Project
Illumina Body Map 2: averaged replicates
Navigation
Downloads
Results for AR_NR3C2
Z-value: 2.49
Transcription factors associated with AR_NR3C2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
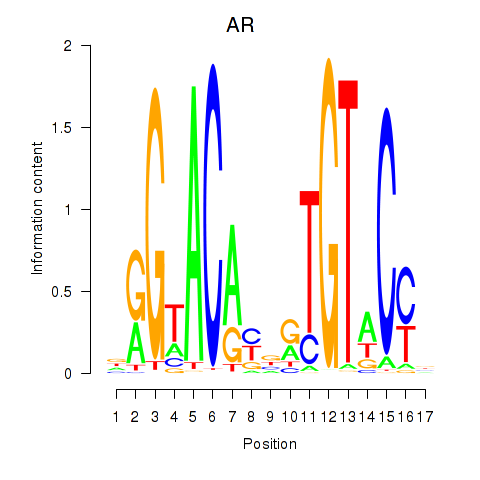
AR
|
ENSG00000169083.11 | androgen receptor |
|
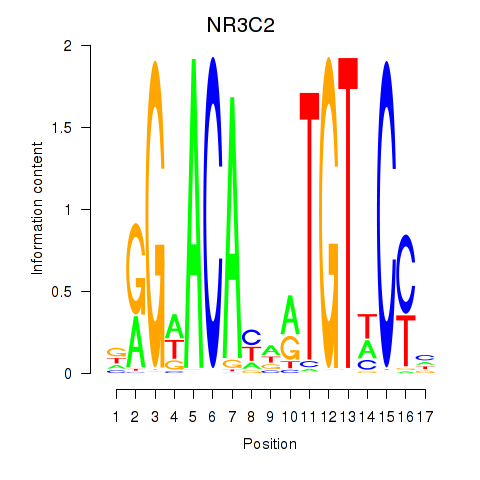
NR3C2
|
ENSG00000151623.10 | nuclear receptor subfamily 3 group C member 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| AR | hg19_v2_chrX_+_66764375_66764465 | 0.57 | 6.1e-04 | Click! |
| NR3C2 | hg19_v2_chr4_-_149363376_149363492 | -0.36 | 4.4e-02 | Click! |
Activity profile of AR_NR3C2 motif
Sorted Z-values of AR_NR3C2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_+_18287721 | 16.48 |
ENST00000356524.4
|
SAA1
|
serum amyloid A1 |
| chr11_+_18287801 | 16.47 |
ENST00000532858.1
ENST00000405158.2 |
SAA1
|
serum amyloid A1 |
| chr12_+_53443963 | 11.95 |
ENST00000546602.1
ENST00000552570.1 ENST00000549700.1 |
TENC1
|
tensin like C1 domain containing phosphatase (tensin 2) |
| chr12_+_53443680 | 11.91 |
ENST00000314250.6
ENST00000451358.1 |
TENC1
|
tensin like C1 domain containing phosphatase (tensin 2) |
| chr16_+_56666563 | 11.40 |
ENST00000570233.1
|
MT1M
|
metallothionein 1M |
| chr16_+_56716336 | 8.54 |
ENST00000394485.4
ENST00000562939.1 |
MT1X
|
metallothionein 1X |
| chr13_-_40924439 | 7.98 |
ENST00000400432.3
|
RP11-172E9.2
|
RP11-172E9.2 |
| chr1_+_22979474 | 7.90 |
ENST00000509305.1
|
C1QB
|
complement component 1, q subcomponent, B chain |
| chr10_+_60759378 | 7.75 |
ENST00000432535.1
|
LINC00844
|
long intergenic non-protein coding RNA 844 |
| chr18_-_48346415 | 7.59 |
ENST00000431965.2
ENST00000436348.2 |
MRO
|
maestro |
| chr19_+_35630628 | 7.46 |
ENST00000588715.1
ENST00000588607.1 |
FXYD1
|
FXYD domain containing ion transport regulator 1 |
| chr1_+_22979676 | 7.33 |
ENST00000432749.2
ENST00000314933.6 |
C1QB
|
complement component 1, q subcomponent, B chain |
| chr2_+_238395879 | 6.73 |
ENST00000445024.2
ENST00000338530.4 ENST00000409373.1 |
MLPH
|
melanophilin |
| chr3_-_58563094 | 6.47 |
ENST00000464064.1
|
FAM107A
|
family with sequence similarity 107, member A |
| chr19_+_35630926 | 6.43 |
ENST00000588081.1
ENST00000589121.1 |
FXYD1
|
FXYD domain containing ion transport regulator 1 |
| chr19_+_35630344 | 5.28 |
ENST00000455515.2
|
FXYD1
|
FXYD domain containing ion transport regulator 1 |
| chr16_+_56659687 | 5.20 |
ENST00000568293.1
ENST00000330439.6 |
MT1E
|
metallothionein 1E |
| chr1_+_1846519 | 5.06 |
ENST00000378604.3
|
CALML6
|
calmodulin-like 6 |
| chr18_-_48346298 | 5.05 |
ENST00000398439.3
|
MRO
|
maestro |
| chr22_+_30477000 | 4.58 |
ENST00000403975.1
|
HORMAD2
|
HORMA domain containing 2 |
| chr18_-_48346130 | 3.91 |
ENST00000592966.1
|
MRO
|
maestro |
| chr16_+_56672571 | 3.68 |
ENST00000290705.8
|
MT1A
|
metallothionein 1A |
| chr16_+_56642489 | 3.66 |
ENST00000561491.1
|
MT2A
|
metallothionein 2A |
| chr19_-_42916499 | 3.63 |
ENST00000601189.1
ENST00000599211.1 |
LIPE
|
lipase, hormone-sensitive |
| chr2_+_102953608 | 3.62 |
ENST00000311734.2
ENST00000409584.1 |
IL1RL1
|
interleukin 1 receptor-like 1 |
| chr14_-_21493884 | 3.56 |
ENST00000556974.1
ENST00000554419.1 ENST00000298687.5 ENST00000397858.1 ENST00000360463.3 ENST00000350792.3 ENST00000397847.2 |
NDRG2
|
NDRG family member 2 |
| chrX_+_8432871 | 3.51 |
ENST00000381032.1
ENST00000453306.1 ENST00000444481.1 |
VCX3B
|
variable charge, X-linked 3B |
| chr8_+_19759228 | 3.48 |
ENST00000520959.1
|
LPL
|
lipoprotein lipase |
| chr9_-_35563896 | 3.47 |
ENST00000399742.2
|
FAM166B
|
family with sequence similarity 166, member B |
| chr16_+_56642041 | 3.35 |
ENST00000245185.5
|
MT2A
|
metallothionein 2A |
| chr18_+_47087055 | 3.25 |
ENST00000577628.1
|
LIPG
|
lipase, endothelial |
| chr17_-_19290483 | 3.24 |
ENST00000395592.2
ENST00000299610.4 |
MFAP4
|
microfibrillar-associated protein 4 |
| chr3_+_125985620 | 3.16 |
ENST00000511512.1
ENST00000512435.1 |
RP11-71E19.1
|
RP11-71E19.1 |
| chr13_+_113777105 | 3.14 |
ENST00000409306.1
ENST00000375551.3 ENST00000375559.3 |
F10
|
coagulation factor X |
| chr17_-_19290117 | 3.12 |
ENST00000497081.2
|
MFAP4
|
microfibrillar-associated protein 4 |
| chr1_-_112903150 | 2.96 |
ENST00000427290.1
|
RP5-965F6.2
|
RP5-965F6.2 |
| chr1_+_162335197 | 2.93 |
ENST00000431696.1
|
RP11-565P22.6
|
Uncharacterized protein |
| chr14_-_21493649 | 2.82 |
ENST00000553442.1
ENST00000555869.1 ENST00000556457.1 ENST00000397844.2 ENST00000554415.1 |
NDRG2
|
NDRG family member 2 |
| chr15_+_42694573 | 2.74 |
ENST00000397200.4
ENST00000569827.1 |
CAPN3
|
calpain 3, (p94) |
| chr14_-_23623577 | 2.60 |
ENST00000422941.2
ENST00000453702.1 |
SLC7A8
|
solute carrier family 7 (amino acid transporter light chain, L system), member 8 |
| chr14_+_105190514 | 2.59 |
ENST00000330877.2
|
ADSSL1
|
adenylosuccinate synthase like 1 |
| chr4_+_75310851 | 2.58 |
ENST00000395748.3
ENST00000264487.2 |
AREG
|
amphiregulin |
| chr8_-_49834299 | 2.50 |
ENST00000396822.1
|
SNAI2
|
snail family zinc finger 2 |
| chrX_+_8433376 | 2.48 |
ENST00000440654.2
ENST00000381029.4 |
VCX3B
|
variable charge, X-linked 3B |
| chr4_+_75311019 | 2.46 |
ENST00000502307.1
|
AREG
|
amphiregulin |
| chr4_+_75480629 | 2.44 |
ENST00000380846.3
|
AREGB
|
amphiregulin B |
| chr14_+_32414059 | 2.35 |
ENST00000553330.1
|
RP11-187E13.1
|
Uncharacterized protein |
| chr2_-_75788424 | 2.33 |
ENST00000410071.1
|
EVA1A
|
eva-1 homolog A (C. elegans) |
| chr8_+_62200509 | 2.30 |
ENST00000519846.1
ENST00000518592.1 ENST00000325897.4 |
CLVS1
|
clavesin 1 |
| chr8_-_49833978 | 2.20 |
ENST00000020945.1
|
SNAI2
|
snail family zinc finger 2 |
| chrX_-_6453159 | 2.15 |
ENST00000381089.3
ENST00000398729.1 |
VCX3A
|
variable charge, X-linked 3A |
| chr3_-_119396193 | 2.13 |
ENST00000484810.1
ENST00000497116.1 ENST00000261070.2 |
COX17
|
COX17 cytochrome c oxidase copper chaperone |
| chr12_-_53575120 | 2.04 |
ENST00000542115.1
|
CSAD
|
cysteine sulfinic acid decarboxylase |
| chr4_-_69817481 | 2.00 |
ENST00000251566.4
|
UGT2A3
|
UDP glucuronosyltransferase 2 family, polypeptide A3 |
| chr2_-_75788428 | 1.96 |
ENST00000432649.1
|
EVA1A
|
eva-1 homolog A (C. elegans) |
| chr2_+_238395803 | 1.95 |
ENST00000264605.3
|
MLPH
|
melanophilin |
| chrY_-_16098393 | 1.91 |
ENST00000250825.4
|
VCY
|
variable charge, Y-linked |
| chrY_+_16168097 | 1.90 |
ENST00000250823.4
|
VCY1B
|
variable charge, Y-linked 1B |
| chr19_-_8408139 | 1.89 |
ENST00000330915.3
ENST00000593649.1 ENST00000595639.1 |
KANK3
|
KN motif and ankyrin repeat domains 3 |
| chr8_+_24298531 | 1.85 |
ENST00000175238.6
|
ADAM7
|
ADAM metallopeptidase domain 7 |
| chr1_-_24469602 | 1.81 |
ENST00000270800.1
|
IL22RA1
|
interleukin 22 receptor, alpha 1 |
| chr8_+_24298438 | 1.78 |
ENST00000441335.2
|
ADAM7
|
ADAM metallopeptidase domain 7 |
| chr18_+_7754957 | 1.76 |
ENST00000400053.4
|
PTPRM
|
protein tyrosine phosphatase, receptor type, M |
| chr8_-_21669826 | 1.69 |
ENST00000517328.1
|
GFRA2
|
GDNF family receptor alpha 2 |
| chr4_-_135122903 | 1.62 |
ENST00000421491.3
ENST00000529122.2 |
PABPC4L
|
poly(A) binding protein, cytoplasmic 4-like |
| chr13_+_114462193 | 1.61 |
ENST00000375353.3
|
TMEM255B
|
transmembrane protein 255B |
| chr7_-_96654133 | 1.59 |
ENST00000486603.2
ENST00000222598.4 |
DLX5
|
distal-less homeobox 5 |
| chr17_+_37824217 | 1.56 |
ENST00000394246.1
|
PNMT
|
phenylethanolamine N-methyltransferase |
| chr8_+_24298597 | 1.56 |
ENST00000380789.1
|
ADAM7
|
ADAM metallopeptidase domain 7 |
| chr7_-_94285402 | 1.52 |
ENST00000428696.2
ENST00000445866.2 |
SGCE
|
sarcoglycan, epsilon |
| chr17_+_79071365 | 1.52 |
ENST00000576756.1
|
BAIAP2
|
BAI1-associated protein 2 |
| chr7_+_74379083 | 1.41 |
ENST00000361825.7
|
GATSL1
|
GATS protein-like 1 |
| chr10_+_695888 | 1.40 |
ENST00000441152.2
|
PRR26
|
proline rich 26 |
| chrX_+_17755563 | 1.38 |
ENST00000380045.3
ENST00000380041.3 ENST00000380043.3 ENST00000398080.1 |
SCML1
|
sex comb on midleg-like 1 (Drosophila) |
| chr5_+_140186647 | 1.38 |
ENST00000512229.2
ENST00000356878.4 ENST00000530339.1 |
PCDHA4
|
protocadherin alpha 4 |
| chrX_+_79270245 | 1.37 |
ENST00000373296.3
ENST00000442340.1 |
TBX22
|
T-box 22 |
| chr16_+_67207838 | 1.37 |
ENST00000566871.1
ENST00000268605.7 |
NOL3
|
nucleolar protein 3 (apoptosis repressor with CARD domain) |
| chr18_+_47087390 | 1.37 |
ENST00000583083.1
|
LIPG
|
lipase, endothelial |
| chr7_-_94285511 | 1.33 |
ENST00000265735.7
|
SGCE
|
sarcoglycan, epsilon |
| chrX_-_80377162 | 1.24 |
ENST00000430960.1
ENST00000447319.1 |
HMGN5
|
high mobility group nucleosome binding domain 5 |
| chr10_-_87551311 | 1.22 |
ENST00000536331.1
|
GRID1
|
glutamate receptor, ionotropic, delta 1 |
| chr9_+_131683174 | 1.22 |
ENST00000372592.3
ENST00000428610.1 |
PHYHD1
|
phytanoyl-CoA dioxygenase domain containing 1 |
| chrX_-_8139308 | 1.15 |
ENST00000317103.4
|
VCX2
|
variable charge, X-linked 2 |
| chr12_+_51318513 | 1.15 |
ENST00000332160.4
|
METTL7A
|
methyltransferase like 7A |
| chr3_-_24207039 | 1.14 |
ENST00000280696.5
|
THRB
|
thyroid hormone receptor, beta |
| chr7_-_4877677 | 1.12 |
ENST00000538469.1
|
RADIL
|
Ras association and DIL domains |
| chr1_+_61547405 | 1.11 |
ENST00000371189.4
|
NFIA
|
nuclear factor I/A |
| chr16_+_67207872 | 1.10 |
ENST00000563258.1
ENST00000568146.1 |
NOL3
|
nucleolar protein 3 (apoptosis repressor with CARD domain) |
| chr10_-_44274273 | 1.07 |
ENST00000419406.1
|
RP11-272J7.4
|
RP11-272J7.4 |
| chrX_-_80377118 | 1.06 |
ENST00000373250.3
|
HMGN5
|
high mobility group nucleosome binding domain 5 |
| chr3_-_134093738 | 1.06 |
ENST00000506107.1
|
AMOTL2
|
angiomotin like 2 |
| chr2_-_242576864 | 1.05 |
ENST00000407315.1
|
THAP4
|
THAP domain containing 4 |
| chr8_-_110615669 | 1.03 |
ENST00000533394.1
|
SYBU
|
syntabulin (syntaxin-interacting) |
| chr12_-_106480587 | 1.00 |
ENST00000548902.1
|
NUAK1
|
NUAK family, SNF1-like kinase, 1 |
| chr16_-_67978016 | 0.99 |
ENST00000264005.5
|
LCAT
|
lecithin-cholesterol acyltransferase |
| chr10_-_375422 | 0.98 |
ENST00000434695.2
|
DIP2C
|
DIP2 disco-interacting protein 2 homolog C (Drosophila) |
| chr8_+_91013577 | 0.96 |
ENST00000220764.2
|
DECR1
|
2,4-dienoyl CoA reductase 1, mitochondrial |
| chr19_-_14945933 | 0.93 |
ENST00000322301.3
|
OR7A5
|
olfactory receptor, family 7, subfamily A, member 5 |
| chr8_+_91013676 | 0.93 |
ENST00000519410.1
ENST00000522161.1 ENST00000517761.1 ENST00000520227.1 |
DECR1
|
2,4-dienoyl CoA reductase 1, mitochondrial |
| chr7_-_94285472 | 0.84 |
ENST00000437425.2
ENST00000447873.1 ENST00000415788.2 |
SGCE
|
sarcoglycan, epsilon |
| chr8_+_9046503 | 0.83 |
ENST00000512942.2
|
RP11-10A14.5
|
RP11-10A14.5 |
| chr8_-_62602327 | 0.81 |
ENST00000445642.3
ENST00000517847.2 ENST00000389204.4 ENST00000517661.1 ENST00000517903.1 ENST00000522603.1 ENST00000522349.1 ENST00000522835.1 ENST00000541428.1 ENST00000518306.1 |
ASPH
|
aspartate beta-hydroxylase |
| chr1_-_209979465 | 0.80 |
ENST00000542854.1
|
IRF6
|
interferon regulatory factor 6 |
| chr6_-_26235206 | 0.80 |
ENST00000244534.5
|
HIST1H1D
|
histone cluster 1, H1d |
| chr3_+_118865028 | 0.80 |
ENST00000460150.1
|
C3orf30
|
chromosome 3 open reading frame 30 |
| chr15_-_28344439 | 0.78 |
ENST00000431101.1
ENST00000445578.1 ENST00000353809.5 ENST00000382996.2 ENST00000354638.3 |
OCA2
|
oculocutaneous albinism II |
| chr12_-_10251603 | 0.77 |
ENST00000457018.2
|
CLEC1A
|
C-type lectin domain family 1, member A |
| chr1_-_204119009 | 0.77 |
ENST00000444817.1
|
ETNK2
|
ethanolamine kinase 2 |
| chr3_-_118864861 | 0.72 |
ENST00000441144.2
|
IGSF11
|
immunoglobulin superfamily, member 11 |
| chr1_+_248651885 | 0.71 |
ENST00000366473.2
|
OR2T5
|
olfactory receptor, family 2, subfamily T, member 5 |
| chr3_+_118864991 | 0.71 |
ENST00000295622.1
|
C3orf30
|
chromosome 3 open reading frame 30 |
| chr11_-_75380165 | 0.70 |
ENST00000304771.3
|
MAP6
|
microtubule-associated protein 6 |
| chrX_-_117119243 | 0.70 |
ENST00000539496.1
ENST00000469946.1 |
KLHL13
|
kelch-like family member 13 |
| chr17_-_79304150 | 0.70 |
ENST00000574093.1
|
TMEM105
|
transmembrane protein 105 |
| chr11_-_114271139 | 0.70 |
ENST00000325636.4
|
C11orf71
|
chromosome 11 open reading frame 71 |
| chr1_-_159832438 | 0.69 |
ENST00000368100.1
|
VSIG8
|
V-set and immunoglobulin domain containing 8 |
| chr16_+_25703274 | 0.69 |
ENST00000331351.5
|
HS3ST4
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 4 |
| chr5_-_173043591 | 0.68 |
ENST00000285908.5
ENST00000480951.1 ENST00000311086.4 |
BOD1
|
biorientation of chromosomes in cell division 1 |
| chr22_+_18632666 | 0.67 |
ENST00000215794.7
|
USP18
|
ubiquitin specific peptidase 18 |
| chr19_+_56187987 | 0.67 |
ENST00000411543.2
|
EPN1
|
epsin 1 |
| chr6_-_111927062 | 0.65 |
ENST00000359831.4
|
TRAF3IP2
|
TRAF3 interacting protein 2 |
| chr2_-_27435390 | 0.61 |
ENST00000428518.1
|
SLC5A6
|
solute carrier family 5 (sodium/multivitamin and iodide cotransporter), member 6 |
| chr6_+_123317116 | 0.61 |
ENST00000275162.5
|
CLVS2
|
clavesin 2 |
| chr12_-_56848426 | 0.61 |
ENST00000257979.4
|
MIP
|
major intrinsic protein of lens fiber |
| chr9_+_139863695 | 0.61 |
ENST00000371629.1
|
C9orf141
|
chromosome 9 open reading frame 141 |
| chr14_+_24616588 | 0.58 |
ENST00000324103.6
ENST00000559260.1 |
RNF31
|
ring finger protein 31 |
| chr2_+_235346970 | 0.56 |
ENST00000418025.1
|
AC097713.3
|
AC097713.3 |
| chr9_+_34957477 | 0.55 |
ENST00000544237.1
|
KIAA1045
|
KIAA1045 |
| chr19_+_35741466 | 0.52 |
ENST00000599658.1
|
LSR
|
lipolysis stimulated lipoprotein receptor |
| chr17_-_14140166 | 0.50 |
ENST00000420162.2
ENST00000431716.2 |
CDRT15
|
CMT1A duplicated region transcript 15 |
| chr1_-_27693349 | 0.48 |
ENST00000374040.3
ENST00000357582.2 ENST00000493901.1 |
MAP3K6
|
mitogen-activated protein kinase kinase kinase 6 |
| chr18_+_45778672 | 0.47 |
ENST00000600091.1
|
AC091150.1
|
HCG1818186; Uncharacterized protein |
| chr19_-_45657028 | 0.45 |
ENST00000429338.1
ENST00000589776.1 |
NKPD1
|
NTPase, KAP family P-loop domain containing 1 |
| chr1_-_10856694 | 0.45 |
ENST00000377022.3
ENST00000344008.5 |
CASZ1
|
castor zinc finger 1 |
| chr11_+_41736067 | 0.44 |
ENST00000528720.1
|
RP11-375D13.2
|
RP11-375D13.2 |
| chr1_+_209545365 | 0.43 |
ENST00000447257.1
|
RP11-372M18.2
|
RP11-372M18.2 |
| chr17_-_61850894 | 0.43 |
ENST00000403162.3
ENST00000582252.1 ENST00000225726.5 |
CCDC47
|
coiled-coil domain containing 47 |
| chr4_+_87928140 | 0.43 |
ENST00000307808.6
|
AFF1
|
AF4/FMR2 family, member 1 |
| chr4_+_186347388 | 0.42 |
ENST00000511138.1
ENST00000511581.1 |
C4orf47
|
chromosome 4 open reading frame 47 |
| chr2_-_27435634 | 0.40 |
ENST00000430186.1
|
SLC5A6
|
solute carrier family 5 (sodium/multivitamin and iodide cotransporter), member 6 |
| chr19_+_8117636 | 0.40 |
ENST00000253451.4
ENST00000315626.4 |
CCL25
|
chemokine (C-C motif) ligand 25 |
| chr22_+_23046750 | 0.35 |
ENST00000390307.2
|
IGLV3-22
|
immunoglobulin lambda variable 3-22 (gene/pseudogene) |
| chr4_-_186347099 | 0.35 |
ENST00000505357.1
ENST00000264689.6 |
UFSP2
|
UFM1-specific peptidase 2 |
| chr6_+_127898312 | 0.33 |
ENST00000329722.7
|
C6orf58
|
chromosome 6 open reading frame 58 |
| chr14_-_24616426 | 0.30 |
ENST00000216802.5
|
PSME2
|
proteasome (prosome, macropain) activator subunit 2 (PA28 beta) |
| chr10_+_48359344 | 0.29 |
ENST00000412534.1
ENST00000444585.1 |
ZNF488
|
zinc finger protein 488 |
| chr11_+_236540 | 0.27 |
ENST00000532097.1
|
PSMD13
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 13 |
| chr4_-_8442438 | 0.25 |
ENST00000356406.5
ENST00000413009.2 |
ACOX3
|
acyl-CoA oxidase 3, pristanoyl |
| chr3_-_118864893 | 0.25 |
ENST00000354673.2
ENST00000425327.2 |
IGSF11
|
immunoglobulin superfamily, member 11 |
| chr10_+_696000 | 0.24 |
ENST00000381489.5
|
PRR26
|
proline rich 26 |
| chr12_+_125549925 | 0.24 |
ENST00000316519.6
|
AACS
|
acetoacetyl-CoA synthetase |
| chr11_+_103907308 | 0.24 |
ENST00000302259.3
|
DDI1
|
DNA-damage inducible 1 homolog 1 (S. cerevisiae) |
| chr22_+_19939026 | 0.22 |
ENST00000406520.3
|
COMT
|
catechol-O-methyltransferase |
| chr14_+_22251210 | 0.22 |
ENST00000390429.3
|
TRAV7
|
T cell receptor alpha variable 7 |
| chr11_-_236326 | 0.21 |
ENST00000525237.1
ENST00000532956.1 ENST00000525319.1 ENST00000524564.1 ENST00000382743.4 |
SIRT3
|
sirtuin 3 |
| chr19_+_8117881 | 0.20 |
ENST00000390669.3
|
CCL25
|
chemokine (C-C motif) ligand 25 |
| chr10_+_26932068 | 0.19 |
ENST00000544033.1
ENST00000454991.1 |
LINC00202-2
RP13-16H11.7
|
long intergenic non-protein coding RNA 202-2 RP13-16H11.7 |
| chr12_+_56615761 | 0.19 |
ENST00000447747.1
ENST00000399713.2 |
NABP2
|
nucleic acid binding protein 2 |
| chr16_-_66584059 | 0.17 |
ENST00000417693.3
ENST00000544898.1 ENST00000569718.1 ENST00000527284.1 ENST00000299697.7 ENST00000451102.2 |
TK2
|
thymidine kinase 2, mitochondrial |
| chr1_+_248524883 | 0.15 |
ENST00000366475.1
|
OR2T4
|
olfactory receptor, family 2, subfamily T, member 4 |
| chr16_-_25269134 | 0.14 |
ENST00000328086.7
|
ZKSCAN2
|
zinc finger with KRAB and SCAN domains 2 |
| chr5_-_68339648 | 0.13 |
ENST00000479830.2
|
CTC-498J12.1
|
CTC-498J12.1 |
| chr19_+_57874835 | 0.12 |
ENST00000543226.1
ENST00000596755.1 ENST00000282282.3 ENST00000597658.1 |
TRAPPC2P1
ZNF547
AC003002.4
|
trafficking protein particle complex 2 pseudogene 1 zinc finger protein 547 Uncharacterized protein |
| chr1_+_76262552 | 0.10 |
ENST00000263187.3
|
MSH4
|
mutS homolog 4 |
| chr2_+_27435179 | 0.06 |
ENST00000606999.1
ENST00000405489.3 |
ATRAID
|
all-trans retinoic acid-induced differentiation factor |
| chr19_+_1261106 | 0.04 |
ENST00000588411.1
|
CIRBP
|
cold inducible RNA binding protein |
| chr16_-_787771 | 0.04 |
ENST00000568545.1
|
NARFL
|
nuclear prelamin A recognition factor-like |
| chr12_+_125549973 | 0.04 |
ENST00000536752.1
ENST00000261686.6 |
AACS
|
acetoacetyl-CoA synthetase |
| chr17_+_79031415 | 0.02 |
ENST00000572073.1
ENST00000573677.1 |
BAIAP2
|
BAI1-associated protein 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of AR_NR3C2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 19.2 | GO:0010732 | protein glutathionylation(GO:0010731) regulation of protein glutathionylation(GO:0010732) negative regulation of protein glutathionylation(GO:0010734) |
| 1.9 | 15.6 | GO:0036018 | cellular response to erythropoietin(GO:0036018) |
| 1.6 | 4.7 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 1.2 | 4.6 | GO:0010983 | positive regulation of high-density lipoprotein particle clearance(GO:0010983) |
| 0.8 | 5.0 | GO:0042695 | thelarche(GO:0042695) mammary gland branching involved in thelarche(GO:0060744) |
| 0.8 | 2.5 | GO:0014876 | response to injury involved in regulation of muscle adaptation(GO:0014876) |
| 0.7 | 3.6 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 0.7 | 33.0 | GO:0050716 | positive regulation of interleukin-1 secretion(GO:0050716) |
| 0.6 | 19.9 | GO:0071294 | cellular response to zinc ion(GO:0071294) |
| 0.5 | 2.6 | GO:0044208 | 'de novo' AMP biosynthetic process(GO:0044208) |
| 0.5 | 3.6 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.5 | 8.7 | GO:0030050 | vesicle transport along actin filament(GO:0030050) |
| 0.5 | 4.6 | GO:0051177 | meiotic sister chromatid cohesion(GO:0051177) |
| 0.5 | 6.4 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.4 | 2.0 | GO:0042412 | taurine biosynthetic process(GO:0042412) |
| 0.4 | 1.6 | GO:0060166 | olfactory pit development(GO:0060166) |
| 0.4 | 3.1 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.4 | 2.7 | GO:0014718 | positive regulation of satellite cell activation involved in skeletal muscle regeneration(GO:0014718) |
| 0.4 | 1.6 | GO:0042418 | epinephrine metabolic process(GO:0042414) epinephrine biosynthetic process(GO:0042418) |
| 0.4 | 3.5 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.4 | 6.4 | GO:0090360 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 0.2 | 1.1 | GO:0008050 | female courtship behavior(GO:0008050) |
| 0.2 | 0.8 | GO:0018197 | peptidyl-aspartic acid modification(GO:0018197) peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.2 | 1.0 | GO:0090107 | regulation of high-density lipoprotein particle assembly(GO:0090107) |
| 0.2 | 1.5 | GO:1905232 | cellular response to L-glutamate(GO:1905232) |
| 0.2 | 1.0 | GO:0015887 | biotin transport(GO:0015878) pantothenate transmembrane transport(GO:0015887) |
| 0.1 | 0.8 | GO:0006726 | eye pigment biosynthetic process(GO:0006726) eye pigment metabolic process(GO:0042441) pigment metabolic process involved in developmental pigmentation(GO:0043324) pigment metabolic process involved in pigmentation(GO:0043474) |
| 0.1 | 0.6 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.1 | 2.1 | GO:0010155 | regulation of proton transport(GO:0010155) mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.1 | 1.7 | GO:0035860 | glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.1 | 0.8 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.1 | 0.5 | GO:1904274 | tricellular tight junction assembly(GO:1904274) |
| 0.1 | 1.0 | GO:0035507 | regulation of myosin-light-chain-phosphatase activity(GO:0035507) |
| 0.1 | 13.2 | GO:0030449 | regulation of complement activation(GO:0030449) |
| 0.1 | 2.0 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.1 | 2.6 | GO:0015695 | organic cation transport(GO:0015695) |
| 0.1 | 0.6 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.1 | 2.3 | GO:0071157 | negative regulation of cell cycle arrest(GO:0071157) |
| 0.0 | 0.2 | GO:0031337 | cellular response to phosphate starvation(GO:0016036) positive regulation of sulfur amino acid metabolic process(GO:0031337) positive regulation of homocysteine metabolic process(GO:0050668) |
| 0.0 | 1.8 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 1.1 | GO:0072189 | ureter development(GO:0072189) |
| 0.0 | 0.4 | GO:0006983 | ER overload response(GO:0006983) |
| 0.0 | 0.2 | GO:0046092 | deoxycytidine metabolic process(GO:0046092) |
| 0.0 | 2.9 | GO:0007040 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.0 | 0.8 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 0.3 | GO:0071394 | response to oleic acid(GO:0034201) cellular response to testosterone stimulus(GO:0071394) |
| 0.0 | 0.8 | GO:0060644 | mammary gland epithelial cell differentiation(GO:0060644) |
| 0.0 | 1.2 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.0 | 2.1 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.0 | 0.6 | GO:0002230 | positive regulation of defense response to virus by host(GO:0002230) |
| 0.0 | 0.6 | GO:0006833 | water transport(GO:0006833) |
| 0.0 | 0.3 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 1.1 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.7 | GO:0060338 | regulation of type I interferon-mediated signaling pathway(GO:0060338) |
| 0.0 | 1.8 | GO:0019221 | cytokine-mediated signaling pathway(GO:0019221) |
| 0.0 | 0.7 | GO:0030201 | heparan sulfate proteoglycan metabolic process(GO:0030201) |
| 0.0 | 0.3 | GO:0048714 | positive regulation of oligodendrocyte differentiation(GO:0048714) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.0 | 15.2 | GO:0005602 | complement component C1 complex(GO:0005602) |
| 1.0 | 33.0 | GO:0071682 | endocytic vesicle lumen(GO:0071682) |
| 0.7 | 19.2 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.6 | 6.4 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.4 | 3.7 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.2 | 4.0 | GO:0042627 | chylomicron(GO:0042627) |
| 0.1 | 0.6 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.1 | 3.1 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.1 | 4.7 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.1 | 0.8 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.1 | 1.0 | GO:0097433 | dense body(GO:0097433) |
| 0.1 | 8.2 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.1 | 0.3 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.0 | 5.0 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 1.7 | GO:0019898 | extrinsic component of membrane(GO:0019898) |
| 0.0 | 4.8 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.8 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 1.0 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 2.5 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.0 | 41.3 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 2.7 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 0.3 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.2 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.0 | 1.5 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 2.1 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 0.8 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.0 | 7.4 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.0 | 7.5 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 0.6 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 23.2 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 2.3 | GO:0000785 | chromatin(GO:0000785) |
| 0.0 | 1.0 | GO:0031526 | brush border membrane(GO:0031526) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 3.6 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 1.2 | 3.6 | GO:0002113 | interleukin-33 binding(GO:0002113) |
| 0.9 | 3.5 | GO:0017129 | triglyceride binding(GO:0017129) |
| 0.9 | 2.6 | GO:0004019 | adenylosuccinate synthase activity(GO:0004019) |
| 0.6 | 33.0 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.5 | 1.6 | GO:0004603 | phenylethanolamine N-methyltransferase activity(GO:0004603) |
| 0.5 | 1.9 | GO:0008670 | 2,4-dienoyl-CoA reductase (NADPH) activity(GO:0008670) |
| 0.4 | 7.0 | GO:0046870 | cadmium ion binding(GO:0046870) |
| 0.4 | 2.0 | GO:0004782 | sulfinoalanine decarboxylase activity(GO:0004782) |
| 0.4 | 2.1 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.4 | 4.6 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.3 | 1.8 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.3 | 0.8 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.2 | 19.2 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.2 | 2.5 | GO:0035877 | death effector domain binding(GO:0035877) |
| 0.2 | 2.6 | GO:0019534 | toxin transporter activity(GO:0019534) |
| 0.2 | 0.8 | GO:0015173 | aromatic amino acid transmembrane transporter activity(GO:0015173) |
| 0.2 | 1.7 | GO:0016167 | glial cell-derived neurotrophic factor receptor activity(GO:0016167) |
| 0.2 | 1.0 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.1 | 0.6 | GO:0031735 | CCR10 chemokine receptor binding(GO:0031735) |
| 0.1 | 2.7 | GO:0031432 | titin binding(GO:0031432) |
| 0.1 | 1.0 | GO:0008523 | sodium-dependent multivitamin transmembrane transporter activity(GO:0008523) |
| 0.1 | 0.7 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.1 | 1.1 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.1 | 8.7 | GO:0017022 | myosin binding(GO:0017022) |
| 0.1 | 0.8 | GO:0004305 | ethanolamine kinase activity(GO:0004305) |
| 0.1 | 2.9 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.1 | 4.2 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 1.6 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 16.4 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 1.8 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 1.5 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 2.3 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 2.0 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 5.2 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.3 | GO:0016402 | pristanoyl-CoA oxidase activity(GO:0016402) |
| 0.0 | 0.6 | GO:0015250 | water channel activity(GO:0015250) |
| 0.0 | 0.3 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 1.2 | GO:0005234 | extracellular-glutamate-gated ion channel activity(GO:0005234) |
| 0.0 | 0.2 | GO:0004137 | deoxycytidine kinase activity(GO:0004137) thymidine kinase activity(GO:0004797) |
| 0.0 | 0.2 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.0 | 4.4 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.0 | 0.3 | GO:0047760 | butyrate-CoA ligase activity(GO:0047760) |
| 0.0 | 1.0 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 23.2 | GO:0008270 | zinc ion binding(GO:0008270) |
| 0.0 | 1.0 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.5 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 1.2 | GO:0051213 | dioxygenase activity(GO:0051213) |
| 0.0 | 0.2 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.0 | 0.8 | GO:0031490 | chromatin DNA binding(GO:0031490) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 33.0 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.1 | 5.0 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.1 | 15.2 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 3.1 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 7.0 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 6.4 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 4.7 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 1.6 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 1.2 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 0.5 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 1.1 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 5.1 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 5.2 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.8 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 1.5 | PID RAC1 PATHWAY | RAC1 signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 15.2 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 1.0 | 33.0 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.2 | 3.1 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.1 | 2.6 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.1 | 3.5 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.1 | 3.6 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.1 | 1.9 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.1 | 1.6 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.1 | 6.4 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 1.0 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.8 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 1.0 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 2.6 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 2.0 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.6 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.7 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.0 | 0.8 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 1.1 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 1.7 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 1.4 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.7 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.7 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |