Project
Illumina Body Map 2: averaged replicates
Navigation
Downloads
Results for ARID3A
Z-value: 1.38
Transcription factors associated with ARID3A
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ARID3A
|
ENSG00000116017.6 | AT-rich interaction domain 3A |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ARID3A | hg19_v2_chr19_+_926000_926046 | -0.20 | 2.8e-01 | Click! |
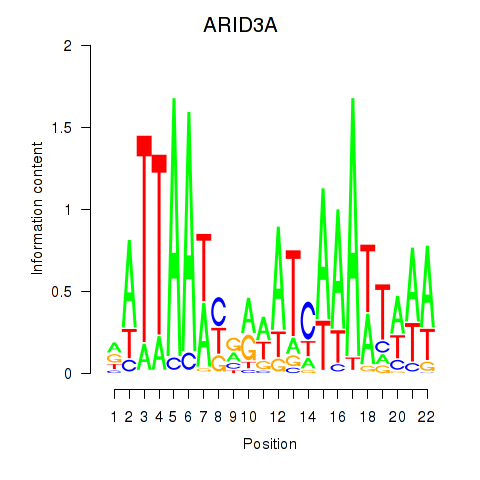
Activity profile of ARID3A motif
Sorted Z-values of ARID3A motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_77333117 | 3.33 |
ENST00000477717.1
|
ST6GALNAC5
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 5 |
| chr5_+_159656437 | 2.98 |
ENST00000402432.3
|
FABP6
|
fatty acid binding protein 6, ileal |
| chr12_+_119772502 | 2.49 |
ENST00000536742.1
ENST00000327554.2 |
CCDC60
|
coiled-coil domain containing 60 |
| chr3_-_172859017 | 2.22 |
ENST00000351008.3
|
SPATA16
|
spermatogenesis associated 16 |
| chrX_-_102531717 | 2.15 |
ENST00000372680.1
|
TCEAL5
|
transcription elongation factor A (SII)-like 5 |
| chr14_-_77737543 | 2.05 |
ENST00000298352.4
|
NGB
|
neuroglobin |
| chrX_+_85969626 | 1.87 |
ENST00000484479.1
|
DACH2
|
dachshund homolog 2 (Drosophila) |
| chr17_-_39041479 | 1.76 |
ENST00000167588.3
|
KRT20
|
keratin 20 |
| chr18_-_25739260 | 1.66 |
ENST00000413878.1
|
CDH2
|
cadherin 2, type 1, N-cadherin (neuronal) |
| chr19_-_46145696 | 1.65 |
ENST00000588172.1
|
EML2
|
echinoderm microtubule associated protein like 2 |
| chrX_-_2882296 | 1.64 |
ENST00000438544.1
ENST00000381134.3 ENST00000545496.1 |
ARSE
|
arylsulfatase E (chondrodysplasia punctata 1) |
| chrX_-_114252193 | 1.60 |
ENST00000243213.1
|
IL13RA2
|
interleukin 13 receptor, alpha 2 |
| chr7_-_92855762 | 1.54 |
ENST00000453812.2
ENST00000394468.2 |
HEPACAM2
|
HEPACAM family member 2 |
| chr10_-_69597915 | 1.53 |
ENST00000225171.2
|
DNAJC12
|
DnaJ (Hsp40) homolog, subfamily C, member 12 |
| chr12_+_41136144 | 1.51 |
ENST00000548005.1
ENST00000552248.1 |
CNTN1
|
contactin 1 |
| chr7_-_100253993 | 1.50 |
ENST00000461605.1
ENST00000160382.5 |
ACTL6B
|
actin-like 6B |
| chr15_-_74658519 | 1.50 |
ENST00000450547.1
ENST00000358632.4 |
CYP11A1
|
cytochrome P450, family 11, subfamily A, polypeptide 1 |
| chr21_-_22175341 | 1.48 |
ENST00000416768.1
ENST00000452561.1 ENST00000419299.1 ENST00000437238.1 |
LINC00320
|
long intergenic non-protein coding RNA 320 |
| chr1_-_39395165 | 1.40 |
ENST00000372985.3
|
RHBDL2
|
rhomboid, veinlet-like 2 (Drosophila) |
| chr3_-_93747425 | 1.39 |
ENST00000315099.2
|
STX19
|
syntaxin 19 |
| chr15_-_74659978 | 1.38 |
ENST00000541301.1
ENST00000416978.1 ENST00000268053.6 |
CYP11A1
|
cytochrome P450, family 11, subfamily A, polypeptide 1 |
| chr6_-_49681235 | 1.36 |
ENST00000339139.4
|
CRISP2
|
cysteine-rich secretory protein 2 |
| chr15_-_74658493 | 1.35 |
ENST00000419019.2
ENST00000569662.1 |
CYP11A1
|
cytochrome P450, family 11, subfamily A, polypeptide 1 |
| chr14_+_24099318 | 1.34 |
ENST00000432832.2
|
DHRS2
|
dehydrogenase/reductase (SDR family) member 2 |
| chr3_+_118892411 | 1.33 |
ENST00000479520.1
ENST00000494855.1 |
UPK1B
|
uroplakin 1B |
| chr3_+_177159695 | 1.33 |
ENST00000442937.1
|
LINC00578
|
long intergenic non-protein coding RNA 578 |
| chr16_-_21289627 | 1.25 |
ENST00000396023.2
ENST00000415987.2 |
CRYM
|
crystallin, mu |
| chr10_-_100995603 | 1.23 |
ENST00000370552.3
ENST00000370549.1 |
HPSE2
|
heparanase 2 |
| chr2_+_79252822 | 1.23 |
ENST00000272324.5
|
REG3G
|
regenerating islet-derived 3 gamma |
| chr14_+_37641012 | 1.22 |
ENST00000556667.1
|
SLC25A21-AS1
|
SLC25A21 antisense RNA 1 |
| chr8_+_85095497 | 1.21 |
ENST00000522455.1
ENST00000521695.1 |
RALYL
|
RALY RNA binding protein-like |
| chr22_+_18721427 | 1.20 |
ENST00000342888.3
|
AC008132.1
|
Uncharacterized protein |
| chr12_-_68696652 | 1.20 |
ENST00000539972.1
|
MDM1
|
Mdm1 nuclear protein homolog (mouse) |
| chr2_+_79252804 | 1.20 |
ENST00000393897.2
|
REG3G
|
regenerating islet-derived 3 gamma |
| chr10_-_69597810 | 1.17 |
ENST00000483798.2
|
DNAJC12
|
DnaJ (Hsp40) homolog, subfamily C, member 12 |
| chr8_-_10336885 | 1.17 |
ENST00000520494.1
|
RP11-981G7.3
|
RP11-981G7.3 |
| chr12_+_18414446 | 1.13 |
ENST00000433979.1
|
PIK3C2G
|
phosphatidylinositol-4-phosphate 3-kinase, catalytic subunit type 2 gamma |
| chr5_+_126988732 | 1.13 |
ENST00000395322.3
|
CTXN3
|
cortexin 3 |
| chr3_-_169487617 | 1.13 |
ENST00000330368.2
|
ACTRT3
|
actin-related protein T3 |
| chr6_-_49989648 | 1.12 |
ENST00000393660.2
ENST00000371148.2 |
DEFB110
|
defensin, beta 110 locus |
| chr22_-_32766972 | 1.11 |
ENST00000382084.4
ENST00000382086.2 |
RFPL3S
|
RFPL3 antisense |
| chr9_-_117420052 | 1.11 |
ENST00000423632.1
|
RP11-402G3.5
|
RP11-402G3.5 |
| chr21_-_22175450 | 1.09 |
ENST00000435279.2
|
LINC00320
|
long intergenic non-protein coding RNA 320 |
| chr12_-_71182695 | 1.08 |
ENST00000342084.4
|
PTPRR
|
protein tyrosine phosphatase, receptor type, R |
| chr3_-_6847096 | 1.06 |
ENST00000454410.2
ENST00000424366.1 ENST00000417482.1 ENST00000412629.1 |
GRM7-AS3
|
GRM7 antisense RNA 3 |
| chrX_-_80457385 | 1.06 |
ENST00000451455.1
ENST00000436386.1 ENST00000358130.2 |
HMGN5
|
high mobility group nucleosome binding domain 5 |
| chr16_-_30064244 | 1.06 |
ENST00000571269.1
ENST00000561666.1 |
FAM57B
|
family with sequence similarity 57, member B |
| chr4_+_69917078 | 1.04 |
ENST00000502942.1
|
UGT2B7
|
UDP glucuronosyltransferase 2 family, polypeptide B7 |
| chr10_-_100995540 | 1.03 |
ENST00000370546.1
ENST00000404542.1 |
HPSE2
|
heparanase 2 |
| chr1_-_31661000 | 1.03 |
ENST00000263693.1
ENST00000398657.2 ENST00000526106.1 |
NKAIN1
|
Na+/K+ transporting ATPase interacting 1 |
| chr2_+_79252834 | 1.02 |
ENST00000409471.1
|
REG3G
|
regenerating islet-derived 3 gamma |
| chr16_-_79263322 | 1.01 |
ENST00000569677.1
|
RP11-679B19.2
|
RP11-679B19.2 |
| chr4_-_163085107 | 1.01 |
ENST00000379164.4
|
FSTL5
|
follistatin-like 5 |
| chr22_-_32767017 | 0.99 |
ENST00000400234.1
|
RFPL3S
|
RFPL3 antisense |
| chr21_+_34442439 | 0.99 |
ENST00000382348.1
ENST00000333063.5 |
OLIG1
|
oligodendrocyte transcription factor 1 |
| chr4_-_163085141 | 0.96 |
ENST00000427802.2
ENST00000306100.5 |
FSTL5
|
follistatin-like 5 |
| chr19_-_12941469 | 0.95 |
ENST00000586969.1
ENST00000589681.1 ENST00000585384.1 ENST00000589808.1 |
RTBDN
|
retbindin |
| chr14_+_38091270 | 0.95 |
ENST00000553443.1
|
TTC6
|
tetratricopeptide repeat domain 6 |
| chr4_-_170948361 | 0.93 |
ENST00000393702.3
|
MFAP3L
|
microfibrillar-associated protein 3-like |
| chr1_+_51567895 | 0.93 |
ENST00000371759.2
|
C1orf185
|
chromosome 1 open reading frame 185 |
| chr5_-_110062349 | 0.93 |
ENST00000511883.2
ENST00000455884.2 |
TMEM232
|
transmembrane protein 232 |
| chr11_+_60260248 | 0.93 |
ENST00000526784.1
ENST00000016913.4 ENST00000537076.1 ENST00000530007.1 |
MS4A12
|
membrane-spanning 4-domains, subfamily A, member 12 |
| chr18_+_616711 | 0.91 |
ENST00000579494.1
|
CLUL1
|
clusterin-like 1 (retinal) |
| chr9_-_19786926 | 0.89 |
ENST00000341998.2
ENST00000286344.3 |
SLC24A2
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 2 |
| chr21_-_15583165 | 0.88 |
ENST00000536861.1
|
LIPI
|
lipase, member I |
| chr18_+_616672 | 0.87 |
ENST00000338387.7
|
CLUL1
|
clusterin-like 1 (retinal) |
| chrX_-_138724677 | 0.87 |
ENST00000370573.4
ENST00000338585.6 ENST00000370576.4 |
MCF2
|
MCF.2 cell line derived transforming sequence |
| chr6_+_31105426 | 0.85 |
ENST00000547221.1
|
PSORS1C1
|
psoriasis susceptibility 1 candidate 1 |
| chr5_-_110062384 | 0.85 |
ENST00000429839.2
|
TMEM232
|
transmembrane protein 232 |
| chr1_+_51567910 | 0.85 |
ENST00000467127.1
|
C1orf185
|
chromosome 1 open reading frame 185 |
| chr16_+_30034655 | 0.84 |
ENST00000300575.2
|
C16orf92
|
chromosome 16 open reading frame 92 |
| chr1_-_205391178 | 0.83 |
ENST00000367153.4
ENST00000367151.2 ENST00000391936.2 ENST00000367149.3 |
LEMD1
|
LEM domain containing 1 |
| chr3_-_179691866 | 0.83 |
ENST00000464614.1
ENST00000476138.1 ENST00000463761.1 |
PEX5L
|
peroxisomal biogenesis factor 5-like |
| chr1_-_72566613 | 0.82 |
ENST00000306821.3
|
NEGR1
|
neuronal growth regulator 1 |
| chr6_-_136847099 | 0.82 |
ENST00000438100.2
|
MAP7
|
microtubule-associated protein 7 |
| chr4_+_69962185 | 0.81 |
ENST00000305231.7
|
UGT2B7
|
UDP glucuronosyltransferase 2 family, polypeptide B7 |
| chrX_+_83116142 | 0.80 |
ENST00000329312.4
|
CYLC1
|
cylicin, basic protein of sperm head cytoskeleton 1 |
| chr16_-_31564881 | 0.80 |
ENST00000567765.1
|
CTD-2014E2.6
|
CTD-2014E2.6 |
| chr4_-_69434245 | 0.80 |
ENST00000317746.2
|
UGT2B17
|
UDP glucuronosyltransferase 2 family, polypeptide B17 |
| chr8_+_85095769 | 0.80 |
ENST00000518566.1
|
RALYL
|
RALY RNA binding protein-like |
| chr18_-_64271363 | 0.79 |
ENST00000262150.2
|
CDH19
|
cadherin 19, type 2 |
| chr14_+_70918874 | 0.79 |
ENST00000603540.1
|
ADAM21
|
ADAM metallopeptidase domain 21 |
| chr10_-_46620012 | 0.77 |
ENST00000508602.1
ENST00000374339.3 ENST00000502254.1 ENST00000437863.1 ENST00000374342.2 ENST00000395722.3 |
PTPN20A
|
protein tyrosine phosphatase, non-receptor type 20A |
| chr13_-_47471155 | 0.77 |
ENST00000543956.1
ENST00000542664.1 |
HTR2A
|
5-hydroxytryptamine (serotonin) receptor 2A, G protein-coupled |
| chr3_-_36781352 | 0.76 |
ENST00000416516.2
|
DCLK3
|
doublecortin-like kinase 3 |
| chr8_-_7673238 | 0.76 |
ENST00000335021.2
|
DEFB107A
|
defensin, beta 107A |
| chr6_+_151561085 | 0.76 |
ENST00000402676.2
|
AKAP12
|
A kinase (PRKA) anchor protein 12 |
| chr8_+_104892639 | 0.75 |
ENST00000436393.2
|
RIMS2
|
regulating synaptic membrane exocytosis 2 |
| chr3_-_195581080 | 0.75 |
ENST00000444346.1
|
AC124944.5
|
AC124944.5 |
| chr14_+_62453803 | 0.75 |
ENST00000446982.2
|
SYT16
|
synaptotagmin XVI |
| chr10_-_69597828 | 0.75 |
ENST00000339758.7
|
DNAJC12
|
DnaJ (Hsp40) homolog, subfamily C, member 12 |
| chr3_+_177159744 | 0.75 |
ENST00000439009.1
|
LINC00578
|
long intergenic non-protein coding RNA 578 |
| chr16_+_76587314 | 0.74 |
ENST00000563764.1
|
RP11-58C22.1
|
Uncharacterized protein |
| chr12_+_51818555 | 0.73 |
ENST00000453097.2
|
SLC4A8
|
solute carrier family 4, sodium bicarbonate cotransporter, member 8 |
| chr8_+_85095553 | 0.73 |
ENST00000521268.1
|
RALYL
|
RALY RNA binding protein-like |
| chr16_-_52061283 | 0.73 |
ENST00000566314.1
|
C16orf97
|
chromosome 16 open reading frame 97 |
| chr16_-_11375179 | 0.71 |
ENST00000312511.3
|
PRM1
|
protamine 1 |
| chr12_-_120765565 | 0.71 |
ENST00000423423.3
ENST00000308366.4 |
PLA2G1B
|
phospholipase A2, group IB (pancreas) |
| chr18_+_61143994 | 0.70 |
ENST00000382771.4
|
SERPINB5
|
serpin peptidase inhibitor, clade B (ovalbumin), member 5 |
| chr15_-_58306295 | 0.70 |
ENST00000559517.1
|
ALDH1A2
|
aldehyde dehydrogenase 1 family, member A2 |
| chrX_-_117119243 | 0.69 |
ENST00000539496.1
ENST00000469946.1 |
KLHL13
|
kelch-like family member 13 |
| chr13_-_20110902 | 0.69 |
ENST00000390680.2
ENST00000382977.4 ENST00000382975.4 ENST00000457266.2 |
TPTE2
|
transmembrane phosphoinositide 3-phosphatase and tensin homolog 2 |
| chr19_-_53794875 | 0.69 |
ENST00000426466.1
|
BIRC8
|
baculoviral IAP repeat containing 8 |
| chr4_+_71200681 | 0.69 |
ENST00000273936.5
|
CABS1
|
calcium-binding protein, spermatid-specific 1 |
| chr3_+_118892362 | 0.68 |
ENST00000497685.1
ENST00000264234.3 |
UPK1B
|
uroplakin 1B |
| chr16_+_19421803 | 0.68 |
ENST00000541464.1
|
TMC5
|
transmembrane channel-like 5 |
| chr4_+_69962212 | 0.67 |
ENST00000508661.1
|
UGT2B7
|
UDP glucuronosyltransferase 2 family, polypeptide B7 |
| chr13_-_102068706 | 0.67 |
ENST00000251127.6
|
NALCN
|
sodium leak channel, non-selective |
| chr8_+_7353368 | 0.67 |
ENST00000355602.2
|
DEFB107B
|
defensin, beta 107B |
| chr10_+_124320156 | 0.66 |
ENST00000338354.3
ENST00000344338.3 ENST00000330163.4 ENST00000368909.3 ENST00000368955.3 ENST00000368956.2 |
DMBT1
|
deleted in malignant brain tumors 1 |
| chr14_+_77425972 | 0.65 |
ENST00000553613.1
|
RP11-7F17.7
|
RP11-7F17.7 |
| chr6_-_52860171 | 0.65 |
ENST00000370963.4
|
GSTA4
|
glutathione S-transferase alpha 4 |
| chr19_-_13617247 | 0.65 |
ENST00000573710.2
|
CACNA1A
|
calcium channel, voltage-dependent, P/Q type, alpha 1A subunit |
| chr15_-_45670924 | 0.64 |
ENST00000396659.3
|
GATM
|
glycine amidinotransferase (L-arginine:glycine amidinotransferase) |
| chr16_-_48281444 | 0.64 |
ENST00000537808.1
ENST00000569991.1 |
ABCC11
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 11 |
| chr1_+_152943122 | 0.64 |
ENST00000328051.2
|
SPRR4
|
small proline-rich protein 4 |
| chr19_+_4153598 | 0.64 |
ENST00000078445.2
ENST00000252587.3 ENST00000595923.1 ENST00000602257.1 ENST00000602147.1 |
CREB3L3
|
cAMP responsive element binding protein 3-like 3 |
| chr6_+_73076432 | 0.64 |
ENST00000414192.2
|
RIMS1
|
regulating synaptic membrane exocytosis 1 |
| chr22_-_40289759 | 0.64 |
ENST00000325157.6
|
ENTHD1
|
ENTH domain containing 1 |
| chr10_-_48806939 | 0.63 |
ENST00000374233.3
ENST00000507417.1 ENST00000512321.1 ENST00000395660.2 ENST00000374235.2 ENST00000395661.3 |
PTPN20B
|
protein tyrosine phosphatase, non-receptor type 20B |
| chr6_+_46761118 | 0.63 |
ENST00000230588.4
|
MEP1A
|
meprin A, alpha (PABA peptide hydrolase) |
| chrY_-_20935572 | 0.62 |
ENST00000382852.1
ENST00000344884.4 ENST00000304790.3 |
HSFY2
|
heat shock transcription factor, Y linked 2 |
| chr19_+_52848659 | 0.62 |
ENST00000327920.8
|
ZNF610
|
zinc finger protein 610 |
| chr4_+_159131630 | 0.61 |
ENST00000504569.1
ENST00000509278.1 ENST00000514558.1 ENST00000503200.1 |
TMEM144
|
transmembrane protein 144 |
| chr21_+_15588499 | 0.60 |
ENST00000400577.3
|
RBM11
|
RNA binding motif protein 11 |
| chr14_+_83108955 | 0.60 |
ENST00000555798.1
ENST00000553760.1 ENST00000555150.1 ENST00000556970.1 |
RP11-406A9.2
|
RP11-406A9.2 |
| chr9_-_21482312 | 0.59 |
ENST00000448696.3
|
IFNE
|
interferon, epsilon |
| chr8_+_18248755 | 0.59 |
ENST00000286479.3
|
NAT2
|
N-acetyltransferase 2 (arylamine N-acetyltransferase) |
| chr5_+_140220769 | 0.58 |
ENST00000531613.1
ENST00000378123.3 |
PCDHA8
|
protocadherin alpha 8 |
| chr4_-_187517928 | 0.57 |
ENST00000512772.1
|
FAT1
|
FAT atypical cadherin 1 |
| chr12_-_81763184 | 0.57 |
ENST00000548670.1
ENST00000541570.2 ENST00000553058.1 |
PPFIA2
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2 |
| chr2_+_210444142 | 0.57 |
ENST00000360351.4
ENST00000361559.4 |
MAP2
|
microtubule-associated protein 2 |
| chr1_-_23520755 | 0.57 |
ENST00000314113.3
|
HTR1D
|
5-hydroxytryptamine (serotonin) receptor 1D, G protein-coupled |
| chrY_+_20708557 | 0.56 |
ENST00000307393.2
ENST00000309834.4 ENST00000382856.2 |
HSFY1
|
heat shock transcription factor, Y-linked 1 |
| chr15_+_54903673 | 0.56 |
ENST00000560537.1
|
UNC13C
|
unc-13 homolog C (C. elegans) |
| chr4_+_175839506 | 0.56 |
ENST00000505141.1
ENST00000359240.3 ENST00000445694.1 |
ADAM29
|
ADAM metallopeptidase domain 29 |
| chr18_-_51884204 | 0.56 |
ENST00000577499.1
ENST00000584040.1 ENST00000581310.1 |
STARD6
|
StAR-related lipid transfer (START) domain containing 6 |
| chr8_-_19102999 | 0.56 |
ENST00000517949.1
|
RP11-1080G15.1
|
RP11-1080G15.1 |
| chr11_-_70507867 | 0.55 |
ENST00000412252.1
ENST00000409161.1 ENST00000409530.1 |
SHANK2
|
SH3 and multiple ankyrin repeat domains 2 |
| chr15_+_26360970 | 0.55 |
ENST00000556159.1
ENST00000557523.1 |
LINC00929
|
long intergenic non-protein coding RNA 929 |
| chr3_-_179692042 | 0.55 |
ENST00000468741.1
|
PEX5L
|
peroxisomal biogenesis factor 5-like |
| chrX_-_138072729 | 0.55 |
ENST00000455663.1
|
FGF13
|
fibroblast growth factor 13 |
| chr1_-_227505289 | 0.54 |
ENST00000366765.3
|
CDC42BPA
|
CDC42 binding protein kinase alpha (DMPK-like) |
| chr1_-_235098861 | 0.54 |
ENST00000458044.1
|
RP11-443B7.1
|
RP11-443B7.1 |
| chr3_+_142442841 | 0.53 |
ENST00000476941.1
ENST00000273482.6 |
TRPC1
|
transient receptor potential cation channel, subfamily C, member 1 |
| chr2_+_20101786 | 0.53 |
ENST00000607190.1
|
RP11-79O8.1
|
RP11-79O8.1 |
| chrX_+_36053908 | 0.53 |
ENST00000378660.2
|
CHDC2
|
calponin homology domain containing 2 |
| chr2_-_135805008 | 0.53 |
ENST00000414343.1
|
MAP3K19
|
mitogen-activated protein kinase kinase kinase 19 |
| chr11_-_5255861 | 0.53 |
ENST00000380299.3
|
HBD
|
hemoglobin, delta |
| chr19_+_45542295 | 0.52 |
ENST00000221455.3
ENST00000391953.4 ENST00000588936.1 |
CLASRP
|
CLK4-associating serine/arginine rich protein |
| chr8_-_40200877 | 0.52 |
ENST00000521030.1
|
CTA-392C11.1
|
CTA-392C11.1 |
| chr4_+_159131596 | 0.52 |
ENST00000512481.1
|
TMEM144
|
transmembrane protein 144 |
| chr20_-_29978383 | 0.51 |
ENST00000339144.3
ENST00000376321.3 |
DEFB119
|
defensin, beta 119 |
| chr1_+_229440129 | 0.51 |
ENST00000366688.3
|
SPHAR
|
S-phase response (cyclin related) |
| chr5_-_10308125 | 0.51 |
ENST00000296658.3
|
CMBL
|
carboxymethylenebutenolidase homolog (Pseudomonas) |
| chr12_+_72061563 | 0.51 |
ENST00000551238.1
|
THAP2
|
THAP domain containing, apoptosis associated protein 2 |
| chr1_+_182419261 | 0.51 |
ENST00000294854.8
ENST00000542961.1 |
RGSL1
|
regulator of G-protein signaling like 1 |
| chr20_-_42355629 | 0.50 |
ENST00000373003.1
|
GTSF1L
|
gametocyte specific factor 1-like |
| chr2_+_210444748 | 0.50 |
ENST00000392194.1
|
MAP2
|
microtubule-associated protein 2 |
| chr2_-_70418032 | 0.50 |
ENST00000425268.1
ENST00000428751.1 ENST00000417203.1 ENST00000417865.1 ENST00000428010.1 ENST00000447804.1 ENST00000264434.2 |
C2orf42
|
chromosome 2 open reading frame 42 |
| chr6_-_52859968 | 0.49 |
ENST00000370959.1
|
GSTA4
|
glutathione S-transferase alpha 4 |
| chr2_-_183106641 | 0.49 |
ENST00000346717.4
|
PDE1A
|
phosphodiesterase 1A, calmodulin-dependent |
| chr11_-_70507901 | 0.49 |
ENST00000449833.2
ENST00000357171.3 ENST00000449116.2 |
SHANK2
|
SH3 and multiple ankyrin repeat domains 2 |
| chr4_+_71108300 | 0.49 |
ENST00000304954.3
|
CSN3
|
casein kappa |
| chr4_+_15341442 | 0.49 |
ENST00000397700.2
ENST00000295297.4 |
C1QTNF7
|
C1q and tumor necrosis factor related protein 7 |
| chr4_+_75174204 | 0.49 |
ENST00000332112.4
ENST00000514968.1 ENST00000503098.1 ENST00000502358.1 ENST00000509145.1 ENST00000505212.1 |
EPGN
|
epithelial mitogen |
| chr9_+_21440440 | 0.48 |
ENST00000276927.1
|
IFNA1
|
interferon, alpha 1 |
| chr9_-_74675521 | 0.48 |
ENST00000377024.3
|
C9orf57
|
chromosome 9 open reading frame 57 |
| chr6_+_36922209 | 0.48 |
ENST00000373674.3
|
PI16
|
peptidase inhibitor 16 |
| chrX_+_80457442 | 0.48 |
ENST00000373212.5
|
SH3BGRL
|
SH3 domain binding glutamic acid-rich protein like |
| chr4_+_5527117 | 0.48 |
ENST00000505296.1
|
C4orf6
|
chromosome 4 open reading frame 6 |
| chr2_+_191002486 | 0.48 |
ENST00000396974.2
|
C2orf88
|
chromosome 2 open reading frame 88 |
| chr1_-_19615744 | 0.47 |
ENST00000361640.4
|
AKR7A3
|
aldo-keto reductase family 7, member A3 (aflatoxin aldehyde reductase) |
| chr14_+_101295638 | 0.47 |
ENST00000523671.2
|
MEG3
|
maternally expressed 3 (non-protein coding) |
| chr19_+_56270380 | 0.47 |
ENST00000434937.2
|
RFPL4A
|
ret finger protein-like 4A |
| chr8_+_18248786 | 0.47 |
ENST00000520116.1
|
NAT2
|
N-acetyltransferase 2 (arylamine N-acetyltransferase) |
| chr12_+_26126681 | 0.46 |
ENST00000542865.1
|
RASSF8
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 8 |
| chr10_-_22498950 | 0.46 |
ENST00000422359.2
|
EBLN1
|
endogenous Bornavirus-like nucleoprotein 1 |
| chr2_+_234875252 | 0.46 |
ENST00000456930.1
|
TRPM8
|
transient receptor potential cation channel, subfamily M, member 8 |
| chr15_+_26360932 | 0.46 |
ENST00000556213.1
|
LINC00929
|
long intergenic non-protein coding RNA 929 |
| chr12_+_103545593 | 0.46 |
ENST00000547418.1
|
RP11-552I14.1
|
Uncharacterized protein |
| chr7_+_6617039 | 0.46 |
ENST00000405731.3
ENST00000396713.2 ENST00000396707.2 ENST00000335965.6 ENST00000396709.1 ENST00000483589.1 ENST00000396706.2 |
ZDHHC4
|
zinc finger, DHHC-type containing 4 |
| chr6_-_142409936 | 0.46 |
ENST00000258042.1
|
NMBR
|
neuromedin B receptor |
| chrX_+_140982452 | 0.46 |
ENST00000544766.1
|
MAGEC3
|
melanoma antigen family C, 3 |
| chr10_-_50143242 | 0.46 |
ENST00000298124.3
|
LRRC18
|
leucine rich repeat containing 18 |
| chrX_+_134975753 | 0.46 |
ENST00000535938.1
|
SAGE1
|
sarcoma antigen 1 |
| chr16_-_85722530 | 0.45 |
ENST00000253462.3
|
GINS2
|
GINS complex subunit 2 (Psf2 homolog) |
| chr4_+_5526883 | 0.45 |
ENST00000195455.2
|
C4orf6
|
chromosome 4 open reading frame 6 |
| chr12_-_74686314 | 0.45 |
ENST00000551210.1
ENST00000515416.2 ENST00000549905.1 |
RP11-81H3.2
|
RP11-81H3.2 |
| chr11_+_71249071 | 0.45 |
ENST00000398534.3
|
KRTAP5-8
|
keratin associated protein 5-8 |
| chr5_-_136834982 | 0.45 |
ENST00000510689.1
ENST00000394945.1 |
SPOCK1
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 1 |
| chr2_+_15840525 | 0.45 |
ENST00000436967.1
|
AC008271.1
|
AC008271.1 |
| chrX_+_134975858 | 0.44 |
ENST00000537770.1
|
SAGE1
|
sarcoma antigen 1 |
| chr3_+_186158134 | 0.44 |
ENST00000414102.1
|
RP11-78H24.1
|
RP11-78H24.1 |
| chr1_-_247887345 | 0.44 |
ENST00000366485.1
|
OR14A2
|
olfactory receptor, family 14, subfamily A, member 2 |
| chr7_+_89783689 | 0.44 |
ENST00000297205.2
|
STEAP1
|
six transmembrane epithelial antigen of the prostate 1 |
| chr5_-_42887494 | 0.44 |
ENST00000514218.1
|
SEPP1
|
selenoprotein P, plasma, 1 |
| chr11_-_5345582 | 0.44 |
ENST00000328813.2
|
OR51B2
|
olfactory receptor, family 51, subfamily B, member 2 |
| chr6_+_46714654 | 0.43 |
ENST00000565422.1
|
ANKRD66
|
ankyrin repeat domain 66 |
| chr3_-_126327398 | 0.43 |
ENST00000383572.2
|
TXNRD3NB
|
thioredoxin reductase 3 neighbor |
Network of associatons between targets according to the STRING database.
First level regulatory network of ARID3A
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.4 | GO:0044278 | cell wall disruption in other organism(GO:0044278) |
| 0.2 | 1.3 | GO:0019355 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.2 | 0.5 | GO:1902283 | negative regulation of primary amine oxidase activity(GO:1902283) |
| 0.2 | 0.5 | GO:0036466 | synaptic vesicle recycling via endosome(GO:0036466) |
| 0.2 | 2.0 | GO:2000809 | positive regulation of synaptic vesicle clustering(GO:2000809) |
| 0.2 | 0.5 | GO:0002590 | regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002589) negative regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002590) positive regulation of iron ion transport(GO:0034758) positive regulation of iron ion transmembrane transport(GO:0034761) regulation of iron ion import(GO:1900390) regulation of ferrous iron import into cell(GO:1903989) positive regulation of ferrous iron import into cell(GO:1903991) regulation of ferrous iron binding(GO:1904432) positive regulation of ferrous iron binding(GO:1904434) regulation of transferrin receptor binding(GO:1904435) positive regulation of transferrin receptor binding(GO:1904437) regulation of ferrous iron import across plasma membrane(GO:1904438) positive regulation of ferrous iron import across plasma membrane(GO:1904440) response to iron ion starvation(GO:1990641) |
| 0.2 | 0.6 | GO:0006601 | creatine biosynthetic process(GO:0006601) |
| 0.2 | 1.6 | GO:0043305 | negative regulation of mast cell degranulation(GO:0043305) |
| 0.2 | 0.8 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.2 | 1.1 | GO:1903385 | regulation of homophilic cell adhesion(GO:1903385) |
| 0.2 | 0.8 | GO:0010513 | positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) |
| 0.1 | 3.5 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.1 | 0.7 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.1 | 1.4 | GO:0016560 | protein import into peroxisome matrix, docking(GO:0016560) |
| 0.1 | 4.1 | GO:0042359 | vitamin D metabolic process(GO:0042359) |
| 0.1 | 3.3 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.1 | 1.7 | GO:0010968 | regulation of microtubule nucleation(GO:0010968) |
| 0.1 | 2.8 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.1 | 2.2 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.1 | 0.5 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.1 | 0.5 | GO:0045196 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.1 | 1.2 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.1 | 0.2 | GO:0006433 | prolyl-tRNA aminoacylation(GO:0006433) |
| 0.1 | 1.2 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.1 | 0.6 | GO:0098706 | ferric iron import(GO:0033216) ferric iron import into cell(GO:0097461) ferric iron import across plasma membrane(GO:0098706) |
| 0.1 | 0.7 | GO:0035799 | ureter maturation(GO:0035799) |
| 0.1 | 0.9 | GO:0060075 | regulation of resting membrane potential(GO:0060075) |
| 0.1 | 0.2 | GO:0042214 | terpene metabolic process(GO:0042214) |
| 0.1 | 0.5 | GO:0050955 | thermoception(GO:0050955) |
| 0.1 | 1.4 | GO:0097151 | positive regulation of inhibitory postsynaptic potential(GO:0097151) modulation of inhibitory postsynaptic potential(GO:0098828) |
| 0.1 | 0.2 | GO:1904204 | regulation of skeletal muscle hypertrophy(GO:1904204) |
| 0.1 | 0.3 | GO:0035105 | sterol regulatory element binding protein import into nucleus(GO:0035105) |
| 0.1 | 0.6 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.1 | 1.8 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.1 | 0.8 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.1 | 2.9 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.1 | 1.4 | GO:0031629 | synaptic vesicle fusion to presynaptic active zone membrane(GO:0031629) vesicle fusion to plasma membrane(GO:0099500) |
| 0.1 | 0.8 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.1 | 0.5 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.0 | 0.5 | GO:0014827 | intestine smooth muscle contraction(GO:0014827) |
| 0.0 | 0.3 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.0 | 2.3 | GO:0060292 | long term synaptic depression(GO:0060292) |
| 0.0 | 0.4 | GO:0061458 | reproductive system development(GO:0061458) |
| 0.0 | 0.3 | GO:0097105 | presynaptic membrane assembly(GO:0097105) |
| 0.0 | 1.3 | GO:0043011 | myeloid dendritic cell differentiation(GO:0043011) |
| 0.0 | 0.3 | GO:0090166 | Golgi disassembly(GO:0090166) |
| 0.0 | 0.3 | GO:0097106 | postsynaptic density organization(GO:0097106) |
| 0.0 | 0.2 | GO:0032354 | response to follicle-stimulating hormone(GO:0032354) response to gonadotropin(GO:0034698) |
| 0.0 | 0.6 | GO:0016446 | somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.0 | 0.5 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.0 | 0.2 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.0 | 0.7 | GO:0015865 | purine nucleotide transport(GO:0015865) |
| 0.0 | 1.7 | GO:0051123 | RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) |
| 0.0 | 0.2 | GO:2001206 | positive regulation of osteoclast development(GO:2001206) |
| 0.0 | 0.2 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.0 | 0.5 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.0 | 0.5 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.0 | 0.3 | GO:0042354 | fucose catabolic process(GO:0019317) L-fucose metabolic process(GO:0042354) L-fucose catabolic process(GO:0042355) |
| 0.0 | 1.1 | GO:0039703 | viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) |
| 0.0 | 0.4 | GO:1990440 | positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990440) |
| 0.0 | 0.1 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.0 | 1.5 | GO:0010765 | positive regulation of sodium ion transport(GO:0010765) |
| 0.0 | 0.2 | GO:1902570 | protein localization to nucleolus(GO:1902570) |
| 0.0 | 0.5 | GO:0032703 | negative regulation of interleukin-2 production(GO:0032703) |
| 0.0 | 0.4 | GO:0045838 | positive regulation of membrane potential(GO:0045838) |
| 0.0 | 0.8 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.0 | 0.6 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.0 | 0.7 | GO:0030261 | chromosome condensation(GO:0030261) |
| 0.0 | 0.9 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.0 | 1.1 | GO:0045599 | negative regulation of fat cell differentiation(GO:0045599) |
| 0.0 | 0.2 | GO:0006987 | activation of signaling protein activity involved in unfolded protein response(GO:0006987) |
| 0.0 | 0.1 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.0 | 0.2 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.0 | 1.0 | GO:0014047 | glutamate secretion(GO:0014047) |
| 0.0 | 0.4 | GO:0071157 | negative regulation of cell cycle arrest(GO:0071157) |
| 0.0 | 1.4 | GO:0006027 | glycosaminoglycan catabolic process(GO:0006027) |
| 0.0 | 0.3 | GO:0032331 | negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.0 | 0.4 | GO:0060512 | prostate gland morphogenesis(GO:0060512) |
| 0.0 | 0.7 | GO:0048663 | neuron fate commitment(GO:0048663) |
| 0.0 | 0.2 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.0 | 1.5 | GO:0021510 | spinal cord development(GO:0021510) |
| 0.0 | 1.7 | GO:0021954 | central nervous system neuron development(GO:0021954) |
| 0.0 | 0.3 | GO:0090036 | regulation of protein kinase C signaling(GO:0090036) |
| 0.0 | 0.8 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.2 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.0 | 1.0 | GO:0002028 | regulation of sodium ion transport(GO:0002028) |
| 0.0 | 0.2 | GO:0001542 | ovulation from ovarian follicle(GO:0001542) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0043159 | cytoskeletal calyx(GO:0033150) acrosomal matrix(GO:0043159) |
| 0.2 | 1.8 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.1 | 0.5 | GO:0000811 | GINS complex(GO:0000811) |
| 0.1 | 0.9 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 0.1 | 1.1 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.1 | 1.7 | GO:0016342 | catenin complex(GO:0016342) |
| 0.1 | 0.6 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.1 | 0.8 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.1 | 1.0 | GO:0005883 | neurofilament(GO:0005883) |
| 0.1 | 0.4 | GO:0044308 | axonal spine(GO:0044308) |
| 0.1 | 0.5 | GO:1990357 | terminal web(GO:1990357) |
| 0.1 | 0.6 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.0 | 0.8 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.0 | 1.5 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 0.8 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.0 | 0.4 | GO:0043196 | varicosity(GO:0043196) |
| 0.0 | 2.4 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 1.1 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.3 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.5 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.0 | 0.2 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.0 | 0.2 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.0 | 1.2 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 1.3 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 0.2 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.0 | 0.8 | GO:0032809 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 0.0 | GO:1990666 | PCSK9-LDLR complex(GO:1990666) |
| 0.0 | 0.1 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.0 | 0.3 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.3 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.0 | 0.4 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.0 | 0.1 | GO:0097125 | cyclin B1-CDK1 complex(GO:0097125) |
| 0.0 | 0.2 | GO:0005687 | U4 snRNP(GO:0005687) |
| 0.0 | 2.7 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 1.7 | GO:0072686 | mitotic spindle(GO:0072686) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 4.2 | GO:0008386 | cholesterol monooxygenase (side-chain-cleaving) activity(GO:0008386) |
| 0.6 | 2.3 | GO:0030305 | heparanase activity(GO:0030305) |
| 0.3 | 3.3 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.3 | 1.2 | GO:0016639 | oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 0.3 | 1.3 | GO:0004090 | carbonyl reductase (NADPH) activity(GO:0004090) |
| 0.3 | 1.1 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.2 | 0.6 | GO:0015068 | amidinotransferase activity(GO:0015067) glycine amidinotransferase activity(GO:0015068) |
| 0.2 | 1.3 | GO:0000309 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) |
| 0.2 | 3.7 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.2 | 3.4 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.2 | 0.7 | GO:0051800 | phosphatidylinositol-3,4-bisphosphate 3-phosphatase activity(GO:0051800) |
| 0.2 | 1.0 | GO:1902444 | riboflavin binding(GO:1902444) |
| 0.2 | 2.2 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.1 | 1.4 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.1 | 0.5 | GO:0052596 | tryptamine:oxygen oxidoreductase (deaminating) activity(GO:0052593) aminoacetone:oxygen oxidoreductase(deaminating) activity(GO:0052594) aliphatic-amine oxidase activity(GO:0052595) phenethylamine:oxygen oxidoreductase (deaminating) activity(GO:0052596) |
| 0.1 | 2.8 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.1 | 1.0 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.1 | 0.9 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.1 | 1.1 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.1 | 1.3 | GO:0051378 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.1 | 1.7 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.1 | 0.6 | GO:0008823 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.1 | 0.4 | GO:0051431 | corticotropin-releasing hormone receptor 2 binding(GO:0051431) |
| 0.1 | 0.2 | GO:0004827 | proline-tRNA ligase activity(GO:0004827) |
| 0.1 | 0.6 | GO:0032138 | single base insertion or deletion binding(GO:0032138) |
| 0.1 | 3.5 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.1 | 0.5 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.1 | 1.6 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.1 | 1.7 | GO:0045294 | alpha-catenin binding(GO:0045294) gamma-catenin binding(GO:0045295) |
| 0.1 | 0.2 | GO:0002113 | interleukin-33 binding(GO:0002113) |
| 0.1 | 1.1 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.1 | 0.2 | GO:0010348 | lithium:proton antiporter activity(GO:0010348) |
| 0.1 | 1.8 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.1 | 0.2 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.1 | 0.4 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.1 | 0.2 | GO:0031493 | nucleosomal histone binding(GO:0031493) |
| 0.0 | 0.2 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 1.6 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 0.7 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.0 | 0.4 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.0 | 0.8 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.6 | GO:0043225 | anion transmembrane-transporting ATPase activity(GO:0043225) |
| 0.0 | 2.2 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.9 | GO:0022840 | leak channel activity(GO:0022840) narrow pore channel activity(GO:0022842) |
| 0.0 | 0.4 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.5 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 1.1 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.3 | GO:0005497 | androgen binding(GO:0005497) |
| 0.0 | 0.5 | GO:1990459 | beta-2-microglobulin binding(GO:0030881) transferrin receptor binding(GO:1990459) |
| 0.0 | 0.8 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.1 | GO:0010309 | acireductone dioxygenase [iron(II)-requiring] activity(GO:0010309) |
| 0.0 | 0.8 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.7 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.5 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 0.5 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.0 | 0.6 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 1.3 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.1 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.0 | 0.4 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 1.1 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.4 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.5 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.1 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.6 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 3.6 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.8 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.2 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.2 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.0 | 0.4 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.3 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.0 | 0.6 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 1.1 | GO:1901476 | carbohydrate transmembrane transporter activity(GO:0015144) carbohydrate transporter activity(GO:1901476) |
| 0.0 | 0.5 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.4 | GO:0004033 | aldo-keto reductase (NADP) activity(GO:0004033) |
| 0.0 | 0.3 | GO:0016289 | CoA hydrolase activity(GO:0016289) |
| 0.0 | 0.4 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.6 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.1 | 0.3 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.0 | 1.7 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 2.0 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 1.2 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 1.5 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 0.5 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 3.8 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 3.0 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.0 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.1 | 3.0 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.1 | 3.3 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.1 | 1.6 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.1 | 1.3 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.1 | 2.3 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.1 | 1.8 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 1.6 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.0 | 0.7 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 1.7 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.6 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.0 | 1.3 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 0.5 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.9 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.0 | 0.6 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.0 | 0.2 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.0 | 1.3 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.3 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.4 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 0.6 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 0.5 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |