Project
Illumina Body Map 2: averaged replicates
Navigation
Downloads
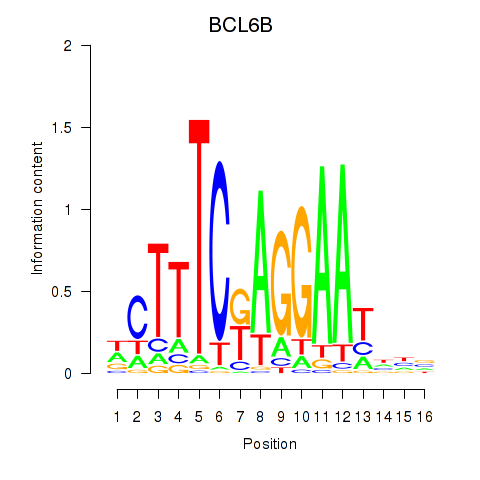
Results for BCL6B
Z-value: 0.99
Transcription factors associated with BCL6B
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
BCL6B
|
ENSG00000161940.6 | BCL6B transcription repressor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| BCL6B | hg19_v2_chr17_+_6926339_6926377 | 0.59 | 4.0e-04 | Click! |
Activity profile of BCL6B motif
Sorted Z-values of BCL6B motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_91574142 | 2.82 |
ENST00000547937.1
|
DCN
|
decorin |
| chr2_+_120189422 | 2.75 |
ENST00000306406.4
|
TMEM37
|
transmembrane protein 37 |
| chr16_-_66952779 | 2.49 |
ENST00000570262.1
ENST00000394055.3 ENST00000299752.4 |
CDH16
|
cadherin 16, KSP-cadherin |
| chr14_+_95078714 | 2.49 |
ENST00000393078.3
ENST00000393080.4 ENST00000467132.1 |
SERPINA3
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 3 |
| chr3_-_112329110 | 2.34 |
ENST00000479368.1
|
CCDC80
|
coiled-coil domain containing 80 |
| chr2_+_169659121 | 2.25 |
ENST00000397206.2
ENST00000397209.2 ENST00000421711.2 |
NOSTRIN
|
nitric oxide synthase trafficking |
| chr16_-_66952742 | 2.22 |
ENST00000565235.2
ENST00000568632.1 ENST00000565796.1 |
CDH16
|
cadherin 16, KSP-cadherin |
| chr2_+_239756671 | 2.17 |
ENST00000448943.2
|
TWIST2
|
twist family bHLH transcription factor 2 |
| chr3_-_64673289 | 2.01 |
ENST00000295903.4
|
ADAMTS9
|
ADAM metallopeptidase with thrombospondin type 1 motif, 9 |
| chr18_+_6834472 | 1.80 |
ENST00000581099.1
ENST00000419673.2 ENST00000531294.1 |
ARHGAP28
|
Rho GTPase activating protein 28 |
| chr20_+_56136136 | 1.76 |
ENST00000319441.4
ENST00000543666.1 |
PCK1
|
phosphoenolpyruvate carboxykinase 1 (soluble) |
| chr1_-_168698433 | 1.70 |
ENST00000367817.3
|
DPT
|
dermatopontin |
| chr11_+_27062860 | 1.70 |
ENST00000528583.1
|
BBOX1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1 |
| chr17_+_77021702 | 1.66 |
ENST00000392445.2
ENST00000354124.3 |
C1QTNF1
|
C1q and tumor necrosis factor related protein 1 |
| chr2_-_188419078 | 1.65 |
ENST00000437725.1
ENST00000409676.1 ENST00000339091.4 ENST00000420747.1 |
TFPI
|
tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor) |
| chr5_+_140501581 | 1.58 |
ENST00000194152.1
|
PCDHB4
|
protocadherin beta 4 |
| chr2_-_188419200 | 1.52 |
ENST00000233156.3
ENST00000426055.1 ENST00000453013.1 ENST00000417013.1 |
TFPI
|
tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor) |
| chr2_+_169658928 | 1.48 |
ENST00000317647.7
ENST00000445023.2 |
NOSTRIN
|
nitric oxide synthase trafficking |
| chr11_+_26495447 | 1.47 |
ENST00000531568.1
|
ANO3
|
anoctamin 3 |
| chr19_-_460996 | 1.46 |
ENST00000264554.6
|
SHC2
|
SHC (Src homology 2 domain containing) transforming protein 2 |
| chr6_+_150690028 | 1.45 |
ENST00000229447.5
ENST00000344419.3 |
IYD
|
iodotyrosine deiodinase |
| chr8_-_91095099 | 1.45 |
ENST00000265431.3
|
CALB1
|
calbindin 1, 28kDa |
| chr6_+_150690089 | 1.44 |
ENST00000392256.2
|
IYD
|
iodotyrosine deiodinase |
| chr5_+_127039075 | 1.36 |
ENST00000514853.2
|
CTC-228N24.1
|
CTC-228N24.1 |
| chr14_-_30396948 | 1.35 |
ENST00000331968.5
|
PRKD1
|
protein kinase D1 |
| chr6_-_28304152 | 1.30 |
ENST00000435857.1
|
ZSCAN31
|
zinc finger and SCAN domain containing 31 |
| chr13_-_36705425 | 1.27 |
ENST00000255448.4
ENST00000360631.3 ENST00000379892.4 |
DCLK1
|
doublecortin-like kinase 1 |
| chr3_-_123512688 | 1.21 |
ENST00000475616.1
|
MYLK
|
myosin light chain kinase |
| chr4_+_70796784 | 1.21 |
ENST00000246891.4
ENST00000444405.3 |
CSN1S1
|
casein alpha s1 |
| chr14_-_80678512 | 1.17 |
ENST00000553968.1
|
DIO2
|
deiodinase, iodothyronine, type II |
| chr1_-_230850043 | 1.11 |
ENST00000366667.4
|
AGT
|
angiotensinogen (serpin peptidase inhibitor, clade A, member 8) |
| chr9_-_117880477 | 1.09 |
ENST00000534839.1
ENST00000340094.3 ENST00000535648.1 ENST00000346706.3 ENST00000345230.3 ENST00000350763.4 |
TNC
|
tenascin C |
| chr5_+_140579162 | 1.04 |
ENST00000536699.1
ENST00000354757.3 |
PCDHB11
|
protocadherin beta 11 |
| chr6_-_87804815 | 1.00 |
ENST00000369582.2
|
CGA
|
glycoprotein hormones, alpha polypeptide |
| chr4_-_184241927 | 0.98 |
ENST00000323319.5
|
CLDN22
|
claudin 22 |
| chr8_+_67039278 | 0.95 |
ENST00000276573.7
ENST00000350034.4 |
TRIM55
|
tripartite motif containing 55 |
| chr6_-_28973037 | 0.94 |
ENST00000377179.3
|
ZNF311
|
zinc finger protein 311 |
| chr6_+_125540951 | 0.92 |
ENST00000524679.1
|
TPD52L1
|
tumor protein D52-like 1 |
| chr8_-_107782463 | 0.92 |
ENST00000311955.3
|
ABRA
|
actin-binding Rho activating protein |
| chr16_+_14396121 | 0.91 |
ENST00000570945.1
|
RP11-65J21.3
|
RP11-65J21.3 |
| chr12_+_19358228 | 0.90 |
ENST00000424268.1
ENST00000543806.1 |
PLEKHA5
|
pleckstrin homology domain containing, family A member 5 |
| chr12_-_15082050 | 0.88 |
ENST00000540097.1
|
ERP27
|
endoplasmic reticulum protein 27 |
| chr3_+_155860751 | 0.85 |
ENST00000471742.1
|
KCNAB1
|
potassium voltage-gated channel, shaker-related subfamily, beta member 1 |
| chr5_+_140480083 | 0.83 |
ENST00000231130.2
|
PCDHB3
|
protocadherin beta 3 |
| chr17_+_79761997 | 0.83 |
ENST00000400723.3
ENST00000570996.1 |
GCGR
|
glucagon receptor |
| chr2_-_216257849 | 0.83 |
ENST00000456923.1
|
FN1
|
fibronectin 1 |
| chr3_+_46618727 | 0.83 |
ENST00000296145.5
|
TDGF1
|
teratocarcinoma-derived growth factor 1 |
| chr6_-_136788001 | 0.82 |
ENST00000544465.1
|
MAP7
|
microtubule-associated protein 7 |
| chr6_+_150690133 | 0.82 |
ENST00000392255.3
ENST00000500320.3 |
IYD
|
iodotyrosine deiodinase |
| chr3_+_102153859 | 0.81 |
ENST00000306176.1
ENST00000466937.1 |
ZPLD1
|
zona pellucida-like domain containing 1 |
| chr5_+_80529104 | 0.79 |
ENST00000254035.4
ENST00000511719.1 ENST00000437669.1 ENST00000424301.2 ENST00000505060.1 |
CKMT2
|
creatine kinase, mitochondrial 2 (sarcomeric) |
| chr8_+_67039131 | 0.79 |
ENST00000315962.4
ENST00000353317.5 |
TRIM55
|
tripartite motif containing 55 |
| chr7_+_134464414 | 0.77 |
ENST00000361901.2
|
CALD1
|
caldesmon 1 |
| chr12_+_104324112 | 0.77 |
ENST00000299767.5
|
HSP90B1
|
heat shock protein 90kDa beta (Grp94), member 1 |
| chr17_-_42907564 | 0.77 |
ENST00000592524.1
|
GJC1
|
gap junction protein, gamma 1, 45kDa |
| chr19_-_44860820 | 0.76 |
ENST00000354340.4
ENST00000337401.4 ENST00000587909.1 |
ZNF112
|
zinc finger protein 112 |
| chr7_+_134464376 | 0.76 |
ENST00000454108.1
ENST00000361675.2 |
CALD1
|
caldesmon 1 |
| chrX_-_139866723 | 0.75 |
ENST00000370532.2
|
CDR1
|
cerebellar degeneration-related protein 1, 34kDa |
| chr8_-_81787006 | 0.75 |
ENST00000327835.3
|
ZNF704
|
zinc finger protein 704 |
| chr4_+_95972822 | 0.74 |
ENST00000509540.1
ENST00000440890.2 |
BMPR1B
|
bone morphogenetic protein receptor, type IB |
| chr12_+_19358192 | 0.72 |
ENST00000538305.1
|
PLEKHA5
|
pleckstrin homology domain containing, family A member 5 |
| chr6_-_28303901 | 0.71 |
ENST00000439158.1
ENST00000446474.1 ENST00000414431.1 ENST00000344279.6 ENST00000453745.1 |
ZSCAN31
|
zinc finger and SCAN domain containing 31 |
| chr1_-_153521714 | 0.71 |
ENST00000368713.3
|
S100A3
|
S100 calcium binding protein A3 |
| chr1_-_31845914 | 0.67 |
ENST00000373713.2
|
FABP3
|
fatty acid binding protein 3, muscle and heart (mammary-derived growth inhibitor) |
| chr4_-_99578776 | 0.64 |
ENST00000515287.1
|
TSPAN5
|
tetraspanin 5 |
| chr15_+_65843130 | 0.62 |
ENST00000569894.1
|
PTPLAD1
|
protein tyrosine phosphatase-like A domain containing 1 |
| chr6_+_7541808 | 0.61 |
ENST00000379802.3
|
DSP
|
desmoplakin |
| chr3_-_123603137 | 0.60 |
ENST00000360304.3
ENST00000359169.1 ENST00000346322.5 ENST00000360772.3 |
MYLK
|
myosin light chain kinase |
| chr9_+_97766409 | 0.59 |
ENST00000425634.2
|
C9orf3
|
chromosome 9 open reading frame 3 |
| chr9_+_97766469 | 0.58 |
ENST00000433691.2
|
C9orf3
|
chromosome 9 open reading frame 3 |
| chr14_-_21567009 | 0.58 |
ENST00000556174.1
ENST00000554478.1 ENST00000553980.1 ENST00000421093.2 |
ZNF219
|
zinc finger protein 219 |
| chr2_-_31361543 | 0.58 |
ENST00000349752.5
|
GALNT14
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 14 (GalNAc-T14) |
| chr22_-_50699701 | 0.58 |
ENST00000395780.1
|
MAPK12
|
mitogen-activated protein kinase 12 |
| chr3_+_35682913 | 0.57 |
ENST00000449196.1
|
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa |
| chr12_+_16500037 | 0.55 |
ENST00000536371.1
ENST00000010404.2 |
MGST1
|
microsomal glutathione S-transferase 1 |
| chr1_-_74663825 | 0.53 |
ENST00000370911.3
ENST00000370909.2 ENST00000354431.4 |
LRRIQ3
|
leucine-rich repeats and IQ motif containing 3 |
| chr6_+_7541845 | 0.52 |
ENST00000418664.2
|
DSP
|
desmoplakin |
| chr14_+_21566980 | 0.52 |
ENST00000418511.2
ENST00000554329.2 |
TMEM253
|
transmembrane protein 253 |
| chr22_-_31503490 | 0.51 |
ENST00000400299.2
|
SELM
|
Selenoprotein M |
| chr12_-_11175219 | 0.51 |
ENST00000390673.2
|
TAS2R19
|
taste receptor, type 2, member 19 |
| chr20_-_4229721 | 0.50 |
ENST00000379453.4
|
ADRA1D
|
adrenoceptor alpha 1D |
| chr12_-_62997214 | 0.50 |
ENST00000408887.2
|
C12orf61
|
chromosome 12 open reading frame 61 |
| chr3_-_23958506 | 0.49 |
ENST00000425478.2
|
NKIRAS1
|
NFKB inhibitor interacting Ras-like 1 |
| chr6_+_44194762 | 0.47 |
ENST00000371708.1
|
SLC29A1
|
solute carrier family 29 (equilibrative nucleoside transporter), member 1 |
| chr3_-_23958402 | 0.46 |
ENST00000415901.2
ENST00000416026.2 ENST00000412028.1 ENST00000388759.3 ENST00000437230.1 |
NKIRAS1
|
NFKB inhibitor interacting Ras-like 1 |
| chr16_+_58283814 | 0.46 |
ENST00000443128.2
ENST00000219299.4 |
CCDC113
|
coiled-coil domain containing 113 |
| chr19_-_4717835 | 0.43 |
ENST00000599248.1
|
DPP9
|
dipeptidyl-peptidase 9 |
| chr14_-_100046444 | 0.42 |
ENST00000554996.1
|
CCDC85C
|
coiled-coil domain containing 85C |
| chr7_-_122840015 | 0.40 |
ENST00000194130.2
|
SLC13A1
|
solute carrier family 13 (sodium/sulfate symporter), member 1 |
| chr1_-_153521597 | 0.39 |
ENST00000368712.1
|
S100A3
|
S100 calcium binding protein A3 |
| chr4_-_99578789 | 0.39 |
ENST00000511651.1
ENST00000505184.1 |
TSPAN5
|
tetraspanin 5 |
| chr3_+_35683651 | 0.38 |
ENST00000187397.4
|
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa |
| chr10_-_70287231 | 0.37 |
ENST00000609923.1
|
SLC25A16
|
solute carrier family 25 (mitochondrial carrier; Graves disease autoantigen), member 16 |
| chr8_-_141810634 | 0.37 |
ENST00000521986.1
ENST00000523539.1 ENST00000538769.1 |
PTK2
|
protein tyrosine kinase 2 |
| chr4_+_86396265 | 0.37 |
ENST00000395184.1
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chr3_+_158787041 | 0.35 |
ENST00000471575.1
ENST00000476809.1 ENST00000485419.1 |
IQCJ-SCHIP1
|
IQCJ-SCHIP1 readthrough |
| chr9_+_70971815 | 0.34 |
ENST00000396392.1
ENST00000396396.1 |
PGM5
|
phosphoglucomutase 5 |
| chr6_+_39760129 | 0.34 |
ENST00000274867.4
|
DAAM2
|
dishevelled associated activator of morphogenesis 2 |
| chr11_-_18258342 | 0.33 |
ENST00000278222.4
|
SAA4
|
serum amyloid A4, constitutive |
| chr10_-_70287172 | 0.32 |
ENST00000539557.1
|
SLC25A16
|
solute carrier family 25 (mitochondrial carrier; Graves disease autoantigen), member 16 |
| chr4_-_185820602 | 0.32 |
ENST00000515864.1
ENST00000507183.1 |
RP11-701P16.4
|
long intergenic non-protein coding RNA 1093 |
| chr2_+_173686303 | 0.31 |
ENST00000397087.3
|
RAPGEF4
|
Rap guanine nucleotide exchange factor (GEF) 4 |
| chr16_+_16481598 | 0.30 |
ENST00000327792.5
|
NPIPA7
|
nuclear pore complex interacting protein family, member A7 |
| chr11_+_48346472 | 0.29 |
ENST00000319856.4
|
OR4C3
|
olfactory receptor, family 4, subfamily C, member 3 |
| chr17_+_4853442 | 0.29 |
ENST00000522301.1
|
ENO3
|
enolase 3 (beta, muscle) |
| chr1_+_209878182 | 0.29 |
ENST00000367027.3
|
HSD11B1
|
hydroxysteroid (11-beta) dehydrogenase 1 |
| chr12_+_31812121 | 0.29 |
ENST00000395763.3
|
METTL20
|
methyltransferase like 20 |
| chr17_-_42908155 | 0.29 |
ENST00000426548.1
ENST00000590758.1 ENST00000591424.1 |
GJC1
|
gap junction protein, gamma 1, 45kDa |
| chr4_-_13546632 | 0.28 |
ENST00000382438.5
|
NKX3-2
|
NK3 homeobox 2 |
| chr5_-_145214893 | 0.28 |
ENST00000394450.2
|
PRELID2
|
PRELI domain containing 2 |
| chr12_-_122238913 | 0.28 |
ENST00000537157.1
|
AC084018.1
|
AC084018.1 |
| chr13_+_34922173 | 0.28 |
ENST00000605909.1
|
RP11-16D22.2
|
RP11-16D22.2 |
| chr5_-_38845812 | 0.27 |
ENST00000513480.1
ENST00000512519.1 |
CTD-2127H9.1
|
CTD-2127H9.1 |
| chr15_+_26147507 | 0.27 |
ENST00000383019.2
ENST00000557171.1 |
RP11-1084I9.1
|
RP11-1084I9.1 |
| chr3_+_156799587 | 0.27 |
ENST00000469196.1
|
RP11-6F2.5
|
RP11-6F2.5 |
| chr7_+_872107 | 0.25 |
ENST00000405266.1
ENST00000401592.1 ENST00000403868.1 ENST00000425407.2 |
SUN1
|
Sad1 and UNC84 domain containing 1 |
| chrX_-_54209640 | 0.23 |
ENST00000375180.2
ENST00000328235.4 ENST00000477084.1 |
FAM120C
|
family with sequence similarity 120C |
| chr2_-_31030277 | 0.23 |
ENST00000534090.2
ENST00000295055.8 |
CAPN13
|
calpain 13 |
| chr19_-_38183201 | 0.23 |
ENST00000590008.1
ENST00000358582.4 |
ZNF781
|
zinc finger protein 781 |
| chr12_+_8975061 | 0.22 |
ENST00000299698.7
|
A2ML1
|
alpha-2-macroglobulin-like 1 |
| chr4_+_86699834 | 0.20 |
ENST00000395183.2
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chr2_+_27760247 | 0.20 |
ENST00000447166.1
|
AC109829.1
|
Uncharacterized protein |
| chr18_+_72167096 | 0.20 |
ENST00000324301.8
|
CNDP2
|
CNDP dipeptidase 2 (metallopeptidase M20 family) |
| chr19_-_7812397 | 0.19 |
ENST00000593660.1
ENST00000354397.6 ENST00000593821.1 ENST00000602261.1 ENST00000315591.8 ENST00000394161.5 ENST00000204801.8 ENST00000601256.1 ENST00000601951.1 ENST00000315599.7 |
CD209
|
CD209 molecule |
| chr5_-_145214848 | 0.19 |
ENST00000505416.1
ENST00000334744.4 ENST00000358004.2 ENST00000511435.1 |
PRELID2
|
PRELI domain containing 2 |
| chr8_+_104427581 | 0.19 |
ENST00000521716.1
ENST00000521971.1 ENST00000519682.1 |
DCAF13
|
DDB1 and CUL4 associated factor 13 |
| chr3_+_35683775 | 0.18 |
ENST00000452563.1
|
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa |
| chr20_+_43990576 | 0.18 |
ENST00000372727.1
ENST00000414310.1 |
SYS1
|
SYS1 Golgi-localized integral membrane protein homolog (S. cerevisiae) |
| chr18_-_52989217 | 0.18 |
ENST00000570287.2
|
TCF4
|
transcription factor 4 |
| chr11_+_123986069 | 0.18 |
ENST00000456829.2
ENST00000361352.5 ENST00000449321.1 ENST00000392748.1 ENST00000360334.4 ENST00000392744.4 |
VWA5A
|
von Willebrand factor A domain containing 5A |
| chr17_-_54893250 | 0.17 |
ENST00000397862.2
|
C17orf67
|
chromosome 17 open reading frame 67 |
| chr12_+_93964746 | 0.17 |
ENST00000536696.2
|
SOCS2
|
suppressor of cytokine signaling 2 |
| chr6_+_31588478 | 0.17 |
ENST00000376007.4
ENST00000376033.2 |
PRRC2A
|
proline-rich coiled-coil 2A |
| chr19_-_13227514 | 0.16 |
ENST00000587487.1
ENST00000592814.1 |
TRMT1
|
tRNA methyltransferase 1 homolog (S. cerevisiae) |
| chr17_+_76183432 | 0.16 |
ENST00000591256.1
ENST00000589256.1 ENST00000588800.1 ENST00000591952.1 ENST00000327898.5 ENST00000586542.1 ENST00000586731.1 |
AFMID
|
arylformamidase |
| chr12_+_20522179 | 0.16 |
ENST00000359062.3
|
PDE3A
|
phosphodiesterase 3A, cGMP-inhibited |
| chr2_-_84686552 | 0.16 |
ENST00000393868.2
|
SUCLG1
|
succinate-CoA ligase, alpha subunit |
| chr12_+_16064106 | 0.16 |
ENST00000428559.2
|
DERA
|
deoxyribose-phosphate aldolase (putative) |
| chr1_-_76398077 | 0.15 |
ENST00000284142.6
|
ASB17
|
ankyrin repeat and SOCS box containing 17 |
| chr19_-_13227534 | 0.15 |
ENST00000588229.1
ENST00000357720.4 |
TRMT1
|
tRNA methyltransferase 1 homolog (S. cerevisiae) |
| chr5_+_140535577 | 0.14 |
ENST00000539533.1
|
PCDHB17
|
Protocadherin-psi1; Uncharacterized protein |
| chr13_+_44596471 | 0.14 |
ENST00000592085.1
ENST00000591799.1 |
LINC00284
|
long intergenic non-protein coding RNA 284 |
| chr15_-_94614049 | 0.14 |
ENST00000556447.1
ENST00000555772.1 |
CTD-3049M7.1
|
CTD-3049M7.1 |
| chr3_+_44690211 | 0.14 |
ENST00000396056.2
ENST00000432115.2 ENST00000415571.2 ENST00000399560.2 ENST00000296092.3 ENST00000542250.1 ENST00000453164.1 |
ZNF35
|
zinc finger protein 35 |
| chr4_+_76995855 | 0.13 |
ENST00000355810.4
ENST00000349321.3 |
ART3
|
ADP-ribosyltransferase 3 |
| chr13_+_73356197 | 0.13 |
ENST00000326291.6
|
PIBF1
|
progesterone immunomodulatory binding factor 1 |
| chr18_-_52989525 | 0.13 |
ENST00000457482.3
|
TCF4
|
transcription factor 4 |
| chr3_+_185000729 | 0.13 |
ENST00000448876.1
ENST00000446828.1 ENST00000447637.1 ENST00000424227.1 ENST00000454237.1 |
MAP3K13
|
mitogen-activated protein kinase kinase kinase 13 |
| chrX_-_135338503 | 0.13 |
ENST00000370663.5
|
MAP7D3
|
MAP7 domain containing 3 |
| chr3_+_48413709 | 0.12 |
ENST00000296438.5
ENST00000436231.1 ENST00000445170.1 ENST00000415155.1 |
FBXW12
|
F-box and WD repeat domain containing 12 |
| chr7_+_2636522 | 0.12 |
ENST00000423196.1
|
IQCE
|
IQ motif containing E |
| chr3_+_185046676 | 0.12 |
ENST00000428617.1
ENST00000443863.1 |
MAP3K13
|
mitogen-activated protein kinase kinase kinase 13 |
| chr19_-_49314269 | 0.12 |
ENST00000545387.2
ENST00000316273.6 ENST00000402551.1 ENST00000598162.1 ENST00000599246.1 |
BCAT2
|
branched chain amino-acid transaminase 2, mitochondrial |
| chr2_-_55237484 | 0.11 |
ENST00000394609.2
|
RTN4
|
reticulon 4 |
| chr2_-_157186630 | 0.11 |
ENST00000406048.2
|
NR4A2
|
nuclear receptor subfamily 4, group A, member 2 |
| chr1_+_244227632 | 0.11 |
ENST00000598000.1
|
AL590483.1
|
Uncharacterized protein; cDNA FLJ42623 fis, clone BRACE3015894 |
| chr6_-_83775442 | 0.11 |
ENST00000369745.5
|
UBE3D
|
ubiquitin protein ligase E3D |
| chr6_-_88001706 | 0.11 |
ENST00000369576.2
|
GJB7
|
gap junction protein, beta 7, 25kDa |
| chr4_-_1670632 | 0.11 |
ENST00000461064.1
|
FAM53A
|
family with sequence similarity 53, member A |
| chr1_+_235530675 | 0.11 |
ENST00000366601.3
ENST00000406207.1 ENST00000543662.1 |
TBCE
|
tubulin folding cofactor E |
| chr3_-_50649192 | 0.10 |
ENST00000443053.2
ENST00000348721.3 |
CISH
|
cytokine inducible SH2-containing protein |
| chr9_-_113018835 | 0.10 |
ENST00000374517.5
|
TXN
|
thioredoxin |
| chr18_-_73967160 | 0.10 |
ENST00000579714.1
|
RP11-94B19.7
|
RP11-94B19.7 |
| chr5_+_72251793 | 0.10 |
ENST00000430046.2
ENST00000341845.6 |
FCHO2
|
FCH domain only 2 |
| chr14_-_69262789 | 0.10 |
ENST00000557022.1
|
ZFP36L1
|
ZFP36 ring finger protein-like 1 |
| chr19_-_7812446 | 0.10 |
ENST00000394173.4
ENST00000301357.8 |
CD209
|
CD209 molecule |
| chr17_+_76183398 | 0.10 |
ENST00000409257.5
|
AFMID
|
arylformamidase |
| chr11_-_128775592 | 0.10 |
ENST00000310799.3
|
C11orf45
|
chromosome 11 open reading frame 45 |
| chr10_-_73976025 | 0.10 |
ENST00000342444.4
ENST00000533958.1 ENST00000527593.1 ENST00000394915.3 ENST00000530461.1 ENST00000317168.6 ENST00000524829.1 |
ASCC1
|
activating signal cointegrator 1 complex subunit 1 |
| chr15_+_74610894 | 0.10 |
ENST00000558821.1
ENST00000268082.4 |
CCDC33
|
coiled-coil domain containing 33 |
| chr2_+_27805880 | 0.09 |
ENST00000379717.1
ENST00000355467.4 ENST00000556601.1 ENST00000416005.2 |
ZNF512
|
zinc finger protein 512 |
| chr12_-_42877726 | 0.09 |
ENST00000548696.1
|
PRICKLE1
|
prickle homolog 1 (Drosophila) |
| chr19_+_45147098 | 0.09 |
ENST00000425690.3
ENST00000344956.4 ENST00000403059.4 |
PVR
|
poliovirus receptor |
| chr3_+_44771088 | 0.09 |
ENST00000396048.2
|
ZNF501
|
zinc finger protein 501 |
| chr7_+_23719749 | 0.09 |
ENST00000409192.3
ENST00000344962.4 ENST00000409653.1 ENST00000409994.3 |
FAM221A
|
family with sequence similarity 221, member A |
| chr6_-_116479897 | 0.08 |
ENST00000418500.1
|
COL10A1
|
collagen, type X, alpha 1 |
| chr16_-_18441131 | 0.08 |
ENST00000339303.5
|
NPIPA8
|
nuclear pore complex interacting protein family, member A8 |
| chr14_-_94970559 | 0.08 |
ENST00000556881.1
|
SERPINA12
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 12 |
| chr20_-_18447667 | 0.07 |
ENST00000262547.5
ENST00000329494.5 ENST00000357236.4 |
DZANK1
|
double zinc ribbon and ankyrin repeat domains 1 |
| chr17_+_66539369 | 0.07 |
ENST00000600820.1
|
AC079210.1
|
Uncharacterized protein; cDNA FLJ45097 fis, clone BRAWH3031054 |
| chr22_-_30642728 | 0.07 |
ENST00000403987.3
|
LIF
|
leukemia inhibitory factor |
| chr13_+_53191605 | 0.07 |
ENST00000342657.3
ENST00000398039.1 |
HNRNPA1L2
|
heterogeneous nuclear ribonucleoprotein A1-like 2 |
| chr17_+_34312621 | 0.06 |
ENST00000591669.1
|
CTB-186H2.3
|
Uncharacterized protein |
| chr19_-_40023450 | 0.06 |
ENST00000326282.4
|
EID2B
|
EP300 interacting inhibitor of differentiation 2B |
| chr3_-_152058779 | 0.06 |
ENST00000408960.3
|
TMEM14E
|
transmembrane protein 14E |
| chr11_+_120081475 | 0.06 |
ENST00000328965.4
|
OAF
|
OAF homolog (Drosophila) |
| chr1_-_108735440 | 0.06 |
ENST00000370041.4
|
SLC25A24
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 24 |
| chr17_-_76183111 | 0.05 |
ENST00000405273.1
ENST00000590862.1 ENST00000590430.1 ENST00000586613.1 |
TK1
|
thymidine kinase 1, soluble |
| chr11_+_18230727 | 0.05 |
ENST00000527059.1
|
RP11-113D6.10
|
Putative mitochondrial carrier protein LOC494141 |
| chr11_-_123865868 | 0.05 |
ENST00000307002.3
|
OR10G6
|
olfactory receptor, family 10, subfamily G, member 6 |
| chr21_+_44394620 | 0.05 |
ENST00000291547.5
|
PKNOX1
|
PBX/knotted 1 homeobox 1 |
| chr3_+_45017722 | 0.04 |
ENST00000265564.7
|
EXOSC7
|
exosome component 7 |
| chr13_+_52436111 | 0.04 |
ENST00000242819.4
|
CCDC70
|
coiled-coil domain containing 70 |
| chr10_-_73976884 | 0.04 |
ENST00000317126.4
ENST00000545550.1 |
ASCC1
|
activating signal cointegrator 1 complex subunit 1 |
| chr4_+_89378261 | 0.04 |
ENST00000264350.3
|
HERC5
|
HECT and RLD domain containing E3 ubiquitin protein ligase 5 |
| chr1_-_247876105 | 0.04 |
ENST00000302084.2
|
OR6F1
|
olfactory receptor, family 6, subfamily F, member 1 |
| chr3_+_35683816 | 0.03 |
ENST00000438577.1
|
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa |
| chr1_-_150669604 | 0.03 |
ENST00000427665.1
ENST00000540514.1 |
GOLPH3L
|
golgi phosphoprotein 3-like |
| chr11_-_4945637 | 0.03 |
ENST00000321961.2
|
OR51G1
|
olfactory receptor, family 51, subfamily G, member 1 |
| chr6_-_32083106 | 0.03 |
ENST00000442721.1
|
TNXB
|
tenascin XB |
Network of associatons between targets according to the STRING database.
First level regulatory network of BCL6B
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.8 | GO:1904425 | negative regulation of GTP binding(GO:1904425) |
| 0.4 | 1.8 | GO:0006114 | glycerol biosynthetic process(GO:0006114) |
| 0.4 | 3.2 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.4 | 1.1 | GO:0034226 | lysine import(GO:0034226) L-lysine import(GO:0061461) L-lysine import into cell(GO:1903410) |
| 0.3 | 0.8 | GO:1904235 | regulation of substrate-dependent cell migration, cell attachment to substrate(GO:1904235) positive regulation of substrate-dependent cell migration, cell attachment to substrate(GO:1904237) |
| 0.3 | 0.8 | GO:0031247 | actin rod assembly(GO:0031247) |
| 0.2 | 3.7 | GO:0006570 | tyrosine metabolic process(GO:0006570) |
| 0.2 | 1.1 | GO:0060447 | bud outgrowth involved in lung branching(GO:0060447) |
| 0.2 | 1.7 | GO:2000857 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.2 | 1.8 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.2 | 2.8 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.2 | 0.5 | GO:0071449 | cellular response to lipid hydroperoxide(GO:0071449) |
| 0.2 | 1.1 | GO:0086021 | SA node cell to atrial cardiac muscle cell communication by electrical coupling(GO:0086021) |
| 0.2 | 8.2 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.2 | 1.5 | GO:0035502 | metanephric part of ureteric bud development(GO:0035502) |
| 0.1 | 1.3 | GO:1901727 | positive regulation of endothelial cell chemotaxis by VEGF-activated vascular endothelial growth factor receptor signaling pathway(GO:0038033) protein kinase D signaling(GO:0089700) positive regulation of histone deacetylase activity(GO:1901727) |
| 0.1 | 1.5 | GO:0061588 | calcium activated phospholipid scrambling(GO:0061588) calcium activated phosphatidylcholine scrambling(GO:0061590) calcium activated galactosylceramide scrambling(GO:0061591) |
| 0.1 | 0.7 | GO:2001245 | regulation of phosphatidylcholine biosynthetic process(GO:2001245) |
| 0.1 | 1.7 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.1 | 0.5 | GO:0001994 | norepinephrine-epinephrine vasoconstriction involved in regulation of systemic arterial blood pressure(GO:0001994) |
| 0.1 | 1.1 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.1 | 0.7 | GO:0048165 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.1 | 0.3 | GO:0002940 | tRNA N2-guanine methylation(GO:0002940) |
| 0.1 | 0.5 | GO:0015862 | uridine transport(GO:0015862) |
| 0.1 | 2.5 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.1 | 1.2 | GO:0001514 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.1 | 0.3 | GO:0046968 | peptide antigen transport(GO:0046968) |
| 0.1 | 0.9 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.1 | 2.0 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.0 | 0.8 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.0 | 0.8 | GO:0008595 | tripartite regional subdivision(GO:0007351) anterior/posterior axis specification, embryo(GO:0008595) |
| 0.0 | 0.1 | GO:0060370 | susceptibility to T cell mediated cytotoxicity(GO:0060370) |
| 0.0 | 1.7 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.0 | 2.2 | GO:0045668 | negative regulation of osteoblast differentiation(GO:0045668) |
| 0.0 | 0.1 | GO:0006549 | isoleucine metabolic process(GO:0006549) isoleucine catabolic process(GO:0006550) |
| 0.0 | 0.1 | GO:0021986 | epithalamus development(GO:0021538) habenula development(GO:0021986) |
| 0.0 | 0.1 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.0 | 1.0 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.2 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.0 | 0.4 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.0 | 0.6 | GO:0006975 | DNA damage induced protein phosphorylation(GO:0006975) |
| 0.0 | 0.2 | GO:0097240 | meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.0 | 2.0 | GO:0050999 | regulation of nitric-oxide synthase activity(GO:0050999) |
| 0.0 | 0.1 | GO:1901545 | cellular response to raffinose(GO:0097403) response to raffinose(GO:1901545) |
| 0.0 | 0.2 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.0 | 0.3 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.0 | 0.1 | GO:2001279 | regulation of prostaglandin biosynthetic process(GO:0031392) regulation of unsaturated fatty acid biosynthetic process(GO:2001279) |
| 0.0 | 0.9 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.0 | 0.3 | GO:0060576 | intestinal epithelial cell development(GO:0060576) |
| 0.0 | 0.4 | GO:1902358 | sulfate transmembrane transport(GO:1902358) |
| 0.0 | 0.1 | GO:0060708 | lung vasculature development(GO:0060426) spongiotrophoblast differentiation(GO:0060708) |
| 0.0 | 0.7 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.0 | 0.8 | GO:0070873 | regulation of glycogen metabolic process(GO:0070873) |
| 0.0 | 0.5 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.3 | GO:0006704 | glucocorticoid biosynthetic process(GO:0006704) |
| 0.0 | 0.1 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.8 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.1 | 3.4 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 0.9 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.1 | 1.5 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 1.1 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 0.6 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 1.2 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.3 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.8 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 1.3 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 2.5 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 0.3 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 4.1 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 1.8 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.6 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 1.6 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 5.5 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 3.5 | GO:0030666 | endocytic vesicle membrane(GO:0030666) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.8 | GO:0004613 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 0.5 | 3.7 | GO:0004447 | iodide peroxidase activity(GO:0004447) |
| 0.4 | 1.7 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.2 | 0.7 | GO:0070538 | oleic acid binding(GO:0070538) |
| 0.2 | 0.8 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 0.2 | 1.3 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.2 | 1.8 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.2 | 1.1 | GO:0086020 | gap junction channel activity involved in SA node cell-atrial cardiac muscle cell electrical coupling(GO:0086020) |
| 0.2 | 0.5 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.1 | 1.2 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.1 | 1.1 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.1 | 0.3 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.1 | 1.1 | GO:0046790 | virion binding(GO:0046790) |
| 0.1 | 0.5 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.1 | 1.1 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 0.1 | 1.6 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.1 | 0.8 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.1 | 0.7 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.1 | 1.5 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.1 | 0.2 | GO:0004119 | cGMP-inhibited cyclic-nucleotide phosphodiesterase activity(GO:0004119) |
| 0.1 | 2.3 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.1 | 0.2 | GO:0004139 | deoxyribose-phosphate aldolase activity(GO:0004139) |
| 0.1 | 1.1 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.0 | 0.3 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.0 | 0.3 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.0 | 1.5 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.6 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.1 | GO:0005136 | interleukin-4 receptor binding(GO:0005136) |
| 0.0 | 0.2 | GO:0102008 | cytosolic dipeptidase activity(GO:0102008) |
| 0.0 | 5.3 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 6.0 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 1.3 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 0.3 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.2 | GO:0004775 | succinate-CoA ligase (ADP-forming) activity(GO:0004775) |
| 0.0 | 0.9 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.0 | 0.5 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.0 | 0.2 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.0 | 0.5 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.1 | GO:0005146 | leukemia inhibitory factor receptor binding(GO:0005146) |
| 0.0 | 0.1 | GO:0052656 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.0 | 0.4 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) |
| 0.0 | 0.4 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 0.3 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 0.5 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.0 | 2.8 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.0 | 0.6 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 1.5 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 0.1 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.0 | 1.8 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 5.5 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 0.8 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 2.8 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 1.1 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 1.5 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.0 | 1.8 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.6 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 1.8 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 1.1 | ST GA13 PATHWAY | G alpha 13 Pathway |
| 0.0 | 0.8 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 1.1 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 4.5 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 5.9 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.1 | 1.8 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.1 | 3.7 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.1 | 2.8 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.1 | 1.5 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.1 | 0.6 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.1 | 2.9 | REACTOME FORMATION OF FIBRIN CLOT CLOTTING CASCADE | Genes involved in Formation of Fibrin Clot (Clotting Cascade) |
| 0.1 | 0.8 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.1 | 3.3 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 1.1 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 1.2 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 1.2 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.0 | 0.7 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 1.3 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.8 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 1.0 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.4 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.7 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.8 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.0 | 0.9 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 1.4 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.5 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |