Project
Illumina Body Map 2: averaged replicates
Navigation
Downloads
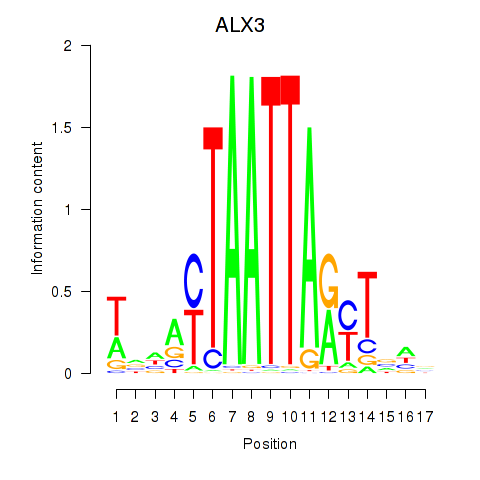
Results for ALX3
Z-value: 1.58
Transcription factors associated with ALX3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ALX3
|
ENSG00000156150.6 | ALX homeobox 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ALX3 | hg19_v2_chr1_-_110613276_110613322 | 0.04 | 8.4e-01 | Click! |
Activity profile of ALX3 motif
Sorted Z-values of ALX3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_89161432 | 15.23 |
ENST00000390242.2
|
IGKJ1
|
immunoglobulin kappa joining 1 |
| chr2_-_89161064 | 15.01 |
ENST00000390241.2
|
IGKJ2
|
immunoglobulin kappa joining 2 |
| chr2_-_89160770 | 11.46 |
ENST00000390240.2
|
IGKJ3
|
immunoglobulin kappa joining 3 |
| chr4_-_25865159 | 4.81 |
ENST00000502949.1
ENST00000264868.5 ENST00000513691.1 ENST00000514872.1 |
SEL1L3
|
sel-1 suppressor of lin-12-like 3 (C. elegans) |
| chr7_-_87342564 | 4.20 |
ENST00000265724.3
ENST00000416177.1 |
ABCB1
|
ATP-binding cassette, sub-family B (MDR/TAP), member 1 |
| chr2_+_90077680 | 4.04 |
ENST00000390270.2
|
IGKV3D-20
|
immunoglobulin kappa variable 3D-20 |
| chr14_-_106494587 | 3.83 |
ENST00000390597.2
|
IGHV2-5
|
immunoglobulin heavy variable 2-5 |
| chr14_-_107049312 | 3.72 |
ENST00000390627.2
|
IGHV3-53
|
immunoglobulin heavy variable 3-53 |
| chr14_-_106668095 | 3.66 |
ENST00000390606.2
|
IGHV3-20
|
immunoglobulin heavy variable 3-20 |
| chr13_-_46742630 | 3.65 |
ENST00000416500.1
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin) |
| chr19_+_17638059 | 3.61 |
ENST00000599164.1
ENST00000449408.2 ENST00000600871.1 ENST00000599124.1 |
FAM129C
|
family with sequence similarity 129, member C |
| chr2_-_89385283 | 3.58 |
ENST00000390252.2
|
IGKV3-15
|
immunoglobulin kappa variable 3-15 |
| chr2_-_89476644 | 3.57 |
ENST00000484817.1
|
IGKV2-24
|
immunoglobulin kappa variable 2-24 |
| chr14_+_22670455 | 3.40 |
ENST00000390460.1
|
TRAV26-2
|
T cell receptor alpha variable 26-2 |
| chr2_+_90211643 | 3.29 |
ENST00000390277.2
|
IGKV3D-11
|
immunoglobulin kappa variable 3D-11 |
| chr2_+_135596180 | 3.23 |
ENST00000283054.4
ENST00000392928.1 |
ACMSD
|
aminocarboxymuconate semialdehyde decarboxylase |
| chr7_+_37723336 | 3.22 |
ENST00000450180.1
|
GPR141
|
G protein-coupled receptor 141 |
| chr12_-_10282742 | 3.12 |
ENST00000298523.5
ENST00000396484.2 ENST00000310002.4 |
CLEC7A
|
C-type lectin domain family 7, member A |
| chr16_+_12059050 | 3.04 |
ENST00000396495.3
|
TNFRSF17
|
tumor necrosis factor receptor superfamily, member 17 |
| chr1_-_92952433 | 3.01 |
ENST00000294702.5
|
GFI1
|
growth factor independent 1 transcription repressor |
| chr14_+_22465771 | 2.97 |
ENST00000390445.2
|
TRAV17
|
T cell receptor alpha variable 17 |
| chr20_+_57594309 | 2.88 |
ENST00000217133.1
|
TUBB1
|
tubulin, beta 1 class VI |
| chr14_-_106552755 | 2.86 |
ENST00000390600.2
|
IGHV3-9
|
immunoglobulin heavy variable 3-9 |
| chr7_+_37723450 | 2.80 |
ENST00000447769.1
|
GPR141
|
G protein-coupled receptor 141 |
| chr16_+_72088376 | 2.79 |
ENST00000570083.1
ENST00000355906.5 ENST00000398131.2 ENST00000569639.1 ENST00000564499.1 ENST00000357763.4 ENST00000562526.1 ENST00000565574.1 ENST00000568417.2 ENST00000356967.5 |
HP
HPR
|
haptoglobin haptoglobin-related protein |
| chr12_-_10282836 | 2.79 |
ENST00000304084.8
ENST00000353231.5 ENST00000525605.1 |
CLEC7A
|
C-type lectin domain family 7, member A |
| chr6_+_160542821 | 2.78 |
ENST00000366963.4
|
SLC22A1
|
solute carrier family 22 (organic cation transporter), member 1 |
| chr2_+_90043607 | 2.74 |
ENST00000462693.1
|
IGKV2D-24
|
immunoglobulin kappa variable 2D-24 (non-functional) |
| chr12_-_10282681 | 2.67 |
ENST00000533022.1
|
CLEC7A
|
C-type lectin domain family 7, member A |
| chr5_-_173217916 | 2.66 |
ENST00000523617.1
|
CTB-43E15.4
|
CTB-43E15.4 |
| chr12_+_8666126 | 2.65 |
ENST00000299665.2
|
CLEC4D
|
C-type lectin domain family 4, member D |
| chr15_+_58702742 | 2.62 |
ENST00000356113.6
ENST00000414170.3 |
LIPC
|
lipase, hepatic |
| chr2_+_135596106 | 2.58 |
ENST00000356140.5
|
ACMSD
|
aminocarboxymuconate semialdehyde decarboxylase |
| chr16_-_28634874 | 2.55 |
ENST00000395609.1
ENST00000350842.4 |
SULT1A1
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 1 |
| chr19_+_17638041 | 2.51 |
ENST00000601861.1
|
FAM129C
|
family with sequence similarity 129, member C |
| chr15_+_94899183 | 2.46 |
ENST00000557742.1
|
MCTP2
|
multiple C2 domains, transmembrane 2 |
| chr2_+_90273679 | 2.45 |
ENST00000423080.2
|
IGKV3D-7
|
immunoglobulin kappa variable 3D-7 |
| chr3_-_191000172 | 2.45 |
ENST00000427544.2
|
UTS2B
|
urotensin 2B |
| chr15_-_20193370 | 2.43 |
ENST00000558565.2
|
IGHV3OR15-7
|
immunoglobulin heavy variable 3/OR15-7 (pseudogene) |
| chr9_-_117111222 | 2.39 |
ENST00000374079.4
|
AKNA
|
AT-hook transcription factor |
| chr15_+_96904487 | 2.38 |
ENST00000600790.1
|
AC087477.1
|
Uncharacterized protein |
| chr2_-_89340242 | 2.38 |
ENST00000480492.1
|
IGKV1-12
|
immunoglobulin kappa variable 1-12 |
| chr19_-_51920952 | 2.38 |
ENST00000356298.5
ENST00000339313.5 ENST00000529627.1 ENST00000439889.2 ENST00000353836.5 ENST00000432469.2 |
SIGLEC10
|
sialic acid binding Ig-like lectin 10 |
| chr3_+_121774202 | 2.34 |
ENST00000469710.1
ENST00000493101.1 ENST00000330540.2 ENST00000264468.5 |
CD86
|
CD86 molecule |
| chr2_+_102615416 | 2.28 |
ENST00000393414.2
|
IL1R2
|
interleukin 1 receptor, type II |
| chr2_+_218994002 | 2.26 |
ENST00000428565.1
|
CXCR2
|
chemokine (C-X-C motif) receptor 2 |
| chr3_+_186692745 | 2.26 |
ENST00000438590.1
|
ST6GAL1
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 |
| chr2_-_225811747 | 2.21 |
ENST00000409592.3
|
DOCK10
|
dedicator of cytokinesis 10 |
| chr18_+_29171689 | 2.19 |
ENST00000237014.3
|
TTR
|
transthyretin |
| chr21_-_15918618 | 2.17 |
ENST00000400564.1
ENST00000400566.1 |
SAMSN1
|
SAM domain, SH3 domain and nuclear localization signals 1 |
| chr2_+_68961934 | 2.14 |
ENST00000409202.3
|
ARHGAP25
|
Rho GTPase activating protein 25 |
| chr6_+_161123270 | 2.12 |
ENST00000366924.2
ENST00000308192.9 ENST00000418964.1 |
PLG
|
plasminogen |
| chr9_+_95709733 | 2.10 |
ENST00000375482.3
|
FGD3
|
FYVE, RhoGEF and PH domain containing 3 |
| chr6_-_138833630 | 2.10 |
ENST00000533765.1
|
NHSL1
|
NHS-like 1 |
| chr2_+_68961905 | 2.03 |
ENST00000295381.3
|
ARHGAP25
|
Rho GTPase activating protein 25 |
| chr19_-_52307357 | 2.02 |
ENST00000594900.1
|
FPR1
|
formyl peptide receptor 1 |
| chr6_+_160542870 | 2.00 |
ENST00000324965.4
ENST00000457470.2 |
SLC22A1
|
solute carrier family 22 (organic cation transporter), member 1 |
| chr4_+_86525299 | 1.99 |
ENST00000512201.1
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chr18_-_67624160 | 1.98 |
ENST00000581982.1
ENST00000280200.4 |
CD226
|
CD226 molecule |
| chr20_-_7238861 | 1.97 |
ENST00000428954.1
|
RP11-19D2.1
|
RP11-19D2.1 |
| chr16_-_28608424 | 1.93 |
ENST00000335715.4
|
SULT1A2
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 2 |
| chr16_+_12059091 | 1.87 |
ENST00000562385.1
|
TNFRSF17
|
tumor necrosis factor receptor superfamily, member 17 |
| chr19_-_58864848 | 1.87 |
ENST00000263100.3
|
A1BG
|
alpha-1-B glycoprotein |
| chr17_-_64225508 | 1.81 |
ENST00000205948.6
|
APOH
|
apolipoprotein H (beta-2-glycoprotein I) |
| chr6_+_130339710 | 1.81 |
ENST00000526087.1
ENST00000533560.1 ENST00000361794.2 |
L3MBTL3
|
l(3)mbt-like 3 (Drosophila) |
| chr3_+_151591422 | 1.77 |
ENST00000362032.5
|
SUCNR1
|
succinate receptor 1 |
| chr5_+_53686658 | 1.75 |
ENST00000512618.1
|
LINC01033
|
long intergenic non-protein coding RNA 1033 |
| chr2_+_68962014 | 1.75 |
ENST00000467265.1
|
ARHGAP25
|
Rho GTPase activating protein 25 |
| chr16_+_31271274 | 1.74 |
ENST00000287497.8
ENST00000544665.3 |
ITGAM
|
integrin, alpha M (complement component 3 receptor 3 subunit) |
| chr14_-_52436247 | 1.72 |
ENST00000597846.1
|
AL358333.1
|
HCG2013195; Uncharacterized protein |
| chr14_+_22977587 | 1.63 |
ENST00000390504.1
|
TRAJ33
|
T cell receptor alpha joining 33 |
| chr3_-_101039402 | 1.61 |
ENST00000193391.7
|
IMPG2
|
interphotoreceptor matrix proteoglycan 2 |
| chr4_-_48116540 | 1.59 |
ENST00000506073.1
|
TXK
|
TXK tyrosine kinase |
| chr4_-_36245561 | 1.58 |
ENST00000506189.1
|
ARAP2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
| chr3_+_186353756 | 1.56 |
ENST00000431018.1
ENST00000450521.1 ENST00000539949.1 |
FETUB
|
fetuin B |
| chr1_+_117544366 | 1.56 |
ENST00000256652.4
ENST00000369470.1 |
CD101
|
CD101 molecule |
| chr16_-_28621298 | 1.54 |
ENST00000566189.1
|
SULT1A1
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 1 |
| chr9_+_90112767 | 1.51 |
ENST00000408954.3
|
DAPK1
|
death-associated protein kinase 1 |
| chr18_-_67624412 | 1.45 |
ENST00000580335.1
|
CD226
|
CD226 molecule |
| chr16_-_28621312 | 1.44 |
ENST00000314752.7
|
SULT1A1
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 1 |
| chr18_-_67623906 | 1.41 |
ENST00000583955.1
|
CD226
|
CD226 molecule |
| chr10_-_99030395 | 1.40 |
ENST00000355366.5
ENST00000371027.1 |
ARHGAP19
|
Rho GTPase activating protein 19 |
| chr16_-_28608364 | 1.36 |
ENST00000533150.1
|
SULT1A2
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 2 |
| chr1_-_150738261 | 1.34 |
ENST00000448301.2
ENST00000368985.3 |
CTSS
|
cathepsin S |
| chr16_-_28621353 | 1.34 |
ENST00000567512.1
|
SULT1A1
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 1 |
| chr12_+_16500599 | 1.34 |
ENST00000535309.1
ENST00000540056.1 ENST00000396209.1 ENST00000540126.1 |
MGST1
|
microsomal glutathione S-transferase 1 |
| chr9_+_90112590 | 1.33 |
ENST00000472284.1
|
DAPK1
|
death-associated protein kinase 1 |
| chrX_-_77225135 | 1.32 |
ENST00000458128.1
|
PGAM4
|
phosphoglycerate mutase family member 4 |
| chr9_+_90112741 | 1.31 |
ENST00000469640.2
|
DAPK1
|
death-associated protein kinase 1 |
| chr6_+_26402517 | 1.29 |
ENST00000414912.2
|
BTN3A1
|
butyrophilin, subfamily 3, member A1 |
| chr17_-_57229155 | 1.28 |
ENST00000584089.1
|
SKA2
|
spindle and kinetochore associated complex subunit 2 |
| chr4_-_164534657 | 1.27 |
ENST00000339875.5
|
MARCH1
|
membrane-associated ring finger (C3HC4) 1, E3 ubiquitin protein ligase |
| chr19_-_3557570 | 1.27 |
ENST00000355415.2
|
MFSD12
|
major facilitator superfamily domain containing 12 |
| chr6_+_26402465 | 1.26 |
ENST00000476549.2
ENST00000289361.6 ENST00000450085.2 ENST00000425234.2 ENST00000427334.1 ENST00000506698.1 |
BTN3A1
|
butyrophilin, subfamily 3, member A1 |
| chr5_+_176811431 | 1.25 |
ENST00000512593.1
ENST00000324417.5 |
SLC34A1
|
solute carrier family 34 (type II sodium/phosphate contransporter), member 1 |
| chr4_-_89442940 | 1.24 |
ENST00000527353.1
|
PIGY
|
phosphatidylinositol glycan anchor biosynthesis, class Y |
| chr1_+_84630645 | 1.23 |
ENST00000394839.2
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta |
| chr5_-_173217931 | 1.23 |
ENST00000522731.1
|
CTB-43E15.4
|
CTB-43E15.4 |
| chr12_+_56435637 | 1.23 |
ENST00000356464.5
ENST00000552361.1 |
RPS26
|
ribosomal protein S26 |
| chr2_+_103035102 | 1.20 |
ENST00000264260.2
|
IL18RAP
|
interleukin 18 receptor accessory protein |
| chr12_+_16500571 | 1.19 |
ENST00000543076.1
ENST00000396210.3 |
MGST1
|
microsomal glutathione S-transferase 1 |
| chr2_-_8715616 | 1.19 |
ENST00000418358.1
|
AC011747.3
|
AC011747.3 |
| chr1_+_84630574 | 1.19 |
ENST00000413538.1
ENST00000417530.1 |
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta |
| chr19_-_36304201 | 1.19 |
ENST00000301175.3
|
PRODH2
|
proline dehydrogenase (oxidase) 2 |
| chr14_-_78083112 | 1.17 |
ENST00000216484.2
|
SPTLC2
|
serine palmitoyltransferase, long chain base subunit 2 |
| chr9_-_4666421 | 1.16 |
ENST00000381895.5
|
SPATA6L
|
spermatogenesis associated 6-like |
| chr11_+_118398178 | 1.15 |
ENST00000302783.4
ENST00000539546.1 |
TTC36
|
tetratricopeptide repeat domain 36 |
| chr16_-_30122717 | 1.15 |
ENST00000566613.1
|
GDPD3
|
glycerophosphodiester phosphodiesterase domain containing 3 |
| chr7_-_38315919 | 1.15 |
ENST00000390339.1
|
TRGJP1
|
T cell receptor gamma joining P1 |
| chr16_-_55866997 | 1.14 |
ENST00000360526.3
ENST00000361503.4 |
CES1
|
carboxylesterase 1 |
| chr10_+_696000 | 1.14 |
ENST00000381489.5
|
PRR26
|
proline rich 26 |
| chr19_+_50016411 | 1.12 |
ENST00000426395.3
ENST00000600273.1 ENST00000599988.1 |
FCGRT
|
Fc fragment of IgG, receptor, transporter, alpha |
| chr4_+_37455536 | 1.12 |
ENST00000381980.4
ENST00000508175.1 |
C4orf19
|
chromosome 4 open reading frame 19 |
| chr2_+_166095898 | 1.11 |
ENST00000424833.1
ENST00000375437.2 ENST00000357398.3 |
SCN2A
|
sodium channel, voltage-gated, type II, alpha subunit |
| chr4_-_19458597 | 1.10 |
ENST00000505347.1
|
RP11-3J1.1
|
RP11-3J1.1 |
| chr9_-_75488984 | 1.09 |
ENST00000423171.1
ENST00000449235.1 ENST00000453787.1 |
RP11-151D14.1
|
RP11-151D14.1 |
| chr22_+_42834029 | 1.09 |
ENST00000428765.1
|
CTA-126B4.7
|
CTA-126B4.7 |
| chr10_+_5135981 | 1.07 |
ENST00000380554.3
|
AKR1C3
|
aldo-keto reductase family 1, member C3 |
| chr12_+_16500037 | 1.04 |
ENST00000536371.1
ENST00000010404.2 |
MGST1
|
microsomal glutathione S-transferase 1 |
| chr2_-_169887827 | 1.04 |
ENST00000263817.6
|
ABCB11
|
ATP-binding cassette, sub-family B (MDR/TAP), member 11 |
| chr4_-_185275104 | 1.01 |
ENST00000317596.3
|
RP11-290F5.2
|
RP11-290F5.2 |
| chr7_-_142124565 | 1.01 |
ENST00000390376.2
|
TRBV6-8
|
T cell receptor beta variable 6-8 |
| chr18_-_44181442 | 1.00 |
ENST00000398722.4
|
LOXHD1
|
lipoxygenase homology domains 1 |
| chr10_-_90611566 | 1.00 |
ENST00000371930.4
|
ANKRD22
|
ankyrin repeat domain 22 |
| chr9_+_90112117 | 0.99 |
ENST00000358077.5
|
DAPK1
|
death-associated protein kinase 1 |
| chr9_-_115095123 | 0.99 |
ENST00000458258.1
|
PTBP3
|
polypyrimidine tract binding protein 3 |
| chr4_+_71859156 | 0.98 |
ENST00000286648.5
ENST00000504730.1 ENST00000504952.1 |
DCK
|
deoxycytidine kinase |
| chr2_-_101925055 | 0.97 |
ENST00000295317.3
|
RNF149
|
ring finger protein 149 |
| chr20_-_50722183 | 0.97 |
ENST00000371523.4
|
ZFP64
|
ZFP64 zinc finger protein |
| chr8_+_92261516 | 0.97 |
ENST00000276609.3
ENST00000309536.2 |
SLC26A7
|
solute carrier family 26 (anion exchanger), member 7 |
| chr1_+_186265399 | 0.95 |
ENST00000367486.3
ENST00000367484.3 ENST00000533951.1 ENST00000367482.4 ENST00000367483.4 ENST00000367485.4 ENST00000445192.2 |
PRG4
|
proteoglycan 4 |
| chr11_-_62521614 | 0.95 |
ENST00000527994.1
ENST00000394807.3 |
ZBTB3
|
zinc finger and BTB domain containing 3 |
| chr5_+_136070614 | 0.95 |
ENST00000502421.1
|
CTB-1I21.1
|
CTB-1I21.1 |
| chr12_+_15699286 | 0.94 |
ENST00000442921.2
ENST00000542557.1 ENST00000445537.2 ENST00000544244.1 |
PTPRO
|
protein tyrosine phosphatase, receptor type, O |
| chr19_+_50016610 | 0.93 |
ENST00000596975.1
|
FCGRT
|
Fc fragment of IgG, receptor, transporter, alpha |
| chr8_-_623547 | 0.93 |
ENST00000522893.1
|
ERICH1
|
glutamate-rich 1 |
| chr5_-_98262240 | 0.90 |
ENST00000284049.3
|
CHD1
|
chromodomain helicase DNA binding protein 1 |
| chr11_+_327171 | 0.89 |
ENST00000534483.1
ENST00000524824.1 ENST00000531076.1 |
RP11-326C3.12
|
RP11-326C3.12 |
| chr13_+_78315295 | 0.89 |
ENST00000351546.3
|
SLAIN1
|
SLAIN motif family, member 1 |
| chr15_+_64680003 | 0.88 |
ENST00000261884.3
|
TRIP4
|
thyroid hormone receptor interactor 4 |
| chr7_-_144435985 | 0.88 |
ENST00000549981.1
|
TPK1
|
thiamin pyrophosphokinase 1 |
| chr13_+_78315348 | 0.87 |
ENST00000441784.1
|
SLAIN1
|
SLAIN motif family, member 1 |
| chr20_-_33735070 | 0.87 |
ENST00000374491.3
ENST00000542871.1 ENST00000374492.3 |
EDEM2
|
ER degradation enhancer, mannosidase alpha-like 2 |
| chr1_+_225600404 | 0.87 |
ENST00000366845.2
|
AC092811.1
|
AC092811.1 |
| chr16_-_28937027 | 0.86 |
ENST00000358201.4
|
RABEP2
|
rabaptin, RAB GTPase binding effector protein 2 |
| chr4_+_80584903 | 0.86 |
ENST00000506460.1
|
RP11-452C8.1
|
RP11-452C8.1 |
| chr11_+_35201826 | 0.86 |
ENST00000531873.1
|
CD44
|
CD44 molecule (Indian blood group) |
| chr1_-_24126051 | 0.86 |
ENST00000445705.1
|
GALE
|
UDP-galactose-4-epimerase |
| chr6_-_32095968 | 0.85 |
ENST00000375203.3
ENST00000375201.4 |
ATF6B
|
activating transcription factor 6 beta |
| chr7_-_142251148 | 0.85 |
ENST00000390360.3
|
TRBV6-4
|
T cell receptor beta variable 6-4 |
| chr5_+_150639360 | 0.84 |
ENST00000523004.1
|
GM2A
|
GM2 ganglioside activator |
| chr8_-_93978216 | 0.84 |
ENST00000517751.1
ENST00000524107.1 |
TRIQK
|
triple QxxK/R motif containing |
| chr1_+_115572415 | 0.84 |
ENST00000256592.1
|
TSHB
|
thyroid stimulating hormone, beta |
| chr6_-_7313381 | 0.83 |
ENST00000489567.1
ENST00000479365.1 ENST00000462112.1 ENST00000397511.2 ENST00000534851.1 ENST00000474597.1 ENST00000244763.4 |
SSR1
|
signal sequence receptor, alpha |
| chr1_+_66820058 | 0.82 |
ENST00000480109.2
|
PDE4B
|
phosphodiesterase 4B, cAMP-specific |
| chr14_+_22947861 | 0.80 |
ENST00000390482.1
|
TRAJ57
|
T cell receptor alpha joining 57 |
| chr4_+_95128748 | 0.78 |
ENST00000359052.4
|
SMARCAD1
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 |
| chr6_-_36515177 | 0.78 |
ENST00000229812.7
|
STK38
|
serine/threonine kinase 38 |
| chr3_+_45927994 | 0.78 |
ENST00000357632.2
ENST00000395963.2 |
CCR9
|
chemokine (C-C motif) receptor 9 |
| chr6_+_78400375 | 0.76 |
ENST00000602452.2
|
MEI4
|
meiosis-specific 4 homolog (S. cerevisiae) |
| chr14_+_55494323 | 0.75 |
ENST00000339298.2
|
SOCS4
|
suppressor of cytokine signaling 4 |
| chr8_-_93978309 | 0.75 |
ENST00000517858.1
ENST00000378861.5 |
TRIQK
|
triple QxxK/R motif containing |
| chr18_+_29027696 | 0.75 |
ENST00000257189.4
|
DSG3
|
desmoglein 3 |
| chr5_-_20575959 | 0.74 |
ENST00000507958.1
|
CDH18
|
cadherin 18, type 2 |
| chr8_-_93978357 | 0.73 |
ENST00000522925.1
ENST00000522903.1 ENST00000537541.1 ENST00000518748.1 ENST00000519069.1 ENST00000521988.1 |
TRIQK
|
triple QxxK/R motif containing |
| chr3_+_138340049 | 0.72 |
ENST00000464668.1
|
FAIM
|
Fas apoptotic inhibitory molecule |
| chr15_+_58724184 | 0.72 |
ENST00000433326.2
|
LIPC
|
lipase, hepatic |
| chr6_+_26440700 | 0.72 |
ENST00000494393.1
ENST00000482451.1 ENST00000244519.2 ENST00000339789.4 ENST00000471353.1 ENST00000361232.3 ENST00000487627.1 ENST00000496719.1 ENST00000490254.1 ENST00000487272.1 |
BTN3A3
|
butyrophilin, subfamily 3, member A3 |
| chr2_+_102953608 | 0.71 |
ENST00000311734.2
ENST00000409584.1 |
IL1RL1
|
interleukin 1 receptor-like 1 |
| chr15_+_48483736 | 0.71 |
ENST00000417307.2
ENST00000559641.1 |
CTXN2
SLC12A1
|
cortexin 2 solute carrier family 12 (sodium/potassium/chloride transporter), member 1 |
| chr5_-_94417314 | 0.71 |
ENST00000505208.1
|
MCTP1
|
multiple C2 domains, transmembrane 1 |
| chr8_-_93978333 | 0.71 |
ENST00000524037.1
ENST00000520430.1 ENST00000521617.1 |
TRIQK
|
triple QxxK/R motif containing |
| chr12_-_10022735 | 0.70 |
ENST00000228438.2
|
CLEC2B
|
C-type lectin domain family 2, member B |
| chr17_-_38821373 | 0.70 |
ENST00000394052.3
|
KRT222
|
keratin 222 |
| chr5_-_94417186 | 0.70 |
ENST00000312216.8
ENST00000512425.1 |
MCTP1
|
multiple C2 domains, transmembrane 1 |
| chr1_-_24126023 | 0.69 |
ENST00000429356.1
|
GALE
|
UDP-galactose-4-epimerase |
| chr4_+_86699834 | 0.69 |
ENST00000395183.2
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chr12_+_66582919 | 0.68 |
ENST00000545837.1
ENST00000457197.2 |
IRAK3
|
interleukin-1 receptor-associated kinase 3 |
| chr7_-_111424506 | 0.68 |
ENST00000450156.1
ENST00000494651.2 |
DOCK4
|
dedicator of cytokinesis 4 |
| chr4_+_26324474 | 0.67 |
ENST00000514675.1
|
RBPJ
|
recombination signal binding protein for immunoglobulin kappa J region |
| chr4_-_120243545 | 0.67 |
ENST00000274024.3
|
FABP2
|
fatty acid binding protein 2, intestinal |
| chr3_+_138340067 | 0.65 |
ENST00000479848.1
|
FAIM
|
Fas apoptotic inhibitory molecule |
| chr14_+_39736582 | 0.65 |
ENST00000556148.1
ENST00000348007.3 |
CTAGE5
|
CTAGE family, member 5 |
| chr5_+_96840389 | 0.64 |
ENST00000504012.1
|
RP11-1E3.1
|
RP11-1E3.1 |
| chr17_-_39191107 | 0.64 |
ENST00000344363.5
|
KRTAP1-3
|
keratin associated protein 1-3 |
| chr1_+_28261533 | 0.63 |
ENST00000411604.1
ENST00000373888.4 |
SMPDL3B
|
sphingomyelin phosphodiesterase, acid-like 3B |
| chr7_+_150020363 | 0.63 |
ENST00000359623.4
ENST00000493307.1 |
LRRC61
|
leucine rich repeat containing 61 |
| chr5_+_66300464 | 0.63 |
ENST00000436277.1
|
MAST4
|
microtubule associated serine/threonine kinase family member 4 |
| chr17_-_295730 | 0.63 |
ENST00000329099.4
|
FAM101B
|
family with sequence similarity 101, member B |
| chr9_-_115095229 | 0.61 |
ENST00000210227.4
|
PTBP3
|
polypyrimidine tract binding protein 3 |
| chr6_+_28317685 | 0.60 |
ENST00000252211.2
ENST00000341464.5 ENST00000377255.3 |
ZKSCAN3
|
zinc finger with KRAB and SCAN domains 3 |
| chr11_-_71823266 | 0.59 |
ENST00000538919.1
ENST00000539395.1 ENST00000542531.1 |
ANAPC15
|
anaphase promoting complex subunit 15 |
| chr8_-_93978346 | 0.58 |
ENST00000523580.1
|
TRIQK
|
triple QxxK/R motif containing |
| chr11_+_46193466 | 0.57 |
ENST00000533793.1
|
RP11-702F3.3
|
RP11-702F3.3 |
| chrX_+_56100757 | 0.57 |
ENST00000433279.1
|
AL353698.1
|
Uncharacterized protein |
| chrX_+_108779004 | 0.54 |
ENST00000218004.1
|
NXT2
|
nuclear transport factor 2-like export factor 2 |
| chrX_+_107288280 | 0.54 |
ENST00000458383.1
|
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chr12_-_118796910 | 0.53 |
ENST00000541186.1
ENST00000539872.1 |
TAOK3
|
TAO kinase 3 |
| chr10_-_74283694 | 0.52 |
ENST00000398763.4
ENST00000418483.2 ENST00000489666.2 |
MICU1
|
mitochondrial calcium uptake 1 |
| chr12_+_104337515 | 0.52 |
ENST00000550595.1
|
HSP90B1
|
heat shock protein 90kDa beta (Grp94), member 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ALX3
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 3.6 | GO:0071449 | cellular response to lipid hydroperoxide(GO:0071449) |
| 1.0 | 3.0 | GO:0070105 | positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 0.9 | 2.8 | GO:2000296 | negative regulation of hydrogen peroxide catabolic process(GO:2000296) |
| 0.8 | 5.8 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.8 | 4.8 | GO:0002729 | positive regulation of natural killer cell cytokine production(GO:0002729) |
| 0.8 | 2.3 | GO:0002644 | negative regulation of tolerance induction(GO:0002644) positive regulation of interleukin-4 biosynthetic process(GO:0045404) |
| 0.7 | 2.1 | GO:0002416 | IgG immunoglobulin transcytosis in epithelial cells mediated by FcRn immunoglobulin receptor(GO:0002416) |
| 0.7 | 4.8 | GO:0048241 | epinephrine transport(GO:0048241) |
| 0.6 | 5.4 | GO:1903961 | positive regulation of anion transmembrane transport(GO:1903961) |
| 0.6 | 2.3 | GO:0032690 | negative regulation of interleukin-1 alpha production(GO:0032690) negative regulation of interleukin-1 alpha secretion(GO:0050712) |
| 0.5 | 2.1 | GO:0052047 | interaction with other organism via secreted substance involved in symbiotic interaction(GO:0052047) |
| 0.4 | 8.6 | GO:0009756 | carbohydrate mediated signaling(GO:0009756) |
| 0.4 | 1.2 | GO:0006667 | sphinganine metabolic process(GO:0006667) |
| 0.4 | 2.3 | GO:0038112 | interleukin-8-mediated signaling pathway(GO:0038112) |
| 0.4 | 10.2 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.4 | 1.1 | GO:2000224 | sesquiterpenoid metabolic process(GO:0006714) sesquiterpenoid catabolic process(GO:0016107) farnesol metabolic process(GO:0016487) farnesol catabolic process(GO:0016488) regulation of testosterone biosynthetic process(GO:2000224) |
| 0.3 | 1.3 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.3 | 0.9 | GO:0036058 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.3 | 1.8 | GO:0034196 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.3 | 1.1 | GO:0090119 | vesicle-mediated cholesterol transport(GO:0090119) |
| 0.2 | 2.4 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.2 | 1.6 | GO:0060332 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.2 | 3.4 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.2 | 3.3 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.2 | 0.8 | GO:0002304 | gamma-delta intraepithelial T cell differentiation(GO:0002304) CD8-positive, gamma-delta intraepithelial T cell differentiation(GO:0002305) |
| 0.2 | 0.9 | GO:0046092 | deoxycytidine metabolic process(GO:0046092) |
| 0.2 | 5.2 | GO:0071447 | cellular response to hydroperoxide(GO:0071447) |
| 0.2 | 0.9 | GO:0042357 | thiamine diphosphate metabolic process(GO:0042357) |
| 0.2 | 1.0 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.2 | 0.5 | GO:0031247 | actin rod assembly(GO:0031247) |
| 0.2 | 0.9 | GO:0036512 | trimming of terminal mannose on B branch(GO:0036509) trimming of first mannose on A branch(GO:0036511) trimming of second mannose on A branch(GO:0036512) |
| 0.2 | 1.2 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.2 | 2.2 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.2 | 0.5 | GO:0072356 | chromosome passenger complex localization to kinetochore(GO:0072356) |
| 0.2 | 26.5 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.1 | 2.0 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.1 | 3.6 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.1 | 0.7 | GO:0034759 | regulation of iron ion transport(GO:0034756) regulation of iron ion transmembrane transport(GO:0034759) |
| 0.1 | 0.8 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.1 | 2.1 | GO:0097475 | motor neuron migration(GO:0097475) |
| 0.1 | 2.2 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.1 | 0.7 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
| 0.1 | 1.7 | GO:0010668 | ectodermal cell differentiation(GO:0010668) |
| 0.1 | 0.7 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.1 | 0.9 | GO:1900625 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.1 | 0.4 | GO:0036367 | adaptation of rhodopsin mediated signaling(GO:0016062) light adaption(GO:0036367) |
| 0.1 | 1.5 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.1 | 2.3 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.1 | 0.7 | GO:0060847 | arterial endothelial cell fate commitment(GO:0060844) blood vessel endothelial cell fate commitment(GO:0060846) endothelial cell fate specification(GO:0060847) blood vessel endothelial cell fate specification(GO:0097101) positive regulation of ephrin receptor signaling pathway(GO:1901189) positive regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901297) positive regulation of canonical Wnt signaling pathway involved in heart development(GO:1905068) |
| 0.1 | 0.3 | GO:0051097 | negative regulation of helicase activity(GO:0051097) |
| 0.1 | 2.6 | GO:0072643 | interferon-gamma secretion(GO:0072643) |
| 0.1 | 0.4 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.1 | 0.4 | GO:0061760 | antifungal innate immune response(GO:0061760) |
| 0.1 | 0.8 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.1 | 1.1 | GO:0008627 | intrinsic apoptotic signaling pathway in response to osmotic stress(GO:0008627) |
| 0.1 | 0.9 | GO:0043923 | positive regulation by host of viral transcription(GO:0043923) |
| 0.1 | 0.8 | GO:0001780 | neutrophil homeostasis(GO:0001780) |
| 0.1 | 0.8 | GO:1990440 | positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990440) |
| 0.1 | 0.5 | GO:0035502 | metanephric part of ureteric bud development(GO:0035502) |
| 0.1 | 1.3 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.1 | 35.2 | GO:0002250 | adaptive immune response(GO:0002250) |
| 0.1 | 1.0 | GO:0009157 | deoxyribonucleoside monophosphate biosynthetic process(GO:0009157) |
| 0.1 | 0.7 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.1 | 1.3 | GO:1902358 | sulfate transmembrane transport(GO:1902358) |
| 0.1 | 0.3 | GO:0030311 | poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.1 | 1.0 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.1 | 0.5 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.1 | 0.4 | GO:0070889 | platelet alpha granule organization(GO:0070889) |
| 0.1 | 0.4 | GO:0050955 | thermoception(GO:0050955) |
| 0.1 | 0.4 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.1 | 0.6 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.4 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.0 | 0.6 | GO:2000773 | negative regulation of cellular senescence(GO:2000773) |
| 0.0 | 1.0 | GO:1903504 | regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.0 | 0.5 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.0 | 1.0 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.0 | 2.7 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.0 | 0.3 | GO:0014901 | regulation of satellite cell activation involved in skeletal muscle regeneration(GO:0014717) satellite cell activation involved in skeletal muscle regeneration(GO:0014901) |
| 0.0 | 1.2 | GO:0033119 | negative regulation of RNA splicing(GO:0033119) |
| 0.0 | 0.7 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.0 | 0.8 | GO:0033189 | response to vitamin A(GO:0033189) |
| 0.0 | 1.6 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.0 | 0.4 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 0.4 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.0 | 0.8 | GO:0070933 | histone H4 deacetylation(GO:0070933) |
| 0.0 | 0.4 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 1.2 | GO:0006505 | GPI anchor metabolic process(GO:0006505) GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.7 | GO:0050776 | regulation of immune response(GO:0050776) |
| 0.0 | 0.1 | GO:0061358 | negative regulation of Wnt protein secretion(GO:0061358) |
| 0.0 | 0.5 | GO:0035455 | response to interferon-alpha(GO:0035455) |
| 0.0 | 1.6 | GO:0002763 | positive regulation of myeloid leukocyte differentiation(GO:0002763) |
| 0.0 | 0.5 | GO:0061099 | negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.0 | 2.9 | GO:0051225 | spindle assembly(GO:0051225) |
| 0.0 | 2.1 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.0 | 0.1 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.0 | 1.0 | GO:0050982 | detection of mechanical stimulus(GO:0050982) |
| 0.0 | 0.4 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.0 | 0.7 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.0 | 0.1 | GO:1990167 | protein K27-linked deubiquitination(GO:1990167) protein K33-linked deubiquitination(GO:1990168) |
| 0.0 | 0.2 | GO:0030299 | intestinal cholesterol absorption(GO:0030299) intestinal lipid absorption(GO:0098856) |
| 0.0 | 0.2 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.0 | 0.2 | GO:0090160 | Golgi to lysosome transport(GO:0090160) |
| 0.0 | 0.1 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.0 | 0.4 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.0 | 0.5 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.8 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.2 | 2.1 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.2 | 1.7 | GO:0034688 | integrin alphaM-beta2 complex(GO:0034688) |
| 0.2 | 0.8 | GO:0005889 | hydrogen:potassium-exchanging ATPase complex(GO:0005889) |
| 0.2 | 1.6 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 0.2 | 1.3 | GO:0036021 | endolysosome lumen(GO:0036021) |
| 0.1 | 6.1 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.1 | 3.6 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.1 | 0.9 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.1 | 5.2 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.1 | 0.4 | GO:0097635 | extrinsic component of autophagosome membrane(GO:0097635) |
| 0.1 | 1.2 | GO:0031211 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.1 | 2.4 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 0.5 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.1 | 1.0 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.1 | 1.2 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.1 | 1.3 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.1 | 0.7 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.1 | 2.3 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 5.5 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.0 | 1.0 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 1.3 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.0 | 0.8 | GO:0000800 | lateral element(GO:0000800) |
| 0.0 | 0.3 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.0 | 1.0 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.5 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.0 | 5.1 | GO:0015629 | actin cytoskeleton(GO:0015629) |
| 0.0 | 0.8 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 0.7 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 4.8 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 4.9 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 0.4 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.0 | 0.5 | GO:0071682 | endocytic vesicle lumen(GO:0071682) |
| 0.0 | 0.6 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 0.7 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 1.4 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 2.6 | GO:0070821 | tertiary granule membrane(GO:0070821) |
| 0.0 | 0.2 | GO:1990357 | terminal web(GO:1990357) |
| 0.0 | 0.4 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 3.9 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 0.9 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.0 | 2.2 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.0 | 2.3 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 1.2 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.6 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 0.1 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.0 | 1.1 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.4 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.3 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 2.0 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.0 | 1.0 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 0.3 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 1.1 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 10.2 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) |
| 1.2 | 4.8 | GO:0005277 | acetylcholine transmembrane transporter activity(GO:0005277) secondary active organic cation transmembrane transporter activity(GO:0008513) acetate ester transmembrane transporter activity(GO:1901375) |
| 0.7 | 2.1 | GO:1904854 | proteasome core complex binding(GO:1904854) |
| 0.7 | 4.2 | GO:0090554 | phosphatidylcholine-translocating ATPase activity(GO:0090554) |
| 0.5 | 2.8 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.5 | 2.3 | GO:0004910 | interleukin-1, Type II, blocking receptor activity(GO:0004910) |
| 0.4 | 2.1 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.4 | 1.5 | GO:0003978 | UDP-N-acetylglucosamine 4-epimerase activity(GO:0003974) UDP-glucose 4-epimerase activity(GO:0003978) |
| 0.4 | 2.3 | GO:0004918 | interleukin-8 receptor activity(GO:0004918) |
| 0.4 | 1.1 | GO:0047017 | geranylgeranyl reductase activity(GO:0045550) prostaglandin-F synthase activity(GO:0047017) |
| 0.3 | 1.0 | GO:0015432 | bile acid-exporting ATPase activity(GO:0015432) |
| 0.3 | 1.3 | GO:0004619 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.3 | 1.9 | GO:0004137 | deoxycytidine kinase activity(GO:0004137) |
| 0.3 | 1.2 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.3 | 1.8 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.3 | 2.0 | GO:0005124 | N-formyl peptide receptor activity(GO:0004982) scavenger receptor binding(GO:0005124) |
| 0.2 | 0.7 | GO:0002113 | interleukin-33 binding(GO:0002113) |
| 0.2 | 0.7 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 0.2 | 1.1 | GO:0047374 | methylumbelliferyl-acetate deacetylase activity(GO:0047374) |
| 0.2 | 2.3 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.2 | 8.6 | GO:0038187 | signaling pattern recognition receptor activity(GO:0008329) pattern recognition receptor activity(GO:0038187) |
| 0.2 | 1.6 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.2 | 3.6 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.2 | 3.3 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.1 | 1.2 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.1 | 1.0 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.1 | 29.4 | GO:0003823 | antigen binding(GO:0003823) |
| 0.1 | 8.7 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.1 | 1.7 | GO:0001851 | complement component C3b binding(GO:0001851) |
| 0.1 | 0.5 | GO:0016532 | superoxide dismutase copper chaperone activity(GO:0016532) copper-dependent protein binding(GO:0032767) |
| 0.1 | 2.4 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.1 | 0.8 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.1 | 0.5 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.1 | 5.1 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.1 | 5.8 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.1 | 0.9 | GO:0016778 | diphosphotransferase activity(GO:0016778) |
| 0.1 | 1.2 | GO:0015321 | sodium-dependent phosphate transmembrane transporter activity(GO:0015321) |
| 0.1 | 0.5 | GO:0046790 | virion binding(GO:0046790) |
| 0.1 | 0.5 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.1 | 2.5 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 1.3 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) |
| 0.1 | 0.9 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.1 | 0.3 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.1 | 0.7 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.1 | 1.6 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.5 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.0 | 0.6 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.8 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 0.8 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 1.3 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 2.3 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.0 | 0.3 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.0 | 1.0 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.7 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 0.4 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 1.3 | GO:0042287 | MHC protein binding(GO:0042287) |
| 0.0 | 1.0 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 0.9 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 2.2 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.4 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 0.0 | 0.8 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 2.0 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 0.5 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.1 | GO:0003875 | ADP-ribosylarginine hydrolase activity(GO:0003875) |
| 0.0 | 5.3 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.9 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 2.3 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.8 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.1 | GO:0038047 | beta-endorphin receptor activity(GO:0004979) morphine receptor activity(GO:0038047) |
| 0.0 | 1.6 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.3 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 3.8 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 0.9 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.0 | 1.0 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 0.2 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.0 | 0.4 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.0 | 0.5 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.0 | 0.7 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 0.3 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 0.0 | GO:0015403 | thiamine uptake transmembrane transporter activity(GO:0015403) uptake transmembrane transporter activity(GO:0015563) |
| 0.0 | 5.0 | GO:0051020 | GTPase binding(GO:0051020) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.5 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.1 | 5.9 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.1 | 5.1 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.0 | 1.1 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 2.3 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.0 | 2.4 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 3.0 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 4.2 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 1.9 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.0 | 2.3 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 2.2 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 7.7 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.9 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 1.6 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 3.0 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.5 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 0.8 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 1.1 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.7 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 9.0 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.3 | 10.2 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.3 | 5.1 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.2 | 5.8 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.1 | 1.3 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.1 | 3.3 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.1 | 2.3 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.1 | 3.6 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 2.9 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.1 | 2.4 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.1 | 1.0 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.1 | 2.3 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 2.1 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 1.9 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 0.5 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 3.0 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.9 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.5 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 4.4 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 3.0 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 0.7 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 9.6 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.7 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.6 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 1.1 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 2.1 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 1.2 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 2.2 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.3 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.7 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.3 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 1.2 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 2.8 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.4 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.8 | REACTOME SPHINGOLIPID METABOLISM | Genes involved in Sphingolipid metabolism |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 0.3 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 1.6 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 2.1 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.8 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |