Project
Illumina Body Map 2, young vs old
Navigation
Downloads
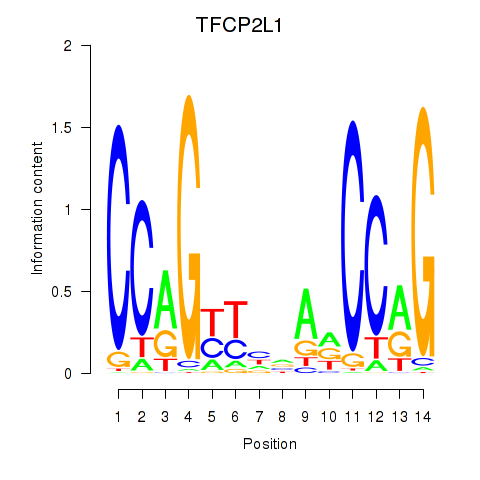
Results for TFCP2L1
Z-value: 0.10
Transcription factors associated with TFCP2L1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TFCP2L1
|
ENSG00000115112.7 | transcription factor CP2 like 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TFCP2L1 | hg19_v2_chr2_-_122042770_122042785 | 0.36 | 4.5e-02 | Click! |
Activity profile of TFCP2L1 motif
Sorted Z-values of TFCP2L1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr20_+_55204351 | 3.69 |
ENST00000201031.2
|
TFAP2C
|
transcription factor AP-2 gamma (activating enhancer binding protein 2 gamma) |
| chr10_+_102505468 | 3.62 |
ENST00000361791.3
ENST00000355243.3 ENST00000428433.1 ENST00000370296.2 |
PAX2
|
paired box 2 |
| chr10_+_102505954 | 3.54 |
ENST00000556085.1
ENST00000427256.1 |
PAX2
|
paired box 2 |
| chr17_-_74023474 | 3.42 |
ENST00000301607.3
|
EVPL
|
envoplakin |
| chr1_-_153538011 | 3.39 |
ENST00000368707.4
|
S100A2
|
S100 calcium binding protein A2 |
| chr17_-_46703826 | 3.32 |
ENST00000550387.1
ENST00000311177.5 |
HOXB9
|
homeobox B9 |
| chr1_+_27668505 | 3.21 |
ENST00000318074.5
|
SYTL1
|
synaptotagmin-like 1 |
| chr20_+_44098385 | 3.17 |
ENST00000217425.5
ENST00000339946.3 |
WFDC2
|
WAP four-disulfide core domain 2 |
| chr17_-_74023291 | 3.15 |
ENST00000586740.1
|
EVPL
|
envoplakin |
| chr20_+_44098346 | 3.14 |
ENST00000372676.3
|
WFDC2
|
WAP four-disulfide core domain 2 |
| chr21_-_31588365 | 3.07 |
ENST00000399899.1
|
CLDN8
|
claudin 8 |
| chr15_+_41136216 | 3.03 |
ENST00000562057.1
ENST00000344051.4 |
SPINT1
|
serine peptidase inhibitor, Kunitz type 1 |
| chr20_-_52790512 | 3.03 |
ENST00000216862.3
|
CYP24A1
|
cytochrome P450, family 24, subfamily A, polypeptide 1 |
| chr11_-_128737259 | 2.98 |
ENST00000440599.2
ENST00000392666.1 ENST00000324036.3 |
KCNJ1
|
potassium inwardly-rectifying channel, subfamily J, member 1 |
| chr1_+_27189631 | 2.94 |
ENST00000339276.4
|
SFN
|
stratifin |
| chr9_-_127269661 | 2.81 |
ENST00000373588.4
|
NR5A1
|
nuclear receptor subfamily 5, group A, member 1 |
| chr1_-_226129083 | 2.78 |
ENST00000420304.2
|
LEFTY2
|
left-right determination factor 2 |
| chr1_-_153538292 | 2.76 |
ENST00000497140.1
ENST00000368708.3 |
S100A2
|
S100 calcium binding protein A2 |
| chr1_-_226129189 | 2.73 |
ENST00000366820.5
|
LEFTY2
|
left-right determination factor 2 |
| chr21_-_31588338 | 2.72 |
ENST00000286809.1
|
CLDN8
|
claudin 8 |
| chr11_-_128737163 | 2.71 |
ENST00000324003.3
ENST00000392665.2 |
KCNJ1
|
potassium inwardly-rectifying channel, subfamily J, member 1 |
| chr1_-_40782347 | 2.66 |
ENST00000417105.1
|
COL9A2
|
collagen, type IX, alpha 2 |
| chr15_+_41136369 | 2.46 |
ENST00000563656.1
|
SPINT1
|
serine peptidase inhibitor, Kunitz type 1 |
| chr22_+_50986462 | 2.42 |
ENST00000395676.2
|
KLHDC7B
|
kelch domain containing 7B |
| chr20_+_9494987 | 2.40 |
ENST00000427562.2
ENST00000246070.2 |
LAMP5
|
lysosomal-associated membrane protein family, member 5 |
| chr6_-_32731243 | 2.23 |
ENST00000427449.1
ENST00000411527.1 |
HLA-DQB2
|
major histocompatibility complex, class II, DQ beta 2 |
| chr15_+_41136263 | 2.23 |
ENST00000568823.1
|
SPINT1
|
serine peptidase inhibitor, Kunitz type 1 |
| chr17_-_27038765 | 2.22 |
ENST00000581289.1
ENST00000301039.2 |
PROCA1
|
protein interacting with cyclin A1 |
| chr6_-_31550192 | 2.19 |
ENST00000429299.2
ENST00000446745.2 |
LTB
|
lymphotoxin beta (TNF superfamily, member 3) |
| chr7_+_142423143 | 2.16 |
ENST00000390399.3
|
TRBV27
|
T cell receptor beta variable 27 |
| chrX_-_51239425 | 2.14 |
ENST00000375992.3
|
NUDT11
|
nudix (nucleoside diphosphate linked moiety X)-type motif 11 |
| chr14_-_107170409 | 2.13 |
ENST00000390633.2
|
IGHV1-69
|
immunoglobulin heavy variable 1-69 |
| chr6_-_32731299 | 2.10 |
ENST00000435145.2
ENST00000437316.2 |
HLA-DQB2
|
major histocompatibility complex, class II, DQ beta 2 |
| chr1_-_207206092 | 2.05 |
ENST00000359470.5
ENST00000461135.2 |
C1orf116
|
chromosome 1 open reading frame 116 |
| chr11_+_46403303 | 2.01 |
ENST00000407067.1
ENST00000395565.1 |
MDK
|
midkine (neurite growth-promoting factor 2) |
| chr1_-_111743285 | 2.00 |
ENST00000357640.4
|
DENND2D
|
DENN/MADD domain containing 2D |
| chr7_-_142240014 | 1.95 |
ENST00000390363.2
|
TRBV9
|
T cell receptor beta variable 9 |
| chr12_-_48398104 | 1.94 |
ENST00000337299.6
ENST00000380518.3 |
COL2A1
|
collagen, type II, alpha 1 |
| chr7_+_87563557 | 1.94 |
ENST00000439864.1
ENST00000412441.1 ENST00000398201.4 ENST00000265727.7 ENST00000315984.7 ENST00000398209.3 |
ADAM22
|
ADAM metallopeptidase domain 22 |
| chr19_-_12251130 | 1.93 |
ENST00000418866.1
ENST00000600335.1 |
ZNF20
|
zinc finger protein 20 |
| chr7_+_87563458 | 1.85 |
ENST00000398204.4
|
ADAM22
|
ADAM metallopeptidase domain 22 |
| chr19_-_6591113 | 1.83 |
ENST00000423145.3
ENST00000245903.3 |
CD70
|
CD70 molecule |
| chr5_-_180018540 | 1.79 |
ENST00000292641.3
|
SCGB3A1
|
secretoglobin, family 3A, member 1 |
| chr2_+_241544834 | 1.78 |
ENST00000319838.5
ENST00000403859.1 ENST00000438013.2 |
GPR35
|
G protein-coupled receptor 35 |
| chr7_-_127671674 | 1.77 |
ENST00000478726.1
|
LRRC4
|
leucine rich repeat containing 4 |
| chr2_-_20212422 | 1.75 |
ENST00000421259.2
ENST00000407540.3 |
MATN3
|
matrilin 3 |
| chr10_-_61469837 | 1.73 |
ENST00000395348.3
|
SLC16A9
|
solute carrier family 16, member 9 |
| chr11_+_46402744 | 1.71 |
ENST00000533952.1
|
MDK
|
midkine (neurite growth-promoting factor 2) |
| chrX_+_51075658 | 1.67 |
ENST00000356450.2
|
NUDT10
|
nudix (nucleoside diphosphate linked moiety X)-type motif 10 |
| chr10_+_91589261 | 1.66 |
ENST00000448963.1
|
LINC00865
|
long intergenic non-protein coding RNA 865 |
| chr5_+_52285144 | 1.66 |
ENST00000296585.5
|
ITGA2
|
integrin, alpha 2 (CD49B, alpha 2 subunit of VLA-2 receptor) |
| chr19_-_46916805 | 1.64 |
ENST00000307522.3
|
CCDC8
|
coiled-coil domain containing 8 |
| chr17_-_27038063 | 1.60 |
ENST00000439862.3
|
PROCA1
|
protein interacting with cyclin A1 |
| chr2_+_219125714 | 1.59 |
ENST00000522678.1
ENST00000519574.1 ENST00000521462.1 |
GPBAR1
|
G protein-coupled bile acid receptor 1 |
| chr11_+_46403194 | 1.57 |
ENST00000395569.4
ENST00000395566.4 |
MDK
|
midkine (neurite growth-promoting factor 2) |
| chr2_-_89385283 | 1.56 |
ENST00000390252.2
|
IGKV3-15
|
immunoglobulin kappa variable 3-15 |
| chr19_+_1071203 | 1.55 |
ENST00000543365.1
|
HMHA1
|
histocompatibility (minor) HA-1 |
| chr16_-_65155833 | 1.54 |
ENST00000566827.1
ENST00000394156.3 ENST00000562998.1 |
CDH11
|
cadherin 11, type 2, OB-cadherin (osteoblast) |
| chr1_-_114696472 | 1.54 |
ENST00000393296.1
ENST00000369547.1 ENST00000610222.1 |
SYT6
|
synaptotagmin VI |
| chr1_-_155959280 | 1.54 |
ENST00000495070.2
|
ARHGEF2
|
Rho/Rac guanine nucleotide exchange factor (GEF) 2 |
| chr1_+_20617362 | 1.53 |
ENST00000375079.2
ENST00000289815.8 ENST00000375083.4 ENST00000289825.4 |
VWA5B1
|
von Willebrand factor A domain containing 5B1 |
| chr17_+_7792101 | 1.53 |
ENST00000358181.4
ENST00000330494.7 |
CHD3
|
chromodomain helicase DNA binding protein 3 |
| chr19_-_12251202 | 1.52 |
ENST00000334213.5
|
ZNF20
|
zinc finger protein 20 |
| chr2_-_31361543 | 1.51 |
ENST00000349752.5
|
GALNT14
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 14 (GalNAc-T14) |
| chr17_-_1532106 | 1.50 |
ENST00000301335.5
ENST00000382147.4 |
SLC43A2
|
solute carrier family 43 (amino acid system L transporter), member 2 |
| chr4_-_25865159 | 1.49 |
ENST00000502949.1
ENST00000264868.5 ENST00000513691.1 ENST00000514872.1 |
SEL1L3
|
sel-1 suppressor of lin-12-like 3 (C. elegans) |
| chr17_-_27278445 | 1.49 |
ENST00000268756.3
ENST00000584685.1 |
PHF12
|
PHD finger protein 12 |
| chr2_-_99757876 | 1.47 |
ENST00000539964.1
ENST00000393482.3 |
TSGA10
|
testis specific, 10 |
| chr1_-_155959853 | 1.46 |
ENST00000462460.2
ENST00000368316.1 |
ARHGEF2
|
Rho/Rac guanine nucleotide exchange factor (GEF) 2 |
| chr16_+_31366536 | 1.45 |
ENST00000562522.1
|
ITGAX
|
integrin, alpha X (complement component 3 receptor 4 subunit) |
| chr11_-_17555421 | 1.44 |
ENST00000526181.1
|
USH1C
|
Usher syndrome 1C (autosomal recessive, severe) |
| chr10_-_121296045 | 1.43 |
ENST00000392865.1
|
RGS10
|
regulator of G-protein signaling 10 |
| chr14_-_106994333 | 1.41 |
ENST00000390624.2
|
IGHV3-48
|
immunoglobulin heavy variable 3-48 |
| chr20_-_57089934 | 1.40 |
ENST00000439429.1
ENST00000371149.3 |
APCDD1L
|
adenomatosis polyposis coli down-regulated 1-like |
| chr5_-_169626104 | 1.37 |
ENST00000520275.1
ENST00000506431.2 |
CTB-27N1.1
|
CTB-27N1.1 |
| chrX_+_69674943 | 1.37 |
ENST00000542398.1
|
DLG3
|
discs, large homolog 3 (Drosophila) |
| chr22_-_37545972 | 1.36 |
ENST00000216223.5
|
IL2RB
|
interleukin 2 receptor, beta |
| chr2_-_99757977 | 1.30 |
ENST00000355053.4
|
TSGA10
|
testis specific, 10 |
| chr12_+_56324933 | 1.29 |
ENST00000549629.1
ENST00000555218.1 |
DGKA
|
diacylglycerol kinase, alpha 80kDa |
| chr4_+_40195262 | 1.28 |
ENST00000503941.1
|
RHOH
|
ras homolog family member H |
| chr19_+_12175535 | 1.28 |
ENST00000550826.1
|
ZNF844
|
zinc finger protein 844 |
| chr22_-_20138302 | 1.27 |
ENST00000540078.1
ENST00000439765.2 |
AC006547.14
|
uncharacterized protein LOC388849 |
| chr2_-_31361362 | 1.26 |
ENST00000430167.1
|
GALNT14
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 14 (GalNAc-T14) |
| chr17_+_15848231 | 1.26 |
ENST00000304222.2
|
ADORA2B
|
adenosine A2b receptor |
| chr12_+_56324756 | 1.25 |
ENST00000331886.5
ENST00000555090.1 |
DGKA
|
diacylglycerol kinase, alpha 80kDa |
| chr17_-_19619917 | 1.25 |
ENST00000325411.5
ENST00000350657.5 ENST00000433844.2 |
SLC47A2
|
solute carrier family 47 (multidrug and toxin extrusion), member 2 |
| chr11_+_1856034 | 1.25 |
ENST00000341958.3
|
SYT8
|
synaptotagmin VIII |
| chr22_-_37823468 | 1.23 |
ENST00000402918.2
|
ELFN2
|
extracellular leucine-rich repeat and fibronectin type III domain containing 2 |
| chr9_+_116111794 | 1.22 |
ENST00000374183.4
|
BSPRY
|
B-box and SPRY domain containing |
| chr12_+_41582103 | 1.22 |
ENST00000402685.2
|
PDZRN4
|
PDZ domain containing ring finger 4 |
| chr10_-_99771079 | 1.20 |
ENST00000309155.3
|
CRTAC1
|
cartilage acidic protein 1 |
| chr3_-_46930171 | 1.20 |
ENST00000593391.1
|
AC109583.1
|
Uncharacterized protein |
| chr11_-_7904464 | 1.20 |
ENST00000502624.2
ENST00000527565.1 |
RP11-35J10.5
|
RP11-35J10.5 |
| chr19_+_20188830 | 1.19 |
ENST00000418063.2
|
ZNF90
|
zinc finger protein 90 |
| chr12_+_49687425 | 1.18 |
ENST00000257860.4
|
PRPH
|
peripherin |
| chr1_-_1356628 | 1.14 |
ENST00000442470.1
ENST00000537107.1 |
ANKRD65
|
ankyrin repeat domain 65 |
| chr19_-_6481776 | 1.14 |
ENST00000543576.1
ENST00000590173.1 ENST00000381480.2 |
DENND1C
|
DENN/MADD domain containing 1C |
| chr15_+_43886057 | 1.13 |
ENST00000441322.1
ENST00000413657.2 ENST00000453733.1 |
CKMT1B
|
creatine kinase, mitochondrial 1B |
| chr16_+_31366455 | 1.13 |
ENST00000268296.4
|
ITGAX
|
integrin, alpha X (complement component 3 receptor 4 subunit) |
| chr17_-_27278304 | 1.13 |
ENST00000577226.1
|
PHF12
|
PHD finger protein 12 |
| chrX_+_133371077 | 1.13 |
ENST00000517294.1
ENST00000370809.4 |
CCDC160
|
coiled-coil domain containing 160 |
| chr1_+_1361494 | 1.12 |
ENST00000378821.3
|
TMEM88B
|
transmembrane protein 88B |
| chr14_+_59104741 | 1.11 |
ENST00000395153.3
ENST00000335867.4 |
DACT1
|
dishevelled-binding antagonist of beta-catenin 1 |
| chr16_-_86542455 | 1.11 |
ENST00000595886.1
ENST00000597578.1 ENST00000593604.1 |
FENDRR
|
FOXF1 adjacent non-coding developmental regulatory RNA |
| chr5_-_148758839 | 1.11 |
ENST00000261796.3
|
IL17B
|
interleukin 17B |
| chr1_+_26872324 | 1.10 |
ENST00000531382.1
|
RPS6KA1
|
ribosomal protein S6 kinase, 90kDa, polypeptide 1 |
| chr12_+_49208234 | 1.10 |
ENST00000540990.1
|
CACNB3
|
calcium channel, voltage-dependent, beta 3 subunit |
| chr17_-_1531635 | 1.09 |
ENST00000571650.1
|
SLC43A2
|
solute carrier family 43 (amino acid system L transporter), member 2 |
| chr1_-_43751276 | 1.09 |
ENST00000423420.1
|
C1orf210
|
chromosome 1 open reading frame 210 |
| chr1_-_1356719 | 1.09 |
ENST00000520296.1
|
ANKRD65
|
ankyrin repeat domain 65 |
| chr19_-_12405606 | 1.08 |
ENST00000356109.5
|
ZNF44
|
zinc finger protein 44 |
| chr10_+_106400859 | 1.08 |
ENST00000369701.3
|
SORCS3
|
sortilin-related VPS10 domain containing receptor 3 |
| chr20_+_30458431 | 1.06 |
ENST00000375938.4
ENST00000535842.1 ENST00000310998.4 ENST00000375921.2 |
TTLL9
|
tubulin tyrosine ligase-like family, member 9 |
| chr11_-_119293903 | 1.05 |
ENST00000580275.1
|
THY1
|
Thy-1 cell surface antigen |
| chr20_-_30458019 | 1.05 |
ENST00000486996.1
ENST00000398084.2 |
DUSP15
|
dual specificity phosphatase 15 |
| chr12_+_48178706 | 1.01 |
ENST00000599515.1
|
AC004466.1
|
Uncharacterized protein |
| chr19_-_12476443 | 1.00 |
ENST00000242804.4
ENST00000438182.1 |
ZNF442
|
zinc finger protein 442 |
| chr19_-_49843539 | 1.00 |
ENST00000602554.1
ENST00000358234.4 |
CTC-301O7.4
|
CTC-301O7.4 |
| chr2_+_98330009 | 0.99 |
ENST00000264972.5
|
ZAP70
|
zeta-chain (TCR) associated protein kinase 70kDa |
| chr7_+_63774321 | 0.99 |
ENST00000423484.2
|
ZNF736
|
zinc finger protein 736 |
| chr3_-_123411191 | 0.99 |
ENST00000354792.5
ENST00000508240.1 |
MYLK
|
myosin light chain kinase |
| chr16_+_84328252 | 0.98 |
ENST00000219454.5
|
WFDC1
|
WAP four-disulfide core domain 1 |
| chr5_-_131826457 | 0.98 |
ENST00000437654.1
ENST00000245414.4 |
IRF1
|
interferon regulatory factor 1 |
| chr13_-_20806440 | 0.97 |
ENST00000400066.3
ENST00000400065.3 ENST00000356192.6 |
GJB6
|
gap junction protein, beta 6, 30kDa |
| chrX_-_138724677 | 0.96 |
ENST00000370573.4
ENST00000338585.6 ENST00000370576.4 |
MCF2
|
MCF.2 cell line derived transforming sequence |
| chr19_-_12405689 | 0.95 |
ENST00000355684.5
|
ZNF44
|
zinc finger protein 44 |
| chr16_+_84328429 | 0.94 |
ENST00000568638.1
|
WFDC1
|
WAP four-disulfide core domain 1 |
| chr19_+_19779589 | 0.94 |
ENST00000541458.1
|
ZNF101
|
zinc finger protein 101 |
| chr6_+_31555045 | 0.94 |
ENST00000396101.3
ENST00000490742.1 |
LST1
|
leukocyte specific transcript 1 |
| chr9_-_127269488 | 0.93 |
ENST00000455734.1
|
NR5A1
|
nuclear receptor subfamily 5, group A, member 1 |
| chr20_-_30458432 | 0.93 |
ENST00000375966.4
ENST00000278979.3 |
DUSP15
|
dual specificity phosphatase 15 |
| chr19_+_14693888 | 0.92 |
ENST00000547437.1
ENST00000397439.2 ENST00000417570.1 |
CLEC17A
|
C-type lectin domain family 17, member A |
| chr17_-_33775760 | 0.91 |
ENST00000534689.1
ENST00000532210.1 ENST00000526861.1 ENST00000531588.1 ENST00000285013.6 |
SLFN13
|
schlafen family member 13 |
| chr1_-_161600990 | 0.91 |
ENST00000531221.1
|
FCGR3B
|
Fc fragment of IgG, low affinity IIIb, receptor (CD16b) |
| chr12_+_56325231 | 0.89 |
ENST00000549368.1
|
DGKA
|
diacylglycerol kinase, alpha 80kDa |
| chr2_+_202937972 | 0.89 |
ENST00000541917.1
ENST00000295844.3 |
AC079354.1
|
uncharacterized protein KIAA2012 |
| chr1_-_38512450 | 0.88 |
ENST00000373012.2
|
POU3F1
|
POU class 3 homeobox 1 |
| chr11_+_67776012 | 0.88 |
ENST00000539229.1
|
ALDH3B1
|
aldehyde dehydrogenase 3 family, member B1 |
| chr19_-_20844343 | 0.87 |
ENST00000595405.1
|
ZNF626
|
zinc finger protein 626 |
| chr9_-_117150243 | 0.87 |
ENST00000374088.3
|
AKNA
|
AT-hook transcription factor |
| chr2_-_136875712 | 0.86 |
ENST00000241393.3
|
CXCR4
|
chemokine (C-X-C motif) receptor 4 |
| chr11_-_119293872 | 0.86 |
ENST00000524970.1
|
THY1
|
Thy-1 cell surface antigen |
| chrX_-_138724994 | 0.85 |
ENST00000536274.1
|
MCF2
|
MCF.2 cell line derived transforming sequence |
| chr5_-_176923803 | 0.84 |
ENST00000506161.1
|
PDLIM7
|
PDZ and LIM domain 7 (enigma) |
| chr20_-_30458354 | 0.83 |
ENST00000428829.1
|
DUSP15
|
dual specificity phosphatase 15 |
| chr1_+_55107449 | 0.83 |
ENST00000421030.2
ENST00000545244.1 ENST00000339553.5 ENST00000409996.1 ENST00000454855.2 |
MROH7
|
maestro heat-like repeat family member 7 |
| chr20_-_62710832 | 0.82 |
ENST00000395042.1
|
RGS19
|
regulator of G-protein signaling 19 |
| chr21_+_45773515 | 0.82 |
ENST00000397932.2
ENST00000300481.9 |
TRPM2
|
transient receptor potential cation channel, subfamily M, member 2 |
| chr14_+_96671016 | 0.82 |
ENST00000542454.2
ENST00000554311.1 ENST00000306005.3 ENST00000539359.1 ENST00000553811.1 |
BDKRB2
RP11-404P21.8
|
bradykinin receptor B2 Uncharacterized protein |
| chr15_+_74218787 | 0.81 |
ENST00000261921.7
|
LOXL1
|
lysyl oxidase-like 1 |
| chr1_-_46089718 | 0.80 |
ENST00000421127.2
ENST00000343901.2 ENST00000528266.1 |
CCDC17
|
coiled-coil domain containing 17 |
| chr3_-_48956818 | 0.80 |
ENST00000408959.2
|
ARIH2OS
|
ariadne homolog 2 opposite strand |
| chr1_+_205225319 | 0.80 |
ENST00000329800.7
|
TMCC2
|
transmembrane and coiled-coil domain family 2 |
| chr1_-_204329013 | 0.79 |
ENST00000272203.3
ENST00000414478.1 |
PLEKHA6
|
pleckstrin homology domain containing, family A member 6 |
| chr1_-_161600942 | 0.78 |
ENST00000421702.2
|
FCGR3B
|
Fc fragment of IgG, low affinity IIIb, receptor (CD16b) |
| chr5_-_176923846 | 0.78 |
ENST00000506537.1
|
PDLIM7
|
PDZ and LIM domain 7 (enigma) |
| chr12_-_45444873 | 0.76 |
ENST00000332700.6
|
DBX2
|
developing brain homeobox 2 |
| chr1_-_43751230 | 0.76 |
ENST00000523677.1
|
C1orf210
|
chromosome 1 open reading frame 210 |
| chr2_+_241564655 | 0.74 |
ENST00000407714.1
|
GPR35
|
G protein-coupled receptor 35 |
| chrX_-_100183894 | 0.73 |
ENST00000328526.5
ENST00000372956.2 |
XKRX
|
XK, Kell blood group complex subunit-related, X-linked |
| chr5_+_139739772 | 0.72 |
ENST00000506757.2
ENST00000230993.6 ENST00000506545.1 ENST00000432095.2 ENST00000507527.1 |
SLC4A9
|
solute carrier family 4, sodium bicarbonate cotransporter, member 9 |
| chr1_-_1142067 | 0.72 |
ENST00000379268.2
|
TNFRSF18
|
tumor necrosis factor receptor superfamily, member 18 |
| chr1_-_46089639 | 0.72 |
ENST00000445048.2
|
CCDC17
|
coiled-coil domain containing 17 |
| chr15_+_68871569 | 0.72 |
ENST00000566799.1
|
CORO2B
|
coronin, actin binding protein, 2B |
| chr15_-_65715401 | 0.72 |
ENST00000352385.2
|
IGDCC4
|
immunoglobulin superfamily, DCC subclass, member 4 |
| chr18_-_2982869 | 0.72 |
ENST00000584915.1
|
LPIN2
|
lipin 2 |
| chr9_-_117160738 | 0.72 |
ENST00000448674.1
|
RP11-9M16.2
|
RP11-9M16.2 |
| chr4_-_174255536 | 0.71 |
ENST00000446922.2
|
HMGB2
|
high mobility group box 2 |
| chr11_-_62380199 | 0.71 |
ENST00000419857.1
ENST00000394773.2 |
EML3
|
echinoderm microtubule associated protein like 3 |
| chr11_-_117667806 | 0.71 |
ENST00000527706.1
ENST00000321322.6 |
DSCAML1
|
Down syndrome cell adhesion molecule like 1 |
| chr19_-_12662240 | 0.71 |
ENST00000416136.1
ENST00000428311.1 |
ZNF564
ZNF709
|
zinc finger protein 564 ZNF709 |
| chr1_+_2477831 | 0.70 |
ENST00000606645.1
|
RP3-395M20.12
|
RP3-395M20.12 |
| chr12_+_52695617 | 0.70 |
ENST00000293525.5
|
KRT86
|
keratin 86 |
| chr20_+_327413 | 0.70 |
ENST00000609179.1
|
NRSN2
|
neurensin 2 |
| chr1_-_156217829 | 0.70 |
ENST00000356983.2
ENST00000335852.1 ENST00000340183.5 ENST00000540423.1 |
PAQR6
|
progestin and adipoQ receptor family member VI |
| chr1_-_1141927 | 0.69 |
ENST00000328596.6
ENST00000379265.5 |
TNFRSF18
|
tumor necrosis factor receptor superfamily, member 18 |
| chr20_+_34680595 | 0.68 |
ENST00000406771.2
|
EPB41L1
|
erythrocyte membrane protein band 4.1-like 1 |
| chr9_-_117150303 | 0.68 |
ENST00000312033.3
|
AKNA
|
AT-hook transcription factor |
| chr19_+_34112850 | 0.68 |
ENST00000591231.1
ENST00000434302.1 ENST00000438847.3 |
CHST8
|
carbohydrate (N-acetylgalactosamine 4-0) sulfotransferase 8 |
| chr11_+_133902667 | 0.67 |
ENST00000533091.1
ENST00000527712.1 |
RP11-713P17.3
|
RP11-713P17.3 |
| chr2_+_71295733 | 0.67 |
ENST00000443938.2
ENST00000244204.6 |
NAGK
|
N-acetylglucosamine kinase |
| chr7_-_2883650 | 0.67 |
ENST00000544127.1
|
GNA12
|
guanine nucleotide binding protein (G protein) alpha 12 |
| chr8_-_54755459 | 0.66 |
ENST00000524234.1
ENST00000521275.1 ENST00000396774.2 |
ATP6V1H
|
ATPase, H+ transporting, lysosomal 50/57kDa, V1 subunit H |
| chr15_+_90234028 | 0.66 |
ENST00000268130.7
ENST00000560294.1 ENST00000558000.1 |
WDR93
|
WD repeat domain 93 |
| chr16_+_680932 | 0.66 |
ENST00000319070.2
|
WFIKKN1
|
WAP, follistatin/kazal, immunoglobulin, kunitz and netrin domain containing 1 |
| chr12_-_120806960 | 0.65 |
ENST00000257552.2
|
MSI1
|
musashi RNA-binding protein 1 |
| chr2_+_71295717 | 0.65 |
ENST00000418807.3
ENST00000443872.2 |
NAGK
|
N-acetylglucosamine kinase |
| chr19_+_41860047 | 0.65 |
ENST00000604123.1
|
TMEM91
|
transmembrane protein 91 |
| chr5_-_160973649 | 0.64 |
ENST00000393959.1
ENST00000517547.1 |
GABRB2
|
gamma-aminobutyric acid (GABA) A receptor, beta 2 |
| chr2_+_71295766 | 0.64 |
ENST00000533981.1
|
NAGK
|
N-acetylglucosamine kinase |
| chr2_-_85890569 | 0.64 |
ENST00000494165.1
|
SFTPB
|
surfactant protein B |
| chr4_+_113970772 | 0.64 |
ENST00000504454.1
ENST00000394537.3 ENST00000357077.4 ENST00000264366.6 |
ANK2
|
ankyrin 2, neuronal |
| chr9_+_108424738 | 0.64 |
ENST00000334077.3
|
TAL2
|
T-cell acute lymphocytic leukemia 2 |
| chr15_+_74466744 | 0.63 |
ENST00000560862.1
ENST00000395118.1 |
ISLR
|
immunoglobulin superfamily containing leucine-rich repeat |
| chr12_-_121342170 | 0.63 |
ENST00000353487.2
|
SPPL3
|
signal peptide peptidase like 3 |
| chr17_+_75369167 | 0.63 |
ENST00000423034.2
|
SEPT9
|
septin 9 |
| chr7_-_64467031 | 0.62 |
ENST00000394323.2
|
ERV3-1
|
endogenous retrovirus group 3, member 1 |
| chr1_-_45272951 | 0.62 |
ENST00000372200.1
|
TCTEX1D4
|
Tctex1 domain containing 4 |
| chr15_-_64665911 | 0.62 |
ENST00000606793.1
ENST00000561349.1 ENST00000560278.1 |
CTD-2116N17.1
|
Uncharacterized protein |
| chr14_-_69445793 | 0.62 |
ENST00000538545.2
ENST00000394419.4 |
ACTN1
|
actinin, alpha 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of TFCP2L1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 7.2 | GO:0035566 | regulation of metanephros size(GO:0035566) |
| 0.9 | 5.3 | GO:0030421 | defecation(GO:0030421) |
| 0.8 | 3.0 | GO:0042369 | vitamin D catabolic process(GO:0042369) |
| 0.7 | 3.7 | GO:0007538 | primary sex determination(GO:0007538) |
| 0.6 | 3.8 | GO:1901909 | diadenosine polyphosphate catabolic process(GO:0015961) diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.6 | 2.9 | GO:0010481 | epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 0.6 | 1.7 | GO:0033341 | regulation of collagen binding(GO:0033341) |
| 0.5 | 1.9 | GO:2000298 | regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000298) |
| 0.4 | 3.5 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.4 | 2.5 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.4 | 2.0 | GO:0006051 | mannosamine metabolic process(GO:0006050) N-acetylmannosamine metabolic process(GO:0006051) |
| 0.4 | 1.4 | GO:1904106 | protein localization to microvillus(GO:1904106) |
| 0.4 | 3.2 | GO:0045084 | positive regulation of interleukin-12 biosynthetic process(GO:0045084) |
| 0.3 | 1.3 | GO:0002545 | chronic inflammatory response to non-antigenic stimulus(GO:0002545) regulation of chronic inflammatory response to non-antigenic stimulus(GO:0002880) |
| 0.3 | 1.5 | GO:0060478 | acrosomal vesicle exocytosis(GO:0060478) |
| 0.3 | 7.7 | GO:0060670 | branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 0.3 | 1.1 | GO:2000176 | regulation of pro-T cell differentiation(GO:2000174) positive regulation of pro-T cell differentiation(GO:2000176) |
| 0.2 | 1.6 | GO:0038183 | bile acid signaling pathway(GO:0038183) |
| 0.2 | 1.4 | GO:0038110 | interleukin-2-mediated signaling pathway(GO:0038110) |
| 0.2 | 0.8 | GO:1902219 | negative regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902219) |
| 0.2 | 0.8 | GO:0018277 | protein deamination(GO:0018277) |
| 0.2 | 0.6 | GO:0097534 | lymphoid lineage cell migration(GO:0097534) lymphoid lineage cell migration into thymus(GO:0097535) regulation of positive thymic T cell selection(GO:1902232) |
| 0.2 | 1.9 | GO:0060174 | limb bud formation(GO:0060174) |
| 0.2 | 1.0 | GO:0043366 | beta selection(GO:0043366) |
| 0.2 | 0.5 | GO:2001037 | tongue muscle cell differentiation(GO:0035981) positive regulation of skeletal muscle fiber differentiation(GO:1902811) regulation of tongue muscle cell differentiation(GO:2001035) positive regulation of tongue muscle cell differentiation(GO:2001037) |
| 0.2 | 0.5 | GO:0006478 | peptidyl-tyrosine sulfation(GO:0006478) |
| 0.2 | 1.1 | GO:0048619 | embryonic hindgut morphogenesis(GO:0048619) |
| 0.2 | 5.8 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.2 | 0.5 | GO:0046103 | adenosine catabolic process(GO:0006154) inosine biosynthetic process(GO:0046103) |
| 0.1 | 1.3 | GO:0046618 | drug export(GO:0046618) |
| 0.1 | 0.6 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.1 | 0.6 | GO:1903412 | response to bile acid(GO:1903412) |
| 0.1 | 1.1 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.1 | 1.0 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.1 | 4.7 | GO:0022011 | myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 0.1 | 0.4 | GO:0071314 | cellular response to cocaine(GO:0071314) |
| 0.1 | 0.5 | GO:1905071 | occluding junction disassembly(GO:1905071) regulation of occluding junction disassembly(GO:1905073) positive regulation of occluding junction disassembly(GO:1905075) |
| 0.1 | 0.3 | GO:0042938 | dipeptide transport(GO:0042938) |
| 0.1 | 0.9 | GO:0039663 | fusion of virus membrane with host plasma membrane(GO:0019064) membrane fusion involved in viral entry into host cell(GO:0039663) multi-organism membrane fusion(GO:0044800) |
| 0.1 | 1.2 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) |
| 0.1 | 5.6 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.1 | 0.2 | GO:1904815 | negative regulation of protein localization to chromosome, telomeric region(GO:1904815) |
| 0.1 | 0.2 | GO:0052151 | positive regulation by symbiont of host apoptotic process(GO:0052151) positive regulation of apoptotic process by virus(GO:0060139) |
| 0.1 | 6.9 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.1 | 1.7 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.1 | 3.5 | GO:0050482 | arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.1 | 1.5 | GO:0021957 | corticospinal tract morphogenesis(GO:0021957) |
| 0.1 | 0.3 | GO:0060166 | olfactory pit development(GO:0060166) |
| 0.1 | 0.6 | GO:0051533 | positive regulation of NFAT protein import into nucleus(GO:0051533) |
| 0.1 | 0.8 | GO:0071502 | cellular response to temperature stimulus(GO:0071502) |
| 0.1 | 3.4 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.1 | 1.1 | GO:0098903 | regulation of membrane repolarization during action potential(GO:0098903) |
| 0.1 | 0.2 | GO:0071205 | protein localization to juxtaparanode region of axon(GO:0071205) |
| 0.1 | 0.1 | GO:0021767 | mammillary body development(GO:0021767) mammillary axonal complex development(GO:0061373) |
| 0.1 | 1.2 | GO:1900121 | negative regulation of receptor binding(GO:1900121) |
| 0.1 | 0.4 | GO:0046985 | positive regulation of hemoglobin biosynthetic process(GO:0046985) |
| 0.1 | 0.4 | GO:0060332 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.1 | 2.8 | GO:0048713 | regulation of oligodendrocyte differentiation(GO:0048713) |
| 0.1 | 1.6 | GO:0097119 | postsynaptic density protein 95 clustering(GO:0097119) |
| 0.1 | 1.1 | GO:1900017 | positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.1 | 0.5 | GO:0000480 | endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.1 | 2.1 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.1 | 0.4 | GO:1902962 | regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) |
| 0.1 | 4.7 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 1.1 | GO:1900452 | regulation of long term synaptic depression(GO:1900452) |
| 0.0 | 0.3 | GO:0030047 | actin modification(GO:0030047) |
| 0.0 | 0.4 | GO:0045602 | negative regulation of endothelial cell differentiation(GO:0045602) |
| 0.0 | 3.0 | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
| 0.0 | 0.4 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.0 | 0.3 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.0 | 0.4 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.0 | 0.4 | GO:0060244 | negative regulation of cell proliferation involved in contact inhibition(GO:0060244) negative regulation of cell growth involved in cardiac muscle cell development(GO:0061052) |
| 0.0 | 2.3 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.0 | 1.4 | GO:0045589 | regulation of regulatory T cell differentiation(GO:0045589) |
| 0.0 | 0.2 | GO:0010847 | regulation of chromatin assembly(GO:0010847) protein auto-ADP-ribosylation(GO:0070213) |
| 0.0 | 0.6 | GO:0021794 | thalamus development(GO:0021794) |
| 0.0 | 0.6 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.0 | 0.3 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.0 | 5.1 | GO:0030449 | regulation of complement activation(GO:0030449) |
| 0.0 | 0.9 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.0 | 4.3 | GO:0031295 | T cell costimulation(GO:0031295) |
| 0.0 | 1.0 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.0 | 1.1 | GO:0043401 | steroid hormone mediated signaling pathway(GO:0043401) |
| 0.0 | 0.3 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.0 | 0.6 | GO:0034776 | response to histamine(GO:0034776) cellular response to histamine(GO:0071420) |
| 0.0 | 0.4 | GO:0070070 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.0 | 0.5 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.0 | 1.5 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 0.2 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.0 | 0.2 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 0.7 | GO:0007035 | vacuolar acidification(GO:0007035) |
| 0.0 | 0.2 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.0 | 0.5 | GO:1990118 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 2.8 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.0 | GO:0003032 | detection of oxygen(GO:0003032) |
| 0.0 | 0.5 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.0 | 0.4 | GO:1902373 | negative regulation of mRNA catabolic process(GO:1902373) |
| 0.0 | 0.3 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.0 | 6.2 | GO:0043542 | endothelial cell migration(GO:0043542) |
| 0.0 | 0.3 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.0 | 3.1 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
| 0.0 | 2.2 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 0.2 | GO:0006294 | nucleotide-excision repair, preincision complex assembly(GO:0006294) |
| 0.0 | 0.1 | GO:0010891 | negative regulation of sequestering of triglyceride(GO:0010891) |
| 0.0 | 1.3 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.0 | 2.8 | GO:1990823 | response to leukemia inhibitory factor(GO:1990823) cellular response to leukemia inhibitory factor(GO:1990830) |
| 0.0 | 0.3 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 0.5 | GO:0097186 | amelogenesis(GO:0097186) |
| 0.0 | 0.8 | GO:0042982 | amyloid precursor protein metabolic process(GO:0042982) |
| 0.0 | 1.9 | GO:0061045 | negative regulation of wound healing(GO:0061045) |
| 0.0 | 1.6 | GO:0045669 | positive regulation of osteoblast differentiation(GO:0045669) |
| 0.0 | 1.4 | GO:0050672 | negative regulation of mononuclear cell proliferation(GO:0032945) negative regulation of lymphocyte proliferation(GO:0050672) |
| 0.0 | 0.5 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.0 | 1.0 | GO:0051602 | response to electrical stimulus(GO:0051602) |
| 0.0 | 1.0 | GO:0090280 | positive regulation of calcium ion import(GO:0090280) |
| 0.0 | 0.5 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 0.7 | GO:0010762 | regulation of fibroblast migration(GO:0010762) |
| 0.0 | 0.5 | GO:0034199 | activation of protein kinase A activity(GO:0034199) |
| 0.0 | 1.8 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.0 | 0.4 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.0 | 0.1 | GO:2000587 | negative regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000587) |
| 0.0 | 2.4 | GO:0030879 | mammary gland development(GO:0030879) |
| 0.0 | 0.4 | GO:0015695 | organic cation transport(GO:0015695) |
| 0.0 | 0.3 | GO:0060334 | regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
| 0.0 | 1.5 | GO:0032508 | DNA duplex unwinding(GO:0032508) |
| 0.0 | 4.4 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 | 0.3 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.0 | 0.5 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 0.0 | 2.3 | GO:0046546 | male gonad development(GO:0008584) development of primary male sexual characteristics(GO:0046546) |
| 0.0 | 0.1 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.0 | 0.2 | GO:0001502 | cartilage condensation(GO:0001502) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.7 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.3 | 3.4 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.3 | 1.7 | GO:0034666 | integrin alpha2-beta1 complex(GO:0034666) |
| 0.2 | 1.6 | GO:1990393 | 3M complex(GO:1990393) |
| 0.2 | 4.3 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.1 | 5.9 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.1 | 0.4 | GO:0097124 | cyclin A2-CDK2 complex(GO:0097124) |
| 0.1 | 5.6 | GO:0030057 | desmosome(GO:0030057) |
| 0.1 | 1.9 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.1 | 0.6 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) |
| 0.1 | 0.6 | GO:0019031 | viral envelope(GO:0019031) viral membrane(GO:0036338) |
| 0.1 | 4.6 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.1 | 0.7 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.1 | 3.2 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.1 | 1.5 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.1 | 2.6 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.1 | 1.9 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.1 | 0.2 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.1 | 0.4 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.1 | 1.1 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.1 | 0.6 | GO:0097486 | alveolar lamellar body(GO:0097208) multivesicular body lumen(GO:0097486) |
| 0.1 | 3.1 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 1.5 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.4 | GO:0031985 | Golgi cisterna(GO:0031985) |
| 0.0 | 1.5 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 4.7 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 0.5 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 6.1 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 0.3 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.0 | 1.0 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.6 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.0 | 2.3 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 2.1 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 1.0 | GO:0031105 | septin complex(GO:0031105) |
| 0.0 | 5.9 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 0.5 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 0.3 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.0 | 1.4 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 0.2 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.0 | 2.6 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.3 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 2.6 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.5 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 3.5 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 0.1 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 0.2 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 0.6 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.1 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
| 0.0 | 0.5 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.3 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 0.2 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 0.4 | GO:0071437 | invadopodium(GO:0071437) |
| 0.0 | 0.5 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 1.4 | GO:0070821 | tertiary granule membrane(GO:0070821) |
| 0.0 | 0.2 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 0.6 | GO:0031430 | M band(GO:0031430) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 6.3 | GO:0019828 | aspartic-type endopeptidase inhibitor activity(GO:0019828) |
| 0.7 | 2.9 | GO:0008426 | protein kinase C inhibitor activity(GO:0008426) |
| 0.6 | 3.8 | GO:0034431 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) inositol diphosphate tetrakisphosphate diphosphatase activity(GO:0052840) inositol bisdiphosphate tetrakisphosphate diphosphatase activity(GO:0052841) inositol diphosphate pentakisphosphate diphosphatase activity(GO:0052842) inositol-1-diphosphate-2,3,4,5,6-pentakisphosphate diphosphatase activity(GO:0052843) inositol-3-diphosphate-1,2,4,5,6-pentakisphosphate diphosphatase activity(GO:0052844) inositol-5-diphosphate-1,2,3,4,6-pentakisphosphate diphosphatase activity(GO:0052845) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 1-diphosphatase activity(GO:0052846) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052847) inositol-3,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052848) |
| 0.6 | 5.7 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.6 | 3.0 | GO:0070576 | vitamin D 24-hydroxylase activity(GO:0070576) |
| 0.5 | 2.0 | GO:0045127 | N-acetylglucosamine kinase activity(GO:0045127) |
| 0.4 | 4.6 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.4 | 6.6 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.3 | 7.2 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.3 | 0.8 | GO:0004947 | bradykinin receptor activity(GO:0004947) |
| 0.3 | 1.4 | GO:0019976 | interleukin-2 receptor activity(GO:0004911) interleukin-2 binding(GO:0019976) |
| 0.3 | 4.3 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.2 | 1.2 | GO:0044378 | non-sequence-specific DNA binding, bending(GO:0044378) |
| 0.2 | 1.6 | GO:0038181 | bile acid receptor activity(GO:0038181) |
| 0.2 | 3.4 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.2 | 1.3 | GO:0015307 | drug:proton antiporter activity(GO:0015307) |
| 0.2 | 1.7 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.2 | 1.9 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.2 | 0.5 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.1 | 2.8 | GO:0019864 | IgG binding(GO:0019864) |
| 0.1 | 1.2 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 0.1 | 0.9 | GO:0042806 | fucose binding(GO:0042806) |
| 0.1 | 0.5 | GO:0052596 | tryptamine:oxygen oxidoreductase (deaminating) activity(GO:0052593) aminoacetone:oxygen oxidoreductase(deaminating) activity(GO:0052594) aliphatic-amine oxidase activity(GO:0052595) phenethylamine:oxygen oxidoreductase (deaminating) activity(GO:0052596) |
| 0.1 | 3.0 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 0.6 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.1 | 0.5 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.1 | 1.1 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.1 | 2.6 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.1 | 1.0 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.1 | 0.6 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) |
| 0.1 | 1.3 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.1 | 4.0 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.1 | 0.5 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.1 | 0.5 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.1 | 1.2 | GO:0004030 | aldehyde dehydrogenase [NAD(P)+] activity(GO:0004030) |
| 0.1 | 2.8 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.1 | 3.8 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.1 | 10.3 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 2.3 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.1 | 0.5 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.1 | 0.3 | GO:0015563 | uptake transmembrane transporter activity(GO:0015563) |
| 0.1 | 0.6 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.1 | 0.4 | GO:0030345 | extracellular matrix structural constituent conferring compression resistance(GO:0030021) structural constituent of tooth enamel(GO:0030345) |
| 0.0 | 2.2 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.6 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 4.0 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.3 | GO:0004906 | interferon-gamma receptor activity(GO:0004906) |
| 0.0 | 3.5 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 2.6 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.0 | 1.1 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 1.5 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 1.3 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 2.8 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.4 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.5 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 0.8 | GO:0016641 | oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor(GO:0016641) |
| 0.0 | 4.9 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.0 | 1.4 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 0.4 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.0 | 0.5 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) potassium ion antiporter activity(GO:0022821) |
| 0.0 | 2.1 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.1 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 3.0 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.8 | GO:0099604 | calcium-release channel activity(GO:0015278) ligand-gated calcium channel activity(GO:0099604) |
| 0.0 | 2.4 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.0 | 1.5 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.0 | 0.2 | GO:0001537 | N-acetylgalactosamine 4-O-sulfotransferase activity(GO:0001537) |
| 0.0 | 0.7 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.5 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.0 | 0.4 | GO:0015101 | organic cation transmembrane transporter activity(GO:0015101) |
| 0.0 | 0.7 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.2 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.0 | 0.6 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 5.0 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.0 | 1.0 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.5 | GO:0001055 | RNA polymerase I activity(GO:0001054) RNA polymerase II activity(GO:0001055) |
| 0.0 | 0.1 | GO:0019826 | oxygen sensor activity(GO:0019826) |
| 0.0 | 0.3 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.7 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 4.7 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 0.3 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.7 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 3.8 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 1.1 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 2.9 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.0 | 0.5 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.1 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.0 | 0.6 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.5 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 1.7 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.1 | GO:0086089 | voltage-gated potassium channel activity involved in atrial cardiac muscle cell action potential repolarization(GO:0086089) |
| 0.0 | 0.4 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.3 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 1.8 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.9 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 1.8 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.3 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.0 | 1.8 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 1.8 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.3 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.2 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 16.9 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 7.9 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.1 | 4.7 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.1 | 4.6 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.1 | 3.2 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.1 | 2.9 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 1.2 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 0.5 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.4 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.0 | 1.8 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 1.9 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 1.0 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 1.1 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.0 | 2.9 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 1.2 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 1.2 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 1.6 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 0.4 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.0 | 1.4 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 0.5 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 1.8 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 1.0 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.0 | 0.2 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 1.0 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.6 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 5.8 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 1.5 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 0.8 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 1.3 | PID CMYB PATHWAY | C-MYB transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 5.5 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 0.9 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.1 | 5.8 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.1 | 1.7 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.1 | 5.8 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.1 | 6.3 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 3.4 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.1 | 3.0 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.1 | 1.3 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 1.2 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.1 | 5.5 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 1.1 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 1.4 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 1.5 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 1.0 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.0 | 0.8 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.0 | 1.0 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 2.6 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 1.7 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 1.2 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.0 | 1.2 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.0 | 0.8 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 3.1 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 1.1 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 2.6 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 1.0 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 2.4 | REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | Genes involved in Cell surface interactions at the vascular wall |
| 0.0 | 1.4 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.5 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.0 | 0.5 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 1.1 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.0 | 8.9 | REACTOME GENERIC TRANSCRIPTION PATHWAY | Genes involved in Generic Transcription Pathway |
| 0.0 | 0.6 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.7 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 0.5 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 0.2 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.5 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 1.2 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.0 | 0.3 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.2 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 1.0 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 2.8 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.3 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 0.4 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.3 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.4 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 1.2 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |