Project
Illumina Body Map 2, young vs old
Navigation
Downloads
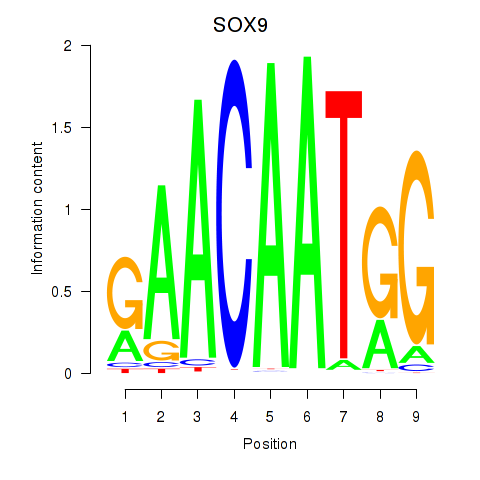
Results for SOX9
Z-value: 0.30
Transcription factors associated with SOX9
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SOX9
|
ENSG00000125398.5 | SRY-box transcription factor 9 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SOX9 | hg19_v2_chr17_+_70117153_70117174 | 0.37 | 3.9e-02 | Click! |
Activity profile of SOX9 motif
Sorted Z-values of SOX9 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_119530428 | 2.56 |
ENST00000369429.3
|
TBX15
|
T-box 15 |
| chr4_-_186732048 | 2.37 |
ENST00000448662.2
ENST00000439049.1 ENST00000420158.1 ENST00000431808.1 ENST00000319471.9 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr8_+_144295067 | 2.32 |
ENST00000330824.2
|
GPIHBP1
|
glycosylphosphatidylinositol anchored high density lipoprotein binding protein 1 |
| chr6_-_139613269 | 2.25 |
ENST00000358430.3
|
TXLNB
|
taxilin beta |
| chr3_-_46904946 | 2.21 |
ENST00000292327.4
|
MYL3
|
myosin, light chain 3, alkali; ventricular, skeletal, slow |
| chr8_+_19759228 | 2.16 |
ENST00000520959.1
|
LPL
|
lipoprotein lipase |
| chr3_-_46904918 | 2.14 |
ENST00000395869.1
|
MYL3
|
myosin, light chain 3, alkali; ventricular, skeletal, slow |
| chr12_-_6233828 | 2.03 |
ENST00000572068.1
ENST00000261405.5 |
VWF
|
von Willebrand factor |
| chr19_-_17375527 | 1.89 |
ENST00000431146.2
ENST00000594190.1 |
USHBP1
|
Usher syndrome 1C binding protein 1 |
| chr8_-_82395461 | 1.88 |
ENST00000256104.4
|
FABP4
|
fatty acid binding protein 4, adipocyte |
| chr12_-_8815215 | 1.88 |
ENST00000544889.1
ENST00000543369.1 |
MFAP5
|
microfibrillar associated protein 5 |
| chrX_-_45060135 | 1.83 |
ENST00000398000.2
ENST00000377934.4 |
CXorf36
|
chromosome X open reading frame 36 |
| chr13_+_110958124 | 1.75 |
ENST00000400163.2
|
COL4A2
|
collagen, type IV, alpha 2 |
| chr14_-_38725573 | 1.73 |
ENST00000342213.2
|
CLEC14A
|
C-type lectin domain family 14, member A |
| chr10_-_97321112 | 1.60 |
ENST00000607232.1
ENST00000371227.4 ENST00000371249.2 ENST00000371247.2 ENST00000371246.2 ENST00000393949.1 ENST00000353505.5 ENST00000347291.4 |
SORBS1
|
sorbin and SH3 domain containing 1 |
| chr11_+_131781290 | 1.59 |
ENST00000425719.2
ENST00000374784.1 |
NTM
|
neurotrimin |
| chr10_-_97321165 | 1.58 |
ENST00000306402.6
|
SORBS1
|
sorbin and SH3 domain containing 1 |
| chr12_-_8815299 | 1.58 |
ENST00000535336.1
|
MFAP5
|
microfibrillar associated protein 5 |
| chr17_-_39743139 | 1.55 |
ENST00000167586.6
|
KRT14
|
keratin 14 |
| chr1_+_156589051 | 1.53 |
ENST00000255039.1
|
HAPLN2
|
hyaluronan and proteoglycan link protein 2 |
| chr5_+_140593509 | 1.51 |
ENST00000341948.4
|
PCDHB13
|
protocadherin beta 13 |
| chr18_+_7946839 | 1.51 |
ENST00000578916.1
|
PTPRM
|
protein tyrosine phosphatase, receptor type, M |
| chr18_+_7946941 | 1.49 |
ENST00000444013.1
|
PTPRM
|
protein tyrosine phosphatase, receptor type, M |
| chr3_-_112329110 | 1.48 |
ENST00000479368.1
|
CCDC80
|
coiled-coil domain containing 80 |
| chr16_+_82660560 | 1.48 |
ENST00000268613.10
ENST00000565636.1 ENST00000431540.3 ENST00000428848.3 |
CDH13
|
cadherin 13 |
| chr10_+_24497704 | 1.39 |
ENST00000376456.4
ENST00000458595.1 |
KIAA1217
|
KIAA1217 |
| chr5_+_148521454 | 1.38 |
ENST00000508983.1
|
ABLIM3
|
actin binding LIM protein family, member 3 |
| chr12_-_18243119 | 1.38 |
ENST00000538724.1
ENST00000229002.2 |
RERGL
|
RERG/RAS-like |
| chr6_-_75915757 | 1.37 |
ENST00000322507.8
|
COL12A1
|
collagen, type XII, alpha 1 |
| chr4_+_41614909 | 1.35 |
ENST00000509454.1
ENST00000396595.3 ENST00000381753.4 |
LIMCH1
|
LIM and calponin homology domains 1 |
| chr12_+_53443680 | 1.32 |
ENST00000314250.6
ENST00000451358.1 |
TENC1
|
tensin like C1 domain containing phosphatase (tensin 2) |
| chr3_-_149051194 | 1.30 |
ENST00000470080.1
|
TM4SF18
|
transmembrane 4 L six family member 18 |
| chr17_+_8213590 | 1.27 |
ENST00000361926.3
|
ARHGEF15
|
Rho guanine nucleotide exchange factor (GEF) 15 |
| chr10_+_24498060 | 1.26 |
ENST00000376454.3
ENST00000376452.3 |
KIAA1217
|
KIAA1217 |
| chr5_-_146833485 | 1.25 |
ENST00000398514.3
|
DPYSL3
|
dihydropyrimidinase-like 3 |
| chr12_+_53443963 | 1.24 |
ENST00000546602.1
ENST00000552570.1 ENST00000549700.1 |
TENC1
|
tensin like C1 domain containing phosphatase (tensin 2) |
| chr3_-_185826855 | 1.23 |
ENST00000306376.5
|
ETV5
|
ets variant 5 |
| chr17_+_1666108 | 1.23 |
ENST00000570731.1
|
SERPINF1
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 1 |
| chr3_-_100565249 | 1.22 |
ENST00000495591.1
ENST00000383691.4 ENST00000466947.1 |
ABI3BP
|
ABI family, member 3 (NESH) binding protein |
| chr10_+_24528108 | 1.19 |
ENST00000438429.1
|
KIAA1217
|
KIAA1217 |
| chr10_+_102106829 | 1.16 |
ENST00000370355.2
|
SCD
|
stearoyl-CoA desaturase (delta-9-desaturase) |
| chr12_-_8815477 | 1.16 |
ENST00000433590.2
|
MFAP5
|
microfibrillar associated protein 5 |
| chr7_-_131241361 | 1.14 |
ENST00000378555.3
ENST00000322985.9 ENST00000541194.1 ENST00000537928.1 |
PODXL
|
podocalyxin-like |
| chr4_+_41614720 | 1.14 |
ENST00000509277.1
|
LIMCH1
|
LIM and calponin homology domains 1 |
| chr5_-_146833803 | 1.13 |
ENST00000512722.1
|
DPYSL3
|
dihydropyrimidinase-like 3 |
| chr3_-_149051444 | 1.13 |
ENST00000296059.2
|
TM4SF18
|
transmembrane 4 L six family member 18 |
| chr19_-_17375541 | 1.12 |
ENST00000252597.3
|
USHBP1
|
Usher syndrome 1C binding protein 1 |
| chr1_-_79472365 | 1.12 |
ENST00000370742.3
|
ELTD1
|
EGF, latrophilin and seven transmembrane domain containing 1 |
| chr6_+_53976235 | 1.09 |
ENST00000502396.1
ENST00000358276.5 |
MLIP
|
muscular LMNA-interacting protein |
| chr1_+_185703513 | 1.08 |
ENST00000271588.4
ENST00000367492.2 |
HMCN1
|
hemicentin 1 |
| chr9_-_124989804 | 1.08 |
ENST00000373755.2
ENST00000373754.2 |
LHX6
|
LIM homeobox 6 |
| chr22_-_19512893 | 1.06 |
ENST00000403084.1
ENST00000413119.2 |
CLDN5
|
claudin 5 |
| chr10_+_6244829 | 1.05 |
ENST00000317350.4
ENST00000379785.1 ENST00000379782.3 ENST00000360521.2 ENST00000379775.4 |
PFKFB3
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 |
| chrX_-_38080077 | 1.04 |
ENST00000378533.3
ENST00000544439.1 ENST00000432886.2 ENST00000538295.1 |
SRPX
|
sushi-repeat containing protein, X-linked |
| chr17_-_73505961 | 1.04 |
ENST00000433559.2
|
CASKIN2
|
CASK interacting protein 2 |
| chr5_+_140602904 | 1.04 |
ENST00000515856.2
ENST00000239449.4 |
PCDHB14
|
protocadherin beta 14 |
| chr6_+_53976211 | 1.02 |
ENST00000503951.1
|
MLIP
|
muscular LMNA-interacting protein |
| chr9_+_27109392 | 1.02 |
ENST00000406359.4
|
TEK
|
TEK tyrosine kinase, endothelial |
| chr2_-_151344172 | 1.02 |
ENST00000375734.2
ENST00000263895.4 ENST00000454202.1 |
RND3
|
Rho family GTPase 3 |
| chr9_+_27109440 | 1.02 |
ENST00000519080.1
|
TEK
|
TEK tyrosine kinase, endothelial |
| chr6_+_148663729 | 1.01 |
ENST00000367467.3
|
SASH1
|
SAM and SH3 domain containing 1 |
| chr12_-_71003568 | 1.00 |
ENST00000547715.1
ENST00000451516.2 ENST00000538708.1 ENST00000550857.1 ENST00000261266.5 |
PTPRB
|
protein tyrosine phosphatase, receptor type, B |
| chr9_+_124461603 | 1.00 |
ENST00000373782.3
|
DAB2IP
|
DAB2 interacting protein |
| chr3_-_65583561 | 1.00 |
ENST00000460329.2
|
MAGI1
|
membrane associated guanylate kinase, WW and PDZ domain containing 1 |
| chr5_+_15500280 | 0.99 |
ENST00000504595.1
|
FBXL7
|
F-box and leucine-rich repeat protein 7 |
| chr2_-_158182410 | 0.99 |
ENST00000419116.2
ENST00000410096.1 |
ERMN
|
ermin, ERM-like protein |
| chr9_+_27109133 | 0.96 |
ENST00000519097.1
ENST00000380036.4 |
TEK
|
TEK tyrosine kinase, endothelial |
| chr17_+_8213539 | 0.96 |
ENST00000583529.1
|
ARHGEF15
|
Rho guanine nucleotide exchange factor (GEF) 15 |
| chrX_+_114874727 | 0.96 |
ENST00000543070.1
|
PLS3
|
plastin 3 |
| chr9_+_2622085 | 0.96 |
ENST00000382099.2
|
VLDLR
|
very low density lipoprotein receptor |
| chr12_-_111358372 | 0.94 |
ENST00000548438.1
ENST00000228841.8 |
MYL2
|
myosin, light chain 2, regulatory, cardiac, slow |
| chr16_+_82660635 | 0.94 |
ENST00000567445.1
ENST00000446376.2 |
CDH13
|
cadherin 13 |
| chr13_+_113656092 | 0.93 |
ENST00000397024.1
|
MCF2L
|
MCF.2 cell line derived transforming sequence-like |
| chr1_-_27339317 | 0.93 |
ENST00000289166.5
|
FAM46B
|
family with sequence similarity 46, member B |
| chr15_+_71184931 | 0.92 |
ENST00000560369.1
ENST00000260382.5 |
LRRC49
|
leucine rich repeat containing 49 |
| chr16_+_66400533 | 0.92 |
ENST00000341529.3
|
CDH5
|
cadherin 5, type 2 (vascular endothelium) |
| chr19_-_8408139 | 0.92 |
ENST00000330915.3
ENST00000593649.1 ENST00000595639.1 |
KANK3
|
KN motif and ankyrin repeat domains 3 |
| chr8_-_38326139 | 0.92 |
ENST00000335922.5
ENST00000532791.1 ENST00000397091.5 |
FGFR1
|
fibroblast growth factor receptor 1 |
| chr3_+_85008089 | 0.90 |
ENST00000383699.3
|
CADM2
|
cell adhesion molecule 2 |
| chr6_-_46293378 | 0.89 |
ENST00000330430.6
|
RCAN2
|
regulator of calcineurin 2 |
| chr5_-_158526693 | 0.89 |
ENST00000380654.4
|
EBF1
|
early B-cell factor 1 |
| chr5_-_146833222 | 0.88 |
ENST00000534907.1
|
DPYSL3
|
dihydropyrimidinase-like 3 |
| chr4_-_182186182 | 0.88 |
ENST00000512547.1
|
RP11-665C14.2
|
RP11-665C14.2 |
| chr17_-_56605341 | 0.88 |
ENST00000583114.1
|
SEPT4
|
septin 4 |
| chr11_+_60691924 | 0.88 |
ENST00000544065.1
ENST00000453848.2 ENST00000005286.4 |
TMEM132A
|
transmembrane protein 132A |
| chr7_+_107110488 | 0.86 |
ENST00000304402.4
|
GPR22
|
G protein-coupled receptor 22 |
| chr1_+_43766642 | 0.86 |
ENST00000372476.3
|
TIE1
|
tyrosine kinase with immunoglobulin-like and EGF-like domains 1 |
| chr5_-_141338627 | 0.85 |
ENST00000231484.3
|
PCDH12
|
protocadherin 12 |
| chr20_+_61147651 | 0.85 |
ENST00000370527.3
ENST00000370524.2 |
C20orf166
|
chromosome 20 open reading frame 166 |
| chr6_+_53976285 | 0.85 |
ENST00000514433.1
|
MLIP
|
muscular LMNA-interacting protein |
| chr7_-_107968921 | 0.84 |
ENST00000442580.1
|
NRCAM
|
neuronal cell adhesion molecule |
| chr10_-_105845674 | 0.83 |
ENST00000353479.5
ENST00000369733.3 |
COL17A1
|
collagen, type XVII, alpha 1 |
| chr12_-_88974236 | 0.82 |
ENST00000228280.5
ENST00000552044.1 ENST00000357116.4 |
KITLG
|
KIT ligand |
| chr5_-_83680192 | 0.82 |
ENST00000380138.3
|
EDIL3
|
EGF-like repeats and discoidin I-like domains 3 |
| chr8_-_38326119 | 0.81 |
ENST00000356207.5
ENST00000326324.6 |
FGFR1
|
fibroblast growth factor receptor 1 |
| chr5_-_158526756 | 0.81 |
ENST00000313708.6
ENST00000517373.1 |
EBF1
|
early B-cell factor 1 |
| chr1_+_43766668 | 0.80 |
ENST00000441333.2
ENST00000538015.1 |
TIE1
|
tyrosine kinase with immunoglobulin-like and EGF-like domains 1 |
| chr17_-_14683517 | 0.80 |
ENST00000379640.1
|
AC005863.1
|
AC005863.1 |
| chr7_-_27219849 | 0.80 |
ENST00000396344.4
|
HOXA10
|
homeobox A10 |
| chr4_-_186732241 | 0.80 |
ENST00000421639.1
|
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr2_-_55277436 | 0.78 |
ENST00000354474.6
|
RTN4
|
reticulon 4 |
| chr5_-_83680603 | 0.78 |
ENST00000296591.5
|
EDIL3
|
EGF-like repeats and discoidin I-like domains 3 |
| chr2_-_1748214 | 0.78 |
ENST00000433670.1
ENST00000425171.1 ENST00000252804.4 |
PXDN
|
peroxidasin homolog (Drosophila) |
| chr4_-_186732892 | 0.78 |
ENST00000451958.1
ENST00000439914.1 ENST00000428330.1 ENST00000429056.1 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr5_+_140557371 | 0.77 |
ENST00000239444.2
|
PCDHB8
|
protocadherin beta 8 |
| chr1_-_217311090 | 0.77 |
ENST00000493603.1
ENST00000366940.2 |
ESRRG
|
estrogen-related receptor gamma |
| chr3_+_89156674 | 0.77 |
ENST00000336596.2
|
EPHA3
|
EPH receptor A3 |
| chr1_+_156589198 | 0.77 |
ENST00000456112.1
|
HAPLN2
|
hyaluronan and proteoglycan link protein 2 |
| chr14_+_63671105 | 0.75 |
ENST00000316754.3
|
RHOJ
|
ras homolog family member J |
| chr7_-_11871815 | 0.74 |
ENST00000423059.4
|
THSD7A
|
thrombospondin, type I, domain containing 7A |
| chr9_+_112403059 | 0.73 |
ENST00000374531.2
|
PALM2
|
paralemmin 2 |
| chr2_-_55277692 | 0.72 |
ENST00000394611.2
|
RTN4
|
reticulon 4 |
| chr9_-_140196703 | 0.71 |
ENST00000356628.2
|
NRARP
|
NOTCH-regulated ankyrin repeat protein |
| chr17_+_16945820 | 0.71 |
ENST00000577514.1
|
MPRIP
|
myosin phosphatase Rho interacting protein |
| chr4_-_110723335 | 0.70 |
ENST00000394634.2
|
CFI
|
complement factor I |
| chr16_+_2083265 | 0.69 |
ENST00000565855.1
ENST00000566198.1 |
SLC9A3R2
|
solute carrier family 9, subfamily A (NHE3, cation proton antiporter 3), member 3 regulator 2 |
| chr2_-_55277654 | 0.69 |
ENST00000337526.6
ENST00000317610.7 ENST00000357732.4 |
RTN4
|
reticulon 4 |
| chr2_-_55277512 | 0.69 |
ENST00000402434.2
|
RTN4
|
reticulon 4 |
| chr2_-_158182322 | 0.69 |
ENST00000420719.2
ENST00000409216.1 |
ERMN
|
ermin, ERM-like protein |
| chr1_-_85930246 | 0.69 |
ENST00000426972.3
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1 |
| chr19_+_45971246 | 0.69 |
ENST00000585836.1
ENST00000417353.2 ENST00000353609.3 ENST00000591858.1 ENST00000443841.2 ENST00000590335.1 |
FOSB
|
FBJ murine osteosarcoma viral oncogene homolog B |
| chr12_+_95611536 | 0.69 |
ENST00000549002.1
|
VEZT
|
vezatin, adherens junctions transmembrane protein |
| chr4_-_110723194 | 0.68 |
ENST00000394635.3
|
CFI
|
complement factor I |
| chr2_+_30569506 | 0.68 |
ENST00000421976.2
|
AC109642.1
|
AC109642.1 |
| chr5_+_140739537 | 0.67 |
ENST00000522605.1
|
PCDHGB2
|
protocadherin gamma subfamily B, 2 |
| chr12_-_24715478 | 0.67 |
ENST00000456299.2
ENST00000540811.1 |
RP11-444D3.1
|
RP11-444D3.1 |
| chr3_-_185826718 | 0.67 |
ENST00000413301.1
ENST00000421809.1 |
ETV5
|
ets variant 5 |
| chr5_+_172068232 | 0.66 |
ENST00000520919.1
ENST00000522853.1 ENST00000369800.5 |
NEURL1B
|
neuralized E3 ubiquitin protein ligase 1B |
| chr17_+_36584662 | 0.66 |
ENST00000431231.2
ENST00000437668.3 |
ARHGAP23
|
Rho GTPase activating protein 23 |
| chr11_-_65640071 | 0.66 |
ENST00000526624.1
|
EFEMP2
|
EGF containing fibulin-like extracellular matrix protein 2 |
| chr2_-_175869936 | 0.65 |
ENST00000409900.3
|
CHN1
|
chimerin 1 |
| chr8_+_70378852 | 0.65 |
ENST00000525061.1
ENST00000458141.2 ENST00000260128.4 |
SULF1
|
sulfatase 1 |
| chr5_+_140729649 | 0.65 |
ENST00000523390.1
|
PCDHGB1
|
protocadherin gamma subfamily B, 1 |
| chr10_-_61122934 | 0.65 |
ENST00000512919.1
|
FAM13C
|
family with sequence similarity 13, member C |
| chr10_+_73724123 | 0.65 |
ENST00000373115.4
|
CHST3
|
carbohydrate (chondroitin 6) sulfotransferase 3 |
| chr11_+_77532233 | 0.65 |
ENST00000525409.1
|
AAMDC
|
adipogenesis associated, Mth938 domain containing |
| chr7_-_22233442 | 0.64 |
ENST00000401957.2
|
RAPGEF5
|
Rap guanine nucleotide exchange factor (GEF) 5 |
| chr2_+_220309379 | 0.64 |
ENST00000451076.1
|
SPEG
|
SPEG complex locus |
| chr8_-_93115445 | 0.64 |
ENST00000523629.1
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr5_+_140797296 | 0.63 |
ENST00000398594.2
|
PCDHGB7
|
protocadherin gamma subfamily B, 7 |
| chr9_+_112852477 | 0.63 |
ENST00000480388.1
|
AKAP2
|
A kinase (PRKA) anchor protein 2 |
| chr14_-_95236551 | 0.63 |
ENST00000238558.3
|
GSC
|
goosecoid homeobox |
| chr1_+_164528866 | 0.63 |
ENST00000420696.2
|
PBX1
|
pre-B-cell leukemia homeobox 1 |
| chr6_+_13272904 | 0.63 |
ENST00000379335.3
ENST00000379329.1 |
PHACTR1
|
phosphatase and actin regulator 1 |
| chr19_+_36359341 | 0.62 |
ENST00000221891.4
|
APLP1
|
amyloid beta (A4) precursor-like protein 1 |
| chr8_-_42358742 | 0.62 |
ENST00000517366.1
|
SLC20A2
|
solute carrier family 20 (phosphate transporter), member 2 |
| chr5_-_146889619 | 0.62 |
ENST00000343218.5
|
DPYSL3
|
dihydropyrimidinase-like 3 |
| chr22_+_38864041 | 0.62 |
ENST00000216014.4
ENST00000409006.3 |
KDELR3
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 3 |
| chr11_+_77532155 | 0.62 |
ENST00000532481.1
ENST00000526415.1 ENST00000393427.2 ENST00000527134.1 ENST00000304716.8 |
AAMDC
|
adipogenesis associated, Mth938 domain containing |
| chr1_-_182360498 | 0.61 |
ENST00000417584.2
|
GLUL
|
glutamate-ammonia ligase |
| chr1_+_209602156 | 0.61 |
ENST00000429156.1
ENST00000366437.3 ENST00000603283.1 ENST00000431096.1 |
MIR205HG
|
MIR205 host gene (non-protein coding) |
| chr9_-_85882145 | 0.61 |
ENST00000328788.1
|
FRMD3
|
FERM domain containing 3 |
| chr21_+_17442799 | 0.61 |
ENST00000602580.1
ENST00000458468.1 ENST00000602935.1 |
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr7_+_116312411 | 0.60 |
ENST00000456159.1
ENST00000397752.3 ENST00000318493.6 |
MET
|
met proto-oncogene |
| chrM_+_4431 | 0.60 |
ENST00000361453.3
|
MT-ND2
|
mitochondrially encoded NADH dehydrogenase 2 |
| chr11_-_76155618 | 0.60 |
ENST00000530759.1
|
RP11-111M22.3
|
RP11-111M22.3 |
| chr1_+_93913713 | 0.60 |
ENST00000604705.1
ENST00000370253.2 |
FNBP1L
|
formin binding protein 1-like |
| chr7_-_107643567 | 0.60 |
ENST00000393559.1
ENST00000439976.1 ENST00000393560.1 |
LAMB1
|
laminin, beta 1 |
| chr3_+_69985734 | 0.60 |
ENST00000314557.6
ENST00000394351.3 |
MITF
|
microphthalmia-associated transcription factor |
| chr18_-_21891460 | 0.60 |
ENST00000357041.4
|
OSBPL1A
|
oxysterol binding protein-like 1A |
| chrX_+_103028638 | 0.60 |
ENST00000434483.1
|
PLP1
|
proteolipid protein 1 |
| chr4_-_110723134 | 0.60 |
ENST00000510800.1
ENST00000512148.1 |
CFI
|
complement factor I |
| chr15_+_101402041 | 0.59 |
ENST00000558475.1
ENST00000558641.1 ENST00000559673.1 |
RP11-66B24.1
|
RP11-66B24.1 |
| chr4_-_87028478 | 0.59 |
ENST00000515400.1
ENST00000395157.3 |
MAPK10
|
mitogen-activated protein kinase 10 |
| chr18_-_53177984 | 0.59 |
ENST00000543082.1
|
TCF4
|
transcription factor 4 |
| chr7_-_112635675 | 0.58 |
ENST00000447785.1
ENST00000451962.1 |
AC018464.3
|
AC018464.3 |
| chr16_+_15528332 | 0.58 |
ENST00000566490.1
|
C16orf45
|
chromosome 16 open reading frame 45 |
| chr8_-_38325219 | 0.58 |
ENST00000533668.1
ENST00000413133.2 ENST00000397108.4 ENST00000526742.1 ENST00000525001.1 ENST00000425967.3 ENST00000529552.1 ENST00000397113.2 |
FGFR1
|
fibroblast growth factor receptor 1 |
| chr14_-_21516590 | 0.57 |
ENST00000555026.1
|
NDRG2
|
NDRG family member 2 |
| chrX_-_17878827 | 0.57 |
ENST00000360011.1
|
RAI2
|
retinoic acid induced 2 |
| chr20_-_1317555 | 0.56 |
ENST00000537552.1
|
AL136531.1
|
HCG2043693; Uncharacterized protein |
| chr11_+_68080077 | 0.56 |
ENST00000294304.7
|
LRP5
|
low density lipoprotein receptor-related protein 5 |
| chr11_-_76155700 | 0.56 |
ENST00000572035.1
|
RP11-111M22.3
|
RP11-111M22.3 |
| chr2_-_175629164 | 0.56 |
ENST00000409323.1
ENST00000261007.5 ENST00000348749.5 |
CHRNA1
|
cholinergic receptor, nicotinic, alpha 1 (muscle) |
| chr3_+_69985792 | 0.55 |
ENST00000531774.1
|
MITF
|
microphthalmia-associated transcription factor |
| chr6_-_127840048 | 0.55 |
ENST00000467753.1
|
SOGA3
|
SOGA family member 3 |
| chr19_+_48972459 | 0.55 |
ENST00000427476.1
|
CYTH2
|
cytohesin 2 |
| chr1_-_205904950 | 0.55 |
ENST00000340781.4
|
SLC26A9
|
solute carrier family 26 (anion exchanger), member 9 |
| chr3_-_48470838 | 0.54 |
ENST00000358459.4
ENST00000358536.4 |
PLXNB1
|
plexin B1 |
| chr11_-_44971702 | 0.54 |
ENST00000533940.1
ENST00000533937.1 |
TP53I11
|
tumor protein p53 inducible protein 11 |
| chr10_-_105845536 | 0.54 |
ENST00000393211.3
|
COL17A1
|
collagen, type XVII, alpha 1 |
| chr7_-_107643674 | 0.54 |
ENST00000222399.6
|
LAMB1
|
laminin, beta 1 |
| chr2_+_223536428 | 0.53 |
ENST00000446656.3
|
MOGAT1
|
monoacylglycerol O-acyltransferase 1 |
| chr16_-_46782221 | 0.53 |
ENST00000394809.4
|
MYLK3
|
myosin light chain kinase 3 |
| chr17_-_10372875 | 0.52 |
ENST00000255381.2
|
MYH4
|
myosin, heavy chain 4, skeletal muscle |
| chr15_-_37392703 | 0.52 |
ENST00000382766.2
ENST00000444725.1 |
MEIS2
|
Meis homeobox 2 |
| chr12_-_24715376 | 0.52 |
ENST00000446891.2
ENST00000429944.2 |
RP11-444D3.1
|
RP11-444D3.1 |
| chr2_+_168725458 | 0.52 |
ENST00000392690.3
|
B3GALT1
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 1 |
| chr7_+_134464376 | 0.52 |
ENST00000454108.1
ENST00000361675.2 |
CALD1
|
caldesmon 1 |
| chr12_+_130646999 | 0.51 |
ENST00000539839.1
ENST00000229030.4 |
FZD10
|
frizzled family receptor 10 |
| chr11_+_110001723 | 0.51 |
ENST00000528673.1
|
ZC3H12C
|
zinc finger CCCH-type containing 12C |
| chr1_+_164529004 | 0.51 |
ENST00000559240.1
ENST00000367897.1 ENST00000540236.1 ENST00000401534.1 |
PBX1
|
pre-B-cell leukemia homeobox 1 |
| chr5_-_16936340 | 0.51 |
ENST00000507288.1
ENST00000513610.1 |
MYO10
|
myosin X |
| chr5_-_141338377 | 0.50 |
ENST00000510041.1
|
PCDH12
|
protocadherin 12 |
| chr21_+_41239243 | 0.50 |
ENST00000328619.5
|
PCP4
|
Purkinje cell protein 4 |
| chr13_+_98795505 | 0.50 |
ENST00000319562.6
|
FARP1
|
FERM, RhoGEF (ARHGEF) and pleckstrin domain protein 1 (chondrocyte-derived) |
| chr10_+_70748487 | 0.50 |
ENST00000361983.4
|
KIAA1279
|
KIAA1279 |
| chr7_+_128379449 | 0.50 |
ENST00000479257.1
|
CALU
|
calumenin |
| chr12_+_95611569 | 0.50 |
ENST00000261219.6
ENST00000551472.1 ENST00000552821.1 |
VEZT
|
vezatin, adherens junctions transmembrane protein |
| chr7_+_94537542 | 0.50 |
ENST00000433881.1
|
PPP1R9A
|
protein phosphatase 1, regulatory subunit 9A |
Network of associatons between targets according to the STRING database.
First level regulatory network of SOX9
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.3 | GO:0007037 | vacuolar phosphate transport(GO:0007037) positive regulation of mitotic cell cycle DNA replication(GO:1903465) positive regulation of parathyroid hormone secretion(GO:2000830) |
| 0.4 | 1.9 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 0.4 | 1.2 | GO:1903964 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.4 | 2.3 | GO:0008065 | establishment of blood-nerve barrier(GO:0008065) |
| 0.4 | 3.0 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.3 | 2.3 | GO:0071503 | positive regulation of lipoprotein particle clearance(GO:0010986) response to heparin(GO:0071503) |
| 0.3 | 0.6 | GO:0014034 | neural crest cell fate commitment(GO:0014034) |
| 0.3 | 4.6 | GO:0055095 | lipoprotein particle mediated signaling(GO:0055095) low-density lipoprotein particle mediated signaling(GO:0055096) |
| 0.3 | 1.2 | GO:0097156 | fasciculation of motor neuron axon(GO:0097156) |
| 0.3 | 3.3 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.3 | 1.1 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.3 | 0.8 | GO:0033023 | mast cell homeostasis(GO:0033023) mast cell apoptotic process(GO:0033024) regulation of mast cell apoptotic process(GO:0033025) regulation of mast cell proliferation(GO:0070666) positive regulation of mast cell proliferation(GO:0070668) |
| 0.3 | 1.1 | GO:0007499 | ectoderm and mesoderm interaction(GO:0007499) |
| 0.3 | 1.6 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 0.2 | 0.7 | GO:0016340 | calcium-dependent cell-matrix adhesion(GO:0016340) |
| 0.2 | 1.1 | GO:0021812 | neuronal-glial interaction involved in cerebral cortex radial glia guided migration(GO:0021812) |
| 0.2 | 2.0 | GO:0038026 | reelin-mediated signaling pathway(GO:0038026) |
| 0.2 | 1.9 | GO:0042078 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.2 | 0.7 | GO:1904049 | negative regulation of spontaneous neurotransmitter secretion(GO:1904049) |
| 0.2 | 0.9 | GO:0030382 | sperm mitochondrion organization(GO:0030382) |
| 0.2 | 1.2 | GO:0071279 | cellular response to cobalt ion(GO:0071279) |
| 0.2 | 0.5 | GO:0034164 | negative regulation of toll-like receptor 9 signaling pathway(GO:0034164) |
| 0.2 | 0.3 | GO:0032972 | regulation of muscle filament sliding speed(GO:0032972) |
| 0.2 | 1.1 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.2 | 0.5 | GO:0032242 | regulation of nucleoside transport(GO:0032242) |
| 0.1 | 0.4 | GO:0001579 | medium-chain fatty acid transport(GO:0001579) |
| 0.1 | 0.6 | GO:0061304 | retinal blood vessel morphogenesis(GO:0061304) |
| 0.1 | 1.0 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.1 | 1.0 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.1 | 0.6 | GO:0071874 | cellular response to norepinephrine stimulus(GO:0071874) |
| 0.1 | 1.0 | GO:0061299 | retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.1 | 1.0 | GO:0048630 | skeletal muscle tissue growth(GO:0048630) |
| 0.1 | 0.5 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.1 | 0.6 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.1 | 0.7 | GO:0070086 | ubiquitin-dependent endocytosis(GO:0070086) |
| 0.1 | 0.7 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.1 | 0.5 | GO:0002528 | regulation of vascular permeability involved in acute inflammatory response(GO:0002528) |
| 0.1 | 1.9 | GO:0071285 | cellular response to lithium ion(GO:0071285) |
| 0.1 | 1.0 | GO:0060244 | negative regulation of cell proliferation involved in contact inhibition(GO:0060244) |
| 0.1 | 4.1 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.1 | 1.2 | GO:1901552 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.1 | 1.0 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.1 | 0.4 | GO:0090149 | mitochondrial membrane fission(GO:0090149) |
| 0.1 | 4.5 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.1 | 3.2 | GO:0045725 | positive regulation of glycogen biosynthetic process(GO:0045725) |
| 0.1 | 1.0 | GO:0070779 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.1 | 4.6 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.1 | 0.5 | GO:1900220 | semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.1 | 5.3 | GO:0002026 | regulation of the force of heart contraction(GO:0002026) |
| 0.1 | 0.5 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.1 | 0.3 | GO:0060489 | orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) |
| 0.1 | 0.3 | GO:1904617 | negative regulation of actin filament binding(GO:1904530) negative regulation of actin binding(GO:1904617) |
| 0.1 | 0.4 | GO:0090285 | negative regulation of protein glycosylation in Golgi(GO:0090285) |
| 0.1 | 0.8 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 0.1 | 0.9 | GO:0044539 | long-chain fatty acid import(GO:0044539) |
| 0.1 | 0.3 | GO:0051710 | cytolysis by symbiont of host cells(GO:0001897) regulation of cytolysis in other organism(GO:0051710) |
| 0.1 | 3.9 | GO:0003301 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.1 | 1.1 | GO:1904936 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.1 | 0.4 | GO:0071603 | endothelial cell-cell adhesion(GO:0071603) |
| 0.1 | 0.6 | GO:1903976 | negative regulation of glial cell migration(GO:1903976) |
| 0.1 | 0.4 | GO:1900827 | maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.1 | 0.2 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.1 | 0.3 | GO:1900063 | regulation of peroxisome organization(GO:1900063) |
| 0.1 | 1.8 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.1 | 2.0 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.1 | 0.2 | GO:0072387 | flavin adenine dinucleotide metabolic process(GO:0072387) |
| 0.1 | 0.4 | GO:1904207 | regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904207) positive regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904209) |
| 0.1 | 0.3 | GO:0009236 | cobalamin biosynthetic process(GO:0009236) |
| 0.1 | 2.3 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.1 | 0.4 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.1 | 1.0 | GO:0098870 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.1 | 0.4 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.1 | 0.7 | GO:0045054 | constitutive secretory pathway(GO:0045054) |
| 0.1 | 0.2 | GO:1904428 | negative regulation of tubulin deacetylation(GO:1904428) |
| 0.1 | 0.2 | GO:0045645 | regulation of eosinophil differentiation(GO:0045643) positive regulation of eosinophil differentiation(GO:0045645) |
| 0.1 | 0.5 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.1 | 0.7 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.1 | 0.5 | GO:0038031 | non-canonical Wnt signaling pathway via JNK cascade(GO:0038031) |
| 0.1 | 0.7 | GO:0098722 | asymmetric stem cell division(GO:0098722) |
| 0.1 | 0.8 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.1 | 0.6 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.1 | 0.2 | GO:0018315 | molybdenum incorporation into molybdenum-molybdopterin complex(GO:0018315) metal incorporation into metallo-molybdopterin complex(GO:0042040) glycine receptor clustering(GO:0072579) |
| 0.1 | 0.3 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.0 | 1.1 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.6 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.0 | 0.4 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.0 | 2.2 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.0 | 0.4 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.0 | 0.5 | GO:0032808 | lacrimal gland development(GO:0032808) |
| 0.0 | 0.6 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 1.0 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.0 | 0.3 | GO:0003185 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.0 | 0.3 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.0 | 0.3 | GO:1902952 | regulation of postsynaptic density protein 95 clustering(GO:1902897) positive regulation of dendritic spine maintenance(GO:1902952) |
| 0.0 | 0.4 | GO:2000465 | regulation of glycogen (starch) synthase activity(GO:2000465) |
| 0.0 | 0.7 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.0 | 0.5 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.0 | 1.4 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.1 | GO:0016240 | autophagosome docking(GO:0016240) |
| 0.0 | 0.1 | GO:0032900 | viral protein processing(GO:0019082) negative regulation of neurotrophin production(GO:0032900) regulation of nerve growth factor production(GO:0032903) negative regulation of nerve growth factor production(GO:0032904) dibasic protein processing(GO:0090472) |
| 0.0 | 0.1 | GO:0021530 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.0 | 0.6 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.0 | 1.4 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.0 | 0.2 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.0 | 0.2 | GO:0090263 | positive regulation of canonical Wnt signaling pathway(GO:0090263) |
| 0.0 | 0.6 | GO:0090360 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 0.0 | 6.1 | GO:0048706 | embryonic skeletal system development(GO:0048706) |
| 0.0 | 0.7 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 | 0.2 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.0 | 0.2 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 0.0 | 0.2 | GO:0009597 | detection of virus(GO:0009597) |
| 0.0 | 0.7 | GO:1902358 | sulfate transmembrane transport(GO:1902358) |
| 0.0 | 1.7 | GO:0006536 | glutamate metabolic process(GO:0006536) |
| 0.0 | 1.7 | GO:0045026 | plasma membrane fusion(GO:0045026) |
| 0.0 | 0.3 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
| 0.0 | 0.3 | GO:0032887 | regulation of spindle elongation(GO:0032887) regulation of mitotic spindle elongation(GO:0032888) anastral spindle assembly(GO:0055048) protein localization to spindle pole body(GO:0071988) regulation of protein localization to spindle pole body(GO:1902363) positive regulation of protein localization to spindle pole body(GO:1902365) positive regulation of mitotic spindle elongation(GO:1902846) |
| 0.0 | 0.2 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.0 | 0.5 | GO:0061436 | establishment of skin barrier(GO:0061436) |
| 0.0 | 0.8 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.0 | 0.4 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 | 0.1 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.0 | 0.9 | GO:0070884 | regulation of calcineurin-NFAT signaling cascade(GO:0070884) |
| 0.0 | 0.3 | GO:0035860 | glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.0 | 0.1 | GO:0016260 | selenocysteine biosynthetic process(GO:0016260) |
| 0.0 | 0.8 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.0 | 1.2 | GO:0030318 | melanocyte differentiation(GO:0030318) |
| 0.0 | 4.2 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.2 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.0 | 0.1 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.0 | 0.4 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.0 | 0.2 | GO:0097116 | gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 0.0 | 0.1 | GO:0042823 | pyridoxal phosphate biosynthetic process(GO:0042823) |
| 0.0 | 0.1 | GO:1903935 | response to sodium arsenite(GO:1903935) cellular response to sodium arsenite(GO:1903936) |
| 0.0 | 4.3 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 | 0.7 | GO:0051412 | response to corticosterone(GO:0051412) |
| 0.0 | 0.6 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.0 | 0.9 | GO:0008038 | neuron recognition(GO:0008038) |
| 0.0 | 0.3 | GO:0035873 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) |
| 0.0 | 0.3 | GO:0021895 | cerebral cortex neuron differentiation(GO:0021895) |
| 0.0 | 0.1 | GO:0071651 | positive regulation of chemokine (C-C motif) ligand 5 production(GO:0071651) |
| 0.0 | 0.8 | GO:0042744 | hydrogen peroxide catabolic process(GO:0042744) |
| 0.0 | 0.2 | GO:0036091 | positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 0.0 | 0.1 | GO:0009098 | branched-chain amino acid biosynthetic process(GO:0009082) leucine biosynthetic process(GO:0009098) valine biosynthetic process(GO:0009099) |
| 0.0 | 0.3 | GO:0006782 | protoporphyrinogen IX biosynthetic process(GO:0006782) |
| 0.0 | 0.6 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.1 | GO:2000588 | positive regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000588) |
| 0.0 | 0.1 | GO:0060179 | male mating behavior(GO:0060179) |
| 0.0 | 0.1 | GO:0051935 | amino acid neurotransmitter reuptake(GO:0051933) glutamate reuptake(GO:0051935) |
| 0.0 | 0.7 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 0.2 | GO:0071798 | response to prostaglandin D(GO:0071798) cellular response to prostaglandin D stimulus(GO:0071799) |
| 0.0 | 0.1 | GO:0061470 | T follicular helper cell differentiation(GO:0061470) |
| 0.0 | 0.9 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.0 | 0.2 | GO:0061303 | cornea development in camera-type eye(GO:0061303) |
| 0.0 | 0.0 | GO:0031204 | posttranslational protein targeting to membrane, translocation(GO:0031204) |
| 0.0 | 0.2 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.0 | 0.1 | GO:0055014 | atrial cardiac muscle cell differentiation(GO:0055011) atrial cardiac muscle cell development(GO:0055014) |
| 0.0 | 2.0 | GO:0030449 | regulation of complement activation(GO:0030449) regulation of protein activation cascade(GO:2000257) |
| 0.0 | 0.1 | GO:0014067 | negative regulation of phosphatidylinositol 3-kinase signaling(GO:0014067) |
| 0.0 | 1.3 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.0 | 0.7 | GO:0033275 | muscle filament sliding(GO:0030049) actin-myosin filament sliding(GO:0033275) |
| 0.0 | 0.5 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.0 | 0.3 | GO:0021681 | cerebellar granular layer development(GO:0021681) |
| 0.0 | 0.1 | GO:0048311 | mitochondrion distribution(GO:0048311) |
| 0.0 | 0.1 | GO:0021781 | glial cell fate commitment(GO:0021781) |
| 0.0 | 1.0 | GO:0032981 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.3 | GO:0036148 | phosphatidylglycerol acyl-chain remodeling(GO:0036148) |
| 0.0 | 0.3 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.1 | GO:0046898 | response to cycloheximide(GO:0046898) |
| 0.0 | 0.1 | GO:0010226 | response to lithium ion(GO:0010226) |
| 0.0 | 0.1 | GO:0042407 | cristae formation(GO:0042407) |
| 0.0 | 0.4 | GO:0014002 | astrocyte development(GO:0014002) |
| 0.0 | 0.2 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.2 | GO:0015671 | oxygen transport(GO:0015671) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:0005595 | collagen type XII trimer(GO:0005595) |
| 0.5 | 3.2 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.4 | 2.6 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.4 | 1.1 | GO:0043257 | laminin-8 complex(GO:0043257) |
| 0.3 | 1.0 | GO:1990032 | parallel fiber(GO:1990032) |
| 0.2 | 1.8 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.2 | 4.6 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.2 | 0.5 | GO:0036457 | keratohyalin granule(GO:0036457) |
| 0.2 | 0.3 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.1 | 2.1 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.1 | 0.6 | GO:1990851 | Wnt-Frizzled-LRP5/6 complex(GO:1990851) |
| 0.1 | 0.4 | GO:0071665 | gamma-catenin-TCF7L2 complex(GO:0071665) |
| 0.1 | 2.0 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.1 | 4.9 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.1 | 2.3 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.1 | 1.1 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.1 | 3.3 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.1 | 2.7 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 3.1 | GO:0034385 | very-low-density lipoprotein particle(GO:0034361) triglyceride-rich lipoprotein particle(GO:0034385) |
| 0.1 | 1.4 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.1 | 0.7 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 0.1 | 0.3 | GO:0097629 | extrinsic component of omegasome membrane(GO:0097629) |
| 0.1 | 0.3 | GO:0060187 | cell pole(GO:0060187) |
| 0.1 | 4.0 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.1 | 0.7 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.1 | 1.0 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.1 | 3.7 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.1 | 0.9 | GO:0030478 | actin cap(GO:0030478) |
| 0.1 | 0.7 | GO:0016011 | dystroglycan complex(GO:0016011) |
| 0.1 | 0.9 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.1 | 1.2 | GO:0043203 | axon hillock(GO:0043203) |
| 0.0 | 0.6 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.0 | 1.0 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.0 | 0.9 | GO:0097227 | sperm annulus(GO:0097227) |
| 0.0 | 0.8 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.5 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 0.2 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.0 | 1.0 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 1.8 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.2 | GO:1990730 | VCP-NSFL1C complex(GO:1990730) |
| 0.0 | 0.5 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.5 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.0 | 0.3 | GO:0055028 | cortical microtubule(GO:0055028) |
| 0.0 | 0.5 | GO:0035748 | myelin sheath abaxonal region(GO:0035748) |
| 0.0 | 0.6 | GO:0097386 | glial cell projection(GO:0097386) |
| 0.0 | 0.3 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 9.4 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 0.5 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 1.1 | GO:0070160 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 0.3 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 1.9 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.5 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 0.7 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.5 | GO:0042641 | actomyosin(GO:0042641) |
| 0.0 | 0.1 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 4.4 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 1.3 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.5 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 1.8 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.2 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.0 | 0.2 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.0 | 0.2 | GO:0097433 | dense body(GO:0097433) |
| 0.0 | 0.1 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 1.4 | GO:0032432 | actin filament bundle(GO:0032432) |
| 0.0 | 0.1 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.0 | 1.1 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 0.2 | GO:0001939 | female pronucleus(GO:0001939) |
| 0.0 | 0.3 | GO:0005845 | mRNA cap binding complex(GO:0005845) |
| 0.0 | 0.1 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.0 | 0.2 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.2 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.3 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 4.3 | GO:0032038 | myosin II heavy chain binding(GO:0032038) |
| 0.5 | 2.2 | GO:0017129 | triglyceride binding(GO:0017129) |
| 0.5 | 2.4 | GO:0055100 | adiponectin binding(GO:0055100) |
| 0.3 | 2.3 | GO:0035473 | lipase binding(GO:0035473) |
| 0.3 | 4.4 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.3 | 1.7 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.2 | 1.7 | GO:0047820 | D-glutamate cyclase activity(GO:0047820) |
| 0.2 | 1.0 | GO:0034189 | very-low-density lipoprotein particle binding(GO:0034189) |
| 0.2 | 1.5 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.2 | 0.8 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.2 | 2.3 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.2 | 0.6 | GO:1904928 | coreceptor activity involved in canonical Wnt signaling pathway(GO:1904928) |
| 0.2 | 0.5 | GO:0047275 | glucosaminylgalactosylglucosylceramide beta-galactosyltransferase activity(GO:0047275) |
| 0.2 | 0.8 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 0.2 | 0.6 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 0.1 | 0.4 | GO:0071633 | dihydroceramidase activity(GO:0071633) |
| 0.1 | 1.0 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.1 | 1.4 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.1 | 1.0 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.1 | 1.2 | GO:0016215 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) |
| 0.1 | 0.9 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.1 | 1.7 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.1 | 0.5 | GO:0016880 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.1 | 0.4 | GO:0005199 | structural constituent of cell wall(GO:0005199) |
| 0.1 | 1.1 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.1 | 4.2 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.1 | 0.4 | GO:0004803 | transposase activity(GO:0004803) |
| 0.1 | 1.0 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.1 | 0.7 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.1 | 0.8 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.1 | 0.5 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 0.1 | 0.4 | GO:0004113 | 2',3'-cyclic-nucleotide 3'-phosphodiesterase activity(GO:0004113) |
| 0.1 | 3.9 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.1 | 1.3 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.1 | 0.3 | GO:0047389 | glycerophosphocholine phosphodiesterase activity(GO:0047389) |
| 0.1 | 0.3 | GO:0047192 | 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 0.1 | 10.5 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 0.6 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.1 | 0.7 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.1 | 0.5 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.1 | 1.2 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.1 | 0.8 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 0.6 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.1 | 0.6 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.1 | 1.1 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.1 | 0.3 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.1 | 0.5 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.1 | 2.0 | GO:0019865 | immunoglobulin binding(GO:0019865) |
| 0.1 | 0.2 | GO:0061599 | molybdopterin adenylyltransferase activity(GO:0061598) molybdopterin molybdotransferase activity(GO:0061599) |
| 0.1 | 0.2 | GO:0004958 | prostaglandin F receptor activity(GO:0004958) |
| 0.1 | 5.3 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.1 | 0.7 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 0.6 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 3.2 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 0.5 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.0 | 0.1 | GO:0004487 | methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 0.0 | 0.5 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.1 | GO:0061752 | telomeric repeat-containing RNA binding(GO:0061752) |
| 0.0 | 1.0 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 1.0 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 1.0 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.0 | 0.2 | GO:0036435 | K48-linked polyubiquitin binding(GO:0036435) |
| 0.0 | 0.2 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.0 | 1.0 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 0.6 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.5 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.0 | 1.4 | GO:0004467 | long-chain fatty acid-CoA ligase activity(GO:0004467) |
| 0.0 | 1.5 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 0.9 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.4 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.5 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.5 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.3 | GO:1903763 | gap junction channel activity involved in cell communication by electrical coupling(GO:1903763) |
| 0.0 | 0.7 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) |
| 0.0 | 0.3 | GO:0016167 | glial cell-derived neurotrophic factor receptor activity(GO:0016167) |
| 0.0 | 1.3 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.6 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 0.3 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 0.8 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.7 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.3 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.1 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.0 | 0.2 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.0 | 0.6 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.0 | 1.7 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 0.1 | GO:0016781 | selenide, water dikinase activity(GO:0004756) phosphotransferase activity, paired acceptors(GO:0016781) |
| 0.0 | 0.1 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.2 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.5 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.4 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.7 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 2.0 | GO:0017022 | myosin binding(GO:0017022) |
| 0.0 | 0.3 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 2.0 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 1.2 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 0.2 | GO:0035005 | lipid kinase activity(GO:0001727) 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 3.8 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.2 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.0 | 1.1 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.0 | 0.1 | GO:0052655 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.0 | 1.7 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.5 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.0 | 0.1 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
| 0.0 | 0.3 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.1 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.0 | 0.3 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 0.1 | GO:0033592 | RNA strand annealing activity(GO:0033592) |
| 0.0 | 0.6 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 0.2 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.3 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.0 | 0.1 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 0.2 | GO:0015194 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.4 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 0.0 | GO:0000992 | polymerase III regulatory region sequence-specific DNA binding(GO:0000992) RNA polymerase III type 1 promoter sequence-specific DNA binding(GO:0001002) RNA polymerase III type 2 promoter sequence-specific DNA binding(GO:0001003) |
| 0.0 | 0.2 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.0 | 0.6 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.8 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.4 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.6 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 0.2 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 2.1 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 0.1 | GO:0004609 | phosphatidylserine decarboxylase activity(GO:0004609) |
| 0.0 | 0.1 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 0.3 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.3 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.2 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.0 | 0.2 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 1.1 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.1 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 0.5 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.1 | GO:0043426 | MRF binding(GO:0043426) |
| 0.0 | 0.1 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.0 | 0.1 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.4 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.1 | 4.6 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.1 | 3.0 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.1 | 0.7 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.1 | 4.3 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 2.5 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 1.6 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 2.9 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 2.5 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 3.3 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.0 | 4.0 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 0.9 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.0 | 0.6 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 0.4 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 1.9 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 2.6 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 2.0 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 1.5 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 0.6 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 0.7 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 2.0 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 2.7 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 0.9 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.2 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 1.0 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 0.2 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 0.6 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 0.7 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.0 | 0.8 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.6 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.5 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 1.7 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.5 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 0.5 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 1.0 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.0 | 0.1 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 1.0 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.3 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.1 | 3.7 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.1 | 2.0 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.1 | 4.7 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 5.8 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 3.8 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 2.2 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.1 | 2.9 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.1 | 1.0 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.1 | 1.8 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.1 | 4.6 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 1.9 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 1.6 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 2.1 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.8 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.0 | 0.8 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 0.9 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.9 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.6 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 1.1 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 1.2 | REACTOME SEMA4D IN SEMAPHORIN SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 1.0 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.6 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.7 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 1.2 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 3.0 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.0 | 0.6 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.0 | 0.3 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.0 | 0.8 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 1.0 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.3 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.0 | 0.3 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 0.3 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 1.4 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |
| 0.0 | 0.2 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.5 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 2.3 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.3 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 0.3 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |