Project
Illumina Body Map 2: averaged replicates
Navigation
Downloads
Results for BACH2
Z-value: 1.10
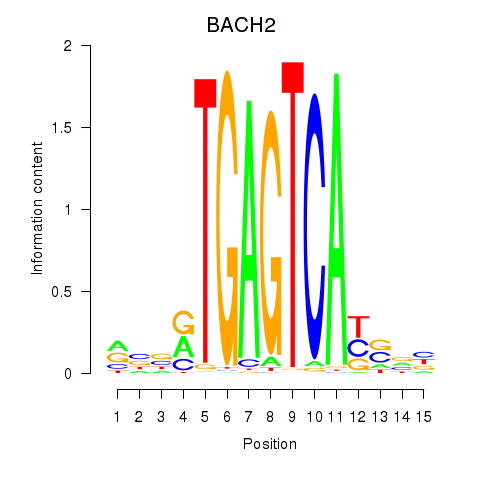
Transcription factors associated with BACH2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
BACH2
|
ENSG00000112182.10 | BTB domain and CNC homolog 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| BACH2 | hg19_v2_chr6_-_91006461_91006517 | 0.34 | 6.0e-02 | Click! |
Activity profile of BACH2 motif
Sorted Z-values of BACH2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_95010147 | 2.07 |
ENST00000548918.1
|
TMCC3
|
transmembrane and coiled-coil domain family 3 |
| chr16_+_30077098 | 1.91 |
ENST00000395240.3
ENST00000566846.1 |
ALDOA
|
aldolase A, fructose-bisphosphate |
| chr2_-_220173685 | 1.85 |
ENST00000423636.2
ENST00000442029.1 ENST00000412847.1 |
PTPRN
|
protein tyrosine phosphatase, receptor type, N |
| chr16_+_30077055 | 1.72 |
ENST00000564595.2
ENST00000569798.1 |
ALDOA
|
aldolase A, fructose-bisphosphate |
| chr12_-_95009837 | 1.66 |
ENST00000551457.1
|
TMCC3
|
transmembrane and coiled-coil domain family 3 |
| chr2_+_169926047 | 1.46 |
ENST00000428522.1
ENST00000450153.1 ENST00000421653.1 |
DHRS9
|
dehydrogenase/reductase (SDR family) member 9 |
| chr12_+_7023735 | 1.46 |
ENST00000538763.1
ENST00000544774.1 ENST00000545045.2 |
ENO2
|
enolase 2 (gamma, neuronal) |
| chr1_+_36621174 | 1.44 |
ENST00000429533.2
|
MAP7D1
|
MAP7 domain containing 1 |
| chr17_-_7123021 | 1.41 |
ENST00000399510.2
|
DLG4
|
discs, large homolog 4 (Drosophila) |
| chr12_+_7023491 | 1.35 |
ENST00000541477.1
ENST00000229277.1 |
ENO2
|
enolase 2 (gamma, neuronal) |
| chr4_+_74606223 | 1.27 |
ENST00000307407.3
ENST00000401931.1 |
IL8
|
interleukin 8 |
| chr3_+_187930491 | 1.26 |
ENST00000443217.1
|
LPP
|
LIM domain containing preferred translocation partner in lipoma |
| chr3_-_71834207 | 1.26 |
ENST00000295619.3
|
PROK2
|
prokineticin 2 |
| chr3_-_98241598 | 1.20 |
ENST00000513287.1
ENST00000514537.1 ENST00000508071.1 ENST00000507944.1 |
CLDND1
|
claudin domain containing 1 |
| chr1_+_36621529 | 1.17 |
ENST00000316156.4
|
MAP7D1
|
MAP7 domain containing 1 |
| chr8_+_22844995 | 1.16 |
ENST00000524077.1
|
RHOBTB2
|
Rho-related BTB domain containing 2 |
| chr5_-_176923846 | 1.15 |
ENST00000506537.1
|
PDLIM7
|
PDZ and LIM domain 7 (enigma) |
| chr5_+_175288631 | 1.14 |
ENST00000509837.1
|
CPLX2
|
complexin 2 |
| chr10_+_88428206 | 1.13 |
ENST00000429277.2
ENST00000458213.2 ENST00000352360.5 |
LDB3
|
LIM domain binding 3 |
| chr1_+_154378049 | 1.08 |
ENST00000512471.1
|
IL6R
|
interleukin 6 receptor |
| chr3_+_35685113 | 1.07 |
ENST00000419330.1
|
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa |
| chr5_-_176923803 | 1.06 |
ENST00000506161.1
|
PDLIM7
|
PDZ and LIM domain 7 (enigma) |
| chr13_-_36429763 | 1.06 |
ENST00000379893.1
|
DCLK1
|
doublecortin-like kinase 1 |
| chr12_-_76478686 | 1.04 |
ENST00000261182.8
|
NAP1L1
|
nucleosome assembly protein 1-like 1 |
| chr11_-_66104237 | 1.04 |
ENST00000530056.1
|
RIN1
|
Ras and Rab interactor 1 |
| chr1_+_36621697 | 1.03 |
ENST00000373150.4
ENST00000373151.2 |
MAP7D1
|
MAP7 domain containing 1 |
| chr12_-_71182695 | 1.02 |
ENST00000342084.4
|
PTPRR
|
protein tyrosine phosphatase, receptor type, R |
| chr6_-_24877490 | 1.01 |
ENST00000540914.1
ENST00000378023.4 |
FAM65B
|
family with sequence similarity 65, member B |
| chr1_+_154377669 | 1.01 |
ENST00000368485.3
ENST00000344086.4 |
IL6R
|
interleukin 6 receptor |
| chr1_-_40157345 | 0.99 |
ENST00000372844.3
|
HPCAL4
|
hippocalcin like 4 |
| chr11_-_62457371 | 0.99 |
ENST00000317449.4
|
LRRN4CL
|
LRRN4 C-terminal like |
| chr20_+_44034804 | 0.96 |
ENST00000357275.2
ENST00000372720.3 |
DBNDD2
|
dysbindin (dystrobrevin binding protein 1) domain containing 2 |
| chr19_-_18995029 | 0.96 |
ENST00000596048.1
|
CERS1
|
ceramide synthase 1 |
| chr5_+_169758393 | 0.95 |
ENST00000521471.1
ENST00000518357.1 ENST00000436248.3 |
CTB-114C7.3
|
CTB-114C7.3 |
| chr11_+_10476851 | 0.95 |
ENST00000396553.2
|
AMPD3
|
adenosine monophosphate deaminase 3 |
| chr9_-_114246332 | 0.94 |
ENST00000602978.1
|
KIAA0368
|
KIAA0368 |
| chr19_+_38794797 | 0.94 |
ENST00000301246.5
ENST00000588605.1 |
C19orf33
|
chromosome 19 open reading frame 33 |
| chr22_+_42372764 | 0.93 |
ENST00000396426.3
ENST00000406029.1 |
SEPT3
|
septin 3 |
| chr15_+_51633826 | 0.91 |
ENST00000335449.6
|
GLDN
|
gliomedin |
| chr11_+_10477733 | 0.91 |
ENST00000528723.1
|
AMPD3
|
adenosine monophosphate deaminase 3 |
| chr3_+_11178779 | 0.91 |
ENST00000438284.2
|
HRH1
|
histamine receptor H1 |
| chr16_+_89988259 | 0.90 |
ENST00000554444.1
ENST00000556565.1 |
TUBB3
|
Tubulin beta-3 chain |
| chr19_+_10959043 | 0.89 |
ENST00000397820.4
|
C19orf38
|
chromosome 19 open reading frame 38 |
| chr8_+_22844913 | 0.89 |
ENST00000519685.1
|
RHOBTB2
|
Rho-related BTB domain containing 2 |
| chr11_-_66103932 | 0.89 |
ENST00000311320.4
|
RIN1
|
Ras and Rab interactor 1 |
| chr18_+_32290218 | 0.88 |
ENST00000348997.5
ENST00000588949.1 ENST00000597599.1 |
DTNA
|
dystrobrevin, alpha |
| chr6_+_56819895 | 0.87 |
ENST00000370748.3
|
BEND6
|
BEN domain containing 6 |
| chr21_+_30672433 | 0.87 |
ENST00000451655.1
|
BACH1
|
BTB and CNC homology 1, basic leucine zipper transcription factor 1 |
| chr7_+_22766766 | 0.86 |
ENST00000426291.1
ENST00000401651.1 ENST00000258743.5 ENST00000420258.2 ENST00000407492.1 ENST00000401630.3 ENST00000406575.1 |
IL6
|
interleukin 6 (interferon, beta 2) |
| chr22_+_42372931 | 0.86 |
ENST00000328414.8
ENST00000396425.3 |
SEPT3
|
septin 3 |
| chr3_+_187930429 | 0.86 |
ENST00000420410.1
|
LPP
|
LIM domain containing preferred translocation partner in lipoma |
| chr10_+_88428370 | 0.85 |
ENST00000372066.3
ENST00000263066.6 ENST00000372056.4 ENST00000310944.6 ENST00000361373.4 ENST00000542786.1 |
LDB3
|
LIM domain binding 3 |
| chr5_-_41794663 | 0.83 |
ENST00000510634.1
|
OXCT1
|
3-oxoacid CoA transferase 1 |
| chr17_-_18161870 | 0.81 |
ENST00000579294.1
ENST00000545457.2 ENST00000379450.4 ENST00000578558.1 |
FLII
|
flightless I homolog (Drosophila) |
| chr3_-_98241358 | 0.80 |
ENST00000503004.1
ENST00000506575.1 ENST00000513452.1 ENST00000515620.1 |
CLDND1
|
claudin domain containing 1 |
| chr16_+_58533951 | 0.79 |
ENST00000566192.1
ENST00000565088.1 ENST00000568640.1 ENST00000563978.1 ENST00000569923.1 ENST00000356752.4 ENST00000563799.1 ENST00000562999.1 ENST00000570248.1 ENST00000562731.1 ENST00000568424.1 |
NDRG4
|
NDRG family member 4 |
| chr11_-_66103867 | 0.78 |
ENST00000424433.2
|
RIN1
|
Ras and Rab interactor 1 |
| chr5_+_150020240 | 0.78 |
ENST00000519664.1
|
SYNPO
|
synaptopodin |
| chr12_-_719573 | 0.78 |
ENST00000397265.3
|
NINJ2
|
ninjurin 2 |
| chr17_-_27503770 | 0.78 |
ENST00000533112.1
|
MYO18A
|
myosin XVIIIA |
| chr2_-_219858123 | 0.77 |
ENST00000453769.1
ENST00000295728.2 ENST00000392096.2 |
CRYBA2
|
crystallin, beta A2 |
| chr11_-_62323702 | 0.77 |
ENST00000530285.1
|
AHNAK
|
AHNAK nucleoprotein |
| chr3_+_48507621 | 0.76 |
ENST00000456089.1
|
TREX1
|
three prime repair exonuclease 1 |
| chr11_+_57308979 | 0.75 |
ENST00000457912.1
|
SMTNL1
|
smoothelin-like 1 |
| chr9_-_35112376 | 0.75 |
ENST00000488109.2
|
FAM214B
|
family with sequence similarity 214, member B |
| chr5_+_176237478 | 0.74 |
ENST00000329542.4
|
UNC5A
|
unc-5 homolog A (C. elegans) |
| chr10_+_104155450 | 0.73 |
ENST00000471698.1
ENST00000189444.6 |
NFKB2
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 (p49/p100) |
| chr1_-_158656488 | 0.72 |
ENST00000368147.4
|
SPTA1
|
spectrin, alpha, erythrocytic 1 (elliptocytosis 2) |
| chr5_+_110559603 | 0.72 |
ENST00000512453.1
|
CAMK4
|
calcium/calmodulin-dependent protein kinase IV |
| chr9_-_114246635 | 0.72 |
ENST00000338205.5
|
KIAA0368
|
KIAA0368 |
| chr17_+_79651007 | 0.71 |
ENST00000572392.1
ENST00000577012.1 |
HGS
|
hepatocyte growth factor-regulated tyrosine kinase substrate |
| chr17_-_70417365 | 0.71 |
ENST00000580948.1
|
LINC00511
|
long intergenic non-protein coding RNA 511 |
| chr5_-_41794313 | 0.70 |
ENST00000512084.1
|
OXCT1
|
3-oxoacid CoA transferase 1 |
| chr3_-_187454281 | 0.70 |
ENST00000232014.4
|
BCL6
|
B-cell CLL/lymphoma 6 |
| chr3_-_71834318 | 0.69 |
ENST00000353065.3
|
PROK2
|
prokineticin 2 |
| chr8_+_123875624 | 0.68 |
ENST00000534247.1
|
ZHX2
|
zinc fingers and homeoboxes 2 |
| chr14_+_77425972 | 0.68 |
ENST00000553613.1
|
RP11-7F17.7
|
RP11-7F17.7 |
| chr3_-_98241713 | 0.67 |
ENST00000502288.1
ENST00000512147.1 ENST00000510541.1 ENST00000503621.1 ENST00000511081.1 |
CLDND1
|
claudin domain containing 1 |
| chr8_+_24771265 | 0.67 |
ENST00000518131.1
ENST00000437366.2 |
NEFM
|
neurofilament, medium polypeptide |
| chr3_-_131756559 | 0.67 |
ENST00000505957.1
|
CPNE4
|
copine IV |
| chr6_+_74405501 | 0.64 |
ENST00000437994.2
ENST00000422508.2 |
CD109
|
CD109 molecule |
| chr22_+_42372970 | 0.64 |
ENST00000291236.11
|
SEPT3
|
septin 3 |
| chr1_-_179112189 | 0.64 |
ENST00000512653.1
ENST00000344730.3 |
ABL2
|
c-abl oncogene 2, non-receptor tyrosine kinase |
| chr10_+_86088337 | 0.64 |
ENST00000359979.4
|
CCSER2
|
coiled-coil serine-rich protein 2 |
| chr1_+_223889285 | 0.63 |
ENST00000433674.2
|
CAPN2
|
calpain 2, (m/II) large subunit |
| chr6_+_74405804 | 0.63 |
ENST00000287097.5
|
CD109
|
CD109 molecule |
| chr16_+_15596123 | 0.62 |
ENST00000452191.2
|
C16orf45
|
chromosome 16 open reading frame 45 |
| chr6_-_119031228 | 0.62 |
ENST00000392500.3
ENST00000368488.5 ENST00000434604.1 |
CEP85L
|
centrosomal protein 85kDa-like |
| chr15_-_83316087 | 0.61 |
ENST00000568757.1
|
CPEB1
|
cytoplasmic polyadenylation element binding protein 1 |
| chr12_-_118490217 | 0.61 |
ENST00000542304.1
|
WSB2
|
WD repeat and SOCS box containing 2 |
| chr12_-_6809958 | 0.61 |
ENST00000320591.5
ENST00000534837.1 |
PIANP
|
PILR alpha associated neural protein |
| chr8_-_68658578 | 0.61 |
ENST00000518549.1
ENST00000297770.4 ENST00000297769.4 |
CPA6
|
carboxypeptidase A6 |
| chr19_-_6057282 | 0.61 |
ENST00000592281.1
|
RFX2
|
regulatory factor X, 2 (influences HLA class II expression) |
| chr11_-_128894053 | 0.60 |
ENST00000392657.3
|
ARHGAP32
|
Rho GTPase activating protein 32 |
| chr4_-_100356551 | 0.60 |
ENST00000209665.4
|
ADH7
|
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
| chr13_+_113698946 | 0.59 |
ENST00000397021.1
|
MCF2L
|
MCF.2 cell line derived transforming sequence-like |
| chr3_+_69134080 | 0.58 |
ENST00000273258.3
|
ARL6IP5
|
ADP-ribosylation-like factor 6 interacting protein 5 |
| chr6_+_44214824 | 0.58 |
ENST00000371646.5
ENST00000353801.3 |
HSP90AB1
|
heat shock protein 90kDa alpha (cytosolic), class B member 1 |
| chr12_-_76478446 | 0.57 |
ENST00000393263.3
ENST00000548044.1 ENST00000547704.1 ENST00000431879.3 ENST00000549596.1 ENST00000550934.1 ENST00000551600.1 ENST00000547479.1 ENST00000547773.1 ENST00000544816.1 ENST00000542344.1 ENST00000548273.1 |
NAP1L1
|
nucleosome assembly protein 1-like 1 |
| chr3_+_164924716 | 0.57 |
ENST00000470138.1
ENST00000498616.1 |
RP11-85M11.2
|
RP11-85M11.2 |
| chr3_+_48507210 | 0.57 |
ENST00000433541.1
ENST00000422277.2 ENST00000436480.2 ENST00000444177.1 |
TREX1
|
three prime repair exonuclease 1 |
| chr12_-_118490403 | 0.56 |
ENST00000535496.1
|
WSB2
|
WD repeat and SOCS box containing 2 |
| chr19_+_36630855 | 0.55 |
ENST00000589146.1
|
CAPNS1
|
calpain, small subunit 1 |
| chr17_+_65375082 | 0.55 |
ENST00000584471.1
|
PITPNC1
|
phosphatidylinositol transfer protein, cytoplasmic 1 |
| chr10_+_68685764 | 0.54 |
ENST00000361320.4
|
LRRTM3
|
leucine rich repeat transmembrane neuronal 3 |
| chr7_+_143078379 | 0.54 |
ENST00000449630.1
ENST00000457235.1 |
ZYX
|
zyxin |
| chr3_+_69134124 | 0.54 |
ENST00000478935.1
|
ARL6IP5
|
ADP-ribosylation-like factor 6 interacting protein 5 |
| chr5_+_110559784 | 0.53 |
ENST00000282356.4
|
CAMK4
|
calcium/calmodulin-dependent protein kinase IV |
| chr12_+_10365082 | 0.52 |
ENST00000545859.1
|
GABARAPL1
|
GABA(A) receptor-associated protein like 1 |
| chr12_-_76478386 | 0.52 |
ENST00000535020.2
|
NAP1L1
|
nucleosome assembly protein 1-like 1 |
| chr15_-_72521017 | 0.52 |
ENST00000561609.1
|
PKM
|
pyruvate kinase, muscle |
| chr12_-_76478417 | 0.52 |
ENST00000552342.1
|
NAP1L1
|
nucleosome assembly protein 1-like 1 |
| chr17_+_79650962 | 0.51 |
ENST00000329138.4
|
HGS
|
hepatocyte growth factor-regulated tyrosine kinase substrate |
| chr22_+_20861858 | 0.51 |
ENST00000414658.1
ENST00000432052.1 ENST00000425759.2 ENST00000292733.7 ENST00000542773.1 ENST00000263205.7 ENST00000406969.1 ENST00000382974.2 |
MED15
|
mediator complex subunit 15 |
| chr1_-_235116495 | 0.51 |
ENST00000549744.1
|
RP11-443B7.3
|
RP11-443B7.3 |
| chr17_+_25799008 | 0.51 |
ENST00000583370.1
ENST00000398988.3 ENST00000268763.6 |
KSR1
|
kinase suppressor of ras 1 |
| chr7_-_15014398 | 0.50 |
ENST00000437998.1
|
DGKB
|
diacylglycerol kinase, beta 90kDa |
| chr9_-_34662651 | 0.50 |
ENST00000259631.4
|
CCL27
|
chemokine (C-C motif) ligand 27 |
| chr6_-_56819385 | 0.50 |
ENST00000370754.5
ENST00000449297.2 |
DST
|
dystonin |
| chr4_-_100356844 | 0.50 |
ENST00000437033.2
|
ADH7
|
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
| chr13_+_113699029 | 0.49 |
ENST00000423251.1
|
MCF2L
|
MCF.2 cell line derived transforming sequence-like |
| chr6_+_56820018 | 0.49 |
ENST00000370746.3
|
BEND6
|
BEN domain containing 6 |
| chr10_+_86088381 | 0.49 |
ENST00000224756.8
ENST00000372088.2 |
CCSER2
|
coiled-coil serine-rich protein 2 |
| chr2_+_87754989 | 0.49 |
ENST00000409898.2
ENST00000419680.2 ENST00000414584.1 ENST00000455131.1 |
LINC00152
|
long intergenic non-protein coding RNA 152 |
| chr3_+_164924769 | 0.49 |
ENST00000494915.1
|
RP11-85M11.2
|
RP11-85M11.2 |
| chr2_+_69201705 | 0.49 |
ENST00000377938.2
|
GKN1
|
gastrokine 1 |
| chr9_-_139922726 | 0.49 |
ENST00000265662.5
ENST00000371605.3 |
ABCA2
|
ATP-binding cassette, sub-family A (ABC1), member 2 |
| chr15_+_89182178 | 0.49 |
ENST00000559876.1
|
ISG20
|
interferon stimulated exonuclease gene 20kDa |
| chr2_-_145275109 | 0.48 |
ENST00000431672.2
|
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr12_+_64846129 | 0.47 |
ENST00000540417.1
ENST00000539810.1 |
TBK1
|
TANK-binding kinase 1 |
| chr14_-_25103388 | 0.47 |
ENST00000526004.1
ENST00000415355.3 |
GZMB
|
granzyme B (granzyme 2, cytotoxic T-lymphocyte-associated serine esterase 1) |
| chrX_-_129244336 | 0.47 |
ENST00000434609.1
|
ELF4
|
E74-like factor 4 (ets domain transcription factor) |
| chr1_-_156675368 | 0.47 |
ENST00000368222.3
|
CRABP2
|
cellular retinoic acid binding protein 2 |
| chr15_+_75335604 | 0.47 |
ENST00000563393.1
|
PPCDC
|
phosphopantothenoylcysteine decarboxylase |
| chr17_-_76921459 | 0.46 |
ENST00000262768.7
|
TIMP2
|
TIMP metallopeptidase inhibitor 2 |
| chrX_-_20074895 | 0.46 |
ENST00000543767.1
|
MAP7D2
|
MAP7 domain containing 2 |
| chr5_+_110559312 | 0.45 |
ENST00000508074.1
|
CAMK4
|
calcium/calmodulin-dependent protein kinase IV |
| chr3_+_184534994 | 0.45 |
ENST00000441141.1
ENST00000445089.1 |
VPS8
|
vacuolar protein sorting 8 homolog (S. cerevisiae) |
| chrX_+_150863951 | 0.45 |
ENST00000370354.1
|
PRRG3
|
proline rich Gla (G-carboxyglutamic acid) 3 (transmembrane) |
| chr19_-_36019123 | 0.44 |
ENST00000588674.1
ENST00000452271.2 ENST00000518157.1 |
SBSN
|
suprabasin |
| chr19_+_38880695 | 0.44 |
ENST00000587947.1
ENST00000338502.4 |
SPRED3
|
sprouty-related, EVH1 domain containing 3 |
| chr3_+_174158732 | 0.42 |
ENST00000434257.1
|
NAALADL2
|
N-acetylated alpha-linked acidic dipeptidase-like 2 |
| chr22_+_20862321 | 0.42 |
ENST00000541476.1
ENST00000438962.1 |
MED15
|
mediator complex subunit 15 |
| chr1_+_223889310 | 0.42 |
ENST00000434648.1
|
CAPN2
|
calpain 2, (m/II) large subunit |
| chr19_+_36630961 | 0.41 |
ENST00000587718.1
ENST00000592483.1 ENST00000590874.1 ENST00000588815.1 |
CAPNS1
|
calpain, small subunit 1 |
| chr11_-_46848393 | 0.41 |
ENST00000526496.1
|
CKAP5
|
cytoskeleton associated protein 5 |
| chr6_-_113953705 | 0.40 |
ENST00000452675.1
|
RP11-367G18.1
|
RP11-367G18.1 |
| chr19_+_36630454 | 0.40 |
ENST00000246533.3
|
CAPNS1
|
calpain, small subunit 1 |
| chr1_-_223853425 | 0.40 |
ENST00000366873.2
ENST00000419193.2 |
CAPN8
|
calpain 8 |
| chr12_+_64845864 | 0.40 |
ENST00000538890.1
|
TBK1
|
TANK-binding kinase 1 |
| chr2_+_87754887 | 0.40 |
ENST00000409054.1
ENST00000331944.6 ENST00000409139.1 |
LINC00152
|
long intergenic non-protein coding RNA 152 |
| chr12_+_6644443 | 0.40 |
ENST00000396858.1
|
GAPDH
|
glyceraldehyde-3-phosphate dehydrogenase |
| chr1_-_27816641 | 0.40 |
ENST00000430629.2
|
WASF2
|
WAS protein family, member 2 |
| chr6_+_44215603 | 0.39 |
ENST00000371554.1
|
HSP90AB1
|
heat shock protein 90kDa alpha (cytosolic), class B member 1 |
| chr21_+_30671690 | 0.39 |
ENST00000399921.1
|
BACH1
|
BTB and CNC homology 1, basic leucine zipper transcription factor 1 |
| chrY_+_15016725 | 0.39 |
ENST00000336079.3
|
DDX3Y
|
DEAD (Asp-Glu-Ala-Asp) box helicase 3, Y-linked |
| chr9_+_34992846 | 0.39 |
ENST00000443266.1
|
DNAJB5
|
DnaJ (Hsp40) homolog, subfamily B, member 5 |
| chr14_+_22989368 | 0.39 |
ENST00000390514.1
|
TRAJ23
|
T cell receptor alpha joining 23 |
| chr5_+_179247759 | 0.39 |
ENST00000389805.4
ENST00000504627.1 ENST00000402874.3 ENST00000510187.1 |
SQSTM1
|
sequestosome 1 |
| chr11_-_65667997 | 0.38 |
ENST00000312562.2
ENST00000534222.1 |
FOSL1
|
FOS-like antigen 1 |
| chr6_+_56819773 | 0.38 |
ENST00000370750.2
|
BEND6
|
BEN domain containing 6 |
| chr4_+_123747834 | 0.38 |
ENST00000264498.3
|
FGF2
|
fibroblast growth factor 2 (basic) |
| chr10_+_17270214 | 0.38 |
ENST00000544301.1
|
VIM
|
vimentin |
| chr7_-_98467543 | 0.37 |
ENST00000345589.4
|
TMEM130
|
transmembrane protein 130 |
| chr2_+_36923830 | 0.37 |
ENST00000379242.3
ENST00000389975.3 |
VIT
|
vitrin |
| chr17_+_30469579 | 0.37 |
ENST00000354266.3
ENST00000581094.1 ENST00000394692.2 |
RHOT1
|
ras homolog family member T1 |
| chr14_-_23426322 | 0.37 |
ENST00000555367.1
|
HAUS4
|
HAUS augmin-like complex, subunit 4 |
| chr14_-_25103472 | 0.37 |
ENST00000216341.4
ENST00000382542.1 ENST00000382540.1 |
GZMB
|
granzyme B (granzyme 2, cytotoxic T-lymphocyte-associated serine esterase 1) |
| chr11_-_65667884 | 0.36 |
ENST00000448083.2
ENST00000531493.1 ENST00000532401.1 |
FOSL1
|
FOS-like antigen 1 |
| chr2_-_220117867 | 0.36 |
ENST00000456818.1
ENST00000447205.1 |
TUBA4A
|
tubulin, alpha 4a |
| chr16_-_89043377 | 0.36 |
ENST00000436887.2
ENST00000448839.1 ENST00000360302.2 |
CBFA2T3
|
core-binding factor, runt domain, alpha subunit 2; translocated to, 3 |
| chr3_+_184016986 | 0.36 |
ENST00000417952.1
|
PSMD2
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 2 |
| chr2_-_145275211 | 0.36 |
ENST00000462355.1
|
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr1_-_52344416 | 0.36 |
ENST00000544028.1
|
NRD1
|
nardilysin (N-arginine dibasic convertase) |
| chr15_+_89182156 | 0.35 |
ENST00000379224.5
|
ISG20
|
interferon stimulated exonuclease gene 20kDa |
| chr1_-_6662919 | 0.35 |
ENST00000377658.4
ENST00000377663.3 |
KLHL21
|
kelch-like family member 21 |
| chr14_-_23426337 | 0.35 |
ENST00000342454.8
ENST00000555986.1 ENST00000541587.1 ENST00000554516.1 ENST00000347758.2 ENST00000206474.7 ENST00000555040.1 |
HAUS4
|
HAUS augmin-like complex, subunit 4 |
| chr16_+_30418910 | 0.35 |
ENST00000566625.1
|
ZNF771
|
zinc finger protein 771 |
| chr14_-_23426270 | 0.35 |
ENST00000557591.1
ENST00000397409.4 ENST00000490506.1 ENST00000554406.1 |
HAUS4
|
HAUS augmin-like complex, subunit 4 |
| chr17_+_30469473 | 0.35 |
ENST00000333942.6
ENST00000358365.3 ENST00000583994.1 ENST00000545287.2 |
RHOT1
|
ras homolog family member T1 |
| chr19_+_49467232 | 0.34 |
ENST00000599784.1
ENST00000594305.1 |
CTD-2639E6.9
|
CTD-2639E6.9 |
| chr2_+_231921574 | 0.34 |
ENST00000308696.6
ENST00000373635.4 ENST00000440838.1 ENST00000409643.1 |
PSMD1
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 1 |
| chr4_-_100356291 | 0.34 |
ENST00000476959.1
ENST00000482593.1 |
ADH7
|
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
| chr15_+_38988799 | 0.34 |
ENST00000318792.1
|
C15orf53
|
chromosome 15 open reading frame 53 |
| chr19_+_6740888 | 0.33 |
ENST00000596673.1
|
TRIP10
|
thyroid hormone receptor interactor 10 |
| chrX_-_129244454 | 0.33 |
ENST00000308167.5
|
ELF4
|
E74-like factor 4 (ets domain transcription factor) |
| chr14_-_51863853 | 0.33 |
ENST00000556762.1
|
RP11-255G12.3
|
RP11-255G12.3 |
| chr10_-_23003460 | 0.33 |
ENST00000376573.4
|
PIP4K2A
|
phosphatidylinositol-5-phosphate 4-kinase, type II, alpha |
| chr4_+_123747979 | 0.33 |
ENST00000608478.1
|
FGF2
|
fibroblast growth factor 2 (basic) |
| chrX_-_129244655 | 0.33 |
ENST00000335997.7
|
ELF4
|
E74-like factor 4 (ets domain transcription factor) |
| chr10_-_31288398 | 0.32 |
ENST00000538351.2
|
ZNF438
|
zinc finger protein 438 |
| chr14_+_105147464 | 0.32 |
ENST00000540171.2
|
RP11-982M15.6
|
RP11-982M15.6 |
| chr11_-_47206965 | 0.32 |
ENST00000525725.1
|
PACSIN3
|
protein kinase C and casein kinase substrate in neurons 3 |
| chr1_+_152881014 | 0.32 |
ENST00000368764.3
ENST00000392667.2 |
IVL
|
involucrin |
| chr9_-_139922631 | 0.32 |
ENST00000341511.6
|
ABCA2
|
ATP-binding cassette, sub-family A (ABC1), member 2 |
| chr11_+_46316677 | 0.31 |
ENST00000534787.1
|
CREB3L1
|
cAMP responsive element binding protein 3-like 1 |
| chr17_-_48785216 | 0.31 |
ENST00000285243.6
|
ANKRD40
|
ankyrin repeat domain 40 |
| chr10_-_73611046 | 0.31 |
ENST00000394934.1
ENST00000394936.3 |
PSAP
|
prosaposin |
| chr11_+_59522837 | 0.31 |
ENST00000437946.2
|
STX3
|
syntaxin 3 |
| chr7_-_98467629 | 0.31 |
ENST00000339375.4
|
TMEM130
|
transmembrane protein 130 |
Network of associatons between targets according to the STRING database.
First level regulatory network of BACH2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 3.0 | GO:0002384 | hepatic immune response(GO:0002384) |
| 0.6 | 1.7 | GO:0002372 | myeloid dendritic cell cytokine production(GO:0002372) |
| 0.5 | 1.8 | GO:1990502 | dense core granule maturation(GO:1990502) |
| 0.4 | 1.4 | GO:0010430 | fatty acid omega-oxidation(GO:0010430) |
| 0.3 | 1.0 | GO:0072720 | cellular response to mycotoxin(GO:0036146) response to dithiothreitol(GO:0072720) |
| 0.3 | 1.6 | GO:0000117 | regulation of transcription involved in G2/M transition of mitotic cell cycle(GO:0000117) |
| 0.3 | 2.7 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.3 | 1.9 | GO:0006196 | AMP catabolic process(GO:0006196) |
| 0.3 | 1.1 | GO:1903028 | positive regulation of opsonization(GO:1903028) |
| 0.2 | 1.0 | GO:1903660 | negative regulation of complement-dependent cytotoxicity(GO:1903660) |
| 0.2 | 0.7 | GO:0042660 | positive regulation of cell fate specification(GO:0042660) |
| 0.2 | 0.7 | GO:0033693 | neurofilament bundle assembly(GO:0033693) |
| 0.2 | 1.1 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.2 | 1.5 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.2 | 3.6 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.2 | 0.7 | GO:0002268 | follicular dendritic cell differentiation(GO:0002268) |
| 0.2 | 0.7 | GO:0032764 | negative regulation of mast cell cytokine production(GO:0032764) negative regulation of isotype switching to IgE isotypes(GO:0048294) |
| 0.2 | 1.3 | GO:0072675 | osteoclast fusion(GO:0072675) |
| 0.1 | 1.0 | GO:1903385 | regulation of homophilic cell adhesion(GO:1903385) |
| 0.1 | 0.8 | GO:0008626 | granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 0.1 | 1.5 | GO:0042904 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.1 | 0.4 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.1 | 1.1 | GO:0002036 | regulation of L-glutamate transport(GO:0002036) |
| 0.1 | 0.7 | GO:1902513 | regulation of organelle transport along microtubule(GO:1902513) |
| 0.1 | 0.9 | GO:0044565 | dendritic cell proliferation(GO:0044565) |
| 0.1 | 0.8 | GO:0032377 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.1 | 0.7 | GO:0098886 | modification of dendritic spine(GO:0098886) |
| 0.1 | 1.4 | GO:0097113 | AMPA glutamate receptor clustering(GO:0097113) glutamate receptor clustering(GO:0097688) |
| 0.1 | 0.5 | GO:1903644 | regulation of chaperone-mediated protein folding(GO:1903644) |
| 0.1 | 0.5 | GO:1902728 | positive regulation of skeletal muscle satellite cell proliferation(GO:1902724) positive regulation of growth factor dependent skeletal muscle satellite cell proliferation(GO:1902728) |
| 0.1 | 0.8 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.1 | 0.7 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.1 | 0.7 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.1 | 0.6 | GO:1903976 | negative regulation of glial cell migration(GO:1903976) |
| 0.1 | 0.2 | GO:0060981 | cell migration involved in coronary angiogenesis(GO:0060981) |
| 0.1 | 0.5 | GO:0032487 | regulation of Rap protein signal transduction(GO:0032487) |
| 0.1 | 0.2 | GO:0038178 | complement component C5a signaling pathway(GO:0038178) negative regulation of granulocyte chemotaxis(GO:0071623) negative regulation of neutrophil chemotaxis(GO:0090024) negative regulation of neutrophil migration(GO:1902623) |
| 0.1 | 1.1 | GO:0048245 | eosinophil chemotaxis(GO:0048245) |
| 0.1 | 0.5 | GO:0019933 | cAMP-mediated signaling(GO:0019933) |
| 0.1 | 0.3 | GO:0018262 | isopeptide cross-linking via N6-(L-isoglutamyl)-L-lysine(GO:0018153) isopeptide cross-linking(GO:0018262) |
| 0.1 | 0.3 | GO:1901094 | regulation of protein tetramerization(GO:1901090) negative regulation of protein tetramerization(GO:1901091) regulation of protein homotetramerization(GO:1901093) negative regulation of protein homotetramerization(GO:1901094) |
| 0.1 | 0.7 | GO:0001866 | NK T cell proliferation(GO:0001866) |
| 0.1 | 0.2 | GO:1904428 | negative regulation of tubulin deacetylation(GO:1904428) |
| 0.1 | 1.3 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.1 | 0.5 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 0.1 | 0.5 | GO:0002138 | retinoic acid biosynthetic process(GO:0002138) diterpenoid biosynthetic process(GO:0016102) |
| 0.0 | 1.2 | GO:1903543 | positive regulation of exosomal secretion(GO:1903543) |
| 0.0 | 0.9 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 1.1 | GO:0031915 | positive regulation of synaptic plasticity(GO:0031915) |
| 0.0 | 0.2 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.0 | 0.7 | GO:0045907 | positive regulation of vasoconstriction(GO:0045907) |
| 0.0 | 0.3 | GO:0098881 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.0 | 0.2 | GO:0010807 | regulation of synaptic vesicle priming(GO:0010807) |
| 0.0 | 0.6 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 0.2 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.0 | 0.8 | GO:2001135 | regulation of endocytic recycling(GO:2001135) |
| 0.0 | 0.6 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.8 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.2 | GO:0019242 | methylglyoxal biosynthetic process(GO:0019242) |
| 0.0 | 0.2 | GO:0018101 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.0 | 0.7 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.5 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 0.3 | GO:0070278 | extracellular matrix constituent secretion(GO:0070278) |
| 0.0 | 0.1 | GO:0090362 | positive regulation of platelet-derived growth factor production(GO:0090362) |
| 0.0 | 0.5 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.0 | 0.6 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.0 | 1.4 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 | 0.1 | GO:0015862 | uridine transport(GO:0015862) |
| 0.0 | 0.2 | GO:0070317 | negative regulation of G0 to G1 transition(GO:0070317) |
| 0.0 | 0.2 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.0 | 1.1 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.0 | 0.5 | GO:1902004 | positive regulation of beta-amyloid formation(GO:1902004) |
| 0.0 | 0.3 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.0 | 0.1 | GO:1903347 | negative regulation of bicellular tight junction assembly(GO:1903347) |
| 0.0 | 0.4 | GO:1903715 | regulation of aerobic respiration(GO:1903715) |
| 0.0 | 0.3 | GO:0051549 | positive regulation of keratinocyte migration(GO:0051549) |
| 0.0 | 0.4 | GO:0060020 | Bergmann glial cell differentiation(GO:0060020) |
| 0.0 | 0.3 | GO:0060012 | synaptic transmission, glycinergic(GO:0060012) |
| 0.0 | 0.1 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.0 | 0.3 | GO:0061470 | T follicular helper cell differentiation(GO:0061470) |
| 0.0 | 0.1 | GO:0009438 | methylglyoxal metabolic process(GO:0009438) |
| 0.0 | 0.4 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.0 | 0.1 | GO:0051582 | glial cell-derived neurotrophic factor secretion(GO:0044467) positive regulation of neurotransmitter uptake(GO:0051582) positive regulation of dopamine uptake involved in synaptic transmission(GO:0051586) positive regulation of catecholamine uptake involved in synaptic transmission(GO:0051944) regulation of glial cell-derived neurotrophic factor secretion(GO:1900166) positive regulation of glial cell-derived neurotrophic factor secretion(GO:1900168) |
| 0.0 | 0.4 | GO:0035092 | sperm chromatin condensation(GO:0035092) |
| 0.0 | 0.2 | GO:0071651 | positive regulation of chemokine (C-C motif) ligand 5 production(GO:0071651) |
| 0.0 | 0.1 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.0 | 0.8 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.0 | 0.7 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.0 | 0.3 | GO:0090154 | positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 0.0 | 0.3 | GO:0060742 | epithelial cell differentiation involved in prostate gland development(GO:0060742) |
| 0.0 | 0.2 | GO:0043932 | ossification involved in bone remodeling(GO:0043932) |
| 0.0 | 1.3 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.0 | 0.5 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.2 | GO:0042636 | negative regulation of hair cycle(GO:0042636) |
| 0.0 | 0.1 | GO:0051083 | 'de novo' cotranslational protein folding(GO:0051083) |
| 0.0 | 0.9 | GO:0006779 | porphyrin-containing compound biosynthetic process(GO:0006779) |
| 0.0 | 2.0 | GO:0045669 | positive regulation of osteoblast differentiation(GO:0045669) |
| 0.0 | 0.3 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 | 0.1 | GO:0006409 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.0 | 0.2 | GO:0014722 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) |
| 0.0 | 2.6 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 0.2 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.8 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.0 | 0.5 | GO:0006735 | NADH regeneration(GO:0006735) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.0 | 0.5 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.0 | 0.8 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.0 | 0.1 | GO:0002480 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-independent(GO:0002480) |
| 0.0 | 0.2 | GO:0006986 | response to unfolded protein(GO:0006986) response to topologically incorrect protein(GO:0035966) |
| 0.0 | 0.1 | GO:0090521 | glomerular visceral epithelial cell migration(GO:0090521) |
| 0.0 | 0.2 | GO:0003417 | growth plate cartilage development(GO:0003417) |
| 0.0 | 0.1 | GO:0021853 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.0 | 0.1 | GO:2001171 | sequestering of actin monomers(GO:0042989) positive regulation of ATP biosynthetic process(GO:2001171) |
| 0.0 | 1.6 | GO:0019233 | sensory perception of pain(GO:0019233) |
| 0.0 | 0.5 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 3.0 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.2 | 0.7 | GO:0034515 | proteasome storage granule(GO:0034515) |
| 0.2 | 0.7 | GO:0097444 | spine apparatus(GO:0097444) |
| 0.2 | 0.7 | GO:0033257 | Bcl3/NF-kappaB2 complex(GO:0033257) |
| 0.2 | 1.2 | GO:0033565 | ESCRT-0 complex(GO:0033565) |
| 0.1 | 0.5 | GO:1902912 | pyruvate kinase complex(GO:1902912) |
| 0.1 | 1.0 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.1 | 1.1 | GO:0070033 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) synaptobrevin 2-SNAP-25-syntaxin-3-complexin complex(GO:0070554) |
| 0.1 | 1.4 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.1 | 0.4 | GO:0044753 | amphisome(GO:0044753) |
| 0.1 | 3.3 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.1 | 1.2 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.1 | 0.9 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.1 | 0.5 | GO:0031673 | H zone(GO:0031673) |
| 0.1 | 0.3 | GO:1990796 | photoreceptor cell terminal bouton(GO:1990796) |
| 0.1 | 0.5 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.1 | 0.2 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.1 | 4.4 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.4 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.0 | 0.8 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.0 | 0.5 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.0 | 0.1 | GO:0002947 | tumor necrosis factor receptor superfamily complex(GO:0002947) |
| 0.0 | 0.7 | GO:0005883 | neurofilament(GO:0005883) |
| 0.0 | 0.3 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.0 | 0.9 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 1.3 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.2 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.0 | 0.2 | GO:0090498 | extrinsic component of Golgi membrane(GO:0090498) |
| 0.0 | 0.4 | GO:0030130 | clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.0 | 0.7 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.0 | 4.4 | GO:0042641 | actomyosin(GO:0042641) |
| 0.0 | 0.6 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.1 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.0 | 0.9 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.7 | GO:0032809 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 1.5 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.3 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.2 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.0 | 3.0 | GO:0101002 | ficolin-1-rich granule(GO:0101002) ficolin-1-rich granule lumen(GO:1904813) |
| 0.0 | 1.1 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.4 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 0.1 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.0 | 2.0 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 2.4 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 0.4 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.1 | GO:0044295 | axonal growth cone(GO:0044295) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.1 | GO:0070119 | ciliary neurotrophic factor binding(GO:0070119) |
| 0.4 | 1.5 | GO:0008260 | 3-oxoacid CoA-transferase activity(GO:0008260) |
| 0.4 | 1.1 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 0.4 | 1.4 | GO:0004031 | aldehyde oxidase activity(GO:0004031) |
| 0.4 | 1.4 | GO:0031811 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 0.3 | 3.6 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.2 | 1.0 | GO:0032551 | pyrimidine nucleoside binding(GO:0001884) UTP binding(GO:0002134) sulfonylurea receptor binding(GO:0017098) pyrimidine ribonucleoside binding(GO:0032551) |
| 0.2 | 1.3 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.2 | 1.9 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.1 | 1.5 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.1 | 0.9 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.1 | 0.5 | GO:0031728 | CCR3 chemokine receptor binding(GO:0031728) |
| 0.1 | 0.4 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.1 | 0.5 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.1 | 1.1 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.1 | 0.3 | GO:0047389 | glycerophosphocholine phosphodiesterase activity(GO:0047389) |
| 0.1 | 0.3 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.1 | 1.6 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.1 | 0.3 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
| 0.1 | 0.2 | GO:0004878 | complement component C5a receptor activity(GO:0004878) |
| 0.1 | 1.7 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.1 | 3.2 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.1 | 0.3 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |
| 0.1 | 0.8 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.1 | 0.5 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.1 | 0.2 | GO:0005019 | platelet-derived growth factor beta-receptor activity(GO:0005019) |
| 0.1 | 1.0 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.1 | 0.4 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.1 | 2.0 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.1 | 0.5 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.0 | 0.7 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 0.5 | GO:0008526 | phosphatidylcholine transporter activity(GO:0008525) phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.2 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.0 | 0.5 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.2 | GO:0004347 | glucose-6-phosphate isomerase activity(GO:0004347) |
| 0.0 | 0.9 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.0 | 0.2 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.0 | 0.7 | GO:0044213 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) intronic transcription regulatory region DNA binding(GO:0044213) |
| 0.0 | 0.4 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.0 | 0.3 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.0 | 0.2 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.0 | 0.3 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.0 | 0.2 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.0 | 0.2 | GO:0001602 | pancreatic polypeptide receptor activity(GO:0001602) |
| 0.0 | 1.8 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.1 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.0 | 1.0 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 1.7 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 1.3 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.0 | 0.3 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.0 | 0.5 | GO:0019841 | retinol binding(GO:0019841) |
| 0.0 | 0.3 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.0 | 1.9 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.6 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.4 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.0 | 1.1 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.9 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.7 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 1.2 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.5 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.5 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 1.0 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.6 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.1 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.6 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.1 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.0 | 0.9 | GO:0043531 | ADP binding(GO:0043531) |
| 0.0 | 0.2 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.0 | 0.8 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 0.1 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 0.2 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 0.4 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.4 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.0 | 1.3 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 2.7 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 1.4 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 0.7 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 2.4 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 3.6 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 2.1 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 2.4 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 1.7 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 0.5 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 1.4 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 0.7 | ST GAQ PATHWAY | G alpha q Pathway |
| 0.0 | 0.7 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 0.4 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.7 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 0.9 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 1.0 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 0.3 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.4 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 1.6 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.4 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.1 | 1.4 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.1 | 4.7 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 1.9 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 1.0 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.7 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.9 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.0 | 0.7 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.0 | 1.2 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 1.3 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.0 | 0.8 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.7 | REACTOME TRAF6 MEDIATED NFKB ACTIVATION | Genes involved in TRAF6 mediated NF-kB activation |
| 0.0 | 1.7 | REACTOME CA DEPENDENT EVENTS | Genes involved in Ca-dependent events |
| 0.0 | 1.1 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.5 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.8 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.9 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.4 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.1 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 1.1 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.9 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.3 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.8 | REACTOME REGULATION OF APOPTOSIS | Genes involved in Regulation of Apoptosis |
| 0.0 | 0.3 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.2 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |