Project
Illumina Body Map 2, young vs old
Navigation
Downloads
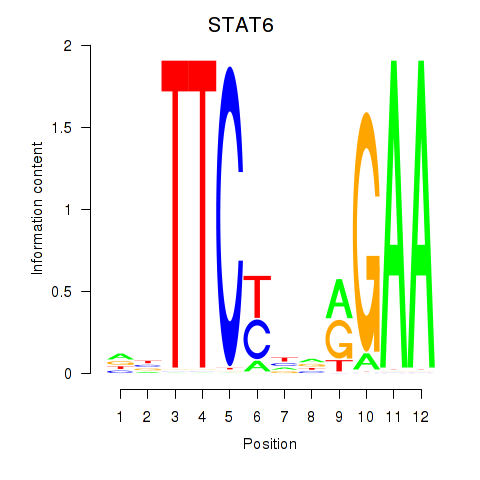
Results for STAT6
Z-value: 0.28
Transcription factors associated with STAT6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
STAT6
|
ENSG00000166888.6 | signal transducer and activator of transcription 6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| STAT6 | hg19_v2_chr12_-_57504975_57505019 | 0.13 | 4.7e-01 | Click! |
Activity profile of STAT6 motif
Sorted Z-values of STAT6 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_160616804 | 2.95 |
ENST00000538290.1
|
SLAMF1
|
signaling lymphocytic activation molecule family member 1 |
| chr14_+_22520762 | 2.45 |
ENST00000390449.3
|
TRAV21
|
T cell receptor alpha variable 21 |
| chr14_-_25078864 | 2.08 |
ENST00000216338.4
ENST00000557220.2 ENST00000382548.4 |
GZMH
|
granzyme H (cathepsin G-like 2, protein h-CCPX) |
| chr5_+_147258266 | 2.03 |
ENST00000296694.4
|
SCGB3A2
|
secretoglobin, family 3A, member 2 |
| chr7_-_38339890 | 1.99 |
ENST00000390341.2
|
TRGV10
|
T cell receptor gamma variable 10 (non-functional) |
| chr17_+_34538310 | 1.83 |
ENST00000444414.1
ENST00000378350.4 ENST00000389068.5 ENST00000588929.1 ENST00000589079.1 ENST00000589336.1 ENST00000400702.4 ENST00000591167.1 ENST00000586598.1 ENST00000591637.1 ENST00000378352.4 ENST00000358756.5 |
CCL4L1
|
chemokine (C-C motif) ligand 4-like 1 |
| chr12_-_55367361 | 1.83 |
ENST00000532804.1
ENST00000531122.1 ENST00000533446.1 |
TESPA1
|
thymocyte expressed, positive selection associated 1 |
| chr6_+_31540056 | 1.79 |
ENST00000418386.2
|
LTA
|
lymphotoxin alpha |
| chr11_-_118083600 | 1.75 |
ENST00000524477.1
|
AMICA1
|
adhesion molecule, interacts with CXADR antigen 1 |
| chr17_+_34640031 | 1.67 |
ENST00000339270.6
ENST00000482104.1 |
CCL4L2
|
chemokine (C-C motif) ligand 4-like 2 |
| chr17_+_34431212 | 1.67 |
ENST00000394495.1
|
CCL4
|
chemokine (C-C motif) ligand 4 |
| chr1_-_203198790 | 1.60 |
ENST00000367229.1
ENST00000255427.3 ENST00000535569.1 |
CHIT1
|
chitinase 1 (chitotriosidase) |
| chr19_-_7764960 | 1.55 |
ENST00000593418.1
|
FCER2
|
Fc fragment of IgE, low affinity II, receptor for (CD23) |
| chr12_-_55367331 | 1.53 |
ENST00000526532.1
ENST00000532757.1 |
TESPA1
|
thymocyte expressed, positive selection associated 1 |
| chr17_+_34639793 | 1.49 |
ENST00000394465.2
ENST00000394463.2 ENST00000378342.4 |
CCL4L2
|
chemokine (C-C motif) ligand 4-like 2 |
| chr5_+_118691706 | 1.47 |
ENST00000415806.2
|
TNFAIP8
|
tumor necrosis factor, alpha-induced protein 8 |
| chr17_-_8770956 | 1.46 |
ENST00000311434.9
|
PIK3R6
|
phosphoinositide-3-kinase, regulatory subunit 6 |
| chr6_-_24877490 | 1.44 |
ENST00000540914.1
ENST00000378023.4 |
FAM65B
|
family with sequence similarity 65, member B |
| chr2_+_191792376 | 1.44 |
ENST00000409428.1
ENST00000409215.1 |
GLS
|
glutaminase |
| chr14_-_25103388 | 1.43 |
ENST00000526004.1
ENST00000415355.3 |
GZMB
|
granzyme B (granzyme 2, cytotoxic T-lymphocyte-associated serine esterase 1) |
| chr14_-_25103472 | 1.39 |
ENST00000216341.4
ENST00000382542.1 ENST00000382540.1 |
GZMB
|
granzyme B (granzyme 2, cytotoxic T-lymphocyte-associated serine esterase 1) |
| chr19_-_7766991 | 1.38 |
ENST00000597921.1
ENST00000346664.5 |
FCER2
|
Fc fragment of IgE, low affinity II, receptor for (CD23) |
| chr2_+_219745020 | 1.32 |
ENST00000258411.3
|
WNT10A
|
wingless-type MMTV integration site family, member 10A |
| chr6_-_41168920 | 1.26 |
ENST00000483722.1
|
TREML2
|
triggering receptor expressed on myeloid cells-like 2 |
| chr14_+_22386325 | 1.19 |
ENST00000390439.2
|
TRAV13-2
|
T cell receptor alpha variable 13-2 |
| chr11_-_58980342 | 1.16 |
ENST00000361050.3
|
MPEG1
|
macrophage expressed 1 |
| chr19_-_54604083 | 1.04 |
ENST00000391761.1
ENST00000356532.3 ENST00000359649.4 ENST00000358375.4 ENST00000391760.1 ENST00000351806.4 |
OSCAR
|
osteoclast associated, immunoglobulin-like receptor |
| chr11_+_118175132 | 1.03 |
ENST00000361763.4
|
CD3E
|
CD3e molecule, epsilon (CD3-TCR complex) |
| chr16_-_29757272 | 1.02 |
ENST00000329410.3
|
C16orf54
|
chromosome 16 open reading frame 54 |
| chr19_+_38924316 | 1.01 |
ENST00000355481.4
ENST00000360985.3 ENST00000359596.3 |
RYR1
|
ryanodine receptor 1 (skeletal) |
| chr6_-_41254403 | 1.00 |
ENST00000589614.1
ENST00000334475.6 ENST00000591620.1 ENST00000244709.4 |
TREM1
|
triggering receptor expressed on myeloid cells 1 |
| chr12_+_9144626 | 0.99 |
ENST00000543895.1
|
KLRG1
|
killer cell lectin-like receptor subfamily G, member 1 |
| chr18_-_24445664 | 0.98 |
ENST00000578776.1
|
AQP4
|
aquaporin 4 |
| chr19_-_52254050 | 0.94 |
ENST00000600815.1
|
FPR1
|
formyl peptide receptor 1 |
| chr1_+_12123414 | 0.89 |
ENST00000263932.2
|
TNFRSF8
|
tumor necrosis factor receptor superfamily, member 8 |
| chr5_-_88119580 | 0.86 |
ENST00000539796.1
|
MEF2C
|
myocyte enhancer factor 2C |
| chr6_+_138188351 | 0.84 |
ENST00000421450.1
|
TNFAIP3
|
tumor necrosis factor, alpha-induced protein 3 |
| chr18_-_67615088 | 0.82 |
ENST00000582621.1
|
CD226
|
CD226 molecule |
| chr10_-_134121438 | 0.81 |
ENST00000298630.3
|
STK32C
|
serine/threonine kinase 32C |
| chr3_+_121311966 | 0.80 |
ENST00000338040.4
|
FBXO40
|
F-box protein 40 |
| chr11_-_128457446 | 0.79 |
ENST00000392668.4
|
ETS1
|
v-ets avian erythroblastosis virus E26 oncogene homolog 1 |
| chr2_-_175462456 | 0.78 |
ENST00000409891.1
ENST00000410117.1 |
WIPF1
|
WAS/WASL interacting protein family, member 1 |
| chr7_-_38305279 | 0.78 |
ENST00000443402.2
|
TRGC1
|
T cell receptor gamma constant 1 |
| chr1_+_12123463 | 0.77 |
ENST00000417814.2
|
TNFRSF8
|
tumor necrosis factor receptor superfamily, member 8 |
| chr1_+_149754227 | 0.76 |
ENST00000444948.1
ENST00000369168.4 |
FCGR1A
|
Fc fragment of IgG, high affinity Ia, receptor (CD64) |
| chr2_+_68961934 | 0.75 |
ENST00000409202.3
|
ARHGAP25
|
Rho GTPase activating protein 25 |
| chr17_-_34207295 | 0.73 |
ENST00000463941.1
ENST00000293272.3 |
CCL5
|
chemokine (C-C motif) ligand 5 |
| chr2_+_68961905 | 0.72 |
ENST00000295381.3
|
ARHGAP25
|
Rho GTPase activating protein 25 |
| chr15_+_77308152 | 0.72 |
ENST00000559859.1
|
PSTPIP1
|
proline-serine-threonine phosphatase interacting protein 1 |
| chr12_-_111358372 | 0.71 |
ENST00000548438.1
ENST00000228841.8 |
MYL2
|
myosin, light chain 2, regulatory, cardiac, slow |
| chr9_-_135996537 | 0.71 |
ENST00000372050.3
ENST00000372047.3 |
RALGDS
|
ral guanine nucleotide dissociation stimulator |
| chr7_+_80254279 | 0.71 |
ENST00000436384.1
|
CD36
|
CD36 molecule (thrombospondin receptor) |
| chr1_-_120935894 | 0.71 |
ENST00000369383.4
ENST00000369384.4 |
FCGR1B
|
Fc fragment of IgG, high affinity Ib, receptor (CD64) |
| chr11_+_65082289 | 0.70 |
ENST00000279249.2
|
CDC42EP2
|
CDC42 effector protein (Rho GTPase binding) 2 |
| chr21_-_35340759 | 0.70 |
ENST00000607953.1
|
AP000569.9
|
AP000569.9 |
| chr2_+_68962014 | 0.70 |
ENST00000467265.1
|
ARHGAP25
|
Rho GTPase activating protein 25 |
| chr8_+_49984894 | 0.69 |
ENST00000522267.1
ENST00000399653.4 ENST00000303202.8 |
C8orf22
|
chromosome 8 open reading frame 22 |
| chr6_+_147527103 | 0.69 |
ENST00000179882.6
|
STXBP5
|
syntaxin binding protein 5 (tomosyn) |
| chr14_-_23285011 | 0.63 |
ENST00000397532.3
|
SLC7A7
|
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7 |
| chr12_-_54779511 | 0.63 |
ENST00000551109.1
ENST00000546970.1 |
ZNF385A
|
zinc finger protein 385A |
| chr13_+_50589390 | 0.62 |
ENST00000360473.4
ENST00000312942.1 |
KCNRG
|
potassium channel regulator |
| chr1_-_36020531 | 0.61 |
ENST00000440579.1
ENST00000494948.1 |
KIAA0319L
|
KIAA0319-like |
| chr6_+_116850174 | 0.58 |
ENST00000416171.2
ENST00000368597.2 ENST00000452373.1 ENST00000405399.1 |
FAM26D
|
family with sequence similarity 26, member D |
| chr4_-_48116540 | 0.58 |
ENST00000506073.1
|
TXK
|
TXK tyrosine kinase |
| chr6_-_32821599 | 0.58 |
ENST00000354258.4
|
TAP1
|
transporter 1, ATP-binding cassette, sub-family B (MDR/TAP) |
| chr18_+_66382428 | 0.56 |
ENST00000578970.1
ENST00000582371.1 ENST00000584775.1 |
CCDC102B
|
coiled-coil domain containing 102B |
| chr5_-_88120083 | 0.56 |
ENST00000509373.1
|
MEF2C
|
myocyte enhancer factor 2C |
| chr3_+_140981456 | 0.55 |
ENST00000504264.1
|
ACPL2
|
acid phosphatase-like 2 |
| chr5_+_36608280 | 0.55 |
ENST00000513646.1
|
SLC1A3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3 |
| chr2_-_109605663 | 0.55 |
ENST00000409271.1
ENST00000258443.2 ENST00000376651.1 |
EDAR
|
ectodysplasin A receptor |
| chr22_-_42342692 | 0.54 |
ENST00000404067.1
ENST00000402338.1 |
CENPM
|
centromere protein M |
| chr12_-_46663734 | 0.54 |
ENST00000550173.1
|
SLC38A1
|
solute carrier family 38, member 1 |
| chr2_-_26700900 | 0.53 |
ENST00000338581.6
ENST00000339598.3 ENST00000402415.3 |
OTOF
|
otoferlin |
| chr11_-_60719213 | 0.53 |
ENST00000227880.3
|
SLC15A3
|
solute carrier family 15 (oligopeptide transporter), member 3 |
| chrX_+_135251835 | 0.53 |
ENST00000456445.1
|
FHL1
|
four and a half LIM domains 1 |
| chr5_+_126984710 | 0.51 |
ENST00000379445.3
|
CTXN3
|
cortexin 3 |
| chr1_+_114472481 | 0.51 |
ENST00000369555.2
|
HIPK1
|
homeodomain interacting protein kinase 1 |
| chr14_-_23285069 | 0.51 |
ENST00000554758.1
ENST00000397528.4 |
SLC7A7
|
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7 |
| chr22_-_42343117 | 0.50 |
ENST00000407253.3
ENST00000215980.5 |
CENPM
|
centromere protein M |
| chr5_-_88120151 | 0.50 |
ENST00000506716.1
|
MEF2C
|
myocyte enhancer factor 2C |
| chr18_-_24445729 | 0.50 |
ENST00000383168.4
|
AQP4
|
aquaporin 4 |
| chr18_-_53068782 | 0.49 |
ENST00000569012.1
|
TCF4
|
transcription factor 4 |
| chr3_+_10312604 | 0.49 |
ENST00000426850.1
|
TATDN2
|
TatD DNase domain containing 2 |
| chr2_-_197226875 | 0.48 |
ENST00000409111.1
|
HECW2
|
HECT, C2 and WW domain containing E3 ubiquitin protein ligase 2 |
| chr4_+_144312659 | 0.48 |
ENST00000509992.1
|
GAB1
|
GRB2-associated binding protein 1 |
| chrX_+_135252050 | 0.48 |
ENST00000449474.1
ENST00000345434.3 |
FHL1
|
four and a half LIM domains 1 |
| chr1_-_116383738 | 0.47 |
ENST00000320238.3
|
NHLH2
|
nescient helix loop helix 2 |
| chr22_+_44464923 | 0.47 |
ENST00000404989.1
|
PARVB
|
parvin, beta |
| chrX_+_102192200 | 0.46 |
ENST00000218249.5
|
RAB40AL
|
RAB40A, member RAS oncogene family-like |
| chr2_-_16804320 | 0.45 |
ENST00000355549.2
|
FAM49A
|
family with sequence similarity 49, member A |
| chr12_+_51619860 | 0.44 |
ENST00000599633.1
|
AC139768.1
|
Uncharacterized protein |
| chr16_+_30076052 | 0.44 |
ENST00000563987.1
|
ALDOA
|
aldolase A, fructose-bisphosphate |
| chr6_+_138188378 | 0.43 |
ENST00000420009.1
|
TNFAIP3
|
tumor necrosis factor, alpha-induced protein 3 |
| chr3_-_10149907 | 0.43 |
ENST00000450660.2
ENST00000524279.1 |
FANCD2OS
|
FANCD2 opposite strand |
| chr20_-_23066953 | 0.42 |
ENST00000246006.4
|
CD93
|
CD93 molecule |
| chr14_+_61449076 | 0.41 |
ENST00000526105.1
|
SLC38A6
|
solute carrier family 38, member 6 |
| chr2_-_134326009 | 0.41 |
ENST00000409261.1
ENST00000409213.1 |
NCKAP5
|
NCK-associated protein 5 |
| chr7_+_10000640 | 0.41 |
ENST00000451755.1
|
AC006373.1
|
AC006373.1 |
| chrX_+_135251783 | 0.40 |
ENST00000394153.2
|
FHL1
|
four and a half LIM domains 1 |
| chr2_-_203735976 | 0.40 |
ENST00000435143.1
|
ICA1L
|
islet cell autoantigen 1,69kDa-like |
| chr6_+_32821924 | 0.40 |
ENST00000374859.2
ENST00000453265.2 |
PSMB9
|
proteasome (prosome, macropain) subunit, beta type, 9 |
| chr7_+_76091775 | 0.39 |
ENST00000442516.1
|
DTX2
|
deltex homolog 2 (Drosophila) |
| chr12_-_42878101 | 0.39 |
ENST00000552108.1
|
PRICKLE1
|
prickle homolog 1 (Drosophila) |
| chr5_+_93954358 | 0.38 |
ENST00000504099.1
|
ANKRD32
|
ankyrin repeat domain 32 |
| chr12_-_118490217 | 0.38 |
ENST00000542304.1
|
WSB2
|
WD repeat and SOCS box containing 2 |
| chr16_+_30075783 | 0.38 |
ENST00000412304.2
|
ALDOA
|
aldolase A, fructose-bisphosphate |
| chr15_+_89402148 | 0.38 |
ENST00000560601.1
|
ACAN
|
aggrecan |
| chrX_+_36254051 | 0.37 |
ENST00000378657.4
|
CXorf30
|
chromosome X open reading frame 30 |
| chr15_+_91643173 | 0.37 |
ENST00000545111.2
|
SV2B
|
synaptic vesicle glycoprotein 2B |
| chr3_-_49851313 | 0.36 |
ENST00000333486.3
|
UBA7
|
ubiquitin-like modifier activating enzyme 7 |
| chr1_+_66796401 | 0.36 |
ENST00000528771.1
|
PDE4B
|
phosphodiesterase 4B, cAMP-specific |
| chr10_+_98064085 | 0.36 |
ENST00000419175.1
ENST00000371174.2 |
DNTT
|
DNA nucleotidylexotransferase |
| chr15_+_91643442 | 0.35 |
ENST00000394232.1
|
SV2B
|
synaptic vesicle glycoprotein 2B |
| chr12_-_42877764 | 0.35 |
ENST00000455697.1
|
PRICKLE1
|
prickle homolog 1 (Drosophila) |
| chr5_+_35856951 | 0.35 |
ENST00000303115.3
ENST00000343305.4 ENST00000506850.1 ENST00000511982.1 |
IL7R
|
interleukin 7 receptor |
| chr14_-_24732738 | 0.34 |
ENST00000558074.1
ENST00000560226.1 |
TGM1
|
transglutaminase 1 |
| chr11_-_77531752 | 0.34 |
ENST00000440064.2
ENST00000528095.1 |
RSF1
|
remodeling and spacing factor 1 |
| chr8_+_123793633 | 0.33 |
ENST00000314393.4
|
ZHX2
|
zinc fingers and homeoboxes 2 |
| chr2_-_203735484 | 0.33 |
ENST00000420558.1
ENST00000418208.1 |
ICA1L
|
islet cell autoantigen 1,69kDa-like |
| chr6_-_33754778 | 0.32 |
ENST00000508327.1
ENST00000513701.1 |
LEMD2
|
LEM domain containing 2 |
| chr3_+_130162328 | 0.32 |
ENST00000512482.1
|
COL6A5
|
collagen, type VI, alpha 5 |
| chr1_-_45965525 | 0.32 |
ENST00000488405.2
ENST00000490551.3 ENST00000432082.1 |
CCDC163P
|
coiled-coil domain containing 163, pseudogene |
| chr16_+_57728701 | 0.32 |
ENST00000569375.1
ENST00000360716.3 ENST00000569167.1 ENST00000394337.4 ENST00000563126.1 ENST00000336825.8 |
CCDC135
|
coiled-coil domain containing 135 |
| chr14_+_61449197 | 0.31 |
ENST00000533744.2
|
SLC38A6
|
solute carrier family 38, member 6 |
| chr11_-_72463421 | 0.31 |
ENST00000393609.3
|
ARAP1
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1 |
| chr5_-_74162739 | 0.31 |
ENST00000513277.1
|
FAM169A
|
family with sequence similarity 169, member A |
| chr9_-_10612703 | 0.31 |
ENST00000463477.1
|
PTPRD
|
protein tyrosine phosphatase, receptor type, D |
| chr15_+_27111510 | 0.31 |
ENST00000335625.5
|
GABRA5
|
gamma-aminobutyric acid (GABA) A receptor, alpha 5 |
| chr11_+_73358594 | 0.30 |
ENST00000227214.6
ENST00000398494.4 ENST00000543085.1 |
PLEKHB1
|
pleckstrin homology domain containing, family B (evectins) member 1 |
| chr2_+_197504278 | 0.30 |
ENST00000272831.7
ENST00000389175.4 ENST00000472405.2 ENST00000423093.2 |
CCDC150
|
coiled-coil domain containing 150 |
| chr7_+_129015484 | 0.30 |
ENST00000490911.1
|
AHCYL2
|
adenosylhomocysteinase-like 2 |
| chr3_-_179692042 | 0.30 |
ENST00000468741.1
|
PEX5L
|
peroxisomal biogenesis factor 5-like |
| chr4_-_120988229 | 0.29 |
ENST00000296509.6
|
MAD2L1
|
MAD2 mitotic arrest deficient-like 1 (yeast) |
| chr3_+_63898275 | 0.29 |
ENST00000538065.1
|
ATXN7
|
ataxin 7 |
| chr11_-_132813627 | 0.29 |
ENST00000374778.4
|
OPCML
|
opioid binding protein/cell adhesion molecule-like |
| chr7_+_129015671 | 0.28 |
ENST00000466993.1
|
AHCYL2
|
adenosylhomocysteinase-like 2 |
| chr11_-_60720002 | 0.28 |
ENST00000538739.1
|
SLC15A3
|
solute carrier family 15 (oligopeptide transporter), member 3 |
| chr1_+_114472222 | 0.27 |
ENST00000369558.1
ENST00000369561.4 |
HIPK1
|
homeodomain interacting protein kinase 1 |
| chr17_+_28268623 | 0.27 |
ENST00000394835.3
ENST00000320856.5 ENST00000394832.2 ENST00000378738.3 |
EFCAB5
|
EF-hand calcium binding domain 5 |
| chr6_+_26440700 | 0.27 |
ENST00000494393.1
ENST00000482451.1 ENST00000244519.2 ENST00000339789.4 ENST00000471353.1 ENST00000361232.3 ENST00000487627.1 ENST00000496719.1 ENST00000490254.1 ENST00000487272.1 |
BTN3A3
|
butyrophilin, subfamily 3, member A3 |
| chr3_-_57233966 | 0.26 |
ENST00000473921.1
ENST00000295934.3 |
HESX1
|
HESX homeobox 1 |
| chr8_+_107593198 | 0.26 |
ENST00000517686.1
|
OXR1
|
oxidation resistance 1 |
| chr5_+_71475449 | 0.26 |
ENST00000504492.1
|
MAP1B
|
microtubule-associated protein 1B |
| chr11_+_118230287 | 0.26 |
ENST00000252108.3
ENST00000431736.2 |
UBE4A
|
ubiquitination factor E4A |
| chr15_+_101402041 | 0.26 |
ENST00000558475.1
ENST00000558641.1 ENST00000559673.1 |
RP11-66B24.1
|
RP11-66B24.1 |
| chr9_-_3469181 | 0.25 |
ENST00000366116.2
|
AL365202.1
|
Uncharacterized protein |
| chr15_+_30918879 | 0.25 |
ENST00000428041.2
|
ARHGAP11B
|
Rho GTPase activating protein 11B |
| chr3_+_189507432 | 0.25 |
ENST00000354600.5
|
TP63
|
tumor protein p63 |
| chr3_+_141121164 | 0.25 |
ENST00000510338.1
ENST00000504673.1 |
ZBTB38
|
zinc finger and BTB domain containing 38 |
| chr8_+_11666649 | 0.25 |
ENST00000528643.1
ENST00000525777.1 |
FDFT1
|
farnesyl-diphosphate farnesyltransferase 1 |
| chr12_-_118490403 | 0.25 |
ENST00000535496.1
|
WSB2
|
WD repeat and SOCS box containing 2 |
| chr2_-_230786619 | 0.25 |
ENST00000389045.3
ENST00000409677.1 |
TRIP12
|
thyroid hormone receptor interactor 12 |
| chr2_-_202316169 | 0.25 |
ENST00000430254.1
|
TRAK2
|
trafficking protein, kinesin binding 2 |
| chr16_+_30075463 | 0.24 |
ENST00000562168.1
ENST00000569545.1 |
ALDOA
|
aldolase A, fructose-bisphosphate |
| chr6_-_30524951 | 0.24 |
ENST00000376621.3
|
GNL1
|
guanine nucleotide binding protein-like 1 |
| chr4_+_54966198 | 0.24 |
ENST00000326902.2
ENST00000503800.1 |
GSX2
|
GS homeobox 2 |
| chr12_-_42877726 | 0.24 |
ENST00000548696.1
|
PRICKLE1
|
prickle homolog 1 (Drosophila) |
| chr2_-_203735586 | 0.24 |
ENST00000454326.1
ENST00000432273.1 ENST00000450143.1 ENST00000411681.1 |
ICA1L
|
islet cell autoantigen 1,69kDa-like |
| chr12_+_56435637 | 0.24 |
ENST00000356464.5
ENST00000552361.1 |
RPS26
|
ribosomal protein S26 |
| chr4_+_141677577 | 0.24 |
ENST00000609937.1
|
RP11-102N12.3
|
RP11-102N12.3 |
| chr12_-_53097247 | 0.24 |
ENST00000341809.3
ENST00000537195.1 |
KRT77
|
keratin 77 |
| chr20_+_58251716 | 0.24 |
ENST00000355648.4
|
PHACTR3
|
phosphatase and actin regulator 3 |
| chr6_-_27880174 | 0.23 |
ENST00000303324.2
|
OR2B2
|
olfactory receptor, family 2, subfamily B, member 2 |
| chr5_+_110427983 | 0.23 |
ENST00000513710.2
ENST00000505303.1 |
WDR36
|
WD repeat domain 36 |
| chr4_+_71337834 | 0.23 |
ENST00000304887.5
|
MUC7
|
mucin 7, secreted |
| chr2_-_31440377 | 0.23 |
ENST00000444918.2
ENST00000403897.3 |
CAPN14
|
calpain 14 |
| chr11_-_8964580 | 0.23 |
ENST00000325884.1
|
ASCL3
|
achaete-scute family bHLH transcription factor 3 |
| chr1_-_153321301 | 0.23 |
ENST00000368739.3
|
PGLYRP4
|
peptidoglycan recognition protein 4 |
| chr5_+_102201687 | 0.23 |
ENST00000304400.7
|
PAM
|
peptidylglycine alpha-amidating monooxygenase |
| chr22_-_42342733 | 0.21 |
ENST00000402420.1
|
CENPM
|
centromere protein M |
| chr11_-_77531858 | 0.21 |
ENST00000360355.2
|
RSF1
|
remodeling and spacing factor 1 |
| chr18_-_44775554 | 0.21 |
ENST00000425639.1
ENST00000400404.1 |
SKOR2
|
SKI family transcriptional corepressor 2 |
| chr16_-_67694597 | 0.20 |
ENST00000393919.4
ENST00000219251.8 |
ACD
|
adrenocortical dysplasia homolog (mouse) |
| chr5_+_102201722 | 0.20 |
ENST00000274392.9
ENST00000455264.2 |
PAM
|
peptidylglycine alpha-amidating monooxygenase |
| chr1_+_109256067 | 0.20 |
ENST00000271311.2
|
FNDC7
|
fibronectin type III domain containing 7 |
| chr6_+_38690622 | 0.19 |
ENST00000327475.6
|
DNAH8
|
dynein, axonemal, heavy chain 8 |
| chr13_-_74708372 | 0.19 |
ENST00000377666.4
|
KLF12
|
Kruppel-like factor 12 |
| chr16_+_67694849 | 0.18 |
ENST00000602551.1
ENST00000458121.2 ENST00000219255.3 |
PARD6A
|
par-6 family cell polarity regulator alpha |
| chr2_+_145425534 | 0.18 |
ENST00000432608.1
ENST00000597655.1 ENST00000598659.1 ENST00000600679.1 ENST00000601277.1 ENST00000451027.1 ENST00000445791.1 ENST00000596540.1 ENST00000596230.1 ENST00000594471.1 ENST00000598248.1 ENST00000597469.1 ENST00000431734.1 ENST00000595686.1 |
TEX41
|
testis expressed 41 (non-protein coding) |
| chr5_+_138609441 | 0.18 |
ENST00000509990.1
ENST00000506147.1 ENST00000512107.1 |
MATR3
|
matrin 3 |
| chr16_+_30075595 | 0.18 |
ENST00000563060.2
|
ALDOA
|
aldolase A, fructose-bisphosphate |
| chr20_+_42136308 | 0.18 |
ENST00000434666.1
ENST00000427442.2 ENST00000439769.1 ENST00000418998.1 |
L3MBTL1
|
l(3)mbt-like 1 (Drosophila) |
| chr11_+_65101225 | 0.17 |
ENST00000528416.1
ENST00000415073.2 ENST00000252268.4 |
DPF2
|
D4, zinc and double PHD fingers family 2 |
| chr14_+_59100774 | 0.17 |
ENST00000556859.1
ENST00000421793.1 |
DACT1
|
dishevelled-binding antagonist of beta-catenin 1 |
| chr3_+_189507523 | 0.17 |
ENST00000437221.1
ENST00000392463.2 ENST00000392461.3 ENST00000449992.1 ENST00000456148.1 |
TP63
|
tumor protein p63 |
| chr17_+_44588877 | 0.17 |
ENST00000576629.1
|
LRRC37A2
|
leucine rich repeat containing 37, member A2 |
| chr14_+_56127960 | 0.17 |
ENST00000553624.1
|
KTN1
|
kinectin 1 (kinesin receptor) |
| chr20_+_42574317 | 0.17 |
ENST00000358131.5
|
TOX2
|
TOX high mobility group box family member 2 |
| chr1_-_241799232 | 0.17 |
ENST00000366553.1
|
CHML
|
choroideremia-like (Rab escort protein 2) |
| chr1_+_34632484 | 0.16 |
ENST00000373374.3
|
C1orf94
|
chromosome 1 open reading frame 94 |
| chr2_+_21444025 | 0.16 |
ENST00000435237.1
ENST00000457901.1 |
AC067959.1
|
AC067959.1 |
| chr2_+_27440229 | 0.14 |
ENST00000264705.4
ENST00000403525.1 |
CAD
|
carbamoyl-phosphate synthetase 2, aspartate transcarbamylase, and dihydroorotase |
| chr2_+_113885138 | 0.14 |
ENST00000409930.3
|
IL1RN
|
interleukin 1 receptor antagonist |
| chr6_+_30951487 | 0.14 |
ENST00000486149.2
ENST00000376296.3 |
MUC21
|
mucin 21, cell surface associated |
| chr6_+_84222220 | 0.13 |
ENST00000369700.3
|
PRSS35
|
protease, serine, 35 |
| chr1_+_109265033 | 0.13 |
ENST00000445274.1
|
FNDC7
|
fibronectin type III domain containing 7 |
| chr9_-_111619239 | 0.13 |
ENST00000374667.3
|
ACTL7B
|
actin-like 7B |
| chr11_-_77532050 | 0.13 |
ENST00000308488.6
|
RSF1
|
remodeling and spacing factor 1 |
| chr8_-_117886732 | 0.13 |
ENST00000517485.1
|
RAD21
|
RAD21 homolog (S. pombe) |
| chr14_+_56127989 | 0.13 |
ENST00000555573.1
|
KTN1
|
kinectin 1 (kinesin receptor) |
Network of associatons between targets according to the STRING database.
First level regulatory network of STAT6
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 4.2 | GO:2000349 | negative regulation of CD40 signaling pathway(GO:2000349) |
| 0.8 | 4.7 | GO:0002925 | positive regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002925) |
| 0.6 | 1.7 | GO:0045556 | TRAIL biosynthetic process(GO:0045553) regulation of TRAIL biosynthetic process(GO:0045554) positive regulation of TRAIL biosynthetic process(GO:0045556) |
| 0.5 | 3.4 | GO:0010387 | COP9 signalosome assembly(GO:0010387) |
| 0.4 | 2.8 | GO:0008626 | granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 0.3 | 2.4 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.3 | 1.9 | GO:0003172 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.2 | 1.6 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.2 | 1.0 | GO:0002669 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.2 | 1.4 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.2 | 1.0 | GO:0051892 | negative regulation of cardioblast differentiation(GO:0051892) regulation of cardiac muscle cell myoblast differentiation(GO:2000690) negative regulation of cardiac muscle cell myoblast differentiation(GO:2000691) |
| 0.2 | 1.0 | GO:0070945 | neutrophil mediated killing of gram-negative bacterium(GO:0070945) |
| 0.1 | 0.6 | GO:0046967 | cytosol to ER transport(GO:0046967) |
| 0.1 | 0.7 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 0.1 | 0.8 | GO:0002729 | positive regulation of natural killer cell cytokine production(GO:0002729) |
| 0.1 | 1.1 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) |
| 0.1 | 0.3 | GO:0098972 | dendritic transport of mitochondrion(GO:0098939) anterograde dendritic transport of mitochondrion(GO:0098972) |
| 0.1 | 0.8 | GO:1904996 | PML body organization(GO:0030578) positive regulation of leukocyte adhesion to vascular endothelial cell(GO:1904996) |
| 0.1 | 0.6 | GO:0060335 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.1 | 0.5 | GO:0006537 | glutamate biosynthetic process(GO:0006537) |
| 0.1 | 0.6 | GO:1902164 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902162) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.1 | 1.3 | GO:0048733 | sebaceous gland development(GO:0048733) |
| 0.1 | 0.2 | GO:0051714 | positive regulation of cytolysis in other organism(GO:0051714) |
| 0.1 | 1.0 | GO:0071313 | cellular response to caffeine(GO:0071313) |
| 0.1 | 0.6 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.1 | 0.4 | GO:0018032 | protein amidation(GO:0018032) |
| 0.1 | 0.2 | GO:0060380 | regulation of single-stranded telomeric DNA binding(GO:0060380) positive regulation of single-stranded telomeric DNA binding(GO:0060381) |
| 0.1 | 1.2 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.1 | 0.8 | GO:0060235 | lens induction in camera-type eye(GO:0060235) |
| 0.1 | 0.7 | GO:0072564 | blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.1 | 0.2 | GO:0021798 | forebrain dorsal/ventral pattern formation(GO:0021798) |
| 0.1 | 0.7 | GO:0031017 | exocrine pancreas development(GO:0031017) |
| 0.1 | 0.7 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.1 | 0.3 | GO:0030037 | actin filament reorganization involved in cell cycle(GO:0030037) |
| 0.1 | 0.3 | GO:0008204 | ergosterol biosynthetic process(GO:0006696) ergosterol metabolic process(GO:0008204) |
| 0.0 | 0.9 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.0 | 0.1 | GO:0044205 | 'de novo' UMP biosynthetic process(GO:0044205) |
| 0.0 | 0.5 | GO:0071963 | establishment or maintenance of cell polarity regulating cell shape(GO:0071963) |
| 0.0 | 0.7 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.2 | GO:0007499 | ectoderm and mesoderm interaction(GO:0007499) |
| 0.0 | 0.3 | GO:0045229 | cell envelope organization(GO:0043163) external encapsulating structure organization(GO:0045229) |
| 0.0 | 0.3 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.0 | 1.5 | GO:0006833 | water transport(GO:0006833) |
| 0.0 | 2.1 | GO:0019835 | cytolysis(GO:0019835) |
| 0.0 | 0.4 | GO:1901315 | negative regulation of histone ubiquitination(GO:0033183) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) |
| 0.0 | 0.6 | GO:1902260 | negative regulation of delayed rectifier potassium channel activity(GO:1902260) |
| 0.0 | 0.5 | GO:0060662 | tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.0 | 1.3 | GO:0034080 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.0 | 0.3 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.0 | 0.4 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.3 | GO:0070197 | meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.0 | 0.3 | GO:0090267 | positive regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090267) |
| 0.0 | 0.3 | GO:0016560 | protein import into peroxisome matrix, docking(GO:0016560) |
| 0.0 | 0.2 | GO:1904781 | positive regulation of protein localization to centrosome(GO:1904781) |
| 0.0 | 0.3 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 1.0 | GO:0042269 | regulation of natural killer cell mediated cytotoxicity(GO:0042269) |
| 0.0 | 0.2 | GO:0048619 | embryonic hindgut morphogenesis(GO:0048619) |
| 0.0 | 0.4 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.0 | 0.3 | GO:0008038 | neuron recognition(GO:0008038) |
| 0.0 | 1.4 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.0 | 0.1 | GO:0048691 | modulation by virus of host transcription(GO:0019056) positive regulation of sprouting of injured axon(GO:0048687) positive regulation of axon extension involved in regeneration(GO:0048691) modulation by symbiont of host transcription(GO:0052026) |
| 0.0 | 0.1 | GO:0050925 | negative regulation of negative chemotaxis(GO:0050925) apoptotic process involved in luteolysis(GO:0061364) |
| 0.0 | 1.4 | GO:0043268 | positive regulation of potassium ion transport(GO:0043268) |
| 0.0 | 0.2 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.0 | 1.9 | GO:0002479 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-dependent(GO:0002479) |
| 0.0 | 0.1 | GO:2000660 | negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.0 | 0.2 | GO:0045876 | positive regulation of sister chromatid cohesion(GO:0045876) |
| 0.0 | 0.3 | GO:0048853 | forebrain morphogenesis(GO:0048853) |
| 0.0 | 0.1 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.0 | 0.3 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 0.3 | GO:0061339 | establishment of monopolar cell polarity(GO:0061162) establishment or maintenance of monopolar cell polarity(GO:0061339) |
| 0.0 | 0.0 | GO:1901963 | glial cell fate determination(GO:0007403) canonical Wnt signaling pathway involved in positive regulation of cardiac outflow tract cell proliferation(GO:0061324) regulation of cell proliferation involved in outflow tract morphogenesis(GO:1901963) regulation of chromatin-mediated maintenance of transcription(GO:1904499) positive regulation of chromatin-mediated maintenance of transcription(GO:1904501) regulation of euchromatin binding(GO:1904793) |
| 0.0 | 1.0 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.0 | 0.5 | GO:0001504 | neurotransmitter uptake(GO:0001504) |
| 0.0 | 0.5 | GO:0035728 | response to hepatocyte growth factor(GO:0035728) |
| 0.0 | 0.1 | GO:0036079 | GDP-fucose transport(GO:0015783) purine nucleotide-sugar transport(GO:0036079) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.5 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.2 | 1.0 | GO:1990425 | ryanodine receptor complex(GO:1990425) |
| 0.2 | 0.7 | GO:0031213 | RSF complex(GO:0031213) |
| 0.1 | 0.6 | GO:0042825 | TAP complex(GO:0042825) |
| 0.1 | 1.0 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.1 | 0.2 | GO:0097679 | other organism cytoplasm(GO:0097679) |
| 0.1 | 2.0 | GO:0071682 | endocytic vesicle lumen(GO:0071682) |
| 0.0 | 3.4 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.4 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.0 | 3.9 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.0 | 2.8 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.7 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.0 | 0.2 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 0.7 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 0.2 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 0.8 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 3.7 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.0 | 0.2 | GO:0000798 | nuclear cohesin complex(GO:0000798) |
| 0.0 | 1.5 | GO:0030669 | clathrin-coated endocytic vesicle membrane(GO:0030669) |
| 0.0 | 0.2 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.0 | 1.6 | GO:0005764 | lytic vacuole(GO:0000323) lysosome(GO:0005764) |
| 0.0 | 1.4 | GO:0101003 | ficolin-1-rich granule membrane(GO:0101003) |
| 0.0 | 0.1 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.0 | 0.2 | GO:0070187 | telosome(GO:0070187) |
| 0.0 | 5.0 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 0.3 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.0 | 0.4 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 1.6 | GO:0001669 | acrosomal vesicle(GO:0001669) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.4 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.3 | 2.9 | GO:0019863 | IgE binding(GO:0019863) |
| 0.3 | 1.0 | GO:0048763 | calcium-induced calcium release activity(GO:0048763) |
| 0.2 | 1.4 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.1 | 0.8 | GO:0015333 | peptide:proton symporter activity(GO:0015333) proton-dependent peptide secondary active transmembrane transporter activity(GO:0022897) |
| 0.1 | 1.7 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.1 | 0.9 | GO:0005124 | N-formyl peptide receptor activity(GO:0004982) scavenger receptor binding(GO:0005124) |
| 0.1 | 1.2 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.1 | 0.7 | GO:0019763 | immunoglobulin receptor activity(GO:0019763) |
| 0.1 | 0.7 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.1 | 1.6 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.1 | 0.3 | GO:0004917 | interleukin-7 receptor activity(GO:0004917) |
| 0.1 | 1.5 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.1 | 0.6 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.1 | 0.6 | GO:0046979 | TAP2 binding(GO:0046979) |
| 0.1 | 1.5 | GO:0015250 | water channel activity(GO:0015250) |
| 0.1 | 0.4 | GO:0004598 | peptidylglycine monooxygenase activity(GO:0004504) peptidylamidoglycolate lyase activity(GO:0004598) |
| 0.1 | 1.3 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.1 | 0.7 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.1 | 1.0 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.1 | 1.9 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.1 | 0.8 | GO:0019864 | IgG binding(GO:0019864) |
| 0.1 | 0.3 | GO:0051996 | farnesyl-diphosphate farnesyltransferase activity(GO:0004310) squalene synthase activity(GO:0051996) |
| 0.1 | 0.2 | GO:0032093 | SAM domain binding(GO:0032093) |
| 0.1 | 0.2 | GO:0008745 | N-acetylmuramoyl-L-alanine amidase activity(GO:0008745) |
| 0.1 | 0.5 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.0 | 1.1 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.0 | 0.1 | GO:0005150 | interleukin-1, Type I receptor binding(GO:0005150) |
| 0.0 | 0.4 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.2 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.4 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.0 | 0.1 | GO:0004088 | carbamoyl-phosphate synthase (ammonia) activity(GO:0004087) carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity(GO:0004088) |
| 0.0 | 1.4 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 0.7 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.0 | 0.8 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 1.9 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.5 | GO:0005283 | sodium:amino acid symporter activity(GO:0005283) |
| 0.0 | 0.3 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.0 | 0.5 | GO:0016888 | endodeoxyribonuclease activity, producing 5'-phosphomonoesters(GO:0016888) |
| 0.0 | 1.5 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 1.3 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.5 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 0.6 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.1 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 2.3 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.0 | 0.3 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.2 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.3 | GO:0031702 | angiotensin receptor binding(GO:0031701) type 1 angiotensin receptor binding(GO:0031702) |
| 0.0 | 0.4 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.1 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.3 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.2 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.0 | 0.1 | GO:0036080 | GDP-fucose transmembrane transporter activity(GO:0005457) purine nucleotide-sugar transmembrane transporter activity(GO:0036080) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.4 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 1.6 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 1.5 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 4.5 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 0.7 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 1.3 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 2.7 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.0 | 1.7 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 1.0 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 1.8 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 0.5 | PID EPO PATHWAY | EPO signaling pathway |
| 0.0 | 1.8 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.9 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 6.5 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.3 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.4 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 1.2 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.5 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 2.8 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.0 | 1.4 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 1.0 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.0 | 1.9 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 0.7 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 2.4 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 3.0 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 0.7 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.0 | 1.3 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.0 | 3.2 | REACTOME ANTIGEN PROCESSING CROSS PRESENTATION | Genes involved in Antigen processing-Cross presentation |
| 0.0 | 1.5 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 2.6 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 1.2 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.3 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.5 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 0.2 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 1.7 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.7 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |