3
|
ELF1
|
E74-like factor 1 (ets domain transcription factor) |
| chr5_-_65018834 | 3.54 |
ENST00000506816.1
|
SGTB
|
small glutamine-rich tetratricopeptide repeat (TPR)-containing, beta |
| chrX_+_12993336 | 3.53 |
ENST00000380635.1
|
TMSB4X
|
thymosin beta 4, X-linked |
| chr9_-_74383302 | 3.52 |
ENST00000377066.5
|
TMEM2
|
transmembrane protein 2 |
| chr5_-_157002775 | 3.47 |
ENST00000257527.4
|
ADAM19
|
ADAM metallopeptidase domain 19 |
| chr5_-_157002749 | 3.45 |
ENST00000517905.1
ENST00000430702.2 ENST00000394020.1 |
ADAM19
|
ADAM metallopeptidase domain 19 |
| chr8_+_61822605 | 3.29 |
ENST00000526936.1
|
AC022182.1
|
AC022182.1 |
| chr4_-_164534657 | 3.20 |
ENST00000339875.5
|
MARCH1
|
membrane-associated ring finger (C3HC4) 1, E3 ubiquitin protein ligase |
| chr4_-_109089573 | 3.16 |
ENST00000265165.1
|
LEF1
|
lymphoid enhancer-binding factor 1 |
| chr14_-_91884150 | 3.10 |
ENST00000553403.1
|
CCDC88C
|
coiled-coil domain containing 88C |
| chr10_+_11206925 | 2.84 |
ENST00000354440.2
ENST00000315874.4 ENST00000427450.1 |
CELF2
|
CUGBP, Elav-like family member 2 |
| chr6_+_114178512 | 2.77 |
ENST00000368635.4
|
MARCKS
|
myristoylated alanine-rich protein kinase C substrate |
| chr1_+_11751748 | 2.71 |
ENST00000294485.5
|
DRAXIN
|
dorsal inhibitory axon guidance protein |
| chr5_-_65017921 | 2.67 |
ENST00000381007.4
|
SGTB
|
small glutamine-rich tetratricopeptide repeat (TPR)-containing, beta |
| chr8_-_80993010 | 2.65 |
ENST00000537855.1
ENST00000520527.1 ENST00000517427.1 ENST00000448733.2 ENST00000379097.3 |
TPD52
|
tumor protein D52 |
| chr14_+_22309368 | 2.65 |
ENST00000390433.1
|
TRAV12-1
|
T cell receptor alpha variable 12-1 |
| chr14_-_91884115 | 2.61 |
ENST00000389857.6
|
CCDC88C
|
coiled-coil domain containing 88C |
| chr9_-_123476612 | 2.55 |
ENST00000426959.1
|
MEGF9
|
multiple EGF-like-domains 9 |
| chr6_-_30654977 | 2.54 |
ENST00000399199.3
|
PPP1R18
|
protein phosphatase 1, regulatory subunit 18 |
| chr11_+_36317830 | 2.49 |
ENST00000530639.1
|
PRR5L
|
proline rich 5 like |
| chr10_+_11207088 | 2.46 |
ENST00000608830.1
|
CELF2
|
CUGBP, Elav-like family member 2 |
| chr17_+_75447326 | 2.44 |
ENST00000591088.1
|
SEPT9
|
septin 9 |
| chr15_-_52043722 | 2.40 |
ENST00000454181.2
|
LYSMD2
|
LysM, putative peptidoglycan-binding, domain containing 2 |
| chrX_+_37639302 | 2.32 |
ENST00000545017.1
ENST00000536160.1 |
CYBB
|
cytochrome b-245, beta polypeptide |
| chr10_-_99030395 | 2.29 |
ENST00000355366.5
ENST00000371027.1 |
ARHGAP19
|
Rho GTPase activating protein 19 |
| chr12_+_8666126 | 2.27 |
ENST00000299665.2
|
CLEC4D
|
C-type lectin domain family 4, member D |
| chr6_-_130536774 | 2.26 |
ENST00000532763.1
|
SAMD3
|
sterile alpha motif domain containing 3 |
| chr1_-_154155675 | 2.25 |
ENST00000330188.9
ENST00000341485.5 |
TPM3
|
tropomyosin 3 |
| chr22_+_27053422 | 2.24 |
ENST00000413665.1
ENST00000421151.1 ENST00000456129.1 ENST00000430080.1 |
MIAT
|
myocardial infarction associated transcript (non-protein coding) |
| chr12_-_42631529 | 2.24 |
ENST00000548917.1
|
YAF2
|
YY1 associated factor 2 |
| chr12_-_45269769 | 2.23 |
ENST00000548826.1
|
NELL2
|
NEL-like 2 (chicken) |
| chr3_+_39509163 | 2.20 |
ENST00000436143.2
ENST00000441980.2 ENST00000311042.6 |
MOBP
|
myelin-associated oligodendrocyte basic protein |
| chr12_-_45270077 | 2.19 |
ENST00000551601.1
ENST00000549027.1 ENST00000452445.2 |
NELL2
|
NEL-like 2 (chicken) |
| chr17_-_9939854 | 2.15 |
ENST00000584146.2
|
GAS7
|
growth arrest-specific 7 |
| chr2_+_232573208 | 2.15 |
ENST00000409115.3
|
PTMA
|
prothymosin, alpha |
| chr21_+_30672433 | 2.14 |
ENST00000451655.1
|
BACH1
|
BTB and CNC homology 1, basic leucine zipper transcription factor 1 |
| chr5_+_149865838 | 2.12 |
ENST00000519157.1
|
NDST1
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1 |
| chr3_+_39509070 | 2.11 |
ENST00000354668.4
ENST00000428261.1 ENST00000420739.1 ENST00000415443.1 ENST00000447324.1 ENST00000383754.3 |
MOBP
|
myelin-associated oligodendrocyte basic protein |
| chr7_+_29234028 | 2.10 |
ENST00000222792.6
|
CHN2
|
chimerin 2 |
| chr12_-_45270151 | 2.09 |
ENST00000429094.2
|
NELL2
|
NEL-like 2 (chicken) |
| chr4_-_176733377 | 2.08 |
ENST00000505375.1
|
GPM6A
|
glycoprotein M6A |
| chr20_-_4804244 | 2.07 |
ENST00000379400.3
|
RASSF2
|
Ras association (RalGDS/AF-6) domain family member 2 |
| chr16_-_4852915 | 2.06 |
ENST00000322048.7
|
ROGDI
|
rogdi homolog (Drosophila) |
| chr5_-_146435572 | 2.03 |
ENST00000394414.1
|
PPP2R2B
|
protein phosphatase 2, regulatory subunit B, beta |
| chr2_-_158182410 | 2.03 |
ENST00000419116.2
ENST00000410096.1 |
ERMN
|
ermin, ERM-like protein |
| chr12_-_109027643 | 2.01 |
ENST00000388962.3
ENST00000550948.1 |
SELPLG
|
selectin P ligand |
| chr17_+_36861735 | 2.01 |
ENST00000378137.5
ENST00000325718.7 |
MLLT6
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 6 |
| chr2_+_232573222 | 2.00 |
ENST00000341369.7
ENST00000409683.1 |
PTMA
|
prothymosin, alpha |
| chr2_+_120517174 | 1.99 |
ENST00000263708.2
|
PTPN4
|
protein tyrosine phosphatase, non-receptor type 4 (megakaryocyte) |
| chr5_-_175964366 | 1.98 |
ENST00000274811.4
|
RNF44
|
ring finger protein 44 |
| chr5_+_147258266 | 1.91 |
ENST00000296694.4
|
SCGB3A2
|
secretoglobin, family 3A, member 2 |
| chrX_+_129305623 | 1.90 |
ENST00000257017.4
|
RAB33A
|
RAB33A, member RAS oncogene family |
| chr17_-_7297519 | 1.88 |
ENST00000576362.1
ENST00000571078.1 |
TMEM256-PLSCR3
|
TMEM256-PLSCR3 readthrough (NMD candidate) |
| chr7_+_50344289 | 1.88 |
ENST00000413698.1
ENST00000359197.5 ENST00000331340.3 ENST00000357364.4 ENST00000343574.5 ENST00000349824.4 ENST00000346667.4 ENST00000440768.2 |
IKZF1
|
IKAROS family zinc finger 1 (Ikaros) |
| chr17_-_7297833 | 1.87 |
ENST00000571802.1
ENST00000576201.1 ENST00000573213.1 ENST00000324822.11 |
TMEM256-PLSCR3
|
TMEM256-PLSCR3 readthrough (NMD candidate) |
| chr17_+_75283973 | 1.87 |
ENST00000431235.2
ENST00000449803.2 |
SEPT9
|
septin 9 |
| chr22_+_27053567 | 1.83 |
ENST00000449717.1
ENST00000453023.1 |
MIAT
|
myocardial infarction associated transcript (non-protein coding) |
| chrY_+_15016013 | 1.82 |
ENST00000360160.4
ENST00000454054.1 |
DDX3Y
|
DEAD (Asp-Glu-Ala-Asp) box helicase 3, Y-linked |
| chr5_-_146435694 | 1.80 |
ENST00000356826.3
|
PPP2R2B
|
protein phosphatase 2, regulatory subunit B, beta |
| chr17_-_9940058 | 1.80 |
ENST00000585266.1
|
GAS7
|
growth arrest-specific 7 |
| chrX_+_37639264 | 1.80 |
ENST00000378588.4
|
CYBB
|
cytochrome b-245, beta polypeptide |
| chr12_-_24715307 | 1.79 |
ENST00000538905.1
|
RP11-444D3.1
|
RP11-444D3.1 |
| chr1_+_40840320 | 1.79 |
ENST00000372708.1
|
SMAP2
|
small ArfGAP2 |
| chr5_-_39424961 | 1.77 |
ENST00000503513.1
|
DAB2
|
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
| chr2_+_89184868 | 1.76 |
ENST00000390243.2
|
IGKV4-1
|
immunoglobulin kappa variable 4-1 |
| chr15_-_70388943 | 1.76 |
ENST00000559048.1
ENST00000560939.1 ENST00000440567.3 ENST00000557907.1 ENST00000558379.1 ENST00000451782.2 ENST00000559929.1 |
TLE3
|
transducin-like enhancer of split 3 (E(sp1) homolog, Drosophila) |
| chr14_+_22320634 | 1.74 |
ENST00000390435.1
|
TRAV8-3
|
T cell receptor alpha variable 8-3 |
| chr3_-_71353892 | 1.72 |
ENST00000484350.1
|
FOXP1
|
forkhead box P1 |
| chr3_-_18466026 | 1.67 |
ENST00000417717.2
|
SATB1
|
SATB homeobox 1 |
| chr6_-_64029879 | 1.66 |
ENST00000370658.5
ENST00000485906.2 ENST00000370657.4 |
LGSN
|
lengsin, lens protein with glutamine synthetase domain |
| chr20_+_48884002 | 1.64 |
ENST00000425497.1
ENST00000445003.1 |
RP11-290F20.3
|
RP11-290F20.3 |
| chr17_-_76713100 | 1.61 |
ENST00000585509.1
|
CYTH1
|
cytohesin 1 |
| chr2_-_158182322 | 1.61 |
ENST00000420719.2
ENST00000409216.1 |
ERMN
|
ermin, ERM-like protein |
| chr6_-_91006461 | 1.60 |
ENST00000257749.4
ENST00000343122.3 ENST00000406998.2 ENST00000453877.1 |
BACH2
|
BTB and CNC homology 1, basic leucine zipper transcription factor 2 |
| chr5_+_149865377 | 1.60 |
ENST00000522491.1
|
NDST1
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1 |
| chr17_-_9939935 | 1.60 |
ENST00000580043.1
|
GAS7
|
growth arrest-specific 7 |
| chr4_+_26322987 | 1.60 |
ENST00000505958.1
|
RBPJ
|
recombination signal binding protein for immunoglobulin kappa J region |
| chr13_-_99959641 | 1.59 |
ENST00000376414.4
|
GPR183
|
G protein-coupled receptor 183 |
| chrY_+_15815447 | 1.56 |
ENST00000284856.3
|
TMSB4Y
|
thymosin beta 4, Y-linked |
| chr11_+_67007518 | 1.55 |
ENST00000530342.1
ENST00000308783.5 |
KDM2A
|
lysine (K)-specific demethylase 2A |
| chr7_+_29234101 | 1.55 |
ENST00000435288.2
|
CHN2
|
chimerin 2 |
| chr5_-_146435501 | 1.52 |
ENST00000336640.6
|
PPP2R2B
|
protein phosphatase 2, regulatory subunit B, beta |
| chr17_+_37809333 | 1.52 |
ENST00000443521.1
|
STARD3
|
StAR-related lipid transfer (START) domain containing 3 |
| chr1_+_66797687 | 1.52 |
ENST00000371045.5
ENST00000531025.1 ENST00000526197.1 |
PDE4B
|
phosphodiesterase 4B, cAMP-specific |
| chr4_+_118955635 | 1.51 |
ENST00000433996.2
|
NDST3
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 3 |
| chr9_-_35115836 | 1.51 |
ENST00000378566.1
ENST00000378554.2 ENST00000322813.5 |
FAM214B
|
family with sequence similarity 214, member B |
| chr17_-_49124230 | 1.51 |
ENST00000510283.1
ENST00000510855.1 |
SPAG9
|
sperm associated antigen 9 |
| chr22_+_37959647 | 1.49 |
ENST00000415670.1
|
CDC42EP1
|
CDC42 effector protein (Rho GTPase binding) 1 |
| chr7_+_31726822 | 1.48 |
ENST00000409146.3
|
PPP1R17
|
protein phosphatase 1, regulatory subunit 17 |
| chr20_-_39317868 | 1.47 |
ENST00000373313.2
|
MAFB
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog B |
| chr10_-_14613968 | 1.44 |
ENST00000488576.1
ENST00000472095.1 |
FAM107B
|
family with sequence similarity 107, member B |
| chr7_-_105926058 | 1.44 |
ENST00000417537.1
|
NAMPT
|
nicotinamide phosphoribosyltransferase |
| chr10_+_114710211 | 1.44 |
ENST00000349937.2
ENST00000369397.4 |
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr3_-_151102529 | 1.43 |
ENST00000302632.3
|
P2RY12
|
purinergic receptor P2Y, G-protein coupled, 12 |
| chr3_+_152879985 | 1.43 |
ENST00000323534.2
|
RAP2B
|
RAP2B, member of RAS oncogene family |
| chr20_+_35202909 | 1.42 |
ENST00000558530.1
ENST00000558028.1 ENST00000560025.1 |
TGIF2-C20orf24
TGIF2
|
TGIF2-C20orf24 readthrough TGFB-induced factor homeobox 2 |
| chr3_+_151986709 | 1.40 |
ENST00000495875.2
ENST00000493459.1 ENST00000324210.5 ENST00000459747.1 |
MBNL1
|
muscleblind-like splicing regulator 1 |
| chr4_-_140477910 | 1.39 |
ENST00000404104.3
|
SETD7
|
SET domain containing (lysine methyltransferase) 7 |
| chr10_+_99079008 | 1.38 |
ENST00000371021.3
|
FRAT1
|
frequently rearranged in advanced T-cell lymphomas |
| chr22_+_19710468 | 1.37 |
ENST00000366425.3
|
GP1BB
|
glycoprotein Ib (platelet), beta polypeptide |
| chr12_-_12674032 | 1.36 |
ENST00000298573.4
|
DUSP16
|
dual specificity phosphatase 16 |
| chr1_+_200993071 | 1.36 |
ENST00000446333.1
ENST00000458003.1 |
RP11-168O16.1
|
RP11-168O16.1 |
| chr9_-_130712995 | 1.35 |
ENST00000373084.4
|
FAM102A
|
family with sequence similarity 102, member A |
| chr12_+_104359641 | 1.35 |
ENST00000537100.1
|
TDG
|
thymine-DNA glycosylase |
| chrX_-_142722897 | 1.34 |
ENST00000338017.4
|
SLITRK4
|
SLIT and NTRK-like family, member 4 |
| chr9_-_123476719 | 1.33 |
ENST00000373930.3
|
MEGF9
|
multiple EGF-like-domains 9 |
| chr14_+_22984601 | 1.33 |
ENST00000390509.1
|
TRAJ28
|
T cell receptor alpha joining 28 |
| chr12_+_15475462 | 1.33 |
ENST00000543886.1
ENST00000348962.2 |
PTPRO
|
protein tyrosine phosphatase, receptor type, O |
| chr1_+_66458072 | 1.31 |
ENST00000423207.2
|
PDE4B
|
phosphodiesterase 4B, cAMP-specific |
| chr2_+_32288725 | 1.30 |
ENST00000315285.3
|
SPAST
|
spastin |
| chr13_+_111767650 | 1.30 |
ENST00000449979.1
ENST00000370623.3 |
ARHGEF7
|
Rho guanine nucleotide exchange factor (GEF) 7 |
| chr10_-_14614311 | 1.30 |
ENST00000479731.1
ENST00000468492.1 |
FAM107B
|
family with sequence similarity 107, member B |
| chr10_-_14614122 | 1.29 |
ENST00000378465.3
ENST00000452706.2 ENST00000378458.2 |
FAM107B
|
family with sequence similarity 107, member B |
| chr14_+_77228532 | 1.28 |
ENST00000167106.4
ENST00000554237.1 |
VASH1
|
vasohibin 1 |
| chr2_-_166060382 | 1.27 |
ENST00000409101.3
|
SCN3A
|
sodium channel, voltage-gated, type III, alpha subunit |
| chr1_-_204436344 | 1.26 |
ENST00000367184.2
|
PIK3C2B
|
phosphatidylinositol-4-phosphate 3-kinase, catalytic subunit type 2 beta |
| chr2_-_166060552 | 1.26 |
ENST00000283254.7
ENST00000453007.1 |
SCN3A
|
sodium channel, voltage-gated, type III, alpha subunit |
| chr15_-_52944231 | 1.24 |
ENST00000546305.2
|
FAM214A
|
family with sequence similarity 214, member A |
| chr13_+_78315466 | 1.24 |
ENST00000314070.5
ENST00000462234.1 |
SLAIN1
|
SLAIN motif family, member 1 |
| chr18_+_72201829 | 1.23 |
ENST00000582365.1
|
CNDP1
|
carnosine dipeptidase 1 (metallopeptidase M20 family) |
| chr4_+_106067943 | 1.23 |
ENST00000380013.4
ENST00000394764.1 ENST00000413648.2 |
TET2
|
tet methylcytosine dioxygenase 2 |
| chr3_-_62359180 | 1.23 |
ENST00000283268.3
|
FEZF2
|
FEZ family zinc finger 2 |
| chr15_+_52043813 | 1.23 |
ENST00000435126.2
|
TMOD2
|
tropomodulin 2 (neuronal) |
| chr15_+_81293254 | 1.22 |
ENST00000267984.2
|
MESDC1
|
mesoderm development candidate 1 |
| chr10_-_14614095 | 1.22 |
ENST00000482277.1
ENST00000378462.1 |
FAM107B
|
family with sequence similarity 107, member B |
| chr3_+_141105235 | 1.22 |
ENST00000503809.1
|
ZBTB38
|
zinc finger and BTB domain containing 38 |
| chr2_-_37501692 | 1.20 |
ENST00000443977.1
|
PRKD3
|
protein kinase D3 |
| chr13_+_78315348 | 1.20 |
ENST00000441784.1
|
SLAIN1
|
SLAIN motif family, member 1 |
| chr10_-_14646388 | 1.19 |
ENST00000468747.1
ENST00000378467.4 |
FAM107B
|
family with sequence similarity 107, member B |
| chr8_-_133071627 | 1.18 |
ENST00000443356.2
ENST00000603859.1 ENST00000254627.3 |
OC90
OC90
|
Otoconin-90 otoconin 90 |
| chr2_-_37899323 | 1.18 |
ENST00000295324.3
ENST00000457889.1 |
CDC42EP3
|
CDC42 effector protein (Rho GTPase binding) 3 |
| chr14_+_23011142 | 1.18 |
ENST00000390533.1
|
TRAJ4
|
T cell receptor alpha joining 4 |
| chr12_-_49582593 | 1.17 |
ENST00000295766.5
|
TUBA1A
|
tubulin, alpha 1a |
| chr13_+_78315528 | 1.17 |
ENST00000496045.1
|
SLAIN1
|
SLAIN motif family, member 1 |
| chr10_-_99094458 | 1.17 |
ENST00000371019.2
|
FRAT2
|
frequently rearranged in advanced T-cell lymphomas 2 |
| chr3_-_69590486 | 1.16 |
ENST00000497880.1
|
FRMD4B
|
FERM domain containing 4B |
| chr1_+_36621174 | 1.15 |
ENST00000429533.2
|
MAP7D1
|
MAP7 domain containing 1 |
| chr1_-_226926864 | 1.15 |
ENST00000429204.1
ENST00000366784.1 |
ITPKB
|
inositol-trisphosphate 3-kinase B |
| chr2_+_32288657 | 1.14 |
ENST00000345662.1
|
SPAST
|
spastin |
| chr17_-_18950310 | 1.14 |
ENST00000573099.1
|
GRAP
|
GRB2-related adaptor protein |
| chr2_-_166060571 | 1.14 |
ENST00000360093.3
|
SCN3A
|
sodium channel, voltage-gated, type III, alpha subunit |
| chr12_-_49504449 | 1.13 |
ENST00000547675.1
|
LMBR1L
|
limb development membrane protein 1-like |
| chr14_-_67981916 | 1.12 |
ENST00000357461.2
|
TMEM229B
|
transmembrane protein 229B |
| chr22_+_22453093 | 1.12 |
ENST00000390283.2
|
IGLV8-61
|
immunoglobulin lambda variable 8-61 |
| chr11_+_35198118 | 1.11 |
ENST00000525211.1
ENST00000526000.1 ENST00000279452.6 ENST00000527889.1 |
CD44
|
CD44 molecule (Indian blood group) |
| chr14_-_57272366 | 1.10 |
ENST00000554788.1
ENST00000554845.1 ENST00000408990.3 |
OTX2
|
orthodenticle homeobox 2 |
| chr18_+_72201664 | 1.09 |
ENST00000358821.3
|
CNDP1
|
carnosine dipeptidase 1 (metallopeptidase M20 family) |
| chr12_-_31477072 | 1.09 |
ENST00000454658.2
|
FAM60A
|
family with sequence similarity 60, member A |
| chr15_+_52043758 | 1.09 |
ENST00000249700.4
ENST00000539962.2 |
TMOD2
|
tropomodulin 2 (neuronal) |
| chr2_-_158182105 | 1.08 |
ENST00000409925.1
|
ERMN
|
ermin, ERM-like protein |
| chr8_-_20040638 | 1.08 |
ENST00000519026.1
ENST00000276373.5 ENST00000440926.1 ENST00000437980.1 |
SLC18A1
|
solute carrier family 18 (vesicular monoamine transporter), member 1 |
| chr11_-_130184555 | 1.08 |
ENST00000525842.1
|
ZBTB44
|
zinc finger and BTB domain containing 44 |
| chr6_-_127840021 | 1.06 |
ENST00000465909.2
|
SOGA3
|
SOGA family member 3 |
| chr4_-_123377880 | 1.05 |
ENST00000226730.4
|
IL2
|
interleukin 2 |
| chr9_+_137218362 | 1.04 |
ENST00000481739.1
|
RXRA
|
retinoid X receptor, alpha |
| chr13_+_78315295 | 1.04 |
ENST00000351546.3
|
SLAIN1
|
SLAIN motif family, member 1 |
| chr1_-_32801825 | 1.04 |
ENST00000329421.7
|
MARCKSL1
|
MARCKS-like 1 |
| chr15_+_75074915 | 1.04 |
ENST00000567123.1
ENST00000569462.1 |
CSK
|
c-src tyrosine kinase |
| chr12_-_90049878 | 1.03 |
ENST00000359142.3
|
ATP2B1
|
ATPase, Ca++ transporting, plasma membrane 1 |
| chr4_+_146403912 | 1.03 |
ENST00000507367.1
ENST00000394092.2 ENST00000515385.1 |
SMAD1
|
SMAD family member 1 |
| chr4_+_118955500 | 1.03 |
ENST00000296499.5
|
NDST3
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 3 |
| chr7_+_114562909 | 1.02 |
ENST00000423503.1
ENST00000427207.1 |
MDFIC
|
MyoD family inhibitor domain containing |
| chr11_-_130184470 | 1.01 |
ENST00000357899.4
ENST00000397753.1 |
ZBTB44
|
zinc finger and BTB domain containing 44 |
| chrX_-_19689106 | 1.01 |
ENST00000379716.1
|
SH3KBP1
|
SH3-domain kinase binding protein 1 |
| chr17_-_73389737 | 1.01 |
ENST00000392563.1
|
GRB2
|
growth factor receptor-bound protein 2 |
| chr22_+_27053545 | 0.98 |
ENST00000458302.1
ENST00000419237.1 |
MIAT
|
myocardial infarction associated transcript (non-protein coding) |
| chr2_+_149402989 | 0.98 |
ENST00000397424.2
|
EPC2
|
enhancer of polycomb homolog 2 (Drosophila) |
| chr18_-_74207146 | 0.97 |
ENST00000443185.2
|
ZNF516
|
zinc finger protein 516 |
| chr1_+_29241027 | 0.97 |
ENST00000373797.1
|
EPB41
|
erythrocyte membrane protein band 4.1 (elliptocytosis 1, RH-linked) |
| chr5_-_150521192 | 0.97 |
ENST00000523714.1
ENST00000521749.1 |
ANXA6
|
annexin A6 |
| chr8_-_121824374 | 0.96 |
ENST00000517992.1
|
SNTB1
|
syntrophin, beta 1 (dystrophin-associated protein A1, 59kDa, basic component 1) |
| chr9_-_130517309 | 0.96 |
ENST00000414380.1
|
SH2D3C
|
SH2 domain containing 3C |
| chr6_+_108881012 | 0.96 |
ENST00000343882.6
|
FOXO3
|
forkhead box O3 |
| chr6_-_134861089 | 0.95 |
ENST00000606039.1
|
RP11-557H15.4
|
RP11-557H15.4 |
| chr10_-_14596140 | 0.94 |
ENST00000496330.1
|
FAM107B
|
family with sequence similarity 107, member B |
| chr18_-_53019208 | 0.94 |
ENST00000562607.1
|
TCF4
|
transcription factor 4 |
| chr22_-_39150947 | 0.93 |
ENST00000411587.2
ENST00000420859.1 ENST00000452294.1 ENST00000456894.1 |
SUN2
|
Sad1 and UNC84 domain containing 2 |
| chr12_-_90049828 | 0.92 |
ENST00000261173.2
ENST00000348959.3 |
ATP2B1
|
ATPase, Ca++ transporting, plasma membrane 1 |
| chr3_+_133118839 | 0.92 |
ENST00000302334.2
|
BFSP2
|
beaded filament structural protein 2, phakinin |
| chr12_-_14133053 | 0.92 |
ENST00000609686.1
|
GRIN2B
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2B |
| chrX_-_110655306 | 0.91 |
ENST00000371993.2
|
DCX
|
doublecortin |
| chr6_+_64282447 | 0.89 |
ENST00000370650.2
ENST00000578299.1 |
PTP4A1
|
protein tyrosine phosphatase type IVA, member 1 |
| chr17_+_29815013 | 0.89 |
ENST00000394744.2
|
RAB11FIP4
|
RAB11 family interacting protein 4 (class II) |
| chr2_+_162272605 | 0.89 |
ENST00000389554.3
|
TBR1
|
T-box, brain, 1 |
| chr11_-_102323489 | 0.89 |
ENST00000361236.3
|
TMEM123
|
transmembrane protein 123 |
| chr22_-_21984282 | 0.89 |
ENST00000398873.3
ENST00000292778.6 |
YDJC
|
YdjC homolog (bacterial) |
| chr8_-_116681686 | 0.89 |
ENST00000519815.1
|
TRPS1
|
trichorhinophalangeal syndrome I |
| chr15_-_67439270 | 0.88 |
ENST00000558463.1
|
RP11-342M21.2
|
Uncharacterized protein |
| chr17_+_73452545 | 0.88 |
ENST00000314256.7
|
KIAA0195
|
KIAA0195 |
| chr20_-_23066953 | 0.87 |
ENST00000246006.4
|
CD93
|
CD93 molecule |
Network of associatons between targets according to the STRING database.
First level regulatory network of SOX10_SOX15
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.2 | 21.7 | GO:0002728 | negative regulation of natural killer cell cytokine production(GO:0002728) |
| 1.2 | 8.1 | GO:0061152 | trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 1.0 | 6.2 | GO:1904209 | regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904207) positive regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904209) |
| 1.0 | 7.7 | GO:1903026 | negative regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903026) |
| 0.8 | 4.1 | GO:1904844 | response to L-glutamine(GO:1904844) cellular response to L-glutamine(GO:1904845) |
| 0.7 | 2.1 | GO:0035283 | central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.6 | 3.0 | GO:0000117 | regulation of transcription involved in G2/M transition of mitotic cell cycle(GO:0000117) |
| 0.5 | 1.6 | GO:0035700 | astrocyte chemotaxis(GO:0035700) regulation of astrocyte chemotaxis(GO:2000458) |
| 0.5 | 2.0 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.5 | 2.0 | GO:0050902 | leukocyte adhesive activation(GO:0050902) |
| 0.5 | 1.9 | GO:1902544 | regulation of DNA N-glycosylase activity(GO:1902544) |
| 0.5 | 0.5 | GO:0000060 | protein import into nucleus, translocation(GO:0000060) |
| 0.4 | 1.3 | GO:0036058 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.4 | 6.5 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.4 | 1.8 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.3 | 1.0 | GO:0045994 | positive regulation of translational initiation by iron(GO:0045994) |
| 0.3 | 1.4 | GO:0045209 | MAPK phosphatase export from nucleus(GO:0045208) MAPK phosphatase export from nucleus, leptomycin B sensitive(GO:0045209) |
| 0.3 | 6.3 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.3 | 2.9 | GO:0071386 | cellular response to corticosterone stimulus(GO:0071386) |
| 0.3 | 1.3 | GO:1901491 | negative regulation of lymphangiogenesis(GO:1901491) |
| 0.3 | 0.9 | GO:1903450 | regulation of G1 to G0 transition(GO:1903450) positive regulation of G1 to G0 transition(GO:1903452) |
| 0.3 | 1.8 | GO:0035026 | leading edge cell differentiation(GO:0035026) |
| 0.3 | 1.7 | GO:0072619 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.3 | 1.7 | GO:0015842 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) |
| 0.3 | 1.1 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.3 | 2.1 | GO:1905068 | positive regulation of ephrin receptor signaling pathway(GO:1901189) positive regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901297) positive regulation of canonical Wnt signaling pathway involved in heart development(GO:1905068) |
| 0.2 | 2.4 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.2 | 0.7 | GO:0048203 | vesicle targeting, trans-Golgi to endosome(GO:0048203) |
| 0.2 | 3.4 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.2 | 1.7 | GO:1902730 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.2 | 2.8 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.2 | 2.3 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.2 | 1.4 | GO:1904139 | microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) |
| 0.2 | 1.6 | GO:1902527 | positive regulation of protein monoubiquitination(GO:1902527) |
| 0.2 | 1.7 | GO:0010989 | negative regulation of low-density lipoprotein particle clearance(GO:0010989) |
| 0.2 | 0.6 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.2 | 4.2 | GO:0043383 | negative T cell selection(GO:0043383) |
| 0.2 | 1.5 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.2 | 5.4 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.2 | 0.7 | GO:0060620 | regulation of cholesterol import(GO:0060620) regulation of sterol import(GO:2000909) |
| 0.2 | 0.5 | GO:1904586 | response to putrescine(GO:1904585) cellular response to putrescine(GO:1904586) hepatocyte dedifferentiation(GO:1990828) |
| 0.2 | 3.0 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.2 | 1.1 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 0.2 | 0.5 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.1 | 0.6 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.1 | 2.5 | GO:0030050 | vesicle transport along actin filament(GO:0030050) |
| 0.1 | 0.6 | GO:1903300 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.1 | 0.6 | GO:0021849 | neuroblast division in subventricular zone(GO:0021849) |
| 0.1 | 1.2 | GO:0033029 | regulation of neutrophil apoptotic process(GO:0033029) |
| 0.1 | 4.6 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.1 | 0.6 | GO:0034445 | regulation of plasma lipoprotein particle oxidation(GO:0034444) negative regulation of plasma lipoprotein particle oxidation(GO:0034445) |
| 0.1 | 1.2 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.1 | 0.9 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.1 | 2.1 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.1 | 0.6 | GO:0010157 | response to chlorate(GO:0010157) |
| 0.1 | 4.3 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.1 | 0.4 | GO:2000616 | negative regulation of histone H3-K9 acetylation(GO:2000616) |
| 0.1 | 1.5 | GO:0006701 | progesterone biosynthetic process(GO:0006701) |
| 0.1 | 2.0 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.1 | 1.1 | GO:2000543 | positive regulation of gastrulation(GO:2000543) |
| 0.1 | 1.3 | GO:0048298 | positive regulation of isotype switching to IgA isotypes(GO:0048298) |
| 0.1 | 1.8 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.1 | 1.6 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.1 | 3.6 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.1 | 7.8 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) |
| 0.1 | 0.5 | GO:0003292 | cardiac septum cell differentiation(GO:0003292) atrioventricular node cell differentiation(GO:0060922) atrioventricular node cell development(GO:0060928) |
| 0.1 | 0.6 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 0.1 | 0.3 | GO:0036515 | serotonergic neuron axon guidance(GO:0036515) |
| 0.1 | 0.3 | GO:0006286 | base-excision repair, base-free sugar-phosphate removal(GO:0006286) |
| 0.1 | 0.9 | GO:0099515 | actin filament-based transport(GO:0099515) |
| 0.1 | 3.5 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.1 | 0.5 | GO:0072369 | regulation of lipid transport by positive regulation of transcription from RNA polymerase II promoter(GO:0072369) |
| 0.1 | 0.3 | GO:0032203 | telomere formation via telomerase(GO:0032203) |
| 0.1 | 0.6 | GO:1904885 | beta-catenin destruction complex assembly(GO:1904885) |
| 0.1 | 0.4 | GO:0046833 | positive regulation of RNA export from nucleus(GO:0046833) |
| 0.1 | 2.1 | GO:1904886 | beta-catenin destruction complex disassembly(GO:1904886) |
| 0.1 | 0.6 | GO:1903527 | positive regulation of membrane tubulation(GO:1903527) |
| 0.1 | 5.3 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.1 | 6.7 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.1 | 1.7 | GO:0070544 | histone H3-K36 demethylation(GO:0070544) |
| 0.1 | 0.4 | GO:0051097 | negative regulation of helicase activity(GO:0051097) oligodendrocyte apoptotic process(GO:0097252) |
| 0.1 | 0.6 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.1 | 0.2 | GO:0014034 | neural crest cell fate commitment(GO:0014034) |
| 0.1 | 0.5 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.1 | 1.0 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.1 | 0.9 | GO:0070777 | gamma-aminobutyric acid biosynthetic process(GO:0009449) D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.1 | 2.2 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.1 | 0.4 | GO:1900224 | positive regulation of nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900224) |
| 0.1 | 1.0 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.1 | 0.3 | GO:0032233 | positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.1 | 0.8 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) negative regulation of chromatin silencing(GO:0031936) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.1 | 1.7 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.1 | 0.4 | GO:0010814 | substance P catabolic process(GO:0010814) calcitonin catabolic process(GO:0010816) endothelin maturation(GO:0034959) |
| 0.1 | 0.3 | GO:1904693 | midbrain morphogenesis(GO:1904693) |
| 0.1 | 0.4 | GO:0046016 | regulation of transcription from RNA polymerase II promoter by glucose(GO:0000430) positive regulation of transcription from RNA polymerase II promoter by glucose(GO:0000432) positive regulation of transcription by glucose(GO:0046016) |
| 0.1 | 0.6 | GO:0019856 | 'de novo' pyrimidine nucleobase biosynthetic process(GO:0006207) pyrimidine nucleobase biosynthetic process(GO:0019856) |
| 0.1 | 1.4 | GO:0034356 | NAD biosynthesis via nicotinamide riboside salvage pathway(GO:0034356) |
| 0.1 | 1.4 | GO:0080182 | histone H3-K4 trimethylation(GO:0080182) |
| 0.1 | 0.3 | GO:0060392 | negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 0.1 | 0.3 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.1 | 0.2 | GO:0010897 | negative regulation of triglyceride catabolic process(GO:0010897) |
| 0.1 | 0.3 | GO:0071469 | detection of mechanical stimulus involved in sensory perception of touch(GO:0050976) cellular response to alkaline pH(GO:0071469) |
| 0.1 | 0.1 | GO:0021569 | rhombomere 3 development(GO:0021569) |
| 0.1 | 1.7 | GO:0021516 | dorsal spinal cord development(GO:0021516) |
| 0.1 | 1.0 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.1 | 2.9 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.1 | 1.4 | GO:0038092 | nodal signaling pathway(GO:0038092) |
| 0.1 | 0.9 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.1 | 1.2 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.1 | 2.1 | GO:0042475 | odontogenesis of dentin-containing tooth(GO:0042475) |
| 0.1 | 1.0 | GO:1904776 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.1 | 0.2 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.1 | 5.2 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.1 | 0.6 | GO:0034378 | chylomicron assembly(GO:0034378) |
| 0.1 | 0.1 | GO:1902174 | positive regulation of keratinocyte apoptotic process(GO:1902174) |
| 0.1 | 1.3 | GO:2000394 | positive regulation of lamellipodium morphogenesis(GO:2000394) |
| 0.1 | 0.3 | GO:0006127 | glycerophosphate shuttle(GO:0006127) |
| 0.1 | 0.3 | GO:2000620 | positive regulation of histone H4-K16 acetylation(GO:2000620) |
| 0.1 | 1.5 | GO:0070933 | histone H4 deacetylation(GO:0070933) |
| 0.1 | 0.6 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.0 | 0.2 | GO:0042539 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.0 | 0.5 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 0.7 | GO:0033235 | positive regulation of protein sumoylation(GO:0033235) |
| 0.0 | 0.5 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.0 | 0.5 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.0 | 0.2 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.0 | 0.3 | GO:0003402 | planar cell polarity pathway involved in axis elongation(GO:0003402) |
| 0.0 | 0.3 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.0 | 1.3 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 0.1 | GO:1900737 | regulation of proteinase activated receptor activity(GO:1900276) negative regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900737) |
| 0.0 | 0.4 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.0 | 0.4 | GO:0034372 | very-low-density lipoprotein particle remodeling(GO:0034372) |
| 0.0 | 0.3 | GO:0001736 | establishment of planar polarity(GO:0001736) establishment of tissue polarity(GO:0007164) |
| 0.0 | 0.3 | GO:0060154 | cellular process regulating host cell cycle in response to virus(GO:0060154) |
| 0.0 | 0.2 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.0 | 1.4 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.0 | 0.3 | GO:0061737 | leukotriene signaling pathway(GO:0061737) |
| 0.0 | 0.8 | GO:0018023 | peptidyl-lysine trimethylation(GO:0018023) |
| 0.0 | 0.7 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.0 | 0.7 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.0 | 0.1 | GO:0015882 | L-ascorbic acid transport(GO:0015882) transepithelial L-ascorbic acid transport(GO:0070904) |
| 0.0 | 0.5 | GO:0001542 | ovulation from ovarian follicle(GO:0001542) |
| 0.0 | 0.6 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.0 | 0.8 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.0 | 0.4 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 1.9 | GO:2000785 | regulation of autophagosome assembly(GO:2000785) |
| 0.0 | 0.4 | GO:0034128 | negative regulation of MyD88-independent toll-like receptor signaling pathway(GO:0034128) |
| 0.0 | 1.8 | GO:1904837 | beta-catenin-TCF complex assembly(GO:1904837) |
| 0.0 | 0.6 | GO:0030308 | negative regulation of cell growth(GO:0030308) |
| 0.0 | 0.4 | GO:1902915 | negative regulation of histone ubiquitination(GO:0033183) negative regulation of protein K63-linked ubiquitination(GO:1900045) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.0 | 0.4 | GO:1902177 | positive regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902177) |
| 0.0 | 0.2 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.0 | 1.2 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.0 | 0.5 | GO:0002091 | negative regulation of receptor internalization(GO:0002091) |
| 0.0 | 0.5 | GO:0016127 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.0 | 0.8 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.0 | 0.4 | GO:0090204 | protein localization to nuclear pore(GO:0090204) |
| 0.0 | 0.8 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.0 | 0.8 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.0 | 0.1 | GO:0035188 | blastocyst hatching(GO:0001835) hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.0 | 1.2 | GO:1902236 | negative regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway(GO:1902236) |
| 0.0 | 0.6 | GO:0045581 | negative regulation of T cell differentiation(GO:0045581) |
| 0.0 | 0.4 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.0 | 0.8 | GO:1903204 | negative regulation of oxidative stress-induced neuron death(GO:1903204) |
| 0.0 | 0.8 | GO:0060117 | auditory receptor cell development(GO:0060117) |
| 0.0 | 0.4 | GO:0048286 | lung alveolus development(GO:0048286) |
| 0.0 | 0.1 | GO:0036333 | hepatocyte homeostasis(GO:0036333) response to tetrachloromethane(GO:1904772) |
| 0.0 | 1.4 | GO:0070534 | protein K63-linked ubiquitination(GO:0070534) |
| 0.0 | 0.3 | GO:0042373 | vitamin K metabolic process(GO:0042373) |
| 0.0 | 1.1 | GO:0030336 | negative regulation of cell migration(GO:0030336) |
| 0.0 | 2.1 | GO:0003407 | neural retina development(GO:0003407) |
| 0.0 | 1.2 | GO:0050482 | arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.0 | 0.9 | GO:0023014 | signal transduction by protein phosphorylation(GO:0023014) |
| 0.0 | 1.1 | GO:0007257 | activation of JUN kinase activity(GO:0007257) |
| 0.0 | 0.3 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.4 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 0.0 | 1.0 | GO:0051560 | mitochondrial calcium ion homeostasis(GO:0051560) |
| 0.0 | 0.7 | GO:0032786 | positive regulation of DNA-templated transcription, elongation(GO:0032786) |
| 0.0 | 0.4 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.0 | 0.3 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.0 | 0.9 | GO:0050691 | regulation of defense response to virus by host(GO:0050691) |
| 0.0 | 0.2 | GO:0033601 | positive regulation of mammary gland epithelial cell proliferation(GO:0033601) endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.0 | 0.1 | GO:0010593 | negative regulation of lamellipodium assembly(GO:0010593) |
| 0.0 | 0.9 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 2.3 | GO:0006879 | cellular iron ion homeostasis(GO:0006879) |
| 0.0 | 2.2 | GO:0002292 | T cell differentiation involved in immune response(GO:0002292) |
| 0.0 | 0.5 | GO:0090331 | negative regulation of platelet aggregation(GO:0090331) |
| 0.0 | 0.7 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.0 | 2.9 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.0 | 1.1 | GO:0042059 | negative regulation of epidermal growth factor receptor signaling pathway(GO:0042059) |
| 0.0 | 0.8 | GO:0051482 | positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G-protein coupled signaling pathway(GO:0051482) |
| 0.0 | 1.2 | GO:0006301 | postreplication repair(GO:0006301) |
| 0.0 | 0.4 | GO:1902187 | negative regulation of viral release from host cell(GO:1902187) |
| 0.0 | 2.7 | GO:0030183 | B cell differentiation(GO:0030183) |
| 0.0 | 1.1 | GO:0050982 | detection of mechanical stimulus(GO:0050982) |
| 0.0 | 0.1 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 0.0 | 0.1 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.0 | 0.1 | GO:0038026 | reelin-mediated signaling pathway(GO:0038026) |
| 0.0 | 1.0 | GO:0060612 | adipose tissue development(GO:0060612) |
| 0.0 | 0.4 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.0 | 1.1 | GO:0043550 | regulation of lipid kinase activity(GO:0043550) |
| 0.0 | 0.5 | GO:0060285 | cilium-dependent cell motility(GO:0060285) |
| 0.0 | 0.8 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.0 | 0.3 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.7 | GO:2000772 | regulation of cellular senescence(GO:2000772) |
| 0.0 | 0.5 | GO:0007264 | small GTPase mediated signal transduction(GO:0007264) |
| 0.0 | 0.4 | GO:0051291 | protein heterooligomerization(GO:0051291) |
| 0.0 | 0.5 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.0 | 0.4 | GO:0031167 | rRNA methylation(GO:0031167) |
| 0.0 | 0.4 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.0 | 0.8 | GO:0050775 | positive regulation of dendrite morphogenesis(GO:0050775) |
| 0.0 | 1.2 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 0.0 | 0.3 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 0.3 | GO:0022011 | myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 0.0 | 1.5 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.0 | 0.9 | GO:0042116 | macrophage activation(GO:0042116) |
| 0.0 | 1.5 | GO:1902036 | regulation of hematopoietic stem cell differentiation(GO:1902036) |
| 0.0 | 0.9 | GO:0050871 | positive regulation of B cell activation(GO:0050871) |
| 0.0 | 0.1 | GO:1903756 | regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903756) negative regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903758) |
| 0.0 | 0.4 | GO:0031643 | positive regulation of myelination(GO:0031643) |
| 0.0 | 1.3 | GO:0002504 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002504) |
| 0.0 | 1.3 | GO:0030218 | erythrocyte differentiation(GO:0030218) |
| 0.0 | 0.3 | GO:0043651 | linoleic acid metabolic process(GO:0043651) |
| 0.0 | 0.5 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 0.2 | GO:0060009 | Sertoli cell development(GO:0060009) |
| 0.0 | 0.8 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 0.1 | GO:0060218 | hematopoietic stem cell differentiation(GO:0060218) |
| 0.0 | 3.8 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.0 | 0.0 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.0 | 0.7 | GO:0001954 | positive regulation of cell-matrix adhesion(GO:0001954) |
| 0.0 | 0.3 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.0 | 0.1 | GO:0051611 | negative regulation of neurotransmitter uptake(GO:0051581) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.0 | 0.1 | GO:0015727 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 10.9 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.7 | 9.8 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.7 | 2.8 | GO:0042585 | germinal vesicle(GO:0042585) |
| 0.3 | 0.9 | GO:0008623 | CHRAC(GO:0008623) |
| 0.3 | 1.7 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.3 | 2.5 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.2 | 2.4 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.2 | 0.6 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.2 | 1.8 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.2 | 1.0 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.1 | 1.8 | GO:0070081 | clathrin-sculpted monoamine transport vesicle(GO:0070081) clathrin-sculpted monoamine transport vesicle membrane(GO:0070083) |
| 0.1 | 1.1 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.1 | 2.4 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.1 | 0.8 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.1 | 1.0 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.1 | 0.4 | GO:1990015 | mesaxon(GO:0097453) ensheathing process(GO:1990015) |
| 0.1 | 4.6 | GO:0032156 | septin cytoskeleton(GO:0032156) |
| 0.1 | 3.0 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.1 | 2.8 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.1 | 1.9 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.1 | 3.5 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.1 | 4.7 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 1.3 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.1 | 2.0 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.1 | 0.4 | GO:0031302 | intrinsic component of endosome membrane(GO:0031302) |
| 0.1 | 2.3 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.1 | 2.0 | GO:0001931 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.1 | 0.6 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.1 | 0.4 | GO:0071008 | U2-type post-mRNA release spliceosomal complex(GO:0071008) |
| 0.1 | 4.2 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.1 | 0.7 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.1 | 0.5 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.1 | 2.1 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.1 | 1.1 | GO:0090544 | BAF-type complex(GO:0090544) |
| 0.1 | 1.9 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.1 | 0.5 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.1 | 0.4 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.1 | 8.5 | GO:0070821 | tertiary granule membrane(GO:0070821) |
| 0.1 | 1.9 | GO:0071682 | endocytic vesicle lumen(GO:0071682) |
| 0.1 | 5.6 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.1 | 1.7 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.1 | GO:0039713 | viral factory(GO:0039713) cytoplasmic viral factory(GO:0039714) host cell viral assembly compartment(GO:0072517) |
| 0.0 | 1.1 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.0 | 0.7 | GO:0070938 | contractile ring(GO:0070938) |
| 0.0 | 4.9 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.3 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.0 | 1.8 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.0 | 0.3 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.7 | GO:0090543 | Flemming body(GO:0090543) |
| 0.0 | 0.4 | GO:0043219 | lateral loop(GO:0043219) |
| 0.0 | 0.7 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 2.1 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.0 | 0.4 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.4 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 1.7 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.0 | 0.9 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 2.2 | GO:0043596 | nuclear replication fork(GO:0043596) |
| 0.0 | 1.1 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.6 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 0.0 | 1.1 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 0.8 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.9 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 5.3 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.0 | 0.4 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.4 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.3 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 0.3 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 0.6 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.6 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 3.2 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
| 0.0 | 0.5 | GO:0005858 | axonemal dynein complex(GO:0005858) |
| 0.0 | 0.4 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.0 | 2.3 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 2.5 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.5 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 1.2 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 1.0 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 4.5 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 0.3 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.0 | 0.4 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.9 | GO:0099738 | cell cortex region(GO:0099738) |
| 0.0 | 0.2 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 5.2 | GO:0036464 | cytoplasmic ribonucleoprotein granule(GO:0036464) |
| 0.0 | 0.4 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 0.1 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.0 | GO:0070931 | Golgi-associated vesicle lumen(GO:0070931) |
| 0.0 | 0.4 | GO:0031312 | extrinsic component of organelle membrane(GO:0031312) |
| 0.0 | 1.8 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.8 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.9 | GO:0043739 | G/U mismatch-specific uracil-DNA glycosylase activity(GO:0043739) |
| 0.6 | 6.3 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.5 | 10.2 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.5 | 2.0 | GO:0016211 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.5 | 1.4 | GO:0047280 | nicotinamide phosphoribosyltransferase activity(GO:0047280) |
| 0.4 | 1.7 | GO:0015222 | serotonin transmembrane transporter activity(GO:0015222) |
| 0.4 | 8.1 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.4 | 1.1 | GO:0030350 | iron-responsive element binding(GO:0030350) |
| 0.4 | 1.1 | GO:0016652 | NAD(P)+ transhydrogenase activity(GO:0008746) oxidoreductase activity, acting on NAD(P)H, NAD(P) as acceptor(GO:0016652) |
| 0.3 | 1.4 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.3 | 1.2 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.3 | 1.0 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.3 | 3.5 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.2 | 2.4 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.2 | 0.6 | GO:1904713 | beta-catenin destruction complex binding(GO:1904713) |
| 0.2 | 1.2 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) inositol-1,3,4-trisphosphate 4-phosphatase activity(GO:0017161) inositol-3,4-bisphosphate 4-phosphatase activity(GO:0052828) |
| 0.2 | 0.6 | GO:0102007 | lactonohydrolase activity(GO:0046573) acyl-L-homoserine-lactone lactonohydrolase activity(GO:0102007) |
| 0.2 | 0.7 | GO:0035650 | AP-1 adaptor complex binding(GO:0035650) |
| 0.2 | 3.5 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.2 | 2.6 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.2 | 0.6 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.1 | 0.4 | GO:0050146 | nucleoside phosphotransferase activity(GO:0050146) |
| 0.1 | 8.5 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 1.5 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.1 | 1.6 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.1 | 6.2 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.1 | 1.1 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) glycosphingolipid binding(GO:0043208) |
| 0.1 | 1.2 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.1 | 1.2 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.1 | 1.5 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.1 | 2.1 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.1 | 2.7 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.1 | 1.9 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.1 | 0.4 | GO:0016435 | rRNA (guanine) methyltransferase activity(GO:0016435) |
| 0.1 | 0.6 | GO:0005119 | smoothened binding(GO:0005119) |
| 0.1 | 1.7 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.1 | 0.9 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.1 | 0.4 | GO:0043398 | HLH domain binding(GO:0043398) |
| 0.1 | 0.9 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.1 | 1.5 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.1 | 0.3 | GO:0098782 | mechanically-gated potassium channel activity(GO:0098782) |
| 0.1 | 0.2 | GO:1990698 | palmitoleoyltransferase activity(GO:1990698) |
| 0.1 | 2.8 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.1 | 1.7 | GO:0005168 | neurotrophin TRKA receptor binding(GO:0005168) |
| 0.1 | 5.4 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.1 | 0.8 | GO:0070915 | lysophosphatidic acid binding(GO:0035727) lysophosphatidic acid receptor activity(GO:0070915) |
| 0.1 | 1.4 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.1 | 1.4 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.1 | 0.3 | GO:0052590 | sn-glycerol-3-phosphate:ubiquinone oxidoreductase activity(GO:0052590) sn-glycerol-3-phosphate:ubiquinone-8 oxidoreductase activity(GO:0052591) |
| 0.1 | 0.3 | GO:0008309 | double-stranded DNA exodeoxyribonuclease activity(GO:0008309) |
| 0.1 | 0.9 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.1 | 0.3 | GO:0004090 | carbonyl reductase (NADPH) activity(GO:0004090) |
| 0.1 | 1.7 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.1 | 0.2 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.1 | 0.5 | GO:0004748 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.1 | 2.3 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.1 | 1.8 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.1 | 0.9 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.1 | 0.3 | GO:0016213 | linoleoyl-CoA desaturase activity(GO:0016213) |
| 0.1 | 2.3 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.1 | 1.0 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.1 | 0.5 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.1 | 0.6 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 2.0 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.1 | 0.3 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.1 | 0.3 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.1 | 1.4 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.9 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 3.1 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.9 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 4.5 | GO:0000980 | RNA polymerase II distal enhancer sequence-specific DNA binding(GO:0000980) |
| 0.0 | 1.5 | GO:0017127 | cholesterol transporter activity(GO:0017127) |
| 0.0 | 1.1 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 1.5 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 5.6 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 1.8 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.7 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.0 | 3.4 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 5.9 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 2.7 | GO:0042287 | MHC protein binding(GO:0042287) |
| 0.0 | 0.9 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.0 | 2.8 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 6.9 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 5.1 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.0 | 0.1 | GO:0008520 | L-ascorbate:sodium symporter activity(GO:0008520) L-ascorbic acid transporter activity(GO:0015229) sodium-dependent L-ascorbate transmembrane transporter activity(GO:0070890) |
| 0.0 | 1.1 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 1.4 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 0.5 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.9 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.6 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.4 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.3 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.1 | GO:0051021 | GDP-dissociation inhibitor binding(GO:0051021) Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.0 | 0.6 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 1.0 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.0 | 0.5 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.5 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 1.2 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.9 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 1.1 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.8 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 2.7 | GO:0016279 | protein-lysine N-methyltransferase activity(GO:0016279) |
| 0.0 | 1.3 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 1.0 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.3 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.0 | 0.7 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.4 | GO:0000990 | transcription factor activity, core RNA polymerase binding(GO:0000990) |
| 0.0 | 4.1 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 1.1 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.4 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.0 | 1.2 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 1.4 | GO:0061650 | ubiquitin-like protein conjugating enzyme activity(GO:0061650) |
| 0.0 | 1.5 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.0 | 0.6 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.0 | 1.5 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.0 | 0.9 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 1.4 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 1.3 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 0.4 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.0 | 0.6 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.8 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) lysophosphatidic acid acyltransferase activity(GO:0042171) lysophospholipid acyltransferase activity(GO:0071617) |
| 0.0 | 1.4 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.2 | GO:0042054 | histone methyltransferase activity(GO:0042054) |
| 0.0 | 2.2 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.1 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.0 | 0.8 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 1.2 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.0 | 0.6 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 0.6 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.1 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.0 | 0.3 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 4.4 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 0.3 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.0 | 0.6 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.7 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.2 | GO:0000182 | rDNA binding(GO:0000182) |
| 0.0 | 4.9 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.0 | 1.9 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.0 | 0.6 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.3 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.0 | 0.5 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.4 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.0 | 2.9 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 0.3 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 7.3 | GO:0005525 | GTP binding(GO:0005525) |
| 0.0 | 1.6 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 0.0 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 1.5 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.0 | 0.2 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.0 | 0.8 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.1 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.2 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.1 | 6.9 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.1 | 1.0 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.1 | 9.5 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.1 | 0.9 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.1 | 4.6 | PID ATR PATHWAY | ATR signaling pathway |
| 0.1 | 6.7 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.1 | 3.7 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.1 | 4.2 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 1.5 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.0 | 1.0 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.8 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 1.6 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 1.8 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.0 | 2.1 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 1.7 | ST B CELL ANTIGEN RECEPTOR | B Cell Antigen Receptor |
| 0.0 | 2.4 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.0 | 5.3 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 1.4 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 0.9 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 3.4 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 1.2 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 2.7 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 0.1 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 2.2 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 1.8 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 1.6 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 1.3 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 1.3 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 0.9 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.0 | 0.6 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 6.1 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 1.0 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 0.6 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.7 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 1.4 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.3 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.6 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 0.9 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.6 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 0.7 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.8 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.2 | 21.7 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.1 | 2.5 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.1 | 2.0 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.1 | 5.3 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.1 | 2.1 | REACTOME SOS MEDIATED SIGNALLING | Genes involved in SOS-mediated signalling |
| 0.1 | 2.3 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.1 | 1.4 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.1 | 2.5 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.1 | 4.1 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.1 | 1.8 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.1 | 1.7 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.1 | 4.0 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.1 | 4.5 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 1.6 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.1 | 1.2 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.1 | 1.1 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 2.7 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 1.8 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 1.5 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.9 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.0 | 1.0 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 6.6 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 1.1 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.6 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 1.1 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.7 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 1.7 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.4 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 1.0 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 2.7 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 1.1 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.6 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.0 | 1.2 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 1.9 | REACTOME SIGNALING BY NOTCH1 | Genes involved in Signaling by NOTCH1 |
| 0.0 | 0.8 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.4 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.0 | 0.4 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 0.4 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 1.0 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.1 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.0 | 1.6 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 0.8 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.3 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 0.4 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.3 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.3 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 1.7 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.3 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.5 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.3 | REACTOME SIGNALING BY NOTCH | Genes involved in Signaling by NOTCH |
| 0.0 | 0.6 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 1.9 | REACTOME REGULATION OF MRNA STABILITY BY PROTEINS THAT BIND AU RICH ELEMENTS | Genes involved in Regulation of mRNA Stability by Proteins that Bind AU-rich Elements |
| 0.0 | 0.4 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.3 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.3 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |