Project
Illumina Body Map 2, young vs old
Navigation
Downloads
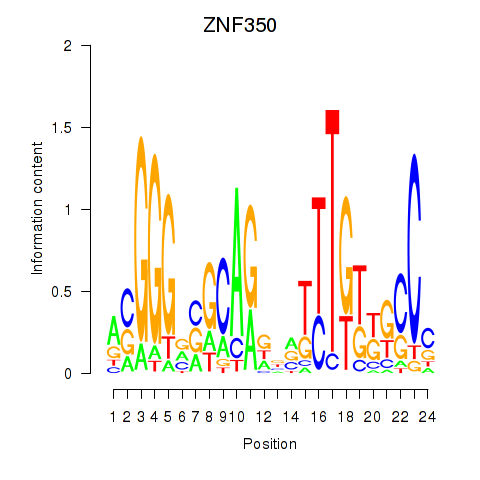
Results for ZNF350
Z-value: 0.45
Transcription factors associated with ZNF350
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZNF350
|
ENSG00000256683.2 | zinc finger protein 350 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZNF350 | hg19_v2_chr19_-_52489923_52490113 | -0.29 | 1.1e-01 | Click! |
Activity profile of ZNF350 motif
Sorted Z-values of ZNF350 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_97175444 | 2.22 |
ENST00000486141.2
|
SORBS1
|
sorbin and SH3 domain containing 1 |
| chr17_+_39969183 | 2.22 |
ENST00000321562.4
|
FKBP10
|
FK506 binding protein 10, 65 kDa |
| chr19_-_17488143 | 2.09 |
ENST00000599426.1
ENST00000252590.4 |
PLVAP
|
plasmalemma vesicle associated protein |
| chr11_-_119217365 | 1.99 |
ENST00000360167.4
ENST00000555262.1 ENST00000449574.2 ENST00000445041.2 |
MFRP
C1QTNF5
|
membrane frizzled-related protein C1q and tumor necrosis factor related protein 5 |
| chr19_+_35532612 | 1.97 |
ENST00000600390.1
ENST00000597419.1 |
HPN
|
hepsin |
| chr1_+_204797749 | 1.91 |
ENST00000367172.4
ENST00000367171.4 ENST00000367170.4 ENST00000338515.6 ENST00000339876.6 ENST00000338586.6 ENST00000539706.1 ENST00000360049.4 ENST00000367169.4 ENST00000446412.1 ENST00000403080.1 |
NFASC
|
neurofascin |
| chr12_+_54519842 | 1.86 |
ENST00000508564.1
|
RP11-834C11.4
|
RP11-834C11.4 |
| chr19_-_49243845 | 1.84 |
ENST00000222145.4
|
RASIP1
|
Ras interacting protein 1 |
| chr12_+_109569155 | 1.77 |
ENST00000539864.1
|
ACACB
|
acetyl-CoA carboxylase beta |
| chr17_+_39968926 | 1.67 |
ENST00000585664.1
ENST00000585922.1 ENST00000429461.1 |
FKBP10
|
FK506 binding protein 10, 65 kDa |
| chr12_+_81471816 | 1.64 |
ENST00000261206.3
|
ACSS3
|
acyl-CoA synthetase short-chain family member 3 |
| chr9_+_124329336 | 1.62 |
ENST00000394340.3
ENST00000436835.1 ENST00000259371.2 |
DAB2IP
|
DAB2 interacting protein |
| chr10_-_15762124 | 1.59 |
ENST00000378076.3
|
ITGA8
|
integrin, alpha 8 |
| chr5_+_42423872 | 1.57 |
ENST00000230882.4
ENST00000357703.3 |
GHR
|
growth hormone receptor |
| chr8_+_144295067 | 1.56 |
ENST00000330824.2
|
GPIHBP1
|
glycosylphosphatidylinositol anchored high density lipoprotein binding protein 1 |
| chr1_+_22970119 | 1.50 |
ENST00000374640.4
ENST00000374639.3 ENST00000374637.1 |
C1QC
|
complement component 1, q subcomponent, C chain |
| chr1_+_228395755 | 1.37 |
ENST00000284548.11
ENST00000570156.2 ENST00000422127.1 ENST00000366707.4 ENST00000366709.4 |
OBSCN
|
obscurin, cytoskeletal calmodulin and titin-interacting RhoGEF |
| chr9_+_34992846 | 1.36 |
ENST00000443266.1
|
DNAJB5
|
DnaJ (Hsp40) homolog, subfamily B, member 5 |
| chr14_+_96671016 | 1.27 |
ENST00000542454.2
ENST00000554311.1 ENST00000306005.3 ENST00000539359.1 ENST00000553811.1 |
BDKRB2
RP11-404P21.8
|
bradykinin receptor B2 Uncharacterized protein |
| chr17_-_73511504 | 1.26 |
ENST00000581870.1
|
CASKIN2
|
CASK interacting protein 2 |
| chr11_-_45687128 | 1.25 |
ENST00000308064.2
|
CHST1
|
carbohydrate (keratan sulfate Gal-6) sulfotransferase 1 |
| chr11_-_7041466 | 1.21 |
ENST00000536068.1
ENST00000278314.4 |
ZNF214
|
zinc finger protein 214 |
| chr2_+_233497931 | 1.20 |
ENST00000264059.3
|
EFHD1
|
EF-hand domain family, member D1 |
| chr6_-_56707943 | 1.18 |
ENST00000370769.4
ENST00000421834.2 ENST00000312431.6 ENST00000361203.3 ENST00000523817.1 |
DST
|
dystonin |
| chr11_-_9113137 | 1.13 |
ENST00000520467.1
ENST00000309263.3 ENST00000457346.2 |
SCUBE2
|
signal peptide, CUB domain, EGF-like 2 |
| chr4_-_76008706 | 1.10 |
ENST00000562355.1
ENST00000563602.1 |
RP11-44F21.5
|
RP11-44F21.5 |
| chr17_+_68071389 | 1.10 |
ENST00000283936.1
ENST00000392671.1 |
KCNJ16
|
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr19_-_4535233 | 1.09 |
ENST00000381848.3
ENST00000588887.1 ENST00000586133.1 |
PLIN5
|
perilipin 5 |
| chr14_-_94254821 | 1.06 |
ENST00000393140.1
|
PRIMA1
|
proline rich membrane anchor 1 |
| chr15_+_42694573 | 0.96 |
ENST00000397200.4
ENST00000569827.1 |
CAPN3
|
calpain 3, (p94) |
| chr2_+_235860690 | 0.94 |
ENST00000416021.1
|
SH3BP4
|
SH3-domain binding protein 4 |
| chr1_-_46089718 | 0.93 |
ENST00000421127.2
ENST00000343901.2 ENST00000528266.1 |
CCDC17
|
coiled-coil domain containing 17 |
| chr9_+_12693336 | 0.92 |
ENST00000381137.2
ENST00000388918.5 |
TYRP1
|
tyrosinase-related protein 1 |
| chr17_+_68071458 | 0.92 |
ENST00000589377.1
|
KCNJ16
|
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr1_-_204654481 | 0.91 |
ENST00000367176.3
|
LRRN2
|
leucine rich repeat neuronal 2 |
| chr11_-_9113093 | 0.91 |
ENST00000450649.2
|
SCUBE2
|
signal peptide, CUB domain, EGF-like 2 |
| chr4_+_75858290 | 0.90 |
ENST00000513238.1
|
PARM1
|
prostate androgen-regulated mucin-like protein 1 |
| chr11_+_118826999 | 0.88 |
ENST00000264031.2
|
UPK2
|
uroplakin 2 |
| chr4_+_75858318 | 0.86 |
ENST00000307428.7
|
PARM1
|
prostate androgen-regulated mucin-like protein 1 |
| chr16_+_31225337 | 0.80 |
ENST00000322122.3
|
TRIM72
|
tripartite motif containing 72 |
| chr4_+_120133791 | 0.77 |
ENST00000274030.6
|
USP53
|
ubiquitin specific peptidase 53 |
| chr11_-_327537 | 0.75 |
ENST00000602735.1
|
IFITM3
|
interferon induced transmembrane protein 3 |
| chr4_+_108910870 | 0.74 |
ENST00000403312.1
ENST00000603302.1 ENST00000309522.3 |
HADH
|
hydroxyacyl-CoA dehydrogenase |
| chr3_-_128212016 | 0.74 |
ENST00000498200.1
ENST00000341105.2 |
GATA2
|
GATA binding protein 2 |
| chr8_+_22414182 | 0.70 |
ENST00000524057.1
|
SORBS3
|
sorbin and SH3 domain containing 3 |
| chrX_-_17878827 | 0.70 |
ENST00000360011.1
|
RAI2
|
retinoic acid induced 2 |
| chr12_+_131438496 | 0.68 |
ENST00000543826.1
|
GPR133
|
G protein-coupled receptor 133 |
| chr2_+_219524379 | 0.65 |
ENST00000443791.1
ENST00000359273.3 ENST00000392109.1 ENST00000392110.2 ENST00000423377.1 ENST00000392111.2 ENST00000412366.1 |
BCS1L
|
BC1 (ubiquinol-cytochrome c reductase) synthesis-like |
| chr3_-_118753716 | 0.64 |
ENST00000393775.2
|
IGSF11
|
immunoglobulin superfamily, member 11 |
| chr4_+_108911036 | 0.63 |
ENST00000505878.1
|
HADH
|
hydroxyacyl-CoA dehydrogenase |
| chr1_+_63788730 | 0.62 |
ENST00000371116.2
|
FOXD3
|
forkhead box D3 |
| chr3_-_118753626 | 0.61 |
ENST00000489689.1
|
IGSF11
|
immunoglobulin superfamily, member 11 |
| chr2_-_157198860 | 0.58 |
ENST00000409572.1
|
NR4A2
|
nuclear receptor subfamily 4, group A, member 2 |
| chr2_+_219524473 | 0.57 |
ENST00000439945.1
ENST00000431802.1 |
BCS1L
|
BC1 (ubiquinol-cytochrome c reductase) synthesis-like |
| chr19_+_5681011 | 0.56 |
ENST00000581893.1
ENST00000411793.2 ENST00000301382.4 ENST00000581773.1 ENST00000423665.2 ENST00000583928.1 ENST00000342970.2 ENST00000422535.2 ENST00000581521.1 ENST00000339423.2 |
HSD11B1L
|
hydroxysteroid (11-beta) dehydrogenase 1-like |
| chr16_+_67282853 | 0.54 |
ENST00000299798.11
|
SLC9A5
|
solute carrier family 9, subfamily A (NHE5, cation proton antiporter 5), member 5 |
| chr18_-_5892103 | 0.50 |
ENST00000383490.2
|
TMEM200C
|
transmembrane protein 200C |
| chr17_-_39968855 | 0.49 |
ENST00000355468.3
ENST00000590496.1 |
LEPREL4
|
leprecan-like 4 |
| chr6_-_28303901 | 0.48 |
ENST00000439158.1
ENST00000446474.1 ENST00000414431.1 ENST00000344279.6 ENST00000453745.1 |
ZSCAN31
|
zinc finger and SCAN domain containing 31 |
| chrX_+_56259316 | 0.48 |
ENST00000468660.1
|
KLF8
|
Kruppel-like factor 8 |
| chr2_+_177025619 | 0.46 |
ENST00000410016.1
|
HOXD3
|
homeobox D3 |
| chr20_+_44035200 | 0.45 |
ENST00000372717.1
ENST00000360981.4 |
DBNDD2
|
dysbindin (dystrobrevin binding protein 1) domain containing 2 |
| chr16_+_69600209 | 0.45 |
ENST00000566899.1
|
NFAT5
|
nuclear factor of activated T-cells 5, tonicity-responsive |
| chr11_+_64323428 | 0.44 |
ENST00000377581.3
|
SLC22A11
|
solute carrier family 22 (organic anion/urate transporter), member 11 |
| chr2_+_235860616 | 0.43 |
ENST00000392011.2
|
SH3BP4
|
SH3-domain binding protein 4 |
| chr16_+_14802801 | 0.41 |
ENST00000526520.1
ENST00000531598.2 |
NPIPA3
|
nuclear pore complex interacting protein family, member A3 |
| chr11_+_64323156 | 0.39 |
ENST00000377585.3
|
SLC22A11
|
solute carrier family 22 (organic anion/urate transporter), member 11 |
| chr3_-_151176497 | 0.39 |
ENST00000282466.3
|
IGSF10
|
immunoglobulin superfamily, member 10 |
| chr11_+_10326918 | 0.37 |
ENST00000528544.1
|
ADM
|
adrenomedullin |
| chr19_-_5680891 | 0.35 |
ENST00000309324.4
|
C19orf70
|
chromosome 19 open reading frame 70 |
| chr19_-_5680499 | 0.34 |
ENST00000587589.1
|
C19orf70
|
chromosome 19 open reading frame 70 |
| chr20_+_44035847 | 0.33 |
ENST00000372712.2
|
DBNDD2
|
dysbindin (dystrobrevin binding protein 1) domain containing 2 |
| chr2_-_219925189 | 0.33 |
ENST00000295731.6
|
IHH
|
indian hedgehog |
| chr11_+_327171 | 0.33 |
ENST00000534483.1
ENST00000524824.1 ENST00000531076.1 |
RP11-326C3.12
|
RP11-326C3.12 |
| chrX_-_134232630 | 0.33 |
ENST00000535837.1
ENST00000433425.2 |
LINC00087
|
long intergenic non-protein coding RNA 87 |
| chr4_+_80748619 | 0.31 |
ENST00000504263.1
|
PCAT4
|
prostate cancer associated transcript 4 (non-protein coding) |
| chr11_-_68671264 | 0.31 |
ENST00000362034.2
|
MRPL21
|
mitochondrial ribosomal protein L21 |
| chr10_-_120938303 | 0.30 |
ENST00000356951.3
ENST00000298510.2 |
PRDX3
|
peroxiredoxin 3 |
| chr1_+_202792385 | 0.29 |
ENST00000549576.1
|
RP11-480I12.4
|
Putative inactive alpha-1,3-mannosyl-glycoprotein 4-beta-N-acetylglucosaminyltransferase-like protein LOC641515 |
| chr12_+_131438443 | 0.29 |
ENST00000261654.5
|
GPR133
|
G protein-coupled receptor 133 |
| chr3_-_155524049 | 0.29 |
ENST00000534941.1
ENST00000340171.2 |
C3orf33
|
chromosome 3 open reading frame 33 |
| chr6_-_28304152 | 0.29 |
ENST00000435857.1
|
ZSCAN31
|
zinc finger and SCAN domain containing 31 |
| chr1_+_173446405 | 0.28 |
ENST00000340385.5
|
PRDX6
|
peroxiredoxin 6 |
| chr11_+_8932828 | 0.28 |
ENST00000530281.1
ENST00000396648.2 ENST00000534147.1 ENST00000529942.1 |
AKIP1
|
A kinase (PRKA) interacting protein 1 |
| chr11_-_68671244 | 0.28 |
ENST00000567045.1
ENST00000450904.2 |
MRPL21
|
mitochondrial ribosomal protein L21 |
| chr11_-_63381823 | 0.28 |
ENST00000323646.5
|
PLA2G16
|
phospholipase A2, group XVI |
| chr11_+_65779283 | 0.27 |
ENST00000312134.2
|
CST6
|
cystatin E/M |
| chr4_+_80748640 | 0.27 |
ENST00000514836.1
|
PCAT4
|
prostate cancer associated transcript 4 (non-protein coding) |
| chr11_-_63381925 | 0.25 |
ENST00000415826.1
|
PLA2G16
|
phospholipase A2, group XVI |
| chr19_+_57050317 | 0.25 |
ENST00000301318.3
ENST00000591844.1 |
ZFP28
|
ZFP28 zinc finger protein |
| chr8_-_12612962 | 0.25 |
ENST00000398246.3
|
LONRF1
|
LON peptidase N-terminal domain and ring finger 1 |
| chr3_-_118753566 | 0.23 |
ENST00000491903.1
|
IGSF11
|
immunoglobulin superfamily, member 11 |
| chr19_+_5681153 | 0.23 |
ENST00000579559.1
ENST00000577222.1 |
HSD11B1L
RPL36
|
hydroxysteroid (11-beta) dehydrogenase 1-like ribosomal protein L36 |
| chr8_-_100905363 | 0.23 |
ENST00000524245.1
|
COX6C
|
cytochrome c oxidase subunit VIc |
| chr14_+_76618242 | 0.23 |
ENST00000557542.1
ENST00000557263.1 ENST00000557207.1 ENST00000312858.5 ENST00000261530.7 |
GPATCH2L
|
G patch domain containing 2-like |
| chr11_+_8932715 | 0.22 |
ENST00000529876.1
ENST00000525005.1 ENST00000524577.1 ENST00000534506.1 |
AKIP1
|
A kinase (PRKA) interacting protein 1 |
| chr1_-_166135952 | 0.22 |
ENST00000354422.3
|
FAM78B
|
family with sequence similarity 78, member B |
| chr12_-_93323013 | 0.19 |
ENST00000322349.8
|
EEA1
|
early endosome antigen 1 |
| chr21_-_30365136 | 0.19 |
ENST00000361371.5
ENST00000389194.2 ENST00000389195.2 |
LTN1
|
listerin E3 ubiquitin protein ligase 1 |
| chr19_+_42829702 | 0.17 |
ENST00000334370.4
|
MEGF8
|
multiple EGF-like-domains 8 |
| chr5_+_176514413 | 0.17 |
ENST00000513166.1
|
FGFR4
|
fibroblast growth factor receptor 4 |
| chr1_-_166136187 | 0.17 |
ENST00000338353.3
|
FAM78B
|
family with sequence similarity 78, member B |
| chr16_+_69600058 | 0.16 |
ENST00000393742.2
|
NFAT5
|
nuclear factor of activated T-cells 5, tonicity-responsive |
| chr11_+_8932654 | 0.15 |
ENST00000299576.5
ENST00000309377.4 ENST00000309357.4 |
AKIP1
|
A kinase (PRKA) interacting protein 1 |
| chr1_-_51425902 | 0.15 |
ENST00000396153.2
|
FAF1
|
Fas (TNFRSF6) associated factor 1 |
| chr15_-_43029252 | 0.13 |
ENST00000563260.1
ENST00000356231.3 |
CDAN1
|
codanin 1 |
| chr3_-_128902729 | 0.13 |
ENST00000451728.2
ENST00000446936.2 ENST00000502976.1 ENST00000500450.2 ENST00000441626.2 |
CNBP
|
CCHC-type zinc finger, nucleic acid binding protein |
| chr11_-_6790286 | 0.13 |
ENST00000338569.2
|
OR2AG2
|
olfactory receptor, family 2, subfamily AG, member 2 |
| chr9_+_19408999 | 0.12 |
ENST00000340967.2
|
ACER2
|
alkaline ceramidase 2 |
| chr21_-_38639813 | 0.11 |
ENST00000309117.6
ENST00000398998.1 |
DSCR3
|
Down syndrome critical region gene 3 |
| chr11_+_68671310 | 0.11 |
ENST00000255078.3
ENST00000539224.1 |
IGHMBP2
|
immunoglobulin mu binding protein 2 |
| chr2_-_21266935 | 0.11 |
ENST00000233242.1
|
APOB
|
apolipoprotein B |
| chr3_-_128902759 | 0.09 |
ENST00000422453.2
ENST00000504813.1 ENST00000512338.1 |
CNBP
|
CCHC-type zinc finger, nucleic acid binding protein |
| chr11_-_112034831 | 0.09 |
ENST00000280357.7
|
IL18
|
interleukin 18 (interferon-gamma-inducing factor) |
| chr11_-_8954491 | 0.09 |
ENST00000526227.1
ENST00000525780.1 ENST00000326053.5 |
C11orf16
|
chromosome 11 open reading frame 16 |
| chr15_+_91445448 | 0.09 |
ENST00000558290.1
ENST00000558853.1 ENST00000559999.1 |
MAN2A2
|
mannosidase, alpha, class 2A, member 2 |
| chr11_-_112034803 | 0.09 |
ENST00000528832.1
|
IL18
|
interleukin 18 (interferon-gamma-inducing factor) |
| chr3_-_118753792 | 0.08 |
ENST00000480431.1
|
IGSF11
|
immunoglobulin superfamily, member 11 |
| chr10_-_12084770 | 0.07 |
ENST00000357604.5
|
UPF2
|
UPF2 regulator of nonsense transcripts homolog (yeast) |
| chr17_+_19030782 | 0.07 |
ENST00000344415.4
ENST00000577213.1 |
GRAPL
|
GRB2-related adaptor protein-like |
| chr17_-_8093471 | 0.07 |
ENST00000389017.4
|
C17orf59
|
chromosome 17 open reading frame 59 |
| chr16_-_46865047 | 0.07 |
ENST00000394806.2
|
C16orf87
|
chromosome 16 open reading frame 87 |
| chr4_-_120133661 | 0.07 |
ENST00000503243.1
ENST00000326780.3 |
RP11-455G16.1
|
Uncharacterized protein |
| chr17_-_39674668 | 0.07 |
ENST00000393981.3
|
KRT15
|
keratin 15 |
| chr11_+_7041677 | 0.06 |
ENST00000299481.4
|
NLRP14
|
NLR family, pyrin domain containing 14 |
| chr10_+_7860460 | 0.05 |
ENST00000344293.5
|
TAF3
|
TAF3 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 140kDa |
| chr15_-_57210769 | 0.05 |
ENST00000559000.1
|
ZNF280D
|
zinc finger protein 280D |
| chr1_-_46089639 | 0.05 |
ENST00000445048.2
|
CCDC17
|
coiled-coil domain containing 17 |
| chr18_+_45778672 | 0.05 |
ENST00000600091.1
|
AC091150.1
|
HCG1818186; Uncharacterized protein |
| chr12_+_50451462 | 0.05 |
ENST00000447966.2
|
ASIC1
|
acid-sensing (proton-gated) ion channel 1 |
| chr11_+_208815 | 0.04 |
ENST00000530889.1
|
RIC8A
|
RIC8 guanine nucleotide exchange factor A |
| chr17_-_18950310 | 0.04 |
ENST00000573099.1
|
GRAP
|
GRB2-related adaptor protein |
| chr19_+_39903185 | 0.03 |
ENST00000409794.3
|
PLEKHG2
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 2 |
| chr17_+_46985823 | 0.03 |
ENST00000508468.2
|
UBE2Z
|
ubiquitin-conjugating enzyme E2Z |
| chr2_-_219157219 | 0.03 |
ENST00000445635.1
ENST00000413976.1 |
TMBIM1
|
transmembrane BAX inhibitor motif containing 1 |
| chr4_+_6784401 | 0.03 |
ENST00000425103.1
ENST00000307659.5 |
KIAA0232
|
KIAA0232 |
| chrX_-_47479246 | 0.03 |
ENST00000295987.7
ENST00000340666.4 |
SYN1
|
synapsin I |
| chr10_+_69644404 | 0.03 |
ENST00000212015.6
|
SIRT1
|
sirtuin 1 |
| chr3_+_14219858 | 0.03 |
ENST00000306024.3
|
LSM3
|
LSM3 homolog, U6 small nuclear RNA associated (S. cerevisiae) |
| chr17_-_39597173 | 0.03 |
ENST00000246646.3
|
KRT38
|
keratin 38 |
| chr17_-_79881408 | 0.02 |
ENST00000392366.3
|
MAFG
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog G |
| chr1_+_204494618 | 0.02 |
ENST00000367180.1
ENST00000391947.2 |
MDM4
|
Mdm4 p53 binding protein homolog (mouse) |
| chr11_-_112034780 | 0.02 |
ENST00000524595.1
|
IL18
|
interleukin 18 (interferon-gamma-inducing factor) |
| chr17_+_7487146 | 0.01 |
ENST00000396501.4
ENST00000584378.1 ENST00000423172.2 ENST00000579445.1 ENST00000585217.1 ENST00000581380.1 |
MPDU1
|
mannose-P-dolichol utilization defect 1 |
| chr2_+_132287237 | 0.01 |
ENST00000467992.2
|
CCDC74A
|
coiled-coil domain containing 74A |
| chr15_+_57210961 | 0.00 |
ENST00000557843.1
|
TCF12
|
transcription factor 12 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ZNF350
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 3.9 | GO:0000412 | histone peptidyl-prolyl isomerization(GO:0000412) |
| 0.5 | 1.6 | GO:2000721 | positive regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000721) |
| 0.5 | 2.0 | GO:0097195 | pilomotor reflex(GO:0097195) |
| 0.4 | 1.1 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 0.3 | 1.6 | GO:0043553 | vascular endothelial growth factor receptor-2 signaling pathway(GO:0036324) negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.3 | 0.9 | GO:0043438 | acetoacetic acid metabolic process(GO:0043438) |
| 0.3 | 1.8 | GO:2001295 | malonyl-CoA biosynthetic process(GO:2001295) |
| 0.2 | 1.4 | GO:0036309 | protein localization to M-band(GO:0036309) |
| 0.2 | 1.6 | GO:0000255 | allantoin metabolic process(GO:0000255) |
| 0.2 | 1.6 | GO:0071503 | positive regulation of lipoprotein particle clearance(GO:0010986) response to heparin(GO:0071503) |
| 0.2 | 2.2 | GO:0045650 | negative regulation of macrophage differentiation(GO:0045650) |
| 0.1 | 1.9 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.1 | 0.6 | GO:0021986 | epithalamus development(GO:0021538) habenula development(GO:0021986) |
| 0.1 | 1.0 | GO:0014718 | positive regulation of satellite cell activation involved in skeletal muscle regeneration(GO:0014718) |
| 0.1 | 1.2 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.1 | 0.3 | GO:0030908 | intein-mediated protein splicing(GO:0016539) protein splicing(GO:0030908) |
| 0.1 | 1.6 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.1 | 2.1 | GO:0002693 | positive regulation of cellular extravasation(GO:0002693) |
| 0.1 | 0.3 | GO:0018171 | peptidyl-cysteine oxidation(GO:0018171) |
| 0.1 | 0.8 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.1 | 0.8 | GO:1902219 | negative regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902219) |
| 0.1 | 1.2 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.1 | 2.2 | GO:0045725 | positive regulation of glycogen biosynthetic process(GO:0045725) |
| 0.1 | 0.5 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.0 | 0.2 | GO:0042231 | interleukin-13 biosynthetic process(GO:0042231) |
| 0.0 | 0.8 | GO:0015747 | urate transport(GO:0015747) |
| 0.0 | 1.2 | GO:0006012 | galactose metabolic process(GO:0006012) |
| 0.0 | 0.5 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.0 | 0.4 | GO:0006701 | progesterone biosynthetic process(GO:0006701) |
| 0.0 | 1.1 | GO:0042135 | neurotransmitter catabolic process(GO:0042135) |
| 0.0 | 0.2 | GO:0097155 | fasciculation of sensory neuron axon(GO:0097155) |
| 0.0 | 1.4 | GO:0043090 | amino acid import(GO:0043090) |
| 0.0 | 1.6 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.0 | 1.9 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.5 | GO:0098719 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.2 | GO:1903412 | response to bile acid(GO:1903412) |
| 0.0 | 0.1 | GO:0090285 | negative regulation of protein glycosylation in Golgi(GO:0090285) |
| 0.0 | 1.8 | GO:0051973 | positive regulation of telomerase activity(GO:0051973) |
| 0.0 | 1.4 | GO:0046676 | negative regulation of insulin secretion(GO:0046676) |
| 0.0 | 0.7 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 0.0 | 0.7 | GO:0010996 | response to auditory stimulus(GO:0010996) |
| 0.0 | 1.8 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.6 | GO:1990032 | parallel fiber(GO:1990032) |
| 0.5 | 1.6 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 0.4 | 1.9 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.3 | 2.2 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.3 | 1.6 | GO:0034678 | integrin alpha8-beta1 complex(GO:0034678) |
| 0.2 | 1.2 | GO:0031673 | H zone(GO:0031673) |
| 0.1 | 1.2 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 1.3 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 0.9 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 0.2 | GO:0044308 | axonal spine(GO:0044308) |
| 0.0 | 3.5 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 1.4 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 2.1 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 1.0 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 0.3 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 1.9 | GO:0009986 | cell surface(GO:0009986) |
| 0.0 | 1.9 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 0.2 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:0004947 | bradykinin receptor activity(GO:0004947) |
| 0.3 | 1.8 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.3 | 2.3 | GO:0035473 | lipase binding(GO:0035473) |
| 0.2 | 1.6 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.2 | 0.9 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.2 | 1.6 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.2 | 1.2 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.1 | 3.7 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.1 | 0.5 | GO:0052740 | 1-acyl-2-lysophosphatidylserine acylhydrolase activity(GO:0052740) |
| 0.1 | 2.3 | GO:0031432 | titin binding(GO:0031432) |
| 0.1 | 1.9 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.1 | 1.9 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.1 | 1.4 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) |
| 0.1 | 2.0 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.0 | 0.8 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 0.6 | GO:0051920 | peroxiredoxin activity(GO:0051920) |
| 0.0 | 0.1 | GO:0071633 | dihydroceramidase activity(GO:0071633) |
| 0.0 | 1.4 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 1.6 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.5 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 2.2 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 1.2 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.3 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 0.6 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.7 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.0 | 1.0 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.1 | GO:0004572 | mannosyl-oligosaccharide 1,3-1,6-alpha-mannosidase activity(GO:0004572) |
| 0.0 | 0.7 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.1 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.0 | 0.2 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.6 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.0 | 2.2 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.0 | 1.6 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 1.2 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.6 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 1.4 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 1.3 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 3.3 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.5 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.1 | 1.4 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 1.6 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 2.9 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 1.8 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 1.5 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 1.2 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.0 | 0.5 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 0.8 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.0 | 1.6 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 1.2 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |