Project
Illumina Body Map 2: averaged replicates
Navigation
Downloads
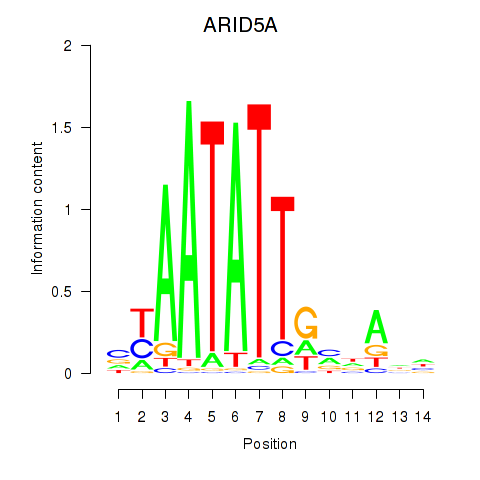
Results for ARID5A
Z-value: 1.65
Transcription factors associated with ARID5A
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ARID5A
|
ENSG00000196843.11 | AT-rich interaction domain 5A |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ARID5A | hg19_v2_chr2_+_97202480_97202499 | 0.36 | 4.1e-02 | Click! |
Activity profile of ARID5A motif
Sorted Z-values of ARID5A motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_217724767 | 4.67 |
ENST00000236979.2
|
TNP1
|
transition protein 1 (during histone to protamine replacement) |
| chr6_-_49834240 | 3.72 |
ENST00000335847.4
|
CRISP1
|
cysteine-rich secretory protein 1 |
| chr6_-_49834209 | 3.63 |
ENST00000507853.1
|
CRISP1
|
cysteine-rich secretory protein 1 |
| chr3_-_150421728 | 3.45 |
ENST00000295910.6
ENST00000491361.1 |
FAM194A
|
family with sequence similarity 194, member A |
| chr4_-_177116772 | 3.34 |
ENST00000280191.2
|
SPATA4
|
spermatogenesis associated 4 |
| chr1_+_40713573 | 3.20 |
ENST00000372766.3
|
TMCO2
|
transmembrane and coiled-coil domains 2 |
| chr8_+_24298531 | 3.16 |
ENST00000175238.6
|
ADAM7
|
ADAM metallopeptidase domain 7 |
| chr7_+_142031986 | 2.99 |
ENST00000547918.2
|
TRBV7-1
|
T cell receptor beta variable 7-1 (non-functional) |
| chr8_+_24298597 | 2.79 |
ENST00000380789.1
|
ADAM7
|
ADAM metallopeptidase domain 7 |
| chr18_-_61329118 | 2.56 |
ENST00000332821.8
ENST00000283752.5 |
SERPINB3
|
serpin peptidase inhibitor, clade B (ovalbumin), member 3 |
| chr2_+_152214098 | 2.50 |
ENST00000243347.3
|
TNFAIP6
|
tumor necrosis factor, alpha-induced protein 6 |
| chr5_+_159848807 | 2.45 |
ENST00000352433.5
|
PTTG1
|
pituitary tumor-transforming 1 |
| chr16_-_11375179 | 2.40 |
ENST00000312511.3
|
PRM1
|
protamine 1 |
| chr5_+_159848854 | 2.40 |
ENST00000517480.1
ENST00000520452.1 ENST00000393964.1 |
PTTG1
|
pituitary tumor-transforming 1 |
| chr11_+_61976137 | 2.36 |
ENST00000244930.4
|
SCGB2A1
|
secretoglobin, family 2A, member 1 |
| chr11_-_26588634 | 2.30 |
ENST00000436318.2
ENST00000281268.8 |
MUC15
|
mucin 15, cell surface associated |
| chrX_+_37850026 | 2.09 |
ENST00000341016.3
|
CXorf27
|
chromosome X open reading frame 27 |
| chr7_-_38339890 | 2.06 |
ENST00000390341.2
|
TRGV10
|
T cell receptor gamma variable 10 (non-functional) |
| chr1_-_76398077 | 2.02 |
ENST00000284142.6
|
ASB17
|
ankyrin repeat and SOCS box containing 17 |
| chrX_+_36053908 | 1.95 |
ENST00000378660.2
|
CHDC2
|
calponin homology domain containing 2 |
| chr3_-_150421752 | 1.89 |
ENST00000498386.1
|
FAM194A
|
family with sequence similarity 194, member A |
| chr10_+_127661942 | 1.88 |
ENST00000417114.1
ENST00000445510.1 ENST00000368691.1 |
FANK1
|
fibronectin type III and ankyrin repeat domains 1 |
| chrX_+_107288239 | 1.86 |
ENST00000217957.5
|
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chr16_+_86229728 | 1.86 |
ENST00000601250.1
|
LINC01082
|
long intergenic non-protein coding RNA 1082 |
| chr8_+_24298438 | 1.85 |
ENST00000441335.2
|
ADAM7
|
ADAM metallopeptidase domain 7 |
| chr20_+_31870927 | 1.83 |
ENST00000253354.1
|
BPIFB1
|
BPI fold containing family B, member 1 |
| chr18_-_69449517 | 1.82 |
ENST00000584810.1
|
RP11-723G8.2
|
Uncharacterized protein |
| chr15_+_45248880 | 1.80 |
ENST00000340827.3
|
C15orf43
|
chromosome 15 open reading frame 43 |
| chr3_-_107596910 | 1.78 |
ENST00000464359.2
ENST00000464823.1 ENST00000466155.1 ENST00000473528.2 ENST00000608306.1 ENST00000488852.1 ENST00000608137.1 ENST00000608307.1 ENST00000609429.1 ENST00000601385.1 ENST00000475362.1 ENST00000600240.1 ENST00000600749.1 |
LINC00635
|
long intergenic non-protein coding RNA 635 |
| chrX_+_27608490 | 1.77 |
ENST00000451261.2
|
DCAF8L2
|
DDB1 and CUL4 associated factor 8-like 2 |
| chr10_-_23528745 | 1.77 |
ENST00000376501.5
|
C10orf115
|
chromosome 10 open reading frame 115 |
| chrX_+_36246735 | 1.76 |
ENST00000378653.3
|
CXorf30
|
chromosome X open reading frame 30 |
| chr5_-_59668541 | 1.74 |
ENST00000514552.1
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chr5_-_131879205 | 1.71 |
ENST00000231454.1
|
IL5
|
interleukin 5 (colony-stimulating factor, eosinophil) |
| chr19_-_42947121 | 1.71 |
ENST00000601181.1
|
CXCL17
|
chemokine (C-X-C motif) ligand 17 |
| chr2_+_79252822 | 1.70 |
ENST00000272324.5
|
REG3G
|
regenerating islet-derived 3 gamma |
| chrX_+_107288280 | 1.69 |
ENST00000458383.1
|
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chr1_-_99999571 | 1.67 |
ENST00000438829.2
|
RP11-413P11.1
|
RP11-413P11.1 |
| chr2_+_74011316 | 1.67 |
ENST00000409561.1
|
C2orf78
|
chromosome 2 open reading frame 78 |
| chr13_+_75126978 | 1.67 |
ENST00000596240.1
ENST00000451336.2 |
LINC00347
|
long intergenic non-protein coding RNA 347 |
| chr2_+_149974684 | 1.66 |
ENST00000450639.1
|
LYPD6B
|
LY6/PLAUR domain containing 6B |
| chr1_+_94798754 | 1.65 |
ENST00000418242.1
|
RP11-148B18.3
|
RP11-148B18.3 |
| chr2_+_79252804 | 1.65 |
ENST00000393897.2
|
REG3G
|
regenerating islet-derived 3 gamma |
| chr9_-_34381536 | 1.64 |
ENST00000379126.3
ENST00000379127.1 ENST00000379133.3 |
C9orf24
|
chromosome 9 open reading frame 24 |
| chr9_-_34381511 | 1.60 |
ENST00000379124.1
|
C9orf24
|
chromosome 9 open reading frame 24 |
| chr10_+_118083919 | 1.60 |
ENST00000333254.3
|
CCDC172
|
coiled-coil domain containing 172 |
| chr6_-_49931818 | 1.59 |
ENST00000322066.3
|
DEFB114
|
defensin, beta 114 |
| chr11_-_102595512 | 1.56 |
ENST00000438475.2
|
MMP8
|
matrix metallopeptidase 8 (neutrophil collagenase) |
| chr5_-_156593273 | 1.56 |
ENST00000302938.4
|
FAM71B
|
family with sequence similarity 71, member B |
| chrX_+_79675965 | 1.56 |
ENST00000308293.5
|
FAM46D
|
family with sequence similarity 46, member D |
| chr10_+_32856764 | 1.54 |
ENST00000375030.2
ENST00000375028.3 |
C10orf68
|
Homo sapiens coiled-coil domain containing 7 (CCDC7), transcript variant 5, mRNA. |
| chr7_-_91808441 | 1.53 |
ENST00000437357.1
ENST00000458448.1 |
LRRD1
|
leucine-rich repeats and death domain containing 1 |
| chr12_+_85430110 | 1.53 |
ENST00000393212.3
ENST00000393217.2 |
LRRIQ1
|
leucine-rich repeats and IQ motif containing 1 |
| chr3_-_167191814 | 1.52 |
ENST00000466903.1
ENST00000264677.4 |
SERPINI2
|
serpin peptidase inhibitor, clade I (pancpin), member 2 |
| chr11_-_125648690 | 1.52 |
ENST00000436890.2
ENST00000358524.3 |
PATE2
|
prostate and testis expressed 2 |
| chr20_-_5426332 | 1.51 |
ENST00000420529.1
|
LINC00658
|
long intergenic non-protein coding RNA 658 |
| chrX_+_55246771 | 1.50 |
ENST00000289619.5
ENST00000374955.3 |
PAGE5
|
P antigen family, member 5 (prostate associated) |
| chr11_+_60102304 | 1.50 |
ENST00000300182.4
|
MS4A6E
|
membrane-spanning 4-domains, subfamily A, member 6E |
| chr9_-_23779367 | 1.50 |
ENST00000440102.1
|
ELAVL2
|
ELAV like neuron-specific RNA binding protein 2 |
| chr1_-_213020991 | 1.47 |
ENST00000332912.3
|
C1orf227
|
chromosome 1 open reading frame 227 |
| chrX_+_107288197 | 1.45 |
ENST00000415430.3
|
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chr11_+_60197040 | 1.45 |
ENST00000300190.2
|
MS4A5
|
membrane-spanning 4-domains, subfamily A, member 5 |
| chr20_+_17680587 | 1.42 |
ENST00000427254.1
ENST00000377805.3 |
BANF2
|
barrier to autointegration factor 2 |
| chr1_+_158815588 | 1.41 |
ENST00000438394.1
|
MNDA
|
myeloid cell nuclear differentiation antigen |
| chrX_+_36065053 | 1.40 |
ENST00000313548.4
|
CHDC2
|
calponin homology domain containing 2 |
| chr10_-_30918669 | 1.40 |
ENST00000375318.2
|
LYZL2
|
lysozyme-like 2 |
| chr6_-_49712147 | 1.39 |
ENST00000433368.2
ENST00000354620.4 |
CRISP3
|
cysteine-rich secretory protein 3 |
| chr8_-_57472154 | 1.39 |
ENST00000499425.1
ENST00000523664.1 ENST00000518943.1 ENST00000524338.1 |
LINC00968
|
long intergenic non-protein coding RNA 968 |
| chrX_+_36254051 | 1.39 |
ENST00000378657.4
|
CXorf30
|
chromosome X open reading frame 30 |
| chr13_+_43355732 | 1.38 |
ENST00000313851.1
|
FAM216B
|
family with sequence similarity 216, member B |
| chr7_+_16566449 | 1.37 |
ENST00000401542.2
|
LRRC72
|
leucine rich repeat containing 72 |
| chr1_-_89736434 | 1.37 |
ENST00000370459.3
|
GBP5
|
guanylate binding protein 5 |
| chr15_+_22368478 | 1.36 |
ENST00000332663.2
|
OR4M2
|
olfactory receptor, family 4, subfamily M, member 2 |
| chr6_+_139135648 | 1.34 |
ENST00000541398.1
|
ECT2L
|
epithelial cell transforming sequence 2 oncogene-like |
| chr4_+_71248795 | 1.34 |
ENST00000304915.3
|
SMR3B
|
submaxillary gland androgen regulated protein 3B |
| chr1_-_85358850 | 1.34 |
ENST00000370611.3
|
LPAR3
|
lysophosphatidic acid receptor 3 |
| chr16_-_52061283 | 1.34 |
ENST00000566314.1
|
C16orf97
|
chromosome 16 open reading frame 97 |
| chr13_+_43355683 | 1.31 |
ENST00000537894.1
|
FAM216B
|
family with sequence similarity 216, member B |
| chr6_+_132455526 | 1.31 |
ENST00000443303.1
|
LINC01013
|
long intergenic non-protein coding RNA 1013 |
| chr19_+_56368803 | 1.30 |
ENST00000587891.1
|
NLRP4
|
NLR family, pyrin domain containing 4 |
| chr4_+_68424434 | 1.29 |
ENST00000265404.2
ENST00000396225.1 |
STAP1
|
signal transducing adaptor family member 1 |
| chr8_-_19102999 | 1.27 |
ENST00000517949.1
|
RP11-1080G15.1
|
RP11-1080G15.1 |
| chr16_-_58328923 | 1.26 |
ENST00000567164.1
ENST00000219301.4 ENST00000569727.1 |
PRSS54
|
protease, serine, 54 |
| chr2_-_69180083 | 1.26 |
ENST00000328895.4
|
GKN2
|
gastrokine 2 |
| chr3_+_26735991 | 1.26 |
ENST00000456208.2
|
LRRC3B
|
leucine rich repeat containing 3B |
| chr8_+_36641842 | 1.25 |
ENST00000523973.1
ENST00000399881.3 |
KCNU1
|
potassium channel, subfamily U, member 1 |
| chr4_+_94125100 | 1.24 |
ENST00000512631.1
|
GRID2
|
glutamate receptor, ionotropic, delta 2 |
| chr12_-_45269430 | 1.23 |
ENST00000395487.2
|
NELL2
|
NEL-like 2 (chicken) |
| chr9_-_24545850 | 1.22 |
ENST00000543880.2
ENST00000418122.1 |
IZUMO3
|
IZUMO family member 3 |
| chr10_-_22498950 | 1.21 |
ENST00000422359.2
|
EBLN1
|
endogenous Bornavirus-like nucleoprotein 1 |
| chr1_+_170904612 | 1.21 |
ENST00000367759.4
ENST00000367758.3 |
MROH9
|
maestro heat-like repeat family member 9 |
| chr3_+_14716606 | 1.18 |
ENST00000253697.3
ENST00000435614.1 ENST00000412910.1 |
C3orf20
|
chromosome 3 open reading frame 20 |
| chr5_-_61031495 | 1.18 |
ENST00000506550.1
ENST00000512882.2 |
CTD-2170G1.2
|
CTD-2170G1.2 |
| chr3_+_94657016 | 1.18 |
ENST00000462219.1
|
LINC00879
|
long intergenic non-protein coding RNA 879 |
| chr10_-_75118471 | 1.17 |
ENST00000340329.3
|
TTC18
|
tetratricopeptide repeat domain 18 |
| chr20_+_55904815 | 1.17 |
ENST00000371263.3
ENST00000345868.4 ENST00000371260.4 ENST00000418127.1 |
SPO11
|
SPO11 meiotic protein covalently bound to DSB |
| chr12_+_8608522 | 1.17 |
ENST00000382073.3
|
CLEC6A
|
C-type lectin domain family 6, member A |
| chr17_+_61473104 | 1.16 |
ENST00000583016.1
|
TANC2
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2 |
| chr17_-_46262541 | 1.16 |
ENST00000579336.1
|
SKAP1
|
src kinase associated phosphoprotein 1 |
| chr6_-_49712072 | 1.15 |
ENST00000423399.2
|
CRISP3
|
cysteine-rich secretory protein 3 |
| chr7_+_142000747 | 1.15 |
ENST00000455382.2
|
TRBV2
|
T cell receptor beta variable 2 |
| chr4_-_122148620 | 1.14 |
ENST00000509841.1
|
TNIP3
|
TNFAIP3 interacting protein 3 |
| chr9_-_107299137 | 1.14 |
ENST00000374781.2
|
OR13C3
|
olfactory receptor, family 13, subfamily C, member 3 |
| chr3_-_157221128 | 1.13 |
ENST00000392833.2
ENST00000362010.2 |
VEPH1
|
ventricular zone expressed PH domain-containing 1 |
| chr20_+_17680599 | 1.13 |
ENST00000246090.5
|
BANF2
|
barrier to autointegration factor 2 |
| chr11_+_60282862 | 1.12 |
ENST00000378186.2
ENST00000378185.2 ENST00000437058.2 ENST00000527948.1 |
MS4A13
|
membrane-spanning 4-domains, subfamily A, member 13 |
| chr15_-_41522889 | 1.11 |
ENST00000458580.2
ENST00000314992.5 ENST00000558396.1 |
EXD1
|
exonuclease 3'-5' domain containing 1 |
| chr6_-_136571400 | 1.10 |
ENST00000418509.2
ENST00000420702.1 ENST00000451457.2 |
MTFR2
|
mitochondrial fission regulator 2 |
| chr19_-_56704421 | 1.10 |
ENST00000358992.3
|
ZSCAN5B
|
zinc finger and SCAN domain containing 5B |
| chr7_-_48068671 | 1.08 |
ENST00000297325.4
|
SUN3
|
Sad1 and UNC84 domain containing 3 |
| chr6_+_15401075 | 1.07 |
ENST00000541660.1
|
JARID2
|
jumonji, AT rich interactive domain 2 |
| chr6_-_130543958 | 1.07 |
ENST00000437477.2
ENST00000439090.2 |
SAMD3
|
sterile alpha motif domain containing 3 |
| chr7_-_16921601 | 1.06 |
ENST00000402239.3
ENST00000310398.2 ENST00000414935.1 |
AGR3
|
anterior gradient 3 |
| chr1_-_149459549 | 1.06 |
ENST00000369175.3
|
FAM72C
|
family with sequence similarity 72, member C |
| chr22_-_50523843 | 1.05 |
ENST00000535444.1
ENST00000431262.2 |
MLC1
|
megalencephalic leukoencephalopathy with subcortical cysts 1 |
| chr3_+_130279178 | 1.04 |
ENST00000358511.6
ENST00000453409.2 |
COL6A6
|
collagen, type VI, alpha 6 |
| chr2_-_166930131 | 1.04 |
ENST00000303395.4
ENST00000409050.1 ENST00000423058.2 ENST00000375405.3 |
SCN1A
|
sodium channel, voltage-gated, type I, alpha subunit |
| chr6_+_132455118 | 1.03 |
ENST00000458028.1
|
LINC01013
|
long intergenic non-protein coding RNA 1013 |
| chr6_-_49712091 | 1.03 |
ENST00000371159.4
|
CRISP3
|
cysteine-rich secretory protein 3 |
| chr5_+_54398463 | 1.02 |
ENST00000274306.6
|
GZMA
|
granzyme A (granzyme 1, cytotoxic T-lymphocyte-associated serine esterase 3) |
| chr11_+_71544246 | 1.01 |
ENST00000328698.1
|
DEFB108B
|
defensin, beta 108B |
| chr12_+_9980113 | 1.01 |
ENST00000537723.1
|
KLRF1
|
killer cell lectin-like receptor subfamily F, member 1 |
| chr12_+_9980069 | 0.99 |
ENST00000354855.3
ENST00000324214.4 ENST00000279544.3 |
KLRF1
|
killer cell lectin-like receptor subfamily F, member 1 |
| chr12_+_10460549 | 0.99 |
ENST00000543420.1
ENST00000543777.1 |
KLRD1
|
killer cell lectin-like receptor subfamily D, member 1 |
| chr16_+_31366536 | 0.99 |
ENST00000562522.1
|
ITGAX
|
integrin, alpha X (complement component 3 receptor 4 subunit) |
| chr1_+_77333117 | 0.99 |
ENST00000477717.1
|
ST6GALNAC5
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 5 |
| chr14_+_39944025 | 0.98 |
ENST00000554328.1
ENST00000556620.1 ENST00000557197.1 |
RP11-111A21.1
|
RP11-111A21.1 |
| chr12_+_81664424 | 0.98 |
ENST00000549161.1
ENST00000550138.1 |
RP11-121G22.3
|
RP11-121G22.3 |
| chr19_+_55014085 | 0.98 |
ENST00000351841.2
|
LAIR2
|
leukocyte-associated immunoglobulin-like receptor 2 |
| chr6_-_49712123 | 0.97 |
ENST00000263045.4
|
CRISP3
|
cysteine-rich secretory protein 3 |
| chr16_-_58328884 | 0.97 |
ENST00000569079.1
|
PRSS54
|
protease, serine, 54 |
| chr12_-_4758159 | 0.97 |
ENST00000545990.2
|
AKAP3
|
A kinase (PRKA) anchor protein 3 |
| chr2_-_69180012 | 0.97 |
ENST00000481498.1
|
GKN2
|
gastrokine 2 |
| chr5_-_131892501 | 0.97 |
ENST00000450655.1
|
IL5
|
interleukin 5 (colony-stimulating factor, eosinophil) |
| chr11_+_60197069 | 0.96 |
ENST00000528905.1
ENST00000528093.1 |
MS4A5
|
membrane-spanning 4-domains, subfamily A, member 5 |
| chrX_+_8432871 | 0.96 |
ENST00000381032.1
ENST00000453306.1 ENST00000444481.1 |
VCX3B
|
variable charge, X-linked 3B |
| chr16_-_58328870 | 0.95 |
ENST00000543437.1
|
PRSS54
|
protease, serine, 54 |
| chr10_+_97759848 | 0.94 |
ENST00000424464.1
ENST00000410012.2 ENST00000344386.3 |
CC2D2B
|
coiled-coil and C2 domain containing 2B |
| chr14_-_25045446 | 0.94 |
ENST00000216336.2
|
CTSG
|
cathepsin G |
| chr10_+_97709725 | 0.94 |
ENST00000472454.2
|
RP11-248J23.6
|
Protein LOC100652732 |
| chr10_+_47746929 | 0.92 |
ENST00000340243.6
ENST00000374277.5 ENST00000449464.2 ENST00000538825.1 ENST00000335083.5 |
ANXA8L2
AL603965.1
|
annexin A8-like 2 Protein LOC100996760 |
| chr2_-_39456673 | 0.90 |
ENST00000378803.1
ENST00000395035.3 |
CDKL4
|
cyclin-dependent kinase-like 4 |
| chr14_+_20295608 | 0.88 |
ENST00000568211.1
ENST00000315947.1 |
OR4N2
|
olfactory receptor, family 4, subfamily N, member 2 |
| chr5_+_57787254 | 0.88 |
ENST00000502276.1
ENST00000396776.2 ENST00000511930.1 |
GAPT
|
GRB2-binding adaptor protein, transmembrane |
| chr10_+_106113515 | 0.88 |
ENST00000369704.3
ENST00000312902.5 |
CCDC147
|
coiled-coil domain containing 147 |
| chrX_+_55246818 | 0.87 |
ENST00000374952.1
|
PAGE5
|
P antigen family, member 5 (prostate associated) |
| chr22_-_32766972 | 0.87 |
ENST00000382084.4
ENST00000382086.2 |
RFPL3S
|
RFPL3 antisense |
| chr8_+_7682694 | 0.87 |
ENST00000335186.2
|
DEFB106A
|
defensin, beta 106A |
| chr6_+_31707725 | 0.87 |
ENST00000375755.3
ENST00000375742.3 ENST00000375750.3 ENST00000425703.1 ENST00000534153.4 ENST00000375703.3 ENST00000375740.3 |
MSH5
|
mutS homolog 5 |
| chr12_+_40787194 | 0.86 |
ENST00000425730.2
ENST00000454784.4 |
MUC19
|
mucin 19, oligomeric |
| chr7_-_38295997 | 0.86 |
ENST00000390334.1
|
TRGJP2
|
T cell receptor gamma joining P2 |
| chr3_-_108672742 | 0.85 |
ENST00000261047.3
|
GUCA1C
|
guanylate cyclase activator 1C |
| chr21_+_43823983 | 0.85 |
ENST00000291535.6
ENST00000450356.1 ENST00000319294.6 ENST00000398367.1 |
UBASH3A
|
ubiquitin associated and SH3 domain containing A |
| chr14_-_21029090 | 0.84 |
ENST00000338904.3
ENST00000554964.1 ENST00000555230.1 ENST00000557068.1 ENST00000404716.3 ENST00000556208.1 ENST00000553541.1 ENST00000429244.2 ENST00000553706.1 ENST00000557209.1 |
RNASE9
|
ribonuclease, RNase A family, 9 (non-active) |
| chr22_-_32767017 | 0.83 |
ENST00000400234.1
|
RFPL3S
|
RFPL3 antisense |
| chr7_+_50135433 | 0.83 |
ENST00000297001.6
|
C7orf72
|
chromosome 7 open reading frame 72 |
| chrX_-_142804516 | 0.83 |
ENST00000370498.1
|
SPANXN2
|
SPANX family, member N2 |
| chrX_-_107682702 | 0.82 |
ENST00000372216.4
|
COL4A6
|
collagen, type IV, alpha 6 |
| chr12_-_45315625 | 0.81 |
ENST00000552993.1
|
NELL2
|
NEL-like 2 (chicken) |
| chrX_+_114423963 | 0.81 |
ENST00000424776.3
|
RBMXL3
|
RNA binding motif protein, X-linked-like 3 |
| chr12_-_15082050 | 0.81 |
ENST00000540097.1
|
ERP27
|
endoplasmic reticulum protein 27 |
| chr2_-_46621306 | 0.81 |
ENST00000418415.1
|
AC016912.3
|
AC016912.3 |
| chr1_+_209602771 | 0.80 |
ENST00000440276.1
|
MIR205HG
|
MIR205 host gene (non-protein coding) |
| chr12_-_45269251 | 0.80 |
ENST00000553120.1
|
NELL2
|
NEL-like 2 (chicken) |
| chr6_-_133055896 | 0.79 |
ENST00000367927.5
ENST00000425515.2 ENST00000207771.3 ENST00000392393.3 ENST00000450865.2 ENST00000392394.2 |
VNN3
|
vanin 3 |
| chr17_-_71223839 | 0.79 |
ENST00000579872.1
ENST00000580032.1 |
FAM104A
|
family with sequence similarity 104, member A |
| chr18_-_19994830 | 0.79 |
ENST00000525417.1
|
CTAGE1
|
cutaneous T-cell lymphoma-associated antigen 1 |
| chr4_+_100737954 | 0.79 |
ENST00000296414.7
ENST00000512369.1 |
DAPP1
|
dual adaptor of phosphotyrosine and 3-phosphoinositides |
| chr5_-_19988339 | 0.78 |
ENST00000382275.1
|
CDH18
|
cadherin 18, type 2 |
| chrX_-_154493791 | 0.78 |
ENST00000369454.3
|
RAB39B
|
RAB39B, member RAS oncogene family |
| chr11_-_26744908 | 0.77 |
ENST00000533617.1
|
SLC5A12
|
solute carrier family 5 (sodium/monocarboxylate cotransporter), member 12 |
| chr11_+_128563652 | 0.76 |
ENST00000527786.2
|
FLI1
|
Fli-1 proto-oncogene, ETS transcription factor |
| chr6_-_133084580 | 0.76 |
ENST00000525270.1
ENST00000530536.1 ENST00000524919.1 |
VNN2
|
vanin 2 |
| chr20_+_15177480 | 0.76 |
ENST00000402914.1
|
MACROD2
|
MACRO domain containing 2 |
| chr2_-_208989225 | 0.75 |
ENST00000264376.4
|
CRYGD
|
crystallin, gamma D |
| chr2_-_118943930 | 0.75 |
ENST00000449075.1
ENST00000414886.1 ENST00000449819.1 |
AC093901.1
|
AC093901.1 |
| chr9_+_71357171 | 0.75 |
ENST00000440050.1
|
PIP5K1B
|
phosphatidylinositol-4-phosphate 5-kinase, type I, beta |
| chr3_+_94657086 | 0.74 |
ENST00000463200.1
|
LINC00879
|
long intergenic non-protein coding RNA 879 |
| chr2_+_90024732 | 0.74 |
ENST00000390268.2
|
IGKV2D-26
|
immunoglobulin kappa variable 2D-26 |
| chr22_+_32754139 | 0.74 |
ENST00000382088.3
|
RFPL3
|
ret finger protein-like 3 |
| chr6_+_26199737 | 0.73 |
ENST00000359985.1
|
HIST1H2BF
|
histone cluster 1, H2bf |
| chr12_+_70219052 | 0.73 |
ENST00000552032.2
ENST00000547771.2 |
MYRFL
|
myelin regulatory factor-like |
| chr12_-_10542617 | 0.72 |
ENST00000240618.6
|
KLRK1
|
killer cell lectin-like receptor subfamily K, member 1 |
| chr19_+_55014013 | 0.71 |
ENST00000301202.2
|
LAIR2
|
leukocyte-associated immunoglobulin-like receptor 2 |
| chr12_-_10607084 | 0.70 |
ENST00000408006.3
ENST00000544822.1 ENST00000536188.1 |
KLRC1
|
killer cell lectin-like receptor subfamily C, member 1 |
| chr14_+_22788560 | 0.70 |
ENST00000390468.1
|
TRAV41
|
T cell receptor alpha variable 41 |
| chr15_-_58571445 | 0.70 |
ENST00000558231.1
|
ALDH1A2
|
aldehyde dehydrogenase 1 family, member A2 |
| chr1_+_152178320 | 0.70 |
ENST00000429352.1
|
RP11-107M16.2
|
RP11-107M16.2 |
| chr7_+_135710494 | 0.70 |
ENST00000440744.2
|
AC024084.1
|
AC024084.1 |
| chr12_-_10562356 | 0.69 |
ENST00000309384.1
|
KLRC4
|
killer cell lectin-like receptor subfamily C, member 4 |
| chr8_+_55528627 | 0.69 |
ENST00000220676.1
|
RP1
|
retinitis pigmentosa 1 (autosomal dominant) |
| chr18_+_61223393 | 0.69 |
ENST00000269491.1
ENST00000382768.1 |
SERPINB12
|
serpin peptidase inhibitor, clade B (ovalbumin), member 12 |
| chr22_-_32860427 | 0.69 |
ENST00000534972.1
ENST00000397450.1 ENST00000397452.1 |
BPIFC
|
BPI fold containing family C |
| chr22_-_23922448 | 0.67 |
ENST00000438703.1
ENST00000330377.2 |
IGLL1
|
immunoglobulin lambda-like polypeptide 1 |
| chr17_-_43339453 | 0.67 |
ENST00000543122.1
|
SPATA32
|
spermatogenesis associated 32 |
| chr2_+_101223490 | 0.67 |
ENST00000414647.1
ENST00000424342.1 |
AC068538.4
|
AC068538.4 |
| chr14_+_56584414 | 0.67 |
ENST00000559044.1
|
PELI2
|
pellino E3 ubiquitin protein ligase family member 2 |
| chr22_-_29457832 | 0.66 |
ENST00000216071.4
|
C22orf31
|
chromosome 22 open reading frame 31 |
| chr7_-_130066571 | 0.66 |
ENST00000492389.1
|
CEP41
|
centrosomal protein 41kDa |
| chr4_-_90756769 | 0.66 |
ENST00000345009.4
ENST00000505199.1 ENST00000502987.1 |
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor) |
Network of associatons between targets according to the STRING database.
First level regulatory network of ARID5A
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.3 | GO:0044278 | cell wall disruption in other organism(GO:0044278) |
| 0.9 | 2.7 | GO:0045645 | regulation of eosinophil differentiation(GO:0045643) positive regulation of eosinophil differentiation(GO:0045645) |
| 0.7 | 4.7 | GO:0007290 | spermatid nucleus elongation(GO:0007290) |
| 0.6 | 2.6 | GO:0035425 | autocrine signaling(GO:0035425) |
| 0.5 | 1.4 | GO:0072616 | interleukin-18 secretion(GO:0072616) |
| 0.4 | 1.3 | GO:1903980 | negative regulation of macrophage colony-stimulating factor signaling pathway(GO:1902227) negative regulation of response to macrophage colony-stimulating factor(GO:1903970) negative regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903973) positive regulation of microglial cell activation(GO:1903980) |
| 0.4 | 2.5 | GO:0061760 | antifungal innate immune response(GO:0061760) |
| 0.4 | 7.8 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.3 | 1.2 | GO:0000706 | meiotic DNA double-strand break processing(GO:0000706) double-strand break repair involved in meiotic recombination(GO:1990918) |
| 0.2 | 0.7 | GO:2000502 | negative regulation of natural killer cell chemotaxis(GO:2000502) |
| 0.2 | 0.7 | GO:0060796 | regulation of transcription involved in primary germ layer cell fate commitment(GO:0060796) |
| 0.2 | 3.4 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.2 | 1.8 | GO:0034144 | negative regulation of toll-like receptor 4 signaling pathway(GO:0034144) |
| 0.2 | 0.9 | GO:0002322 | B cell proliferation involved in immune response(GO:0002322) |
| 0.2 | 0.8 | GO:0060474 | positive regulation of sperm motility involved in capacitation(GO:0060474) |
| 0.2 | 0.5 | GO:0001812 | positive regulation of type I hypersensitivity(GO:0001812) B cell cytokine production(GO:0002368) |
| 0.2 | 0.8 | GO:0000117 | regulation of transcription involved in G2/M transition of mitotic cell cycle(GO:0000117) |
| 0.2 | 4.7 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.2 | 1.6 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.1 | 1.2 | GO:1900454 | positive regulation of long term synaptic depression(GO:1900454) |
| 0.1 | 1.1 | GO:0038195 | urokinase plasminogen activator signaling pathway(GO:0038195) |
| 0.1 | 0.7 | GO:1903285 | negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of glutathione peroxidase activity(GO:1903282) positive regulation of glutathione peroxidase activity(GO:1903284) positive regulation of hydrogen peroxide catabolic process(GO:1903285) positive regulation of peroxidase activity(GO:2000470) |
| 0.1 | 2.1 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.1 | 0.9 | GO:0070942 | neutrophil mediated cytotoxicity(GO:0070942) neutrophil mediated killing of symbiont cell(GO:0070943) neutrophil mediated killing of bacterium(GO:0070944) |
| 0.1 | 0.2 | GO:0032915 | positive regulation of transforming growth factor beta2 production(GO:0032915) |
| 0.1 | 0.6 | GO:1904383 | response to sodium phosphate(GO:1904383) |
| 0.1 | 0.3 | GO:1903926 | signal transduction involved in intra-S DNA damage checkpoint(GO:0072428) response to bisphenol A(GO:1903925) cellular response to bisphenol A(GO:1903926) |
| 0.1 | 1.5 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.1 | 0.9 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.1 | 0.5 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.1 | 0.7 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.1 | 0.8 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.1 | 0.4 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.1 | 1.4 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.1 | 0.6 | GO:0014038 | regulation of Schwann cell differentiation(GO:0014038) |
| 0.1 | 1.3 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.1 | 0.4 | GO:0003409 | optic cup structural organization(GO:0003409) |
| 0.1 | 1.3 | GO:0031284 | positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.1 | 4.8 | GO:2000816 | negative regulation of mitotic sister chromatid separation(GO:2000816) |
| 0.1 | 1.9 | GO:0010759 | positive regulation of macrophage chemotaxis(GO:0010759) |
| 0.1 | 2.5 | GO:0030728 | ovulation(GO:0030728) |
| 0.1 | 1.0 | GO:0032071 | regulation of endodeoxyribonuclease activity(GO:0032071) |
| 0.1 | 0.7 | GO:0035799 | ureter maturation(GO:0035799) |
| 0.1 | 1.1 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.1 | 1.5 | GO:0035090 | maintenance of apical/basal cell polarity(GO:0035090) maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.1 | 0.6 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 0.1 | 0.4 | GO:0060369 | positive regulation of natural killer cell cytokine production(GO:0002729) positive regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060369) |
| 0.1 | 0.2 | GO:0007113 | endomitotic cell cycle(GO:0007113) |
| 0.1 | 1.4 | GO:0030889 | negative regulation of B cell proliferation(GO:0030889) |
| 0.1 | 0.3 | GO:1900041 | negative regulation of interleukin-2 secretion(GO:1900041) |
| 0.1 | 1.1 | GO:0047484 | regulation of response to osmotic stress(GO:0047484) |
| 0.1 | 0.2 | GO:1900275 | negative regulation of phospholipase C activity(GO:1900275) regulation of proteinase activated receptor activity(GO:1900276) negative regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900737) |
| 0.1 | 0.4 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.1 | 0.3 | GO:0007402 | ganglion mother cell fate determination(GO:0007402) |
| 0.1 | 0.7 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.1 | 0.6 | GO:0003350 | pulmonary myocardium development(GO:0003350) |
| 0.0 | 0.1 | GO:0021503 | neural fold bending(GO:0021503) |
| 0.0 | 2.4 | GO:0030261 | chromosome condensation(GO:0030261) |
| 0.0 | 0.2 | GO:2000639 | regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.0 | 0.5 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.0 | 0.7 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.0 | 0.3 | GO:0071461 | cellular response to redox state(GO:0071461) |
| 0.0 | 0.6 | GO:0035873 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) |
| 0.0 | 0.2 | GO:0042357 | thiamine diphosphate metabolic process(GO:0042357) |
| 0.0 | 1.1 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.0 | 1.1 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.0 | 0.3 | GO:0043696 | dedifferentiation(GO:0043696) cell dedifferentiation(GO:0043697) |
| 0.0 | 0.4 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.0 | 0.2 | GO:0050773 | regulation of dendrite development(GO:0050773) |
| 0.0 | 1.0 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.0 | 0.5 | GO:0097105 | presynaptic membrane assembly(GO:0097105) |
| 0.0 | 0.4 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 1.1 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.0 | 0.9 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 | 2.0 | GO:0035036 | sperm-egg recognition(GO:0035036) |
| 0.0 | 0.4 | GO:0048245 | eosinophil chemotaxis(GO:0048245) |
| 0.0 | 0.4 | GO:0060050 | positive regulation of protein glycosylation(GO:0060050) |
| 0.0 | 0.1 | GO:1904844 | response to L-glutamine(GO:1904844) cellular response to L-glutamine(GO:1904845) |
| 0.0 | 0.2 | GO:0010908 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.0 | 0.3 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 0.0 | 2.9 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.4 | GO:1900086 | positive regulation of peptidyl-tyrosine autophosphorylation(GO:1900086) |
| 0.0 | 1.2 | GO:0050832 | defense response to fungus(GO:0050832) |
| 0.0 | 0.4 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.0 | 0.3 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.0 | 0.6 | GO:0033189 | response to vitamin A(GO:0033189) |
| 0.0 | 0.8 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.0 | 1.1 | GO:0034142 | toll-like receptor 4 signaling pathway(GO:0034142) |
| 0.0 | 1.7 | GO:0035987 | endodermal cell differentiation(GO:0035987) |
| 0.0 | 0.2 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.0 | 0.1 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.0 | 1.3 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 0.3 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.0 | 3.1 | GO:0031295 | T cell costimulation(GO:0031295) |
| 0.0 | 0.1 | GO:0045726 | positive regulation of integrin biosynthetic process(GO:0045726) |
| 0.0 | 0.2 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.0 | 0.5 | GO:0060122 | inner ear receptor stereocilium organization(GO:0060122) |
| 0.0 | 0.7 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.0 | 1.5 | GO:0099601 | regulation of neurotransmitter receptor activity(GO:0099601) |
| 0.0 | 2.3 | GO:0030521 | androgen receptor signaling pathway(GO:0030521) |
| 0.0 | 0.5 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.0 | 0.6 | GO:2000401 | regulation of lymphocyte migration(GO:2000401) |
| 0.0 | 4.2 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.0 | GO:0098507 | polynucleotide 5' dephosphorylation(GO:0098507) |
| 0.0 | 0.3 | GO:0071360 | cellular response to exogenous dsRNA(GO:0071360) |
| 0.0 | 0.2 | GO:0019216 | regulation of lipid metabolic process(GO:0019216) |
| 0.0 | 1.2 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.0 | 0.7 | GO:0070306 | lens fiber cell differentiation(GO:0070306) |
| 0.0 | 1.0 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 0.0 | GO:1902173 | negative regulation of keratinocyte apoptotic process(GO:1902173) |
| 0.0 | 0.1 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.0 | 0.1 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 0.0 | 0.1 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.0 | 1.0 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.0 | 0.3 | GO:0086069 | bundle of His cell to Purkinje myocyte communication(GO:0086069) |
| 0.0 | 0.1 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.0 | 0.4 | GO:0007202 | activation of phospholipase C activity(GO:0007202) |
| 0.0 | 0.6 | GO:0006779 | porphyrin-containing compound biosynthetic process(GO:0006779) |
| 0.0 | 0.1 | GO:0046878 | positive regulation of saliva secretion(GO:0046878) |
| 0.0 | 0.2 | GO:0045475 | locomotor rhythm(GO:0045475) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 4.9 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.3 | 1.1 | GO:1990923 | PET complex(GO:1990923) |
| 0.2 | 0.6 | GO:0097679 | other organism cytoplasm(GO:0097679) |
| 0.2 | 3.4 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.1 | 0.9 | GO:1990742 | microvesicle(GO:1990742) |
| 0.1 | 0.6 | GO:0097125 | cyclin B1-CDK1 complex(GO:0097125) |
| 0.1 | 0.9 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.1 | 8.6 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.1 | 0.5 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.1 | 0.7 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.1 | 1.0 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.1 | 0.8 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.1 | 0.2 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 0.1 | 1.6 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.1 | 1.5 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.1 | 2.3 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
| 0.1 | 1.1 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.1 | 0.2 | GO:0072517 | viral factory(GO:0039713) cytoplasmic viral factory(GO:0039714) host cell viral assembly compartment(GO:0072517) |
| 0.1 | 0.9 | GO:0032300 | mismatch repair complex(GO:0032300) |
| 0.0 | 1.0 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 1.1 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.5 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 0.5 | GO:0060091 | kinocilium(GO:0060091) |
| 0.0 | 0.2 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.0 | 1.2 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 0.3 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.0 | 0.2 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 0.1 | GO:1902912 | pyruvate kinase complex(GO:1902912) |
| 0.0 | 0.4 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 0.8 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 0.2 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.0 | 0.2 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 1.6 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 1.1 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 1.8 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 1.3 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.7 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 2.8 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.0 | 0.3 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.0 | 0.2 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.0 | 0.7 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 0.2 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.0 | 0.7 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 0.6 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 0.3 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 0.4 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 1.0 | GO:0045178 | basal part of cell(GO:0045178) |
| 0.0 | 1.4 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 1.2 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.7 | GO:0005137 | interleukin-5 receptor binding(GO:0005137) |
| 0.4 | 2.2 | GO:0023024 | MHC class I protein complex binding(GO:0023024) |
| 0.4 | 2.1 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.3 | 1.2 | GO:0003918 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.2 | 0.7 | GO:0052810 | 1-phosphatidylinositol-5-kinase activity(GO:0052810) |
| 0.2 | 1.1 | GO:0017161 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) inositol-1,3,4-trisphosphate 4-phosphatase activity(GO:0017161) inositol-3,4-bisphosphate 4-phosphatase activity(GO:0052828) |
| 0.2 | 1.3 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.2 | 3.3 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.2 | 0.9 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.2 | 1.3 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.2 | 1.1 | GO:0030377 | urokinase plasminogen activator receptor activity(GO:0030377) |
| 0.1 | 2.5 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.1 | 1.2 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.1 | 1.8 | GO:0032393 | MHC class I receptor activity(GO:0032393) |
| 0.1 | 0.7 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.1 | 1.3 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.1 | 0.3 | GO:0098770 | FBXO family protein binding(GO:0098770) |
| 0.1 | 0.6 | GO:0005539 | glycosaminoglycan binding(GO:0005539) |
| 0.1 | 1.5 | GO:0099602 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.1 | 6.9 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.1 | 0.6 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.1 | 1.5 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.1 | 1.6 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.1 | 7.8 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.1 | 2.5 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 0.4 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.1 | 9.8 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 0.2 | GO:0019781 | NEDD8 activating enzyme activity(GO:0019781) |
| 0.1 | 2.3 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.0 | 0.2 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 0.0 | 0.6 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.5 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.0 | 0.8 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.1 | GO:0004531 | deoxyribonuclease II activity(GO:0004531) |
| 0.0 | 0.2 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.0 | 0.9 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.0 | 0.5 | GO:0015252 | hydrogen ion channel activity(GO:0015252) |
| 0.0 | 0.4 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.0 | 0.2 | GO:0032558 | adenyl deoxyribonucleotide binding(GO:0032558) dATP binding(GO:0032564) |
| 0.0 | 0.5 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.4 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 1.1 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.4 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.4 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 1.0 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 0.6 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 0.3 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.0 | 0.4 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.7 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.8 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.2 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.0 | 0.4 | GO:0016615 | malate dehydrogenase activity(GO:0016615) |
| 0.0 | 0.6 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.3 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.0 | 0.4 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.0 | 0.6 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.2 | GO:0016618 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.0 | 0.8 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 1.7 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 0.3 | GO:0008047 | enzyme activator activity(GO:0008047) |
| 0.0 | 0.3 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.0 | 0.3 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.1 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.0 | 0.7 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.0 | 1.1 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.0 | 2.8 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.2 | GO:0016778 | diphosphotransferase activity(GO:0016778) |
| 0.0 | 0.3 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.0 | 0.4 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 1.2 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.2 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) |
| 0.0 | 0.7 | GO:0005234 | extracellular-glutamate-gated ion channel activity(GO:0005234) |
| 0.0 | 0.2 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.5 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.0 | 0.9 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 1.0 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 4.2 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 0.0 | GO:0004651 | polynucleotide 5'-phosphatase activity(GO:0004651) |
| 0.0 | 0.5 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.2 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.0 | 1.3 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 0.9 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.0 | 0.2 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 0.3 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 1.0 | GO:0008408 | 3'-5' exonuclease activity(GO:0008408) |
| 0.0 | 0.1 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.0 | 0.3 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.0 | 0.2 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 0.0 | 0.1 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.0 | 0.2 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.3 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.1 | 2.2 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 15.2 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 2.3 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.6 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 8.5 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 1.9 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.1 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 1.3 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.0 | 1.3 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.3 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 1.5 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 0.7 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.3 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 1.3 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.0 | 0.2 | PID ATR PATHWAY | ATR signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.0 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.1 | 4.0 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.1 | 2.9 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 1.1 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 0.6 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.6 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.0 | 0.3 | REACTOME P53 INDEPENDENT G1 S DNA DAMAGE CHECKPOINT | Genes involved in p53-Independent G1/S DNA damage checkpoint |
| 0.0 | 2.3 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.7 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 0.1 | REACTOME AUTODEGRADATION OF THE E3 UBIQUITIN LIGASE COP1 | Genes involved in Autodegradation of the E3 ubiquitin ligase COP1 |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 2.6 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 2.8 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 2.5 | REACTOME APC C CDH1 MEDIATED DEGRADATION OF CDC20 AND OTHER APC C CDH1 TARGETED PROTEINS IN LATE MITOSIS EARLY G1 | Genes involved in APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 |
| 0.0 | 1.1 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.0 | 1.4 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.6 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 1.6 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 0.6 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 0.8 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 3.4 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.0 | 0.7 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 1.6 | REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | Genes involved in Cell surface interactions at the vascular wall |
| 0.0 | 0.4 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.5 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |