Project
Illumina Body Map 2: averaged replicates
Navigation
Downloads
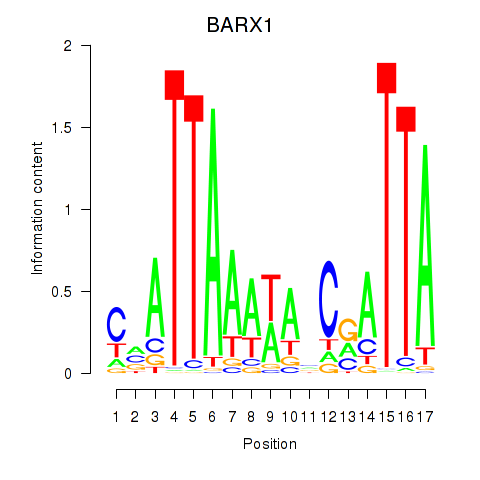
Results for BARX1
Z-value: 2.11
Transcription factors associated with BARX1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
BARX1
|
ENSG00000131668.9 | BARX homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| BARX1 | hg19_v2_chr9_-_96717654_96717666 | 0.52 | 2.4e-03 | Click! |
Activity profile of BARX1 motif
Sorted Z-values of BARX1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_150404904 | 4.97 |
ENST00000521632.1
|
GPX3
|
glutathione peroxidase 3 (plasma) |
| chr4_+_119810134 | 4.73 |
ENST00000434046.2
|
SYNPO2
|
synaptopodin 2 |
| chr3_-_172859017 | 4.62 |
ENST00000351008.3
|
SPATA16
|
spermatogenesis associated 16 |
| chr8_-_124749609 | 4.42 |
ENST00000262219.6
ENST00000419625.1 |
ANXA13
|
annexin A13 |
| chr7_-_16921601 | 4.39 |
ENST00000402239.3
ENST00000310398.2 ENST00000414935.1 |
AGR3
|
anterior gradient 3 |
| chr4_-_177162822 | 4.15 |
ENST00000512254.1
|
ASB5
|
ankyrin repeat and SOCS box containing 5 |
| chr4_+_119809984 | 3.91 |
ENST00000307142.4
ENST00000448416.2 ENST00000429713.2 |
SYNPO2
|
synaptopodin 2 |
| chr5_+_127039075 | 3.65 |
ENST00000514853.2
|
CTC-228N24.1
|
CTC-228N24.1 |
| chr12_-_71551652 | 3.62 |
ENST00000546561.1
|
TSPAN8
|
tetraspanin 8 |
| chr9_-_95166976 | 3.59 |
ENST00000447356.1
|
OGN
|
osteoglycin |
| chr12_+_101988627 | 3.59 |
ENST00000547405.1
ENST00000452455.2 ENST00000441232.1 ENST00000360610.2 ENST00000392934.3 ENST00000547509.1 ENST00000361685.2 ENST00000549145.1 ENST00000553190.1 |
MYBPC1
|
myosin binding protein C, slow type |
| chr15_-_76304731 | 3.26 |
ENST00000394907.3
|
NRG4
|
neuregulin 4 |
| chr11_+_12766583 | 3.26 |
ENST00000361985.2
|
TEAD1
|
TEA domain family member 1 (SV40 transcriptional enhancer factor) |
| chr9_-_95166884 | 3.24 |
ENST00000375561.5
|
OGN
|
osteoglycin |
| chr13_-_38172863 | 3.17 |
ENST00000541481.1
ENST00000379743.4 ENST00000379742.4 ENST00000379749.4 ENST00000541179.1 ENST00000379747.4 |
POSTN
|
periostin, osteoblast specific factor |
| chr9_-_95298254 | 3.15 |
ENST00000444490.2
|
ECM2
|
extracellular matrix protein 2, female organ and adipocyte specific |
| chr15_-_56757329 | 3.09 |
ENST00000260453.3
|
MNS1
|
meiosis-specific nuclear structural 1 |
| chr21_-_43735628 | 3.06 |
ENST00000291525.10
ENST00000518498.1 |
TFF3
|
trefoil factor 3 (intestinal) |
| chr19_-_55660561 | 3.06 |
ENST00000587758.1
ENST00000356783.5 ENST00000291901.8 ENST00000588426.1 ENST00000588147.1 ENST00000536926.1 ENST00000588981.1 |
TNNT1
|
troponin T type 1 (skeletal, slow) |
| chr11_-_102401469 | 3.02 |
ENST00000260227.4
|
MMP7
|
matrix metallopeptidase 7 (matrilysin, uterine) |
| chr13_+_23755054 | 2.96 |
ENST00000218867.3
|
SGCG
|
sarcoglycan, gamma (35kDa dystrophin-associated glycoprotein) |
| chr1_+_160160283 | 2.91 |
ENST00000368079.3
|
CASQ1
|
calsequestrin 1 (fast-twitch, skeletal muscle) |
| chr4_+_41540160 | 2.88 |
ENST00000503057.1
ENST00000511496.1 |
LIMCH1
|
LIM and calponin homology domains 1 |
| chr1_+_160160346 | 2.84 |
ENST00000368078.3
|
CASQ1
|
calsequestrin 1 (fast-twitch, skeletal muscle) |
| chr11_+_61976137 | 2.83 |
ENST00000244930.4
|
SCGB2A1
|
secretoglobin, family 2A, member 1 |
| chr12_-_71551868 | 2.82 |
ENST00000247829.3
|
TSPAN8
|
tetraspanin 8 |
| chr12_+_101988774 | 2.79 |
ENST00000545503.2
ENST00000536007.1 ENST00000541119.1 ENST00000361466.2 ENST00000551300.1 ENST00000550270.1 |
MYBPC1
|
myosin binding protein C, slow type |
| chr12_-_91572278 | 2.67 |
ENST00000425043.1
ENST00000420120.2 ENST00000441303.2 ENST00000456569.2 |
DCN
|
decorin |
| chr20_+_123010 | 2.63 |
ENST00000382398.3
|
DEFB126
|
defensin, beta 126 |
| chr11_+_60467047 | 2.61 |
ENST00000300226.2
|
MS4A8
|
membrane-spanning 4-domains, subfamily A, member 8 |
| chr11_+_100784231 | 2.59 |
ENST00000531183.1
|
ARHGAP42
|
Rho GTPase activating protein 42 |
| chr10_+_124670121 | 2.58 |
ENST00000368894.1
|
FAM24A
|
family with sequence similarity 24, member A |
| chr5_+_140474181 | 2.57 |
ENST00000194155.4
|
PCDHB2
|
protocadherin beta 2 |
| chr13_-_49987885 | 2.50 |
ENST00000409082.1
|
CAB39L
|
calcium binding protein 39-like |
| chr4_-_177190364 | 2.45 |
ENST00000296525.3
|
ASB5
|
ankyrin repeat and SOCS box containing 5 |
| chr9_-_95298927 | 2.44 |
ENST00000395534.2
|
ECM2
|
extracellular matrix protein 2, female organ and adipocyte specific |
| chr6_+_118869452 | 2.41 |
ENST00000357525.5
|
PLN
|
phospholamban |
| chr12_-_91574142 | 2.40 |
ENST00000547937.1
|
DCN
|
decorin |
| chr5_+_137203465 | 2.39 |
ENST00000239926.4
|
MYOT
|
myotilin |
| chr10_+_90354503 | 2.38 |
ENST00000531458.1
|
LIPJ
|
lipase, family member J |
| chr8_+_30244580 | 2.36 |
ENST00000523115.1
ENST00000519647.1 |
RBPMS
|
RNA binding protein with multiple splicing |
| chr10_-_128210005 | 2.36 |
ENST00000284694.7
ENST00000454341.1 ENST00000432642.1 ENST00000392694.1 |
C10orf90
|
chromosome 10 open reading frame 90 |
| chr17_+_68071389 | 2.36 |
ENST00000283936.1
ENST00000392671.1 |
KCNJ16
|
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chrX_-_15511438 | 2.35 |
ENST00000380420.5
|
PIR
|
pirin (iron-binding nuclear protein) |
| chr3_+_107096188 | 2.33 |
ENST00000261058.1
|
CCDC54
|
coiled-coil domain containing 54 |
| chr8_-_120259055 | 2.32 |
ENST00000522828.1
ENST00000523307.1 ENST00000524129.1 ENST00000521048.1 ENST00000522187.1 |
RP11-4K16.2
|
RP11-4K16.2 |
| chr4_+_52917451 | 2.25 |
ENST00000295213.4
ENST00000419395.2 |
SPATA18
|
spermatogenesis associated 18 |
| chr12_+_26348246 | 2.23 |
ENST00000422622.2
|
SSPN
|
sarcospan |
| chr4_+_169575875 | 2.20 |
ENST00000503457.1
|
PALLD
|
palladin, cytoskeletal associated protein |
| chr2_+_15840525 | 2.18 |
ENST00000436967.1
|
AC008271.1
|
AC008271.1 |
| chr1_-_185597619 | 2.18 |
ENST00000608417.1
ENST00000436955.1 |
GS1-204I12.1
|
GS1-204I12.1 |
| chr3_-_149093499 | 2.17 |
ENST00000472441.1
|
TM4SF1
|
transmembrane 4 L six family member 1 |
| chr9_-_95244781 | 2.16 |
ENST00000375544.3
ENST00000375543.1 ENST00000395538.3 ENST00000450139.2 |
ASPN
|
asporin |
| chr12_+_41831485 | 2.14 |
ENST00000539469.2
ENST00000298919.7 |
PDZRN4
|
PDZ domain containing ring finger 4 |
| chr10_-_79398250 | 2.13 |
ENST00000286627.5
|
KCNMA1
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr1_-_153085984 | 2.13 |
ENST00000468739.1
|
SPRR2F
|
small proline-rich protein 2F |
| chrX_+_36246735 | 2.12 |
ENST00000378653.3
|
CXorf30
|
chromosome X open reading frame 30 |
| chr10_-_10836919 | 2.06 |
ENST00000602763.1
ENST00000415590.2 ENST00000434919.2 |
SFTA1P
|
surfactant associated 1, pseudogene |
| chr1_+_36549676 | 2.03 |
ENST00000207457.3
|
TEKT2
|
tektin 2 (testicular) |
| chr19_-_36643329 | 2.01 |
ENST00000589154.1
|
COX7A1
|
cytochrome c oxidase subunit VIIa polypeptide 1 (muscle) |
| chr1_+_167063282 | 2.00 |
ENST00000361200.2
|
DUSP27
|
dual specificity phosphatase 27 (putative) |
| chrX_-_142605301 | 2.00 |
ENST00000370503.2
|
SPANXN3
|
SPANX family, member N3 |
| chr1_+_87012753 | 1.97 |
ENST00000370563.3
|
CLCA4
|
chloride channel accessory 4 |
| chr6_-_76072719 | 1.97 |
ENST00000370020.1
|
FILIP1
|
filamin A interacting protein 1 |
| chr8_+_134029937 | 1.95 |
ENST00000518108.1
|
TG
|
thyroglobulin |
| chr5_+_140552218 | 1.94 |
ENST00000231137.3
|
PCDHB7
|
protocadherin beta 7 |
| chr17_-_57158523 | 1.94 |
ENST00000581468.1
|
TRIM37
|
tripartite motif containing 37 |
| chr8_-_82395461 | 1.93 |
ENST00000256104.4
|
FABP4
|
fatty acid binding protein 4, adipocyte |
| chr7_-_88425025 | 1.92 |
ENST00000297203.2
|
C7orf62
|
chromosome 7 open reading frame 62 |
| chr5_+_140235469 | 1.91 |
ENST00000506939.2
ENST00000307360.5 |
PCDHA10
|
protocadherin alpha 10 |
| chr4_+_9446156 | 1.90 |
ENST00000334879.1
|
DEFB131
|
defensin, beta 131 |
| chr13_-_86373536 | 1.90 |
ENST00000400286.2
|
SLITRK6
|
SLIT and NTRK-like family, member 6 |
| chr10_-_115423792 | 1.88 |
ENST00000369360.3
ENST00000360478.3 ENST00000359988.3 ENST00000369358.4 |
NRAP
|
nebulin-related anchoring protein |
| chr12_+_12223867 | 1.87 |
ENST00000308721.5
|
BCL2L14
|
BCL2-like 14 (apoptosis facilitator) |
| chr2_+_168675182 | 1.85 |
ENST00000305861.1
|
B3GALT1
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 1 |
| chr9_-_95166841 | 1.85 |
ENST00000262551.4
|
OGN
|
osteoglycin |
| chr12_-_10978957 | 1.85 |
ENST00000240619.2
|
TAS2R10
|
taste receptor, type 2, member 10 |
| chr11_+_60467142 | 1.85 |
ENST00000529752.1
|
MS4A8
|
membrane-spanning 4-domains, subfamily A, member 8 |
| chr22_+_18721427 | 1.83 |
ENST00000342888.3
|
AC008132.1
|
Uncharacterized protein |
| chr2_+_158114051 | 1.82 |
ENST00000259056.4
|
GALNT5
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 5 (GalNAc-T5) |
| chr14_-_71001708 | 1.81 |
ENST00000256389.3
|
ADAM20
|
ADAM metallopeptidase domain 20 |
| chr17_-_19265982 | 1.80 |
ENST00000268841.6
ENST00000261499.4 ENST00000575478.1 |
B9D1
|
B9 protein domain 1 |
| chr5_-_16742330 | 1.79 |
ENST00000505695.1
ENST00000427430.2 |
MYO10
|
myosin X |
| chr10_-_69597915 | 1.77 |
ENST00000225171.2
|
DNAJC12
|
DnaJ (Hsp40) homolog, subfamily C, member 12 |
| chr6_-_84419101 | 1.77 |
ENST00000520302.1
ENST00000520213.1 ENST00000439399.2 ENST00000428679.2 ENST00000437520.1 |
SNAP91
|
synaptosomal-associated protein, 91kDa |
| chr5_+_175288631 | 1.76 |
ENST00000509837.1
|
CPLX2
|
complexin 2 |
| chr1_+_172389821 | 1.76 |
ENST00000367727.4
|
C1orf105
|
chromosome 1 open reading frame 105 |
| chr12_+_26274917 | 1.73 |
ENST00000538142.1
|
SSPN
|
sarcospan |
| chr20_+_31805131 | 1.72 |
ENST00000375454.3
ENST00000375452.3 |
BPIFA3
|
BPI fold containing family A, member 3 |
| chr11_-_77734260 | 1.71 |
ENST00000353172.5
|
KCTD14
|
potassium channel tetramerization domain containing 14 |
| chr1_-_72566613 | 1.70 |
ENST00000306821.3
|
NEGR1
|
neuronal growth regulator 1 |
| chr5_+_20616500 | 1.70 |
ENST00000512688.1
|
RP11-774D14.1
|
RP11-774D14.1 |
| chr15_+_86686953 | 1.70 |
ENST00000421325.2
|
AGBL1
|
ATP/GTP binding protein-like 1 |
| chr11_-_114430570 | 1.69 |
ENST00000251921.2
|
NXPE1
|
neurexophilin and PC-esterase domain family, member 1 |
| chr8_-_86253888 | 1.69 |
ENST00000522389.1
ENST00000432364.2 ENST00000517618.1 |
CA1
|
carbonic anhydrase I |
| chrX_-_49965663 | 1.67 |
ENST00000376056.2
ENST00000376058.2 ENST00000358526.2 |
AKAP4
|
A kinase (PRKA) anchor protein 4 |
| chr1_-_20306909 | 1.67 |
ENST00000375111.3
ENST00000400520.3 |
PLA2G2A
|
phospholipase A2, group IIA (platelets, synovial fluid) |
| chr11_+_7110165 | 1.67 |
ENST00000306904.5
|
RBMXL2
|
RNA binding motif protein, X-linked-like 2 |
| chr3_-_146213722 | 1.66 |
ENST00000336685.2
ENST00000489015.1 |
PLSCR2
|
phospholipid scramblase 2 |
| chr3_-_93747425 | 1.65 |
ENST00000315099.2
|
STX19
|
syntaxin 19 |
| chr3_-_164796269 | 1.65 |
ENST00000264382.3
|
SI
|
sucrase-isomaltase (alpha-glucosidase) |
| chr10_+_51549498 | 1.64 |
ENST00000358559.2
ENST00000298239.6 |
MSMB
|
microseminoprotein, beta- |
| chr5_-_135290651 | 1.62 |
ENST00000522943.1
ENST00000514447.2 |
LECT2
|
leukocyte cell-derived chemotaxin 2 |
| chr5_-_42887494 | 1.62 |
ENST00000514218.1
|
SEPP1
|
selenoprotein P, plasma, 1 |
| chr12_-_52911718 | 1.62 |
ENST00000548409.1
|
KRT5
|
keratin 5 |
| chr5_+_121465207 | 1.60 |
ENST00000296600.4
|
ZNF474
|
zinc finger protein 474 |
| chr17_-_66951474 | 1.60 |
ENST00000269080.2
|
ABCA8
|
ATP-binding cassette, sub-family A (ABC1), member 8 |
| chr1_-_153029980 | 1.59 |
ENST00000392653.2
|
SPRR2A
|
small proline-rich protein 2A |
| chr17_-_66951382 | 1.58 |
ENST00000586539.1
|
ABCA8
|
ATP-binding cassette, sub-family A (ABC1), member 8 |
| chr15_-_78913521 | 1.58 |
ENST00000326828.5
|
CHRNA3
|
cholinergic receptor, nicotinic, alpha 3 (neuronal) |
| chr9_-_28670283 | 1.55 |
ENST00000379992.2
|
LINGO2
|
leucine rich repeat and Ig domain containing 2 |
| chrX_-_103499602 | 1.55 |
ENST00000372588.4
|
ESX1
|
ESX homeobox 1 |
| chr6_-_28303901 | 1.55 |
ENST00000439158.1
ENST00000446474.1 ENST00000414431.1 ENST00000344279.6 ENST00000453745.1 |
ZSCAN31
|
zinc finger and SCAN domain containing 31 |
| chr22_-_40289759 | 1.54 |
ENST00000325157.6
|
ENTHD1
|
ENTH domain containing 1 |
| chr15_+_84115868 | 1.54 |
ENST00000427482.2
|
SH3GL3
|
SH3-domain GRB2-like 3 |
| chr4_-_171011323 | 1.53 |
ENST00000509167.1
ENST00000353187.2 ENST00000507375.1 ENST00000515480.1 |
AADAT
|
aminoadipate aminotransferase |
| chr21_+_17566643 | 1.53 |
ENST00000419952.1
ENST00000445461.2 |
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr22_+_26138108 | 1.53 |
ENST00000536101.1
ENST00000335473.7 ENST00000407587.2 |
MYO18B
|
myosin XVIIIB |
| chr9_-_21351377 | 1.52 |
ENST00000380210.1
|
IFNA6
|
interferon, alpha 6 |
| chr4_-_144940477 | 1.51 |
ENST00000513128.1
ENST00000429670.2 ENST00000502664.1 |
GYPB
|
glycophorin B (MNS blood group) |
| chr8_+_134125727 | 1.50 |
ENST00000521107.1
|
TG
|
thyroglobulin |
| chr19_+_56905024 | 1.49 |
ENST00000591172.1
ENST00000589888.1 ENST00000587979.1 ENST00000585659.1 ENST00000593109.1 |
ZNF582-AS1
|
ZNF582 antisense RNA 1 (head to head) |
| chr5_+_121465234 | 1.49 |
ENST00000504912.1
ENST00000505843.1 |
ZNF474
|
zinc finger protein 474 |
| chrX_+_69501943 | 1.49 |
ENST00000509895.1
ENST00000374473.2 ENST00000276066.4 |
RAB41
|
RAB41, member RAS oncogene family |
| chr2_+_74011316 | 1.48 |
ENST00000409561.1
|
C2orf78
|
chromosome 2 open reading frame 78 |
| chr10_-_79397740 | 1.47 |
ENST00000372440.1
ENST00000480683.1 |
KCNMA1
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr1_-_100643765 | 1.47 |
ENST00000370137.1
ENST00000370138.1 ENST00000342895.3 |
LRRC39
|
leucine rich repeat containing 39 |
| chr7_-_100253993 | 1.46 |
ENST00000461605.1
ENST00000160382.5 |
ACTL6B
|
actin-like 6B |
| chr11_+_114168085 | 1.45 |
ENST00000541754.1
|
NNMT
|
nicotinamide N-methyltransferase |
| chr13_+_73629107 | 1.45 |
ENST00000539231.1
|
KLF5
|
Kruppel-like factor 5 (intestinal) |
| chr18_-_47721447 | 1.45 |
ENST00000285039.7
|
MYO5B
|
myosin VB |
| chrX_+_107020963 | 1.44 |
ENST00000509000.2
|
NCBP2L
|
nuclear cap binding protein subunit 2-like |
| chr1_-_145076186 | 1.44 |
ENST00000369348.3
|
PDE4DIP
|
phosphodiesterase 4D interacting protein |
| chr8_-_28347737 | 1.42 |
ENST00000517673.1
ENST00000518734.1 ENST00000346498.2 ENST00000380254.2 |
FBXO16
|
F-box protein 16 |
| chr2_+_210517895 | 1.42 |
ENST00000447185.1
|
MAP2
|
microtubule-associated protein 2 |
| chr10_+_90346519 | 1.41 |
ENST00000371939.3
|
LIPJ
|
lipase, family member J |
| chr5_+_140588269 | 1.39 |
ENST00000541609.1
ENST00000239450.2 |
PCDHB12
|
protocadherin beta 12 |
| chr16_-_20681177 | 1.39 |
ENST00000524149.1
|
ACSM1
|
acyl-CoA synthetase medium-chain family member 1 |
| chr22_-_32767017 | 1.38 |
ENST00000400234.1
|
RFPL3S
|
RFPL3 antisense |
| chr4_-_69817481 | 1.37 |
ENST00000251566.4
|
UGT2A3
|
UDP glucuronosyltransferase 2 family, polypeptide A3 |
| chr3_-_145881432 | 1.36 |
ENST00000469350.1
|
PLOD2
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 2 |
| chr8_-_33457453 | 1.36 |
ENST00000523956.1
ENST00000256261.4 |
DUSP26
|
dual specificity phosphatase 26 (putative) |
| chr3_+_26735991 | 1.35 |
ENST00000456208.2
|
LRRC3B
|
leucine rich repeat containing 3B |
| chr22_+_51176624 | 1.35 |
ENST00000216139.5
ENST00000529621.1 |
ACR
|
acrosin |
| chrX_-_114253536 | 1.33 |
ENST00000371936.1
|
IL13RA2
|
interleukin 13 receptor, alpha 2 |
| chr6_-_55443831 | 1.33 |
ENST00000428842.1
ENST00000358072.5 ENST00000508459.1 |
HMGCLL1
|
3-hydroxymethyl-3-methylglutaryl-CoA lyase-like 1 |
| chrY_+_20708557 | 1.32 |
ENST00000307393.2
ENST00000309834.4 ENST00000382856.2 |
HSFY1
|
heat shock transcription factor, Y-linked 1 |
| chr15_-_74659978 | 1.31 |
ENST00000541301.1
ENST00000416978.1 ENST00000268053.6 |
CYP11A1
|
cytochrome P450, family 11, subfamily A, polypeptide 1 |
| chr12_+_12224331 | 1.31 |
ENST00000396367.1
ENST00000266434.4 ENST00000396369.1 |
BCL2L14
|
BCL2-like 14 (apoptosis facilitator) |
| chr18_+_5748793 | 1.30 |
ENST00000566533.1
ENST00000562452.2 |
RP11-945C19.1
|
RP11-945C19.1 |
| chr1_-_151148442 | 1.30 |
ENST00000441701.1
ENST00000416280.2 |
TMOD4
|
tropomodulin 4 (muscle) |
| chr15_-_72978490 | 1.30 |
ENST00000311755.3
|
HIGD2B
|
HIG1 hypoxia inducible domain family, member 2B |
| chr3_+_8543393 | 1.30 |
ENST00000157600.3
ENST00000415597.1 ENST00000535732.1 |
LMCD1
|
LIM and cysteine-rich domains 1 |
| chr6_-_150212029 | 1.30 |
ENST00000529948.1
ENST00000357183.4 ENST00000367363.3 |
RAET1E
|
retinoic acid early transcript 1E |
| chr12_+_103545593 | 1.30 |
ENST00000547418.1
|
RP11-552I14.1
|
Uncharacterized protein |
| chr14_+_38264418 | 1.28 |
ENST00000267368.7
ENST00000382320.3 |
TTC6
|
tetratricopeptide repeat domain 6 |
| chr15_+_54901540 | 1.28 |
ENST00000539562.2
|
UNC13C
|
unc-13 homolog C (C. elegans) |
| chrX_-_110507098 | 1.28 |
ENST00000541758.1
|
CAPN6
|
calpain 6 |
| chr4_-_168155169 | 1.28 |
ENST00000534949.1
ENST00000535728.1 |
SPOCK3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr3_+_189349162 | 1.28 |
ENST00000264731.3
ENST00000382063.4 ENST00000418709.2 ENST00000320472.5 ENST00000392460.3 ENST00000440651.2 |
TP63
|
tumor protein p63 |
| chr12_-_52604607 | 1.27 |
ENST00000551894.1
ENST00000553017.1 |
C12orf80
|
chromosome 12 open reading frame 80 |
| chr15_-_42448788 | 1.27 |
ENST00000382396.4
ENST00000397272.3 |
PLA2G4F
|
phospholipase A2, group IVF |
| chr22_-_32766972 | 1.27 |
ENST00000382084.4
ENST00000382086.2 |
RFPL3S
|
RFPL3 antisense |
| chr7_-_137028498 | 1.27 |
ENST00000393083.2
|
PTN
|
pleiotrophin |
| chr6_+_28249332 | 1.26 |
ENST00000259883.3
|
PGBD1
|
piggyBac transposable element derived 1 |
| chr6_-_52859046 | 1.26 |
ENST00000457564.1
ENST00000541324.1 ENST00000370960.1 |
GSTA4
|
glutathione S-transferase alpha 4 |
| chr6_-_55443975 | 1.25 |
ENST00000308161.4
ENST00000398661.2 ENST00000274901.4 |
HMGCLL1
|
3-hydroxymethyl-3-methylglutaryl-CoA lyase-like 1 |
| chr1_+_182419261 | 1.25 |
ENST00000294854.8
ENST00000542961.1 |
RGSL1
|
regulator of G-protein signaling like 1 |
| chr7_-_122339162 | 1.24 |
ENST00000340112.2
|
RNF133
|
ring finger protein 133 |
| chr11_-_114430588 | 1.24 |
ENST00000534921.1
|
NXPE1
|
neurexophilin and PC-esterase domain family, member 1 |
| chrX_+_8432871 | 1.23 |
ENST00000381032.1
ENST00000453306.1 ENST00000444481.1 |
VCX3B
|
variable charge, X-linked 3B |
| chr4_-_137842536 | 1.23 |
ENST00000512039.1
|
RP11-138I17.1
|
RP11-138I17.1 |
| chr7_-_117067541 | 1.23 |
ENST00000284629.2
|
ASZ1
|
ankyrin repeat, SAM and basic leucine zipper domain containing 1 |
| chr7_+_134430212 | 1.22 |
ENST00000436461.2
|
CALD1
|
caldesmon 1 |
| chr3_+_155860751 | 1.22 |
ENST00000471742.1
|
KCNAB1
|
potassium voltage-gated channel, shaker-related subfamily, beta member 1 |
| chr1_+_16083154 | 1.22 |
ENST00000375771.1
|
FBLIM1
|
filamin binding LIM protein 1 |
| chr1_-_203320617 | 1.22 |
ENST00000354955.4
|
FMOD
|
fibromodulin |
| chr5_+_169011033 | 1.22 |
ENST00000513795.1
|
SPDL1
|
spindle apparatus coiled-coil protein 1 |
| chr6_+_123100620 | 1.21 |
ENST00000368444.3
|
FABP7
|
fatty acid binding protein 7, brain |
| chr2_+_159651821 | 1.20 |
ENST00000309950.3
ENST00000409042.1 |
DAPL1
|
death associated protein-like 1 |
| chr10_+_19777984 | 1.20 |
ENST00000377265.3
ENST00000455457.2 |
C10orf112
|
MAM and LDL receptor class A domain containing 1 |
| chr14_-_35183886 | 1.19 |
ENST00000298159.6
|
CFL2
|
cofilin 2 (muscle) |
| chr20_-_45179213 | 1.19 |
ENST00000279028.2
|
OCSTAMP
|
osteoclast stimulatory transmembrane protein |
| chr13_-_28562791 | 1.19 |
ENST00000332715.5
|
URAD
|
ureidoimidazoline (2-oxo-4-hydroxy-4-carboxy-5-) decarboxylase |
| chrX_-_114252193 | 1.19 |
ENST00000243213.1
|
IL13RA2
|
interleukin 13 receptor, alpha 2 |
| chr17_-_19265855 | 1.19 |
ENST00000440841.1
ENST00000395615.1 ENST00000461069.2 |
B9D1
|
B9 protein domain 1 |
| chr4_-_157563459 | 1.19 |
ENST00000504237.1
ENST00000511399.1 |
RP11-171N4.2
|
Uncharacterized protein |
| chr22_-_36220420 | 1.17 |
ENST00000473487.2
|
RBFOX2
|
RNA binding protein, fox-1 homolog (C. elegans) 2 |
| chr5_+_140762268 | 1.17 |
ENST00000518325.1
|
PCDHGA7
|
protocadherin gamma subfamily A, 7 |
| chr2_+_102803433 | 1.16 |
ENST00000264257.2
ENST00000441515.2 |
IL1RL2
|
interleukin 1 receptor-like 2 |
| chr19_+_29456034 | 1.16 |
ENST00000589921.1
|
LINC00906
|
long intergenic non-protein coding RNA 906 |
| chr3_-_123512688 | 1.16 |
ENST00000475616.1
|
MYLK
|
myosin light chain kinase |
| chr14_+_85994943 | 1.16 |
ENST00000553678.1
|
RP11-497E19.2
|
Uncharacterized protein |
| chr14_+_101295638 | 1.15 |
ENST00000523671.2
|
MEG3
|
maternally expressed 3 (non-protein coding) |
| chr9_+_105757590 | 1.14 |
ENST00000374798.3
ENST00000487798.1 |
CYLC2
|
cylicin, basic protein of sperm head cytoskeleton 2 |
| chr1_-_68516393 | 1.14 |
ENST00000395201.1
|
DIRAS3
|
DIRAS family, GTP-binding RAS-like 3 |
| chr10_-_29923893 | 1.13 |
ENST00000355867.4
|
SVIL
|
supervillin |
| chr9_+_108463234 | 1.13 |
ENST00000374688.1
|
TMEM38B
|
transmembrane protein 38B |
| chr11_-_13517565 | 1.13 |
ENST00000282091.1
ENST00000529816.1 |
PTH
|
parathyroid hormone |
| chr8_-_86290333 | 1.13 |
ENST00000521846.1
ENST00000523022.1 ENST00000524324.1 ENST00000519991.1 ENST00000520663.1 ENST00000517590.1 ENST00000522579.1 ENST00000522814.1 ENST00000522662.1 ENST00000523858.1 ENST00000519129.1 |
CA1
|
carbonic anhydrase I |
Network of associatons between targets according to the STRING database.
First level regulatory network of BARX1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.4 | GO:1901877 | regulation of calcium ion binding(GO:1901876) negative regulation of calcium ion binding(GO:1901877) regulation of calcium ion import into sarcoplasmic reticulum(GO:1902080) negative regulation of calcium ion import into sarcoplasmic reticulum(GO:1902081) |
| 0.7 | 4.7 | GO:0006982 | response to lipid hydroperoxide(GO:0006982) |
| 0.6 | 1.3 | GO:1904397 | negative regulation of neuromuscular junction development(GO:1904397) |
| 0.6 | 2.3 | GO:0090290 | positive regulation of osteoclast proliferation(GO:0090290) |
| 0.6 | 2.3 | GO:0060084 | synaptic transmission involved in micturition(GO:0060084) |
| 0.5 | 3.2 | GO:0032571 | response to vitamin K(GO:0032571) regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904207) positive regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904209) bone regeneration(GO:1990523) |
| 0.5 | 6.7 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.4 | 2.2 | GO:0070171 | negative regulation of tooth mineralization(GO:0070171) |
| 0.4 | 9.4 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.4 | 4.7 | GO:0034465 | response to carbon monoxide(GO:0034465) |
| 0.4 | 5.7 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.4 | 8.2 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.4 | 1.1 | GO:0002025 | vasodilation by norepinephrine-epinephrine involved in regulation of systemic arterial blood pressure(GO:0002025) |
| 0.4 | 1.1 | GO:1902299 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.3 | 1.0 | GO:0003365 | establishment of cell polarity involved in ameboidal cell migration(GO:0003365) |
| 0.3 | 1.4 | GO:0018874 | benzoate metabolic process(GO:0018874) |
| 0.3 | 3.1 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.3 | 1.4 | GO:0046947 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.3 | 1.4 | GO:1902310 | positive regulation of peptidyl-serine dephosphorylation(GO:1902310) |
| 0.3 | 2.0 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.3 | 1.3 | GO:0031455 | glycine betaine biosynthetic process from choline(GO:0019285) glycine betaine metabolic process(GO:0031455) glycine betaine biosynthetic process(GO:0031456) |
| 0.3 | 2.6 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.3 | 1.9 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.3 | 1.3 | GO:0007499 | ectoderm and mesoderm interaction(GO:0007499) |
| 0.3 | 2.2 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.3 | 0.8 | GO:0042270 | protection from natural killer cell mediated cytotoxicity(GO:0042270) |
| 0.3 | 1.3 | GO:0003277 | apoptotic process involved in endocardial cushion morphogenesis(GO:0003277) intermediate mesoderm morphogenesis(GO:0048390) intermediate mesoderm formation(GO:0048391) intermediate mesodermal cell differentiation(GO:0048392) regulation of cardiac muscle fiber development(GO:0055018) positive regulation of cardiac muscle fiber development(GO:0055020) bud dilation involved in lung branching(GO:0060503) BMP signaling pathway involved in ureter morphogenesis(GO:0061149) renal system segmentation(GO:0061150) BMP signaling pathway involved in renal system segmentation(GO:0061151) pulmonary artery endothelial tube morphogenesis(GO:0061155) regulation of transcription from RNA polymerase II promoter involved in mesonephros development(GO:0061216) BMP signaling pathway involved in nephric duct formation(GO:0071893) negative regulation of branch elongation involved in ureteric bud branching(GO:0072096) negative regulation of branch elongation involved in ureteric bud branching by BMP signaling pathway(GO:0072097) anterior/posterior pattern specification involved in ureteric bud development(GO:0072099) specification of ureteric bud anterior/posterior symmetry(GO:0072100) specification of ureteric bud anterior/posterior symmetry by BMP signaling pathway(GO:0072101) ureter epithelial cell differentiation(GO:0072192) negative regulation of mesenchymal cell proliferation involved in ureter development(GO:0072200) positive regulation of cell proliferation involved in outflow tract morphogenesis(GO:1901964) cardiac jelly development(GO:1905072) regulation of metanephric S-shaped body morphogenesis(GO:2000004) negative regulation of metanephric S-shaped body morphogenesis(GO:2000005) regulation of metanephric comma-shaped body morphogenesis(GO:2000006) negative regulation of metanephric comma-shaped body morphogenesis(GO:2000007) |
| 0.3 | 2.6 | GO:0035609 | C-terminal protein deglutamylation(GO:0035609) |
| 0.3 | 0.8 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.3 | 1.5 | GO:0033512 | L-lysine catabolic process to acetyl-CoA via saccharopine(GO:0033512) L-kynurenine metabolic process(GO:0097052) |
| 0.3 | 2.5 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.3 | 2.5 | GO:0043305 | negative regulation of mast cell degranulation(GO:0043305) |
| 0.2 | 0.7 | GO:0000921 | septin ring assembly(GO:0000921) septin ring organization(GO:0031106) |
| 0.2 | 0.9 | GO:1901350 | cell-cell signaling involved in cell-cell junction organization(GO:1901350) |
| 0.2 | 1.7 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.2 | 3.4 | GO:0015705 | iodide transport(GO:0015705) |
| 0.2 | 1.3 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.2 | 1.1 | GO:0070173 | regulation of enamel mineralization(GO:0070173) |
| 0.2 | 0.7 | GO:0006481 | C-terminal protein methylation(GO:0006481) |
| 0.2 | 0.8 | GO:0086097 | phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) |
| 0.2 | 0.6 | GO:0046603 | negative regulation of mitotic centrosome separation(GO:0046603) |
| 0.2 | 0.6 | GO:0021912 | regulation of transcription from RNA polymerase II promoter involved in spinal cord motor neuron fate specification(GO:0021912) |
| 0.2 | 0.8 | GO:0032053 | ciliary basal body organization(GO:0032053) |
| 0.2 | 0.8 | GO:0036116 | medium-chain fatty-acyl-CoA catabolic process(GO:0036114) long-chain fatty-acyl-CoA catabolic process(GO:0036116) palmitic acid metabolic process(GO:1900533) palmitic acid biosynthetic process(GO:1900535) |
| 0.2 | 1.0 | GO:0046726 | positive regulation by virus of viral protein levels in host cell(GO:0046726) |
| 0.2 | 0.6 | GO:0051758 | homologous chromosome movement towards spindle pole involved in homologous chromosome segregation(GO:0051758) |
| 0.2 | 1.4 | GO:0032439 | endosome localization(GO:0032439) |
| 0.2 | 0.9 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.2 | 1.1 | GO:0061624 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 0.2 | 0.7 | GO:1902725 | negative regulation of skeletal muscle cell proliferation(GO:0014859) negative regulation of skeletal muscle satellite cell proliferation(GO:1902723) negative regulation of satellite cell differentiation(GO:1902725) |
| 0.2 | 0.2 | GO:0050965 | detection of temperature stimulus involved in sensory perception(GO:0050961) detection of temperature stimulus involved in sensory perception of pain(GO:0050965) |
| 0.2 | 1.1 | GO:0000165 | MAPK cascade(GO:0000165) |
| 0.2 | 0.7 | GO:0046490 | isopentenyl diphosphate biosynthetic process(GO:0009240) isopentenyl diphosphate metabolic process(GO:0046490) dimethylallyl diphosphate biosynthetic process(GO:0050992) dimethylallyl diphosphate metabolic process(GO:0050993) |
| 0.2 | 0.9 | GO:0010513 | positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) |
| 0.2 | 1.2 | GO:0000255 | allantoin metabolic process(GO:0000255) |
| 0.2 | 2.2 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.2 | 2.8 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.2 | 2.9 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.2 | 1.1 | GO:0033133 | positive regulation of glucokinase activity(GO:0033133) positive regulation of hexokinase activity(GO:1903301) |
| 0.2 | 0.6 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.2 | 2.5 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 0.2 | 0.9 | GO:0003383 | apical constriction(GO:0003383) |
| 0.2 | 1.5 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.2 | 0.6 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.1 | 4.0 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.1 | 1.0 | GO:0002786 | regulation of antimicrobial peptide production(GO:0002784) regulation of antibacterial peptide production(GO:0002786) |
| 0.1 | 0.4 | GO:0000454 | snoRNA guided rRNA pseudouridine synthesis(GO:0000454) |
| 0.1 | 0.4 | GO:0046707 | IDP metabolic process(GO:0046707) IDP catabolic process(GO:0046709) |
| 0.1 | 0.7 | GO:0038108 | negative regulation of appetite by leptin-mediated signaling pathway(GO:0038108) |
| 0.1 | 1.2 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.1 | 1.7 | GO:0032596 | protein transport into membrane raft(GO:0032596) |
| 0.1 | 1.2 | GO:0072592 | oxygen metabolic process(GO:0072592) |
| 0.1 | 2.9 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.1 | 1.3 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.1 | 1.2 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.1 | 0.5 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.1 | 0.8 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.1 | 0.8 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.1 | 3.3 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.1 | 0.4 | GO:0032594 | protein transport within lipid bilayer(GO:0032594) |
| 0.1 | 0.4 | GO:1901382 | chorionic trophoblast cell proliferation(GO:0097360) regulation of chorionic trophoblast cell proliferation(GO:1901382) |
| 0.1 | 0.7 | GO:0070172 | positive regulation of tooth mineralization(GO:0070172) |
| 0.1 | 1.9 | GO:0071285 | cellular response to lithium ion(GO:0071285) |
| 0.1 | 2.3 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.1 | 2.4 | GO:0036149 | phosphatidylinositol acyl-chain remodeling(GO:0036149) |
| 0.1 | 2.3 | GO:0060391 | positive regulation of SMAD protein import into nucleus(GO:0060391) |
| 0.1 | 0.6 | GO:0002331 | pre-B cell allelic exclusion(GO:0002331) |
| 0.1 | 0.9 | GO:0055011 | atrial cardiac muscle cell differentiation(GO:0055011) atrial cardiac muscle cell development(GO:0055014) |
| 0.1 | 0.6 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.1 | 1.0 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.1 | 2.0 | GO:0044849 | estrous cycle(GO:0044849) |
| 0.1 | 0.4 | GO:0014038 | regulation of Schwann cell differentiation(GO:0014038) |
| 0.1 | 1.1 | GO:0070836 | caveola assembly(GO:0070836) |
| 0.1 | 0.3 | GO:1901253 | negative regulation of intracellular transport of viral material(GO:1901253) |
| 0.1 | 1.3 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.1 | 0.7 | GO:2000117 | negative regulation of cysteine-type endopeptidase activity(GO:2000117) |
| 0.1 | 0.3 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.1 | 0.5 | GO:0070980 | biphenyl catabolic process(GO:0070980) |
| 0.1 | 1.8 | GO:0031915 | positive regulation of synaptic plasticity(GO:0031915) |
| 0.1 | 5.5 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.1 | 0.6 | GO:0060011 | Sertoli cell proliferation(GO:0060011) |
| 0.1 | 1.0 | GO:0042670 | retinal cone cell differentiation(GO:0042670) retinal cone cell development(GO:0046549) |
| 0.1 | 0.9 | GO:0021694 | cerebellar Purkinje cell layer formation(GO:0021694) cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.1 | 8.6 | GO:0032233 | positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.1 | 1.2 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.1 | 3.3 | GO:1902895 | positive regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902895) |
| 0.1 | 0.7 | GO:2000576 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.1 | 0.6 | GO:1900264 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.1 | 0.8 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.1 | 1.4 | GO:0034356 | NAD biosynthesis via nicotinamide riboside salvage pathway(GO:0034356) |
| 0.1 | 1.1 | GO:0032074 | negative regulation of nuclease activity(GO:0032074) |
| 0.1 | 0.4 | GO:0061760 | antifungal innate immune response(GO:0061760) |
| 0.1 | 0.6 | GO:0051673 | membrane disruption in other organism(GO:0051673) |
| 0.1 | 0.6 | GO:0021940 | positive regulation of cerebellar granule cell precursor proliferation(GO:0021940) |
| 0.1 | 0.6 | GO:0070383 | DNA cytosine deamination(GO:0070383) |
| 0.1 | 1.3 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.1 | 0.7 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.1 | 1.7 | GO:0099500 | synaptic vesicle fusion to presynaptic active zone membrane(GO:0031629) vesicle fusion to plasma membrane(GO:0099500) |
| 0.1 | 1.2 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.1 | 1.4 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.1 | 0.7 | GO:0038166 | angiotensin-activated signaling pathway(GO:0038166) |
| 0.1 | 3.0 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.1 | 1.1 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.1 | 1.7 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.1 | 0.4 | GO:1902731 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) negative regulation of chondrocyte proliferation(GO:1902731) |
| 0.1 | 0.4 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.1 | 1.4 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.1 | 1.2 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.1 | 0.9 | GO:0003310 | pancreatic A cell differentiation(GO:0003310) |
| 0.1 | 0.5 | GO:0006578 | amino-acid betaine biosynthetic process(GO:0006578) |
| 0.1 | 0.2 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.1 | 0.5 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 0.1 | 2.1 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 0.8 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.1 | 2.3 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.1 | 0.4 | GO:1902897 | regulation of postsynaptic density protein 95 clustering(GO:1902897) |
| 0.1 | 2.7 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.1 | 1.4 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.1 | 0.8 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.1 | 0.4 | GO:0006987 | activation of signaling protein activity involved in unfolded protein response(GO:0006987) |
| 0.1 | 1.1 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.1 | 0.6 | GO:0044821 | meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.1 | 1.3 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.1 | 0.2 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.1 | 0.9 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.1 | 0.5 | GO:0042256 | mature ribosome assembly(GO:0042256) |
| 0.1 | 0.8 | GO:0007213 | G-protein coupled acetylcholine receptor signaling pathway(GO:0007213) |
| 0.0 | 10.2 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.3 | GO:0010637 | negative regulation of mitochondrial fusion(GO:0010637) |
| 0.0 | 0.2 | GO:0016598 | protein arginylation(GO:0016598) |
| 0.0 | 1.5 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.0 | 0.2 | GO:0061034 | olfactory bulb mitral cell layer development(GO:0061034) |
| 0.0 | 1.6 | GO:0060080 | inhibitory postsynaptic potential(GO:0060080) |
| 0.0 | 0.9 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.0 | 2.8 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.0 | 0.8 | GO:0098719 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 4.1 | GO:0042733 | embryonic digit morphogenesis(GO:0042733) |
| 0.0 | 1.1 | GO:1900364 | negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.0 | 0.6 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.0 | 0.7 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.0 | 1.2 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 0.5 | GO:0051611 | negative regulation of neurotransmitter uptake(GO:0051581) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.0 | 0.6 | GO:0032494 | response to peptidoglycan(GO:0032494) |
| 0.0 | 0.5 | GO:0060087 | relaxation of vascular smooth muscle(GO:0060087) |
| 0.0 | 0.9 | GO:0001501 | skeletal system development(GO:0001501) |
| 0.0 | 0.7 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 | 0.6 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 0.2 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.0 | 0.7 | GO:2000394 | positive regulation of lamellipodium morphogenesis(GO:2000394) |
| 0.0 | 0.2 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 10.3 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.1 | GO:2000078 | glandular epithelial cell maturation(GO:0002071) type B pancreatic cell maturation(GO:0072560) positive regulation of type B pancreatic cell development(GO:2000078) |
| 0.0 | 1.3 | GO:0042359 | vitamin D metabolic process(GO:0042359) |
| 0.0 | 0.1 | GO:0055099 | detection of endogenous stimulus(GO:0009726) response to high density lipoprotein particle(GO:0055099) |
| 0.0 | 1.3 | GO:1901685 | glutathione derivative metabolic process(GO:1901685) glutathione derivative biosynthetic process(GO:1901687) |
| 0.0 | 0.2 | GO:2001106 | regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.0 | 0.4 | GO:1900004 | negative regulation of serine-type endopeptidase activity(GO:1900004) negative regulation of serine-type peptidase activity(GO:1902572) |
| 0.0 | 1.3 | GO:0003301 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 0.1 | GO:1902173 | negative regulation of keratinocyte apoptotic process(GO:1902173) |
| 0.0 | 0.7 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.0 | 2.9 | GO:0030195 | negative regulation of blood coagulation(GO:0030195) negative regulation of hemostasis(GO:1900047) |
| 0.0 | 3.3 | GO:1901185 | negative regulation of ERBB signaling pathway(GO:1901185) |
| 0.0 | 5.5 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 | 1.1 | GO:0006349 | regulation of gene expression by genetic imprinting(GO:0006349) |
| 0.0 | 0.4 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.9 | GO:2000479 | regulation of cAMP-dependent protein kinase activity(GO:2000479) |
| 0.0 | 0.2 | GO:1990481 | mRNA pseudouridine synthesis(GO:1990481) |
| 0.0 | 2.0 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.0 | 0.3 | GO:1904526 | regulation of microtubule binding(GO:1904526) |
| 0.0 | 0.2 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.0 | 0.1 | GO:0090362 | positive regulation of platelet-derived growth factor production(GO:0090362) |
| 0.0 | 0.5 | GO:0050713 | negative regulation of interleukin-1 beta secretion(GO:0050713) |
| 0.0 | 0.7 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 | 0.2 | GO:2000643 | positive regulation of early endosome to late endosome transport(GO:2000643) |
| 0.0 | 0.2 | GO:0090063 | positive regulation of microtubule nucleation(GO:0090063) |
| 0.0 | 0.5 | GO:0032098 | regulation of appetite(GO:0032098) |
| 0.0 | 0.5 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.0 | 0.3 | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 0.0 | 0.2 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.0 | 0.4 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 2.8 | GO:0030521 | androgen receptor signaling pathway(GO:0030521) |
| 0.0 | 0.1 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 0.6 | GO:0042749 | regulation of circadian sleep/wake cycle(GO:0042749) regulation of circadian sleep/wake cycle, sleep(GO:0045187) |
| 0.0 | 0.4 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.0 | 0.6 | GO:0060134 | prepulse inhibition(GO:0060134) |
| 0.0 | 0.8 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.0 | 0.4 | GO:0010886 | positive regulation of cholesterol storage(GO:0010886) |
| 0.0 | 0.5 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.0 | 0.3 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 0.0 | 0.2 | GO:0046784 | viral mRNA export from host cell nucleus(GO:0046784) |
| 0.0 | 0.3 | GO:0010764 | negative regulation of fibroblast migration(GO:0010764) |
| 0.0 | 0.2 | GO:0035986 | senescence-associated heterochromatin focus assembly(GO:0035986) |
| 0.0 | 0.9 | GO:0032508 | DNA duplex unwinding(GO:0032508) |
| 0.0 | 0.7 | GO:0050974 | detection of mechanical stimulus involved in sensory perception(GO:0050974) |
| 0.0 | 1.0 | GO:0030261 | chromosome condensation(GO:0030261) |
| 0.0 | 1.0 | GO:0003382 | epithelial cell morphogenesis(GO:0003382) |
| 0.0 | 0.2 | GO:0034378 | chylomicron assembly(GO:0034378) |
| 0.0 | 0.6 | GO:0010880 | regulation of release of sequestered calcium ion into cytosol by sarcoplasmic reticulum(GO:0010880) |
| 0.0 | 0.3 | GO:0021960 | anterior commissure morphogenesis(GO:0021960) |
| 0.0 | 0.5 | GO:1904778 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.0 | 3.6 | GO:0007586 | digestion(GO:0007586) |
| 0.0 | 3.4 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 0.4 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.3 | GO:0072189 | ureter development(GO:0072189) |
| 0.0 | 0.4 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.8 | GO:0061512 | protein localization to cilium(GO:0061512) |
| 0.0 | 1.6 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 0.3 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.0 | 0.3 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.1 | GO:0006127 | glycerophosphate shuttle(GO:0006127) |
| 0.0 | 0.4 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.9 | GO:0090004 | positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.0 | 0.1 | GO:0010387 | COP9 signalosome assembly(GO:0010387) |
| 0.0 | 1.7 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.0 | 0.3 | GO:1904293 | negative regulation of ERAD pathway(GO:1904293) |
| 0.0 | 6.2 | GO:0006936 | muscle contraction(GO:0006936) |
| 0.0 | 0.3 | GO:0034505 | tooth mineralization(GO:0034505) |
| 0.0 | 0.2 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 0.3 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.0 | 1.3 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.0 | 0.0 | GO:0051311 | meiotic metaphase I plate congression(GO:0043060) meiotic spindle midzone assembly(GO:0051257) meiotic metaphase plate congression(GO:0051311) |
| 0.0 | 0.2 | GO:0001542 | ovulation from ovarian follicle(GO:0001542) |
| 0.0 | 1.5 | GO:0021510 | spinal cord development(GO:0021510) |
| 0.0 | 0.3 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.4 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.0 | 0.8 | GO:0035987 | endodermal cell differentiation(GO:0035987) |
| 0.0 | 0.5 | GO:0015695 | organic cation transport(GO:0015695) |
| 0.0 | 1.5 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 1.2 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.0 | 0.1 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.0 | 2.8 | GO:0031032 | actomyosin structure organization(GO:0031032) |
| 0.0 | 0.3 | GO:0050482 | arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.0 | 1.0 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 0.0 | 1.6 | GO:0055001 | muscle cell development(GO:0055001) |
| 0.0 | 0.6 | GO:0032781 | positive regulation of ATPase activity(GO:0032781) |
| 0.0 | 0.4 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.0 | 0.2 | GO:0030509 | BMP signaling pathway(GO:0030509) |
| 0.0 | 0.9 | GO:0070897 | DNA-templated transcriptional preinitiation complex assembly(GO:0070897) |
| 0.0 | 0.3 | GO:0007617 | mating behavior(GO:0007617) multi-organism reproductive behavior(GO:0044705) |
| 0.0 | 0.1 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.3 | GO:0006474 | N-terminal protein amino acid acetylation(GO:0006474) |
| 0.0 | 0.6 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.0 | 0.4 | GO:0042417 | dopamine metabolic process(GO:0042417) |
| 0.0 | 1.7 | GO:0006024 | glycosaminoglycan biosynthetic process(GO:0006024) |
| 0.0 | 0.2 | GO:0043496 | regulation of protein homodimerization activity(GO:0043496) |
| 0.0 | 0.8 | GO:0019730 | antimicrobial humoral response(GO:0019730) |
| 0.0 | 7.5 | GO:0043687 | post-translational protein modification(GO:0043687) |
| 0.0 | 0.2 | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.0 | 0.3 | GO:0042135 | neurotransmitter catabolic process(GO:0042135) |
| 0.0 | 1.2 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.0 | 0.3 | GO:0036498 | IRE1-mediated unfolded protein response(GO:0036498) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 5.8 | GO:0014802 | terminal cisterna(GO:0014802) |
| 0.6 | 1.1 | GO:0033150 | cytoskeletal calyx(GO:0033150) |
| 0.5 | 3.3 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 0.5 | 1.4 | GO:0043159 | acrosomal matrix(GO:0043159) |
| 0.4 | 5.7 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.4 | 1.1 | GO:0005656 | nuclear pre-replicative complex(GO:0005656) pre-replicative complex(GO:0036387) |
| 0.3 | 1.0 | GO:0098855 | HCN channel complex(GO:0098855) |
| 0.3 | 1.9 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.3 | 3.0 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.3 | 0.9 | GO:1990716 | axonemal central apparatus(GO:1990716) |
| 0.3 | 2.5 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.2 | 1.4 | GO:0005846 | nuclear cap binding complex(GO:0005846) |
| 0.2 | 0.7 | GO:0031021 | interphase microtubule organizing center(GO:0031021) |
| 0.2 | 6.8 | GO:0032982 | myosin filament(GO:0032982) |
| 0.2 | 4.2 | GO:0036038 | MKS complex(GO:0036038) |
| 0.2 | 2.7 | GO:0030478 | actin cap(GO:0030478) |
| 0.2 | 0.7 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.2 | 1.9 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.2 | 1.8 | GO:0070033 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) synaptobrevin 2-SNAP-25-syntaxin-3-complexin complex(GO:0070554) |
| 0.2 | 5.6 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.2 | 0.8 | GO:0009328 | phenylalanine-tRNA ligase complex(GO:0009328) |
| 0.2 | 5.8 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.2 | 1.2 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.1 | 1.0 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.1 | 2.0 | GO:0034464 | BBSome(GO:0034464) |
| 0.1 | 1.7 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.1 | 1.7 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.1 | 3.1 | GO:0005861 | troponin complex(GO:0005861) |
| 0.1 | 0.9 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.1 | 0.8 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.1 | 0.7 | GO:0031673 | H zone(GO:0031673) |
| 0.1 | 1.2 | GO:0071546 | pi-body(GO:0071546) |
| 0.1 | 0.2 | GO:0005683 | U7 snRNP(GO:0005683) |
| 0.1 | 1.0 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.1 | 0.5 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.1 | 2.5 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.1 | 1.4 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.1 | 1.3 | GO:0036379 | striated muscle thin filament(GO:0005865) myofilament(GO:0036379) |
| 0.1 | 1.4 | GO:0045179 | apical cortex(GO:0045179) |
| 0.1 | 0.9 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.1 | 1.2 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.1 | 0.4 | GO:0097125 | cyclin B1-CDK1 complex(GO:0097125) |
| 0.1 | 1.0 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.1 | 1.8 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.1 | 2.4 | GO:0090533 | cation-transporting ATPase complex(GO:0090533) |
| 0.1 | 0.9 | GO:0032797 | SMN complex(GO:0032797) |
| 0.1 | 1.2 | GO:0042641 | actomyosin(GO:0042641) |
| 0.1 | 0.6 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.1 | 0.5 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.1 | 4.0 | GO:0002102 | podosome(GO:0002102) |
| 0.1 | 9.9 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.1 | 0.1 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.1 | 0.4 | GO:0090661 | box H/ACA scaRNP complex(GO:0072589) box H/ACA telomerase RNP complex(GO:0090661) |
| 0.1 | 4.9 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.1 | 9.3 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.1 | 1.1 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.1 | 0.2 | GO:1990723 | cytoplasmic periphery of the nuclear pore complex(GO:1990723) |
| 0.1 | 0.6 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.1 | 0.7 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.1 | 2.3 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.1 | 0.7 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.1 | 1.4 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.1 | 0.3 | GO:0031417 | NatC complex(GO:0031417) |
| 0.0 | 1.5 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 1.7 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.0 | 0.1 | GO:0034515 | proteasome storage granule(GO:0034515) |
| 0.0 | 0.9 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.0 | 0.8 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 0.5 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.0 | 0.8 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.0 | 4.0 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 1.0 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 4.0 | GO:0005930 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.0 | 0.7 | GO:0043220 | Schmidt-Lanterman incisure(GO:0043220) |
| 0.0 | 0.7 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.2 | GO:0071547 | piP-body(GO:0071547) |
| 0.0 | 4.1 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 4.9 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 0.4 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.0 | 0.2 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.0 | 0.5 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.3 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 12.2 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 0.4 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 1.7 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.7 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 1.0 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 0.4 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.0 | 0.9 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.6 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.6 | GO:0044298 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 0.2 | GO:0045240 | dihydrolipoyl dehydrogenase complex(GO:0045240) |
| 0.0 | 0.4 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 0.2 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 1.2 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.5 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 1.3 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 0.9 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.6 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.0 | 4.2 | GO:0030016 | myofibril(GO:0030016) |
| 0.0 | 0.3 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.3 | GO:0005845 | mRNA cap binding complex(GO:0005845) |
| 0.0 | 0.4 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.3 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.0 | 0.0 | GO:0005715 | late recombination nodule(GO:0005715) |
| 0.0 | 0.4 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.2 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.0 | 0.1 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.0 | 1.6 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 0.1 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 1.0 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.2 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.9 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.1 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 0.4 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.9 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.2 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.1 | GO:0032426 | stereocilium tip(GO:0032426) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 5.6 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.7 | 3.3 | GO:0004419 | hydroxymethylglutaryl-CoA lyase activity(GO:0004419) |
| 0.6 | 1.9 | GO:0047275 | glucosaminylgalactosylglucosylceramide beta-galactosyltransferase activity(GO:0047275) |
| 0.6 | 22.8 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.5 | 4.7 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.5 | 1.5 | GO:0047536 | 2-aminoadipate transaminase activity(GO:0047536) |
| 0.5 | 2.4 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.4 | 1.3 | GO:0008386 | cholesterol monooxygenase (side-chain-cleaving) activity(GO:0008386) |
| 0.4 | 1.7 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 0.4 | 4.0 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 0.4 | 1.1 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) |
| 0.4 | 1.4 | GO:0008112 | nicotinamide N-methyltransferase activity(GO:0008112) pyridine N-methyltransferase activity(GO:0030760) |
| 0.4 | 2.8 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.3 | 2.4 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.3 | 1.4 | GO:0033823 | procollagen-lysine 5-dioxygenase activity(GO:0008475) procollagen glucosyltransferase activity(GO:0033823) |
| 0.3 | 2.4 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.3 | 1.3 | GO:0004040 | amidase activity(GO:0004040) |
| 0.3 | 1.3 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.3 | 0.9 | GO:0050571 | 1,5-anhydro-D-fructose reductase activity(GO:0050571) |
| 0.3 | 3.6 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.3 | 0.8 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 0.3 | 1.9 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.2 | 3.2 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.2 | 3.1 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.2 | 0.7 | GO:0046848 | hydroxyapatite binding(GO:0046848) |
| 0.2 | 5.0 | GO:0008430 | selenium binding(GO:0008430) |
| 0.2 | 0.7 | GO:0050211 | procollagen galactosyltransferase activity(GO:0050211) |
| 0.2 | 2.5 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.2 | 0.9 | GO:1904493 | Ac-Asp-Glu binding(GO:1904492) tetrahydrofolyl-poly(glutamate) polymer binding(GO:1904493) |
| 0.2 | 7.3 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.2 | 3.1 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.2 | 0.7 | GO:0003880 | protein C-terminal carboxyl O-methyltransferase activity(GO:0003880) |
| 0.2 | 0.6 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.2 | 1.2 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.2 | 1.0 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.2 | 0.6 | GO:0019781 | NEDD8 activating enzyme activity(GO:0019781) |
| 0.2 | 1.0 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.2 | 1.2 | GO:0004471 | malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.2 | 0.7 | GO:0004452 | isopentenyl-diphosphate delta-isomerase activity(GO:0004452) |
| 0.2 | 1.4 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.2 | 0.8 | GO:0016297 | oleoyl-[acyl-carrier-protein] hydrolase activity(GO:0004320) myristoyl-[acyl-carrier-protein] hydrolase activity(GO:0016295) palmitoyl-[acyl-carrier-protein] hydrolase activity(GO:0016296) acyl-[acyl-carrier-protein] hydrolase activity(GO:0016297) |
| 0.2 | 1.2 | GO:0004909 | interleukin-1, Type I, activating receptor activity(GO:0004909) |
| 0.2 | 0.7 | GO:0004945 | angiotensin receptor activity(GO:0001595) angiotensin type II receptor activity(GO:0004945) |
| 0.2 | 1.8 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.2 | 2.3 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.2 | 2.8 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.2 | 0.6 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 0.1 | 4.7 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.1 | 4.1 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.1 | 0.4 | GO:0019150 | D-ribulokinase activity(GO:0019150) |
| 0.1 | 0.4 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.1 | 2.8 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.1 | 0.4 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.1 | 1.1 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.1 | 3.3 | GO:0001134 | transcription factor activity, transcription factor recruiting(GO:0001134) |
| 0.1 | 1.2 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.1 | 0.3 | GO:0015361 | low-affinity sodium:dicarboxylate symporter activity(GO:0015361) |
| 0.1 | 0.5 | GO:0009383 | rRNA (cytosine-C5-)-methyltransferase activity(GO:0009383) |
| 0.1 | 0.8 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.1 | 0.9 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.1 | 1.4 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.1 | 1.1 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.1 | 2.4 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.1 | 0.5 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.1 | 1.3 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.1 | 1.3 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.1 | 2.1 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.1 | 1.2 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.1 | 0.3 | GO:1904455 | ubiquitin-specific protease activity involved in negative regulation of ERAD pathway(GO:1904455) |
| 0.1 | 0.2 | GO:0010309 | acireductone dioxygenase [iron(II)-requiring] activity(GO:0010309) |
| 0.1 | 1.6 | GO:0070513 | death domain binding(GO:0070513) |
| 0.1 | 2.9 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.1 | 1.3 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.1 | 0.2 | GO:0003826 | alpha-ketoacid dehydrogenase activity(GO:0003826) 3-methyl-2-oxobutanoate dehydrogenase (2-methylpropanoyl-transferring) activity(GO:0003863) |
| 0.1 | 0.5 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.1 | 3.6 | GO:0005549 | odorant binding(GO:0005549) |
| 0.1 | 0.8 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.1 | 0.2 | GO:0030626 | U12 snRNA binding(GO:0030626) |
| 0.1 | 0.2 | GO:0016768 | spermine synthase activity(GO:0016768) |
| 0.1 | 2.8 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.1 | 2.5 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.1 | 1.8 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.1 | 0.3 | GO:0030021 | extracellular matrix structural constituent conferring compression resistance(GO:0030021) structural constituent of tooth enamel(GO:0030345) |
| 0.1 | 0.3 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.1 | 0.4 | GO:0043262 | adenosine-diphosphatase activity(GO:0043262) |
| 0.1 | 2.4 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.1 | 0.5 | GO:0001602 | pancreatic polypeptide receptor activity(GO:0001602) |
| 0.1 | 1.6 | GO:0031005 | filamin binding(GO:0031005) |
| 0.1 | 1.0 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 0.1 | 0.6 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.1 | 0.5 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.1 | 0.2 | GO:0071209 | U7 snRNA binding(GO:0071209) |
| 0.1 | 0.8 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.1 | 1.1 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.1 | 0.2 | GO:0008124 | 4-alpha-hydroxytetrahydrobiopterin dehydratase activity(GO:0008124) |
| 0.1 | 0.4 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.1 | 1.5 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 0.2 | GO:0005148 | prolactin receptor binding(GO:0005148) |
| 0.1 | 13.3 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.0 | 0.2 | GO:0004057 | arginyltransferase activity(GO:0004057) |
| 0.0 | 3.3 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.2 | GO:0034602 | oxoglutarate dehydrogenase (NAD+) activity(GO:0034602) |
| 0.0 | 0.8 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.0 | 0.9 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 1.1 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 1.1 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.0 | 1.1 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.0 | 1.0 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.0 | 1.3 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 7.8 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.2 | GO:0051800 | phosphatidylinositol-3,4-bisphosphate 3-phosphatase activity(GO:0051800) |
| 0.0 | 0.5 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.0 | 0.2 | GO:0001032 | transcription factor activity, RNA polymerase III transcription factor binding(GO:0001007) RNA polymerase III type 3 promoter DNA binding(GO:0001032) |
| 0.0 | 0.7 | GO:0030548 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.0 | 0.4 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 0.2 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 0.0 | 6.9 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 3.6 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 1.0 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.0 | 0.5 | GO:0050694 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.0 | 0.1 | GO:0034041 | sterol-transporting ATPase activity(GO:0034041) |
| 0.0 | 0.8 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 0.4 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.5 | GO:0089720 | caspase binding(GO:0089720) |
| 0.0 | 0.6 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.0 | 1.3 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 1.8 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 1.1 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 2.6 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 1.1 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.2 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.0 | 1.7 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.2 | GO:0009019 | tRNA (guanine-N1-)-methyltransferase activity(GO:0009019) |
| 0.0 | 1.6 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.9 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.0 | 0.2 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 1.0 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.0 | 0.3 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.0 | 1.3 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 5.7 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 0.6 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 5.7 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.7 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 0.1 | GO:0032093 | SAM domain binding(GO:0032093) |
| 0.0 | 0.4 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.0 | 0.5 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.0 | 1.0 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.9 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.9 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.9 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 0.1 | GO:0052591 | sn-glycerol-3-phosphate:ubiquinone oxidoreductase activity(GO:0052590) sn-glycerol-3-phosphate:ubiquinone-8 oxidoreductase activity(GO:0052591) |
| 0.0 | 0.3 | GO:1903136 | cuprous ion binding(GO:1903136) |
| 0.0 | 0.1 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 0.0 | 0.7 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.0 | 0.4 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.5 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.3 | GO:0042805 | actinin binding(GO:0042805) |
| 0.0 | 0.3 | GO:0035242 | protein-arginine omega-N asymmetric methyltransferase activity(GO:0035242) |
| 0.0 | 0.2 | GO:0043855 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.0 | 0.4 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.0 | 0.9 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.5 | GO:0015101 | organic cation transmembrane transporter activity(GO:0015101) |
| 0.0 | 0.4 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.0 | 0.4 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.6 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.0 | 2.3 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 15.9 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 0.6 | GO:0019956 | chemokine binding(GO:0019956) |
| 0.0 | 0.6 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.0 | 0.3 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.5 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.4 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.0 | 0.6 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.3 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.4 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 0.3 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 0.2 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.6 | GO:0003700 | transcription factor activity, sequence-specific DNA binding(GO:0003700) |
| 0.0 | 0.3 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.0 | 0.3 | GO:0004033 | aldo-keto reductase (NADP) activity(GO:0004033) |
| 0.0 | 0.4 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.0 | 0.6 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 0.7 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 0.2 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.4 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 1.3 | GO:0003774 | motor activity(GO:0003774) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 20.1 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.2 | 0.2 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.1 | 1.1 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.1 | 3.3 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.1 | 1.4 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.1 | 2.4 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 3.5 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 0.9 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 4.0 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 1.7 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 2.6 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 2.4 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 4.3 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 1.6 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 1.8 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 8.0 | NABA CORE MATRISOME | Ensemble of genes encoding core extracellular matrix including ECM glycoproteins, collagens and proteoglycans |
| 0.0 | 0.8 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 1.9 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 2.4 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 2.0 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.0 | 0.9 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 0.9 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
| 0.0 | 2.6 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 0.2 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.8 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 1.0 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 0.4 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 9.4 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.2 | 5.7 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.2 | 3.6 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.2 | 2.8 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.1 | 10.4 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 2.3 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.1 | 1.1 | REACTOME SIGNALING BY FGFR | Genes involved in Signaling by FGFR |
| 0.1 | 1.7 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.1 | 2.9 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.1 | 3.8 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.1 | 1.4 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.1 | 3.6 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.1 | 3.9 | REACTOME MUSCLE CONTRACTION | Genes involved in Muscle contraction |
| 0.1 | 4.0 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.1 | 1.4 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.1 | 1.1 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.1 | 1.0 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 2.6 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 1.0 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.6 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 1.0 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.0 | 1.5 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 0.3 | REACTOME INFLUENZA LIFE CYCLE | Genes involved in Influenza Life Cycle |
| 0.0 | 1.5 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 1.0 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 3.2 | REACTOME EXTRACELLULAR MATRIX ORGANIZATION | Genes involved in Extracellular matrix organization |
| 0.0 | 1.6 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 2.9 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 1.5 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.0 | 0.6 | REACTOME POL SWITCHING | Genes involved in Polymerase switching |
| 0.0 | 7.0 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.0 | 1.2 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 1.0 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.6 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.9 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 0.2 | REACTOME APC C CDC20 MEDIATED DEGRADATION OF MITOTIC PROTEINS | Genes involved in APC/C:Cdc20 mediated degradation of mitotic proteins |
| 0.0 | 9.0 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.5 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 2.2 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.8 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 1.2 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.3 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 1.0 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.7 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.4 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.5 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.7 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |