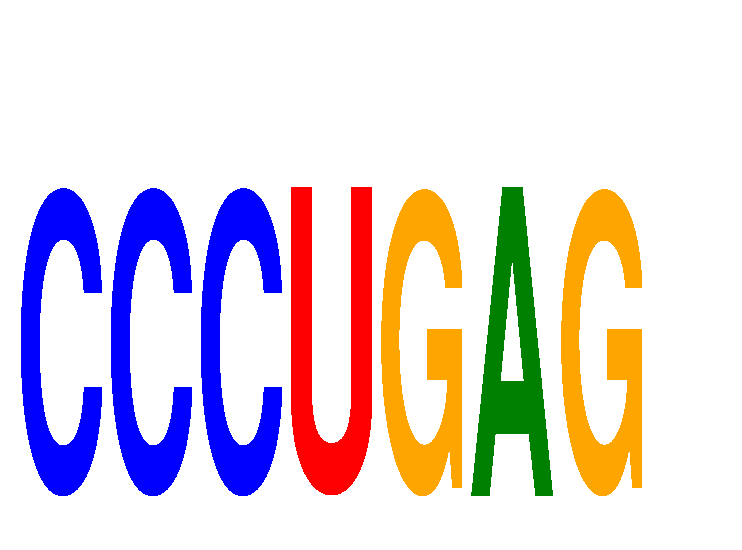Project
Illumina Body Map 2: averaged replicates
Navigation
Downloads
Results for CCCUGAG
Z-value: 1.34
miRNA associated with seed CCCUGAG
| Name | miRBASE accession |
|---|---|
|
hsa-miR-125a-5p
|
MIMAT0000443 |
|
hsa-miR-125b-5p
|
MIMAT0000423 |
|
hsa-miR-4319
|
MIMAT0016870 |
Activity profile of CCCUGAG motif
Sorted Z-values of CCCUGAG motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_-_90358048 | 3.04 |
ENST00000300060.6
ENST00000560137.1 |
ANPEP
|
alanyl (membrane) aminopeptidase |
| chr6_-_24911195 | 2.51 |
ENST00000259698.4
|
FAM65B
|
family with sequence similarity 65, member B |
| chr17_-_61777459 | 2.44 |
ENST00000578993.1
ENST00000583211.1 ENST00000259006.3 |
LIMD2
|
LIM domain containing 2 |
| chr5_-_100238956 | 2.37 |
ENST00000231461.5
|
ST8SIA4
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 4 |
| chr15_+_81489213 | 2.31 |
ENST00000559383.1
ENST00000394660.2 |
IL16
|
interleukin 16 |
| chr17_-_76713100 | 2.27 |
ENST00000585509.1
|
CYTH1
|
cytohesin 1 |
| chr3_-_13461807 | 2.27 |
ENST00000254508.5
|
NUP210
|
nucleoporin 210kDa |
| chr3_+_47021168 | 2.17 |
ENST00000450053.3
ENST00000292309.5 ENST00000383740.2 |
NBEAL2
|
neurobeachin-like 2 |
| chr1_+_12227035 | 2.14 |
ENST00000376259.3
ENST00000536782.1 |
TNFRSF1B
|
tumor necrosis factor receptor superfamily, member 1B |
| chr3_+_186648274 | 2.07 |
ENST00000169298.3
|
ST6GAL1
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 |
| chr11_+_122526383 | 2.03 |
ENST00000284273.5
|
UBASH3B
|
ubiquitin associated and SH3 domain containing B |
| chr6_+_106546808 | 2.03 |
ENST00000369089.3
|
PRDM1
|
PR domain containing 1, with ZNF domain |
| chr6_+_391739 | 1.92 |
ENST00000380956.4
|
IRF4
|
interferon regulatory factor 4 |
| chr1_+_26856236 | 1.91 |
ENST00000374168.2
ENST00000374166.4 |
RPS6KA1
|
ribosomal protein S6 kinase, 90kDa, polypeptide 1 |
| chr19_+_10959043 | 1.82 |
ENST00000397820.4
|
C19orf38
|
chromosome 19 open reading frame 38 |
| chr6_-_16761678 | 1.78 |
ENST00000244769.4
ENST00000436367.1 |
ATXN1
|
ataxin 1 |
| chr15_+_31619013 | 1.56 |
ENST00000307145.3
|
KLF13
|
Kruppel-like factor 13 |
| chr3_-_183273477 | 1.54 |
ENST00000341319.3
|
KLHL6
|
kelch-like family member 6 |
| chr2_-_163695128 | 1.52 |
ENST00000332142.5
|
KCNH7
|
potassium voltage-gated channel, subfamily H (eag-related), member 7 |
| chr17_-_38020392 | 1.49 |
ENST00000346872.3
ENST00000439167.2 ENST00000377945.3 ENST00000394189.2 ENST00000377944.3 ENST00000377958.2 ENST00000535189.1 ENST00000377952.2 |
IKZF3
|
IKAROS family zinc finger 3 (Aiolos) |
| chr19_-_11450249 | 1.47 |
ENST00000222120.3
|
RAB3D
|
RAB3D, member RAS oncogene family |
| chr19_+_926000 | 1.43 |
ENST00000263620.3
|
ARID3A
|
AT rich interactive domain 3A (BRIGHT-like) |
| chr1_-_108507631 | 1.42 |
ENST00000527011.1
ENST00000370056.4 |
VAV3
|
vav 3 guanine nucleotide exchange factor |
| chr11_-_102323489 | 1.39 |
ENST00000361236.3
|
TMEM123
|
transmembrane protein 123 |
| chr9_-_35650900 | 1.32 |
ENST00000259608.3
|
SIT1
|
signaling threshold regulating transmembrane adaptor 1 |
| chr9_-_123476719 | 1.30 |
ENST00000373930.3
|
MEGF9
|
multiple EGF-like-domains 9 |
| chr15_-_61521495 | 1.30 |
ENST00000335670.6
|
RORA
|
RAR-related orphan receptor A |
| chr19_-_4066890 | 1.23 |
ENST00000322357.4
|
ZBTB7A
|
zinc finger and BTB domain containing 7A |
| chr11_-_128392085 | 1.22 |
ENST00000526145.2
ENST00000531611.1 ENST00000319397.6 ENST00000345075.4 ENST00000535549.1 |
ETS1
|
v-ets avian erythroblastosis virus E26 oncogene homolog 1 |
| chr12_-_58240470 | 1.21 |
ENST00000548823.1
ENST00000398073.2 |
CTDSP2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 2 |
| chr3_+_127391769 | 1.20 |
ENST00000393363.3
ENST00000232744.8 ENST00000453791.2 |
ABTB1
|
ankyrin repeat and BTB (POZ) domain containing 1 |
| chr17_-_40540377 | 1.19 |
ENST00000404395.3
ENST00000389272.3 ENST00000585517.1 ENST00000588065.1 |
STAT3
|
signal transducer and activator of transcription 3 (acute-phase response factor) |
| chr10_+_82213904 | 1.17 |
ENST00000429989.3
|
TSPAN14
|
tetraspanin 14 |
| chr14_-_67982146 | 1.16 |
ENST00000557779.1
ENST00000557006.1 |
TMEM229B
|
transmembrane protein 229B |
| chr3_+_5229356 | 1.15 |
ENST00000256497.4
|
EDEM1
|
ER degradation enhancer, mannosidase alpha-like 1 |
| chr15_-_79383102 | 1.12 |
ENST00000558480.2
ENST00000419573.3 |
RASGRF1
|
Ras protein-specific guanine nucleotide-releasing factor 1 |
| chr17_-_66287257 | 1.11 |
ENST00000327268.4
|
SLC16A6
|
solute carrier family 16, member 6 |
| chr11_-_65381643 | 1.11 |
ENST00000309100.3
ENST00000529839.1 ENST00000526293.1 |
MAP3K11
|
mitogen-activated protein kinase kinase kinase 11 |
| chr18_-_19284724 | 1.08 |
ENST00000580981.1
ENST00000289119.2 |
ABHD3
|
abhydrolase domain containing 3 |
| chr12_-_54779511 | 1.07 |
ENST00000551109.1
ENST00000546970.1 |
ZNF385A
|
zinc finger protein 385A |
| chr19_-_1863567 | 1.05 |
ENST00000250916.4
|
KLF16
|
Kruppel-like factor 16 |
| chr3_+_58223228 | 1.04 |
ENST00000478253.1
ENST00000295962.4 |
ABHD6
|
abhydrolase domain containing 6 |
| chr8_-_63998590 | 1.02 |
ENST00000260116.4
|
TTPA
|
tocopherol (alpha) transfer protein |
| chr6_-_33547975 | 1.01 |
ENST00000442998.2
ENST00000360661.5 |
BAK1
|
BCL2-antagonist/killer 1 |
| chr10_-_73533255 | 1.00 |
ENST00000394957.3
|
C10orf54
|
chromosome 10 open reading frame 54 |
| chr8_-_8751068 | 1.00 |
ENST00000276282.6
|
MFHAS1
|
malignant fibrous histiocytoma amplified sequence 1 |
| chr9_+_79074068 | 1.00 |
ENST00000444201.2
ENST00000376730.4 |
GCNT1
|
glucosaminyl (N-acetyl) transferase 1, core 2 |
| chr7_+_142985308 | 0.99 |
ENST00000310447.5
|
CASP2
|
caspase 2, apoptosis-related cysteine peptidase |
| chr5_+_56111361 | 0.99 |
ENST00000399503.3
|
MAP3K1
|
mitogen-activated protein kinase kinase kinase 1, E3 ubiquitin protein ligase |
| chr3_-_72496035 | 0.98 |
ENST00000477973.2
|
RYBP
|
RING1 and YY1 binding protein |
| chr5_-_175964366 | 0.96 |
ENST00000274811.4
|
RNF44
|
ring finger protein 44 |
| chr19_+_13261216 | 0.95 |
ENST00000587885.1
ENST00000292433.3 |
IER2
|
immediate early response 2 |
| chr16_-_85045131 | 0.95 |
ENST00000313732.4
|
ZDHHC7
|
zinc finger, DHHC-type containing 7 |
| chr12_+_83080659 | 0.94 |
ENST00000321196.3
|
TMTC2
|
transmembrane and tetratricopeptide repeat containing 2 |
| chr12_-_110318263 | 0.94 |
ENST00000318348.4
|
GLTP
|
glycolipid transfer protein |
| chr1_-_200379180 | 0.94 |
ENST00000294740.3
|
ZNF281
|
zinc finger protein 281 |
| chr14_+_70078303 | 0.94 |
ENST00000342745.4
|
KIAA0247
|
KIAA0247 |
| chr1_-_111682662 | 0.94 |
ENST00000286692.4
|
DRAM2
|
DNA-damage regulated autophagy modulator 2 |
| chr8_-_141645645 | 0.94 |
ENST00000519980.1
ENST00000220592.5 |
AGO2
|
argonaute RISC catalytic component 2 |
| chr5_+_156887027 | 0.92 |
ENST00000435489.2
ENST00000311946.7 |
NIPAL4
|
NIPA-like domain containing 4 |
| chr8_-_53322303 | 0.92 |
ENST00000276480.7
|
ST18
|
suppression of tumorigenicity 18 (breast carcinoma) (zinc finger protein) |
| chr13_-_53024725 | 0.91 |
ENST00000378060.4
|
VPS36
|
vacuolar protein sorting 36 homolog (S. cerevisiae) |
| chr6_-_32811771 | 0.91 |
ENST00000395339.3
ENST00000374882.3 |
PSMB8
|
proteasome (prosome, macropain) subunit, beta type, 8 |
| chr19_-_19754404 | 0.91 |
ENST00000587205.1
ENST00000445806.2 ENST00000203556.4 |
GMIP
|
GEM interacting protein |
| chr1_+_47901689 | 0.90 |
ENST00000334793.5
|
FOXD2
|
forkhead box D2 |
| chr12_+_111843749 | 0.90 |
ENST00000341259.2
|
SH2B3
|
SH2B adaptor protein 3 |
| chr8_-_66754172 | 0.90 |
ENST00000401827.3
|
PDE7A
|
phosphodiesterase 7A |
| chr12_+_104359576 | 0.89 |
ENST00000392872.3
ENST00000436021.2 |
TDG
|
thymine-DNA glycosylase |
| chr20_-_62610982 | 0.89 |
ENST00000369886.3
ENST00000450107.1 |
SAMD10
|
sterile alpha motif domain containing 10 |
| chr2_+_5832799 | 0.88 |
ENST00000322002.3
|
SOX11
|
SRY (sex determining region Y)-box 11 |
| chr12_+_122150646 | 0.87 |
ENST00000449592.2
|
TMEM120B
|
transmembrane protein 120B |
| chr18_-_43652211 | 0.85 |
ENST00000589328.1
ENST00000409746.5 |
PSTPIP2
|
proline-serine-threonine phosphatase interacting protein 2 |
| chr1_+_89990431 | 0.85 |
ENST00000330947.2
ENST00000358200.4 |
LRRC8B
|
leucine rich repeat containing 8 family, member B |
| chr6_-_90062543 | 0.85 |
ENST00000435041.2
|
UBE2J1
|
ubiquitin-conjugating enzyme E2, J1 |
| chr12_-_56224546 | 0.85 |
ENST00000357606.3
ENST00000547445.1 |
DNAJC14
|
DnaJ (Hsp40) homolog, subfamily C, member 14 |
| chr3_-_52090461 | 0.84 |
ENST00000296483.6
ENST00000495880.1 |
DUSP7
|
dual specificity phosphatase 7 |
| chr2_-_201936302 | 0.84 |
ENST00000453765.1
ENST00000452799.1 ENST00000446678.1 ENST00000418596.3 |
FAM126B
|
family with sequence similarity 126, member B |
| chr12_+_11802753 | 0.83 |
ENST00000396373.4
|
ETV6
|
ets variant 6 |
| chr17_-_27507395 | 0.81 |
ENST00000354329.4
ENST00000527372.1 |
MYO18A
|
myosin XVIIIA |
| chr3_+_141205852 | 0.81 |
ENST00000286364.3
ENST00000452898.1 |
RASA2
|
RAS p21 protein activator 2 |
| chr5_+_74632993 | 0.79 |
ENST00000287936.4
|
HMGCR
|
3-hydroxy-3-methylglutaryl-CoA reductase |
| chr11_+_58939965 | 0.78 |
ENST00000227451.3
|
DTX4
|
deltex homolog 4 (Drosophila) |
| chr1_-_23857698 | 0.78 |
ENST00000361729.2
|
E2F2
|
E2F transcription factor 2 |
| chr15_+_63481668 | 0.77 |
ENST00000321437.4
ENST00000559006.1 ENST00000448330.2 |
RAB8B
|
RAB8B, member RAS oncogene family |
| chr12_+_53774423 | 0.76 |
ENST00000426431.2
|
SP1
|
Sp1 transcription factor |
| chr6_-_30658745 | 0.76 |
ENST00000376420.5
ENST00000376421.5 |
NRM
|
nurim (nuclear envelope membrane protein) |
| chrX_+_153639856 | 0.76 |
ENST00000426834.1
ENST00000369790.4 ENST00000454722.1 ENST00000350743.4 ENST00000299328.5 ENST00000351413.4 |
TAZ
|
tafazzin |
| chr11_-_78052923 | 0.76 |
ENST00000340149.2
|
GAB2
|
GRB2-associated binding protein 2 |
| chr8_+_37887772 | 0.76 |
ENST00000338825.4
|
EIF4EBP1
|
eukaryotic translation initiation factor 4E binding protein 1 |
| chr15_-_23086394 | 0.74 |
ENST00000337435.4
|
NIPA1
|
non imprinted in Prader-Willi/Angelman syndrome 1 |
| chr1_-_21671968 | 0.74 |
ENST00000415912.2
|
ECE1
|
endothelin converting enzyme 1 |
| chr12_+_56521840 | 0.73 |
ENST00000394048.5
|
ESYT1
|
extended synaptotagmin-like protein 1 |
| chr12_-_68553512 | 0.73 |
ENST00000229135.3
|
IFNG
|
interferon, gamma |
| chr3_-_48672859 | 0.73 |
ENST00000395550.2
ENST00000455886.2 ENST00000431739.1 ENST00000426599.1 ENST00000383733.3 ENST00000420764.2 ENST00000337000.8 |
SLC26A6
|
solute carrier family 26 (anion exchanger), member 6 |
| chr9_+_33817461 | 0.73 |
ENST00000263228.3
|
UBE2R2
|
ubiquitin-conjugating enzyme E2R 2 |
| chr11_+_94277017 | 0.73 |
ENST00000358752.2
|
FUT4
|
fucosyltransferase 4 (alpha (1,3) fucosyltransferase, myeloid-specific) |
| chr12_+_49932886 | 0.70 |
ENST00000257981.6
|
KCNH3
|
potassium voltage-gated channel, subfamily H (eag-related), member 3 |
| chr11_+_75526212 | 0.70 |
ENST00000356136.3
|
UVRAG
|
UV radiation resistance associated |
| chr9_-_130829588 | 0.70 |
ENST00000373078.4
|
NAIF1
|
nuclear apoptosis inducing factor 1 |
| chrX_+_78003204 | 0.70 |
ENST00000435339.3
ENST00000514744.1 |
LPAR4
|
lysophosphatidic acid receptor 4 |
| chr2_+_131113580 | 0.70 |
ENST00000175756.5
|
PTPN18
|
protein tyrosine phosphatase, non-receptor type 18 (brain-derived) |
| chr12_-_69326940 | 0.70 |
ENST00000549781.1
ENST00000548262.1 ENST00000551568.1 ENST00000548954.1 |
CPM
|
carboxypeptidase M |
| chr10_+_72575643 | 0.69 |
ENST00000373202.3
|
SGPL1
|
sphingosine-1-phosphate lyase 1 |
| chr5_+_96271141 | 0.69 |
ENST00000231368.5
|
LNPEP
|
leucyl/cystinyl aminopeptidase |
| chrX_-_103401649 | 0.68 |
ENST00000357421.4
|
SLC25A53
|
solute carrier family 25, member 53 |
| chr17_-_40761375 | 0.68 |
ENST00000543197.1
ENST00000309428.5 |
FAM134C
|
family with sequence similarity 134, member C |
| chr7_-_139876812 | 0.67 |
ENST00000397560.2
|
JHDM1D
|
lysine (K)-specific demethylase 7A |
| chr17_+_73512594 | 0.67 |
ENST00000333213.6
|
TSEN54
|
TSEN54 tRNA splicing endonuclease subunit |
| chr11_-_64901978 | 0.67 |
ENST00000294256.8
ENST00000377190.3 |
SYVN1
|
synovial apoptosis inhibitor 1, synoviolin |
| chr10_-_118764862 | 0.66 |
ENST00000260777.10
|
KIAA1598
|
KIAA1598 |
| chr3_+_15468862 | 0.66 |
ENST00000396842.2
|
EAF1
|
ELL associated factor 1 |
| chr1_-_207224307 | 0.66 |
ENST00000315927.4
|
YOD1
|
YOD1 deubiquitinase |
| chr2_-_85555385 | 0.65 |
ENST00000377386.3
|
TGOLN2
|
trans-golgi network protein 2 |
| chr5_+_80256453 | 0.65 |
ENST00000265080.4
|
RASGRF2
|
Ras protein-specific guanine nucleotide-releasing factor 2 |
| chr14_-_73493825 | 0.64 |
ENST00000318876.5
ENST00000556143.1 |
ZFYVE1
|
zinc finger, FYVE domain containing 1 |
| chr16_-_23521710 | 0.64 |
ENST00000562117.1
ENST00000567468.1 ENST00000562944.1 ENST00000309859.4 |
GGA2
|
golgi-associated, gamma adaptin ear containing, ARF binding protein 2 |
| chr2_+_12857015 | 0.64 |
ENST00000155926.4
|
TRIB2
|
tribbles pseudokinase 2 |
| chr17_-_48227877 | 0.64 |
ENST00000316878.6
|
PPP1R9B
|
protein phosphatase 1, regulatory subunit 9B |
| chr12_+_50135588 | 0.64 |
ENST00000423828.1
ENST00000550445.1 |
TMBIM6
|
transmembrane BAX inhibitor motif containing 6 |
| chr1_+_36273743 | 0.64 |
ENST00000373210.3
|
AGO4
|
argonaute RISC catalytic component 4 |
| chr9_+_129622904 | 0.64 |
ENST00000319119.4
|
ZBTB34
|
zinc finger and BTB domain containing 34 |
| chr7_-_143059845 | 0.63 |
ENST00000443739.2
|
FAM131B
|
family with sequence similarity 131, member B |
| chr1_+_203274639 | 0.63 |
ENST00000290551.4
|
BTG2
|
BTG family, member 2 |
| chr12_-_51717922 | 0.63 |
ENST00000452142.2
|
BIN2
|
bridging integrator 2 |
| chr10_-_60027642 | 0.63 |
ENST00000373935.3
|
IPMK
|
inositol polyphosphate multikinase |
| chr17_+_61699766 | 0.62 |
ENST00000579585.1
ENST00000584573.1 ENST00000361733.3 ENST00000361357.3 |
MAP3K3
|
mitogen-activated protein kinase kinase kinase 3 |
| chr17_+_53828381 | 0.62 |
ENST00000576183.1
|
PCTP
|
phosphatidylcholine transfer protein |
| chr12_+_68042495 | 0.62 |
ENST00000344096.3
|
DYRK2
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 2 |
| chr17_-_27224621 | 0.62 |
ENST00000394906.2
ENST00000585169.1 ENST00000394908.4 |
FLOT2
|
flotillin 2 |
| chr11_-_65769594 | 0.62 |
ENST00000532707.1
ENST00000533544.1 ENST00000526451.1 ENST00000312234.2 ENST00000530462.1 ENST00000525767.1 ENST00000529964.1 ENST00000527249.1 |
EIF1AD
|
eukaryotic translation initiation factor 1A domain containing |
| chr8_-_95961578 | 0.62 |
ENST00000448464.2
ENST00000342697.4 |
TP53INP1
|
tumor protein p53 inducible nuclear protein 1 |
| chr2_+_47168313 | 0.60 |
ENST00000319190.5
ENST00000394850.2 ENST00000536057.1 |
TTC7A
|
tetratricopeptide repeat domain 7A |
| chr17_-_79876010 | 0.60 |
ENST00000328666.6
|
SIRT7
|
sirtuin 7 |
| chr22_-_37608325 | 0.60 |
ENST00000328544.3
|
SSTR3
|
somatostatin receptor 3 |
| chr1_+_160051319 | 0.60 |
ENST00000368088.3
|
KCNJ9
|
potassium inwardly-rectifying channel, subfamily J, member 9 |
| chr5_-_134788086 | 0.59 |
ENST00000537858.1
|
TIFAB
|
TRAF-interacting protein with forkhead-associated domain, family member B |
| chr6_-_43596899 | 0.58 |
ENST00000307126.5
ENST00000452781.1 |
GTPBP2
|
GTP binding protein 2 |
| chr5_+_110559784 | 0.58 |
ENST00000282356.4
|
CAMK4
|
calcium/calmodulin-dependent protein kinase IV |
| chr18_+_19321281 | 0.58 |
ENST00000261537.6
|
MIB1
|
mindbomb E3 ubiquitin protein ligase 1 |
| chr4_-_57301748 | 0.57 |
ENST00000264220.2
|
PPAT
|
phosphoribosyl pyrophosphate amidotransferase |
| chr5_-_131347306 | 0.57 |
ENST00000296869.4
ENST00000379249.3 ENST00000379272.2 ENST00000379264.2 |
ACSL6
|
acyl-CoA synthetase long-chain family member 6 |
| chr5_+_177019159 | 0.57 |
ENST00000332598.6
|
TMED9
|
transmembrane emp24 protein transport domain containing 9 |
| chr1_+_178694300 | 0.56 |
ENST00000367635.3
|
RALGPS2
|
Ral GEF with PH domain and SH3 binding motif 2 |
| chr10_-_14590644 | 0.56 |
ENST00000378470.1
|
FAM107B
|
family with sequence similarity 107, member B |
| chr19_+_13906250 | 0.54 |
ENST00000254323.2
|
ZSWIM4
|
zinc finger, SWIM-type containing 4 |
| chr5_+_151151471 | 0.54 |
ENST00000394123.3
ENST00000543466.1 |
G3BP1
|
GTPase activating protein (SH3 domain) binding protein 1 |
| chr16_-_2264779 | 0.53 |
ENST00000333503.7
|
PGP
|
phosphoglycolate phosphatase |
| chr18_+_32558208 | 0.53 |
ENST00000436190.2
|
MAPRE2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr2_-_158485387 | 0.53 |
ENST00000243349.8
|
ACVR1C
|
activin A receptor, type IC |
| chr11_+_45825896 | 0.53 |
ENST00000314134.3
|
SLC35C1
|
solute carrier family 35 (GDP-fucose transporter), member C1 |
| chr12_+_65004292 | 0.53 |
ENST00000542104.1
ENST00000336061.2 |
RASSF3
|
Ras association (RalGDS/AF-6) domain family member 3 |
| chr6_-_31830655 | 0.53 |
ENST00000375631.4
|
NEU1
|
sialidase 1 (lysosomal sialidase) |
| chr5_-_133561752 | 0.53 |
ENST00000519718.1
ENST00000481195.1 |
CTD-2410N18.5
PPP2CA
|
S-phase kinase-associated protein 1 protein phosphatase 2, catalytic subunit, alpha isozyme |
| chr7_-_106301405 | 0.52 |
ENST00000523505.1
|
CCDC71L
|
coiled-coil domain containing 71-like |
| chr1_+_32573636 | 0.52 |
ENST00000373625.3
|
KPNA6
|
karyopherin alpha 6 (importin alpha 7) |
| chr10_-_94003003 | 0.52 |
ENST00000412050.4
|
CPEB3
|
cytoplasmic polyadenylation element binding protein 3 |
| chr16_-_71610985 | 0.51 |
ENST00000355962.4
|
TAT
|
tyrosine aminotransferase |
| chr1_+_26737253 | 0.51 |
ENST00000326279.6
|
LIN28A
|
lin-28 homolog A (C. elegans) |
| chr2_+_61293021 | 0.51 |
ENST00000402291.1
|
KIAA1841
|
KIAA1841 |
| chr5_+_133861790 | 0.50 |
ENST00000395003.1
|
PHF15
|
jade family PHD finger 2 |
| chr8_-_67579418 | 0.50 |
ENST00000310421.4
|
VCPIP1
|
valosin containing protein (p97)/p47 complex interacting protein 1 |
| chr13_-_29292956 | 0.50 |
ENST00000266943.6
|
SLC46A3
|
solute carrier family 46, member 3 |
| chr1_-_179198702 | 0.50 |
ENST00000502732.1
|
ABL2
|
c-abl oncogene 2, non-receptor tyrosine kinase |
| chr2_+_7057523 | 0.50 |
ENST00000320892.6
|
RNF144A
|
ring finger protein 144A |
| chr22_-_37099555 | 0.50 |
ENST00000300105.6
|
CACNG2
|
calcium channel, voltage-dependent, gamma subunit 2 |
| chr17_+_2496971 | 0.50 |
ENST00000397195.5
|
PAFAH1B1
|
platelet-activating factor acetylhydrolase 1b, regulatory subunit 1 (45kDa) |
| chr3_-_39195037 | 0.49 |
ENST00000273153.5
|
CSRNP1
|
cysteine-serine-rich nuclear protein 1 |
| chr19_+_19303008 | 0.49 |
ENST00000353145.1
ENST00000421262.3 ENST00000303088.4 ENST00000456252.3 ENST00000593273.1 |
RFXANK
|
regulatory factor X-associated ankyrin-containing protein |
| chr11_-_67169253 | 0.49 |
ENST00000527663.1
ENST00000312989.7 |
PPP1CA
|
protein phosphatase 1, catalytic subunit, alpha isozyme |
| chr13_+_41363581 | 0.49 |
ENST00000338625.4
|
SLC25A15
|
solute carrier family 25 (mitochondrial carrier; ornithine transporter) member 15 |
| chr8_-_23315190 | 0.48 |
ENST00000356206.6
ENST00000358689.4 ENST00000417069.2 |
ENTPD4
|
ectonucleoside triphosphate diphosphohydrolase 4 |
| chr6_+_18387570 | 0.48 |
ENST00000259939.3
|
RNF144B
|
ring finger protein 144B |
| chr14_-_91976488 | 0.48 |
ENST00000554684.1
ENST00000337238.4 ENST00000428424.2 ENST00000554511.1 |
SMEK1
|
SMEK homolog 1, suppressor of mek1 (Dictyostelium) |
| chr20_-_31071239 | 0.48 |
ENST00000359676.5
|
C20orf112
|
chromosome 20 open reading frame 112 |
| chr18_-_60987220 | 0.47 |
ENST00000398117.1
|
BCL2
|
B-cell CLL/lymphoma 2 |
| chr12_+_50690489 | 0.47 |
ENST00000598429.1
|
AC140061.12
|
Uncharacterized protein |
| chr10_+_72432559 | 0.46 |
ENST00000373208.1
ENST00000373207.1 |
ADAMTS14
|
ADAM metallopeptidase with thrombospondin type 1 motif, 14 |
| chr3_-_53381539 | 0.46 |
ENST00000606822.1
ENST00000294241.6 ENST00000607628.1 |
DCP1A
|
decapping mRNA 1A |
| chr14_+_71374122 | 0.46 |
ENST00000304743.2
ENST00000238570.5 |
PCNX
|
pecanex homolog (Drosophila) |
| chr1_+_165796753 | 0.46 |
ENST00000367879.4
|
UCK2
|
uridine-cytidine kinase 2 |
| chr6_+_35995488 | 0.46 |
ENST00000229795.3
|
MAPK14
|
mitogen-activated protein kinase 14 |
| chr10_+_65281123 | 0.45 |
ENST00000298249.4
ENST00000373758.4 |
REEP3
|
receptor accessory protein 3 |
| chr12_-_117319236 | 0.45 |
ENST00000257572.5
|
HRK
|
harakiri, BCL2 interacting protein (contains only BH3 domain) |
| chr1_-_23495340 | 0.45 |
ENST00000418342.1
|
LUZP1
|
leucine zipper protein 1 |
| chr2_-_97535708 | 0.45 |
ENST00000305476.5
|
SEMA4C
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4C |
| chr9_-_19786926 | 0.45 |
ENST00000341998.2
ENST00000286344.3 |
SLC24A2
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 2 |
| chr1_-_36022979 | 0.45 |
ENST00000469892.1
ENST00000325722.3 |
KIAA0319L
|
KIAA0319-like |
| chr17_+_38375574 | 0.45 |
ENST00000323571.4
ENST00000585043.1 ENST00000394103.3 ENST00000536600.1 |
WIPF2
|
WAS/WASL interacting protein family, member 2 |
| chr4_+_174089904 | 0.45 |
ENST00000265000.4
|
GALNT7
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 7 (GalNAc-T7) |
| chr11_-_36531774 | 0.45 |
ENST00000348124.5
ENST00000526995.1 |
TRAF6
|
TNF receptor-associated factor 6, E3 ubiquitin protein ligase |
| chr2_-_206950781 | 0.44 |
ENST00000403263.1
|
INO80D
|
INO80 complex subunit D |
| chr5_+_137673945 | 0.44 |
ENST00000513056.1
ENST00000511276.1 |
FAM53C
|
family with sequence similarity 53, member C |
| chr1_-_173174681 | 0.44 |
ENST00000367718.1
|
TNFSF4
|
tumor necrosis factor (ligand) superfamily, member 4 |
| chr20_+_62795827 | 0.44 |
ENST00000328439.1
ENST00000536311.1 |
MYT1
|
myelin transcription factor 1 |
| chr9_-_130497565 | 0.43 |
ENST00000336067.6
ENST00000373281.5 ENST00000373284.5 ENST00000458505.3 |
TOR2A
|
torsin family 2, member A |
| chr11_+_61447845 | 0.43 |
ENST00000257215.5
|
DAGLA
|
diacylglycerol lipase, alpha |
| chr8_+_22857048 | 0.43 |
ENST00000251822.6
|
RHOBTB2
|
Rho-related BTB domain containing 2 |
| chr4_+_25235597 | 0.43 |
ENST00000264864.6
|
PI4K2B
|
phosphatidylinositol 4-kinase type 2 beta |
| chr8_-_74791051 | 0.42 |
ENST00000453587.2
ENST00000602969.1 ENST00000602593.1 ENST00000419880.3 ENST00000517608.1 |
UBE2W
|
ubiquitin-conjugating enzyme E2W (putative) |
| chr14_+_60715928 | 0.42 |
ENST00000395076.4
|
PPM1A
|
protein phosphatase, Mg2+/Mn2+ dependent, 1A |
Network of associatons between targets according to the STRING database.
First level regulatory network of CCCUGAG
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.0 | GO:1990654 | sebum secreting cell proliferation(GO:1990654) |
| 0.6 | 1.9 | GO:0045082 | positive regulation of interleukin-10 biosynthetic process(GO:0045082) positive regulation of interleukin-4 biosynthetic process(GO:0045404) |
| 0.3 | 1.0 | GO:0048597 | B cell negative selection(GO:0002352) post-embryonic camera-type eye morphogenesis(GO:0048597) |
| 0.3 | 1.5 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.3 | 0.9 | GO:0061386 | closure of optic fissure(GO:0061386) |
| 0.3 | 1.6 | GO:0036510 | trimming of terminal mannose on C branch(GO:0036510) |
| 0.2 | 0.7 | GO:2000309 | CD8-positive, alpha-beta T cell differentiation involved in immune response(GO:0002302) regulation of CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0032829) positive regulation of CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0032831) positive regulation of vitamin metabolic process(GO:0046136) positive regulation of vitamin D biosynthetic process(GO:0060557) positive regulation of calcidiol 1-monooxygenase activity(GO:0060559) positive regulation of tumor necrosis factor (ligand) superfamily member 11 production(GO:2000309) |
| 0.2 | 0.7 | GO:0042938 | dipeptide transport(GO:0042938) |
| 0.2 | 1.2 | GO:0010730 | negative regulation of hydrogen peroxide biosynthetic process(GO:0010730) positive regulation of skeletal muscle satellite cell proliferation(GO:1902724) positive regulation of growth factor dependent skeletal muscle satellite cell proliferation(GO:1902728) |
| 0.2 | 1.6 | GO:0090625 | mRNA cleavage involved in gene silencing by siRNA(GO:0090625) |
| 0.2 | 0.9 | GO:1902544 | regulation of DNA N-glycosylase activity(GO:1902544) |
| 0.2 | 0.6 | GO:0045081 | negative regulation of interleukin-10 biosynthetic process(GO:0045081) |
| 0.2 | 0.6 | GO:1904761 | negative regulation of myofibroblast differentiation(GO:1904761) |
| 0.2 | 1.0 | GO:0090212 | vitamin E metabolic process(GO:0042360) regulation of establishment of blood-brain barrier(GO:0090210) negative regulation of establishment of blood-brain barrier(GO:0090212) |
| 0.2 | 0.8 | GO:1903028 | positive regulation of opsonization(GO:1903028) |
| 0.2 | 0.6 | GO:0002372 | myeloid dendritic cell cytokine production(GO:0002372) |
| 0.2 | 1.9 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.2 | 1.5 | GO:0071926 | endocannabinoid signaling pathway(GO:0071926) |
| 0.2 | 0.9 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.2 | 0.5 | GO:0006114 | glycerol biosynthetic process(GO:0006114) |
| 0.2 | 0.5 | GO:1901383 | negative regulation of chorionic trophoblast cell proliferation(GO:1901383) |
| 0.2 | 0.5 | GO:1900365 | positive regulation of mRNA polyadenylation(GO:1900365) |
| 0.2 | 0.7 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.2 | 0.7 | GO:1990167 | protein K27-linked deubiquitination(GO:1990167) protein K33-linked deubiquitination(GO:1990168) |
| 0.2 | 0.6 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) |
| 0.2 | 0.8 | GO:1990086 | lens fiber cell apoptotic process(GO:1990086) |
| 0.2 | 1.1 | GO:1902164 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902162) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.2 | 1.2 | GO:0030578 | PML body organization(GO:0030578) |
| 0.2 | 0.6 | GO:0045897 | positive regulation of transcription during mitosis(GO:0045897) |
| 0.2 | 0.5 | GO:0021503 | neural fold bending(GO:0021503) |
| 0.1 | 0.3 | GO:2000646 | positive regulation of receptor catabolic process(GO:2000646) |
| 0.1 | 0.4 | GO:2000525 | regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 0.1 | 1.0 | GO:0035234 | ectopic germ cell programmed cell death(GO:0035234) |
| 0.1 | 0.6 | GO:0048205 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.1 | 1.2 | GO:0046836 | glycolipid transport(GO:0046836) |
| 0.1 | 2.1 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.1 | 0.8 | GO:0036166 | phenotypic switching(GO:0036166) regulation of phenotypic switching(GO:1900239) |
| 0.1 | 0.4 | GO:0090222 | centrosome-templated microtubule nucleation(GO:0090222) |
| 0.1 | 0.4 | GO:1904562 | phosphatidylinositol 5-phosphate metabolic process(GO:1904562) |
| 0.1 | 0.7 | GO:0010814 | substance P catabolic process(GO:0010814) calcitonin catabolic process(GO:0010816) endothelin maturation(GO:0034959) |
| 0.1 | 0.5 | GO:0009138 | pyrimidine nucleoside diphosphate metabolic process(GO:0009138) |
| 0.1 | 0.6 | GO:0008628 | hormone-mediated apoptotic signaling pathway(GO:0008628) somatostatin receptor signaling pathway(GO:0038169) somatostatin signaling pathway(GO:0038170) |
| 0.1 | 0.4 | GO:2000705 | dense core granule biogenesis(GO:0061110) regulation of dense core granule biogenesis(GO:2000705) |
| 0.1 | 0.5 | GO:0043375 | negative regulation of cellular pH reduction(GO:0032848) CD8-positive, alpha-beta T cell lineage commitment(GO:0043375) negative regulation of retinal cell programmed cell death(GO:0046671) |
| 0.1 | 0.4 | GO:0002384 | hepatic immune response(GO:0002384) |
| 0.1 | 0.6 | GO:0021650 | vestibulocochlear nerve formation(GO:0021650) |
| 0.1 | 0.5 | GO:0046035 | CMP salvage(GO:0006238) CMP biosynthetic process(GO:0009224) CMP metabolic process(GO:0046035) |
| 0.1 | 2.4 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.1 | 0.8 | GO:0051461 | positive regulation of corticotropin secretion(GO:0051461) |
| 0.1 | 1.1 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.1 | 2.3 | GO:0045779 | negative regulation of bone resorption(GO:0045779) |
| 0.1 | 0.5 | GO:0036079 | GDP-fucose transport(GO:0015783) purine nucleotide-sugar transport(GO:0036079) |
| 0.1 | 1.6 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.1 | 0.8 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.1 | 1.7 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.1 | 0.6 | GO:1903903 | regulation of establishment of T cell polarity(GO:1903903) |
| 0.1 | 0.7 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.1 | 1.0 | GO:0070862 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.1 | 0.4 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.1 | 0.5 | GO:0036496 | regulation of translational initiation by eIF2 alpha dephosphorylation(GO:0036496) |
| 0.1 | 0.6 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.1 | 0.4 | GO:0021816 | lamellipodium assembly involved in ameboidal cell migration(GO:0003363) extension of a leading process involved in cell motility in cerebral cortex radial glia guided migration(GO:0021816) |
| 0.1 | 1.3 | GO:0021694 | cerebellar Purkinje cell layer formation(GO:0021694) cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.1 | 2.3 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.1 | 0.6 | GO:0031438 | negative regulation of mRNA cleavage(GO:0031438) negative regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904721) |
| 0.1 | 0.5 | GO:0014835 | myoblast differentiation involved in skeletal muscle regeneration(GO:0014835) positive regulation of interleukin-12 secretion(GO:2001184) |
| 0.1 | 1.0 | GO:0060352 | cell adhesion molecule production(GO:0060352) |
| 0.1 | 1.2 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.1 | 1.6 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.1 | 0.8 | GO:1990928 | response to amino acid starvation(GO:1990928) |
| 0.1 | 0.8 | GO:0035965 | cardiolipin acyl-chain remodeling(GO:0035965) |
| 0.1 | 0.4 | GO:0071043 | CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) |
| 0.1 | 0.5 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.1 | 0.6 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.1 | 0.3 | GO:0042351 | 'de novo' GDP-L-fucose biosynthetic process(GO:0042351) |
| 0.1 | 1.6 | GO:0002467 | germinal center formation(GO:0002467) |
| 0.1 | 0.4 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.1 | 0.4 | GO:0035549 | positive regulation of interferon-beta secretion(GO:0035549) |
| 0.1 | 2.1 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.1 | 2.3 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 0.3 | GO:1903722 | negative regulation of exosomal secretion(GO:1903542) regulation of centriole elongation(GO:1903722) |
| 0.1 | 0.9 | GO:0070102 | interleukin-6-mediated signaling pathway(GO:0070102) |
| 0.1 | 0.6 | GO:0032484 | Ral protein signal transduction(GO:0032484) regulation of Ral protein signal transduction(GO:0032485) |
| 0.1 | 0.5 | GO:1903352 | L-ornithine transmembrane transport(GO:1903352) |
| 0.1 | 0.7 | GO:0002480 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-independent(GO:0002480) |
| 0.1 | 0.4 | GO:1903378 | positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 0.1 | 2.4 | GO:0030220 | platelet formation(GO:0030220) |
| 0.1 | 0.9 | GO:0019317 | fucose catabolic process(GO:0019317) L-fucose metabolic process(GO:0042354) L-fucose catabolic process(GO:0042355) |
| 0.1 | 0.2 | GO:0005989 | lactose metabolic process(GO:0005988) lactose biosynthetic process(GO:0005989) apoptotic process involved in mammary gland involution(GO:0060057) positive regulation of apoptotic process involved in mammary gland involution(GO:0060058) positive regulation of apoptotic process involved in morphogenesis(GO:1902339) regulation of mammary gland involution(GO:1903519) positive regulation of mammary gland involution(GO:1903521) positive regulation of apoptotic process involved in development(GO:1904747) |
| 0.1 | 0.2 | GO:0060392 | negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 0.1 | 0.6 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.1 | 0.4 | GO:1904674 | positive regulation of somatic stem cell population maintenance(GO:1904674) |
| 0.1 | 0.8 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.1 | 0.2 | GO:0015920 | lipopolysaccharide transport(GO:0015920) |
| 0.1 | 0.3 | GO:0061528 | negative regulation of gamma-aminobutyric acid secretion(GO:0014053) aspartate secretion(GO:0061528) regulation of aspartate secretion(GO:1904448) positive regulation of aspartate secretion(GO:1904450) |
| 0.1 | 0.5 | GO:0031087 | nuclear-transcribed mRNA catabolic process, deadenylation-independent decay(GO:0031086) deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.1 | 0.3 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.1 | 0.5 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.1 | 0.3 | GO:0090370 | negative regulation of cholesterol efflux(GO:0090370) |
| 0.1 | 0.4 | GO:0045084 | positive regulation of interleukin-12 biosynthetic process(GO:0045084) |
| 0.1 | 0.5 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.1 | 0.2 | GO:1900224 | positive regulation of nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900224) |
| 0.1 | 0.4 | GO:0034670 | chemotaxis to arachidonic acid(GO:0034670) response to arachidonic acid(GO:1904550) |
| 0.1 | 0.4 | GO:0006499 | N-terminal protein myristoylation(GO:0006499) |
| 0.1 | 0.1 | GO:0021699 | cerebellum maturation(GO:0021590) cerebellar Purkinje cell layer maturation(GO:0021691) cerebellar cortex maturation(GO:0021699) |
| 0.1 | 0.3 | GO:0097327 | response to antineoplastic agent(GO:0097327) |
| 0.1 | 0.3 | GO:0071883 | activation of MAPK activity by adrenergic receptor signaling pathway(GO:0071883) regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.0 | 0.4 | GO:0060292 | long term synaptic depression(GO:0060292) |
| 0.0 | 1.1 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 0.0 | 1.0 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.0 | 0.4 | GO:0097411 | hypoxia-inducible factor-1alpha signaling pathway(GO:0097411) |
| 0.0 | 0.1 | GO:0002194 | hepatocyte cell migration(GO:0002194) otic placode formation(GO:0043049) branching involved in pancreas morphogenesis(GO:0061114) acinar cell differentiation(GO:0090425) positive regulation of forebrain neuron differentiation(GO:2000979) |
| 0.0 | 0.7 | GO:0001553 | luteinization(GO:0001553) |
| 0.0 | 1.5 | GO:0045577 | regulation of B cell differentiation(GO:0045577) |
| 0.0 | 0.5 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.0 | 0.2 | GO:0097045 | phosphatidylserine exposure on blood platelet(GO:0097045) |
| 0.0 | 0.4 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.0 | 0.2 | GO:1904781 | positive regulation of protein localization to centrosome(GO:1904781) |
| 0.0 | 0.2 | GO:1903826 | arginine transmembrane transport(GO:1903826) |
| 0.0 | 2.5 | GO:0006409 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.0 | 2.1 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.0 | 0.8 | GO:0043306 | positive regulation of mast cell activation involved in immune response(GO:0033008) positive regulation of mast cell degranulation(GO:0043306) |
| 0.0 | 0.4 | GO:0036089 | cleavage furrow formation(GO:0036089) |
| 0.0 | 0.1 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.0 | 0.3 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.0 | 0.9 | GO:0035162 | embryonic hemopoiesis(GO:0035162) |
| 0.0 | 0.2 | GO:2001270 | regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001270) negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.0 | 0.3 | GO:2000049 | positive regulation of cell-cell adhesion mediated by cadherin(GO:2000049) |
| 0.0 | 0.6 | GO:0060211 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) |
| 0.0 | 0.3 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.0 | 0.9 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.0 | 1.4 | GO:0030890 | positive regulation of B cell proliferation(GO:0030890) |
| 0.0 | 0.3 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.0 | 1.3 | GO:0043029 | T cell homeostasis(GO:0043029) |
| 0.0 | 0.1 | GO:0001579 | medium-chain fatty acid transport(GO:0001579) |
| 0.0 | 0.9 | GO:0010172 | embryonic body morphogenesis(GO:0010172) |
| 0.0 | 0.2 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.0 | 0.1 | GO:1905071 | proepicardium development(GO:0003342) septum transversum development(GO:0003343) occluding junction disassembly(GO:1905071) regulation of occluding junction disassembly(GO:1905073) positive regulation of occluding junction disassembly(GO:1905075) |
| 0.0 | 0.5 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.0 | 0.9 | GO:1904380 | endoplasmic reticulum mannose trimming(GO:1904380) |
| 0.0 | 0.9 | GO:0035518 | histone H2A monoubiquitination(GO:0035518) |
| 0.0 | 1.9 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 0.4 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 0.0 | 0.3 | GO:1901098 | positive regulation of autophagosome maturation(GO:1901098) |
| 0.0 | 0.1 | GO:0070904 | L-ascorbic acid transport(GO:0015882) molecular hydrogen transport(GO:0015993) transepithelial L-ascorbic acid transport(GO:0070904) |
| 0.0 | 0.3 | GO:2001222 | regulation of neuron migration(GO:2001222) |
| 0.0 | 1.2 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.0 | 0.2 | GO:0034128 | negative regulation of MyD88-independent toll-like receptor signaling pathway(GO:0034128) |
| 0.0 | 0.6 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.0 | 0.1 | GO:0031998 | regulation of fatty acid beta-oxidation(GO:0031998) |
| 0.0 | 0.2 | GO:2001138 | regulation of phospholipid transport(GO:2001138) positive regulation of phospholipid transport(GO:2001140) |
| 0.0 | 1.0 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.5 | GO:0032464 | positive regulation of protein homooligomerization(GO:0032464) |
| 0.0 | 0.3 | GO:0006655 | phosphatidylglycerol biosynthetic process(GO:0006655) |
| 0.0 | 0.3 | GO:0034378 | chylomicron assembly(GO:0034378) |
| 0.0 | 0.1 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.0 | 0.4 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 0.2 | GO:1904379 | protein localization to cytosolic proteasome complex(GO:1904327) protein localization to cytosolic proteasome complex involved in ERAD pathway(GO:1904379) |
| 0.0 | 0.4 | GO:0061099 | negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.0 | 0.9 | GO:0045494 | photoreceptor cell maintenance(GO:0045494) |
| 0.0 | 0.4 | GO:0000183 | chromatin silencing at rDNA(GO:0000183) |
| 0.0 | 0.5 | GO:0061157 | mRNA destabilization(GO:0061157) |
| 0.0 | 2.7 | GO:2000134 | negative regulation of G1/S transition of mitotic cell cycle(GO:2000134) |
| 0.0 | 0.7 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.0 | 0.9 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.2 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.0 | 0.3 | GO:0006474 | N-terminal protein amino acid acetylation(GO:0006474) |
| 0.0 | 0.0 | GO:0010360 | negative regulation of anion channel activity(GO:0010360) Golgi to plasma membrane CFTR protein transport(GO:0043000) |
| 0.0 | 0.1 | GO:0034499 | late endosome to Golgi transport(GO:0034499) |
| 0.0 | 0.2 | GO:0070544 | histone H3-K36 demethylation(GO:0070544) |
| 0.0 | 0.2 | GO:0051292 | nuclear pore organization(GO:0006999) nuclear pore complex assembly(GO:0051292) |
| 0.0 | 0.4 | GO:0071218 | cellular response to misfolded protein(GO:0071218) |
| 0.0 | 0.3 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 1.0 | GO:0006656 | phosphatidylcholine biosynthetic process(GO:0006656) |
| 0.0 | 0.1 | GO:0072719 | copper ion import(GO:0015677) cellular response to cisplatin(GO:0072719) |
| 0.0 | 0.1 | GO:0090063 | positive regulation of microtubule nucleation(GO:0090063) |
| 0.0 | 0.1 | GO:2001241 | positive regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001241) |
| 0.0 | 0.1 | GO:0006021 | inositol biosynthetic process(GO:0006021) |
| 0.0 | 0.1 | GO:0001767 | establishment of lymphocyte polarity(GO:0001767) |
| 0.0 | 0.1 | GO:0051532 | regulation of NFAT protein import into nucleus(GO:0051532) |
| 0.0 | 0.2 | GO:0044387 | negative regulation of protein kinase activity by regulation of protein phosphorylation(GO:0044387) |
| 0.0 | 0.1 | GO:0014028 | notochord formation(GO:0014028) |
| 0.0 | 1.0 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.0 | 0.1 | GO:0036155 | acylglycerol acyl-chain remodeling(GO:0036155) |
| 0.0 | 0.6 | GO:0071800 | podosome assembly(GO:0071800) |
| 0.0 | 0.3 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 0.3 | GO:0006974 | cellular response to DNA damage stimulus(GO:0006974) |
| 0.0 | 0.3 | GO:0045589 | regulation of regulatory T cell differentiation(GO:0045589) |
| 0.0 | 0.3 | GO:0071243 | cellular response to arsenic-containing substance(GO:0071243) |
| 0.0 | 0.5 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.0 | 0.2 | GO:0000338 | protein deneddylation(GO:0000338) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0000814 | ESCRT II complex(GO:0000814) |
| 0.3 | 2.6 | GO:0043196 | varicosity(GO:0043196) |
| 0.3 | 1.8 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.2 | 0.7 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.2 | 0.9 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.2 | 0.5 | GO:0097637 | intrinsic component of autophagosome membrane(GO:0097636) integral component of autophagosome membrane(GO:0097637) |
| 0.2 | 0.6 | GO:0097629 | extrinsic component of omegasome membrane(GO:0097629) |
| 0.1 | 0.7 | GO:0031905 | early endosome lumen(GO:0031905) |
| 0.1 | 0.7 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.1 | 0.5 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.1 | 1.6 | GO:0035068 | micro-ribonucleoprotein complex(GO:0035068) |
| 0.1 | 0.9 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.1 | 0.7 | GO:0031302 | intrinsic component of endosome membrane(GO:0031302) |
| 0.1 | 1.9 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.1 | 0.9 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.1 | 1.2 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.1 | 1.1 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.1 | 0.3 | GO:0097196 | Shu complex(GO:0097196) |
| 0.1 | 0.4 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.1 | 0.3 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.1 | 0.2 | GO:0002947 | tumor necrosis factor receptor superfamily complex(GO:0002947) |
| 0.1 | 0.8 | GO:0051286 | cell tip(GO:0051286) |
| 0.1 | 1.5 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.1 | 0.3 | GO:0032144 | 4-aminobutyrate transaminase complex(GO:0032144) |
| 0.1 | 0.4 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.1 | 0.4 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.1 | 1.1 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.4 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.0 | 0.4 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.0 | 0.2 | GO:0031251 | PAN complex(GO:0031251) |
| 0.0 | 0.2 | GO:0042643 | actomyosin, actin portion(GO:0042643) |
| 0.0 | 0.5 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.0 | 1.6 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.0 | 0.7 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.0 | 1.5 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 0.7 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 0.2 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.0 | 0.4 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.0 | 0.7 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.0 | 0.3 | GO:0097443 | sorting endosome(GO:0097443) cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.0 | 0.2 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.0 | 0.4 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 0.5 | GO:0000235 | astral microtubule(GO:0000235) aster(GO:0005818) |
| 0.0 | 0.4 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.0 | 0.4 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.6 | GO:0001931 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 0.1 | GO:0034665 | integrin alpha1-beta1 complex(GO:0034665) |
| 0.0 | 1.1 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 3.2 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.3 | GO:0090543 | Flemming body(GO:0090543) |
| 0.0 | 0.3 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 0.6 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 3.9 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 2.3 | GO:0043657 | host(GO:0018995) host cell(GO:0043657) |
| 0.0 | 0.1 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 0.0 | 0.5 | GO:0046930 | pore complex(GO:0046930) |
| 0.0 | 0.2 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.3 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.0 | 0.6 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.5 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 0.4 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 2.2 | GO:0101003 | ficolin-1-rich granule membrane(GO:0101003) |
| 0.0 | 0.4 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.0 | 0.2 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.0 | 0.1 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.0 | 0.9 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 0.1 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 3.1 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 0.1 | GO:0030663 | COPI vesicle coat(GO:0030126) COPI-coated vesicle membrane(GO:0030663) |
| 0.0 | 0.2 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.0 | 1.4 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 0.6 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.1 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.2 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.9 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 1.0 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.1 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 0.2 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.0 | 0.7 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.1 | GO:0071203 | WASH complex(GO:0071203) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0043739 | G/U mismatch-specific uracil-DNA glycosylase activity(GO:0043739) |
| 0.3 | 1.0 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.3 | 1.0 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.2 | 1.2 | GO:0017089 | glycolipid transporter activity(GO:0017089) |
| 0.2 | 0.9 | GO:0098808 | mRNA cap binding(GO:0098808) |
| 0.2 | 1.6 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.2 | 1.1 | GO:0052740 | 1-acyl-2-lysophosphatidylserine acylhydrolase activity(GO:0052740) |
| 0.2 | 2.4 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.2 | 2.1 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.2 | 1.0 | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0003829) |
| 0.2 | 0.9 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.2 | 0.5 | GO:0038100 | nodal binding(GO:0038100) |
| 0.2 | 0.5 | GO:0080130 | L-phenylalanine aminotransferase activity(GO:0070546) L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.2 | 0.7 | GO:1904455 | ubiquitin-specific protease activity involved in negative regulation of ERAD pathway(GO:1904455) |
| 0.2 | 0.6 | GO:0051765 | inositol tetrakisphosphate kinase activity(GO:0051765) |
| 0.2 | 0.6 | GO:0034739 | histone deacetylase activity (H4-K16 specific)(GO:0034739) |
| 0.1 | 0.7 | GO:0015563 | uptake transmembrane transporter activity(GO:0015563) |
| 0.1 | 0.6 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.1 | 1.2 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.1 | 0.4 | GO:0070119 | ciliary neurotrophic factor binding(GO:0070119) |
| 0.1 | 0.7 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.1 | 1.3 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.1 | 0.3 | GO:0042356 | GDP-4-dehydro-D-rhamnose reductase activity(GO:0042356) GDP-L-fucose synthase activity(GO:0050577) |
| 0.1 | 0.5 | GO:0036080 | GDP-fucose transmembrane transporter activity(GO:0005457) purine nucleotide-sugar transmembrane transporter activity(GO:0036080) |
| 0.1 | 1.6 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.1 | 3.1 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.1 | 0.3 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 0.1 | 0.4 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.1 | 1.2 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.1 | 0.5 | GO:0098519 | nucleotide phosphatase activity, acting on free nucleotides(GO:0098519) |
| 0.1 | 1.5 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.1 | 0.6 | GO:0060698 | endoribonuclease inhibitor activity(GO:0060698) |
| 0.1 | 2.3 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.1 | 4.0 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.1 | 0.4 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.1 | 0.3 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.1 | 0.2 | GO:0043849 | Ras palmitoyltransferase activity(GO:0043849) |
| 0.1 | 0.9 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.1 | 0.3 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.1 | 0.4 | GO:0098626 | methylselenol reductase activity(GO:0098625) methylseleninic acid reductase activity(GO:0098626) |
| 0.1 | 0.5 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.1 | 1.7 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.1 | 0.3 | GO:1990189 | peptide-serine-N-acetyltransferase activity(GO:1990189) |
| 0.1 | 0.7 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.1 | 0.5 | GO:0052796 | exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.1 | 0.6 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.1 | 0.7 | GO:0070915 | lysophosphatidic acid binding(GO:0035727) lysophosphatidic acid receptor activity(GO:0070915) |
| 0.1 | 0.4 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 0.1 | 2.3 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.1 | 0.8 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.1 | 0.2 | GO:0070506 | high-density lipoprotein particle receptor activity(GO:0070506) |
| 0.1 | 0.3 | GO:0003867 | 4-aminobutyrate transaminase activity(GO:0003867) succinate-semialdehyde dehydrogenase binding(GO:0032145) (S)-3-amino-2-methylpropionate transaminase activity(GO:0047298) |
| 0.1 | 0.3 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.1 | 0.4 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.1 | 0.2 | GO:0017057 | 6-phosphogluconolactonase activity(GO:0017057) |
| 0.1 | 0.5 | GO:0045134 | uridine-diphosphatase activity(GO:0045134) |
| 0.1 | 1.7 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.7 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.2 | GO:0010465 | nerve growth factor receptor activity(GO:0010465) |
| 0.0 | 2.1 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 0.2 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.0 | 1.0 | GO:0033549 | MAP kinase phosphatase activity(GO:0033549) |
| 0.0 | 0.4 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 1.1 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.6 | GO:0008525 | phosphatidylcholine transporter activity(GO:0008525) |
| 0.0 | 1.8 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.5 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.1 | GO:0008441 | 3'(2'),5'-bisphosphate nucleotidase activity(GO:0008441) |
| 0.0 | 0.9 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 0.5 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 0.2 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.0 | 1.6 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.0 | 0.2 | GO:0052836 | inositol 5-diphosphate pentakisphosphate 5-kinase activity(GO:0052836) inositol diphosphate tetrakisphosphate kinase activity(GO:0052839) |
| 0.0 | 2.0 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.5 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.0 | 0.8 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.0 | 1.2 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.3 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.0 | 0.6 | GO:0055106 | ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.0 | 1.9 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 1.9 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.2 | GO:0045145 | single-stranded DNA 5'-3' exodeoxyribonuclease activity(GO:0045145) |
| 0.0 | 0.7 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 1.4 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 1.1 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.7 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.4 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 0.5 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.3 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.0 | 0.5 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.0 | 0.2 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 0.0 | 0.4 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.0 | 0.6 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 0.9 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.2 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.0 | 0.1 | GO:0070890 | L-ascorbate:sodium symporter activity(GO:0008520) L-ascorbic acid transporter activity(GO:0015229) sodium-dependent L-ascorbate transmembrane transporter activity(GO:0070890) |
| 0.0 | 1.0 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 0.7 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.0 | 0.8 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.1 | GO:0050252 | retinol O-fatty-acyltransferase activity(GO:0050252) |
| 0.0 | 0.4 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 0.4 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.3 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 1.0 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 2.2 | GO:0035254 | glutamate receptor binding(GO:0035254) |
| 0.0 | 0.9 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.6 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.0 | 0.9 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 1.3 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.2 | GO:0015181 | arginine transmembrane transporter activity(GO:0015181) |
| 0.0 | 1.8 | GO:0016279 | protein-lysine N-methyltransferase activity(GO:0016279) |
| 0.0 | 0.8 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.4 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 2.5 | GO:0051219 | phosphoprotein binding(GO:0051219) |
| 0.0 | 0.1 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) core promoter proximal region sequence-specific DNA binding(GO:0000987) core promoter proximal region DNA binding(GO:0001159) |
| 0.0 | 0.4 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 0.5 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.1 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.0 | 0.1 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.0 | 0.2 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.0 | 0.2 | GO:0034713 | type I transforming growth factor beta receptor binding(GO:0034713) |
| 0.0 | 0.1 | GO:0019828 | aspartic-type endopeptidase inhibitor activity(GO:0019828) |
| 0.0 | 0.1 | GO:0035252 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 0.0 | 0.1 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 0.2 | GO:0001163 | RNA polymerase I regulatory region DNA binding(GO:0001013) RNA polymerase I regulatory region sequence-specific DNA binding(GO:0001163) RNA polymerase I CORE element sequence-specific DNA binding(GO:0001164) |
| 0.0 | 0.2 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.2 | GO:0000182 | rDNA binding(GO:0000182) |
| 0.0 | 0.3 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.0 | 0.8 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 0.5 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.2 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 0.1 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.0 | 1.4 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.6 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.1 | GO:0005114 | transforming growth factor beta receptor activity, type I(GO:0005025) type II transforming growth factor beta receptor binding(GO:0005114) |
| 0.0 | 0.2 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.1 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.0 | 0.3 | GO:0008252 | nucleotidase activity(GO:0008252) 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.2 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 0.1 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.0 | 0.5 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.0 | 0.8 | GO:0043531 | ADP binding(GO:0043531) |
| 0.0 | 0.6 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.0 | 0.2 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.4 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.9 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.3 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.1 | 2.5 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.1 | 1.5 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 1.3 | PID EPO PATHWAY | EPO signaling pathway |
| 0.0 | 1.8 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 1.9 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 2.6 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 2.3 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.0 | 4.4 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 0.8 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 2.3 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 1.1 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 1.2 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 1.2 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 3.2 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 6.6 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 1.7 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 1.0 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
| 0.0 | 0.7 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.0 | 0.6 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 0.2 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.0 | 0.6 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.0 | 0.3 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.6 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.6 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 0.5 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 0.4 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 0.3 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 0.8 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 1.0 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 0.3 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 1.1 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.5 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.7 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.1 | 1.5 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.1 | 2.4 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.1 | 1.6 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.1 | 0.7 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.1 | 2.3 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.1 | 1.0 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.1 | 1.8 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.1 | 0.4 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.1 | 0.3 | REACTOME ARMS MEDIATED ACTIVATION | Genes involved in ARMS-mediated activation |
| 0.0 | 0.9 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 0.0 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 2.5 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.0 | 1.8 | REACTOME TRAF6 MEDIATED NFKB ACTIVATION | Genes involved in TRAF6 mediated NF-kB activation |
| 0.0 | 1.8 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.6 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.0 | 0.9 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.0 | 2.8 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.4 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 1.3 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.8 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 1.2 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.5 | REACTOME DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 0.0 | 1.4 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 0.7 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 1.6 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.0 | 0.6 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.6 | REACTOME POST NMDA RECEPTOR ACTIVATION EVENTS | Genes involved in Post NMDA receptor activation events |
| 0.0 | 0.3 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.0 | 1.5 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.0 | 0.3 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.0 | 0.8 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 2.0 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.4 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 1.8 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.5 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 1.0 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 0.5 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.4 | REACTOME TRANSCRIPTIONAL ACTIVITY OF SMAD2 SMAD3 SMAD4 HETEROTRIMER | Genes involved in Transcriptional activity of SMAD2/SMAD3:SMAD4 heterotrimer |
| 0.0 | 0.7 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.0 | 1.1 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 1.0 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.3 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 1.3 | REACTOME CELL DEATH SIGNALLING VIA NRAGE NRIF AND NADE | Genes involved in Cell death signalling via NRAGE, NRIF and NADE |
| 0.0 | 0.1 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.6 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.5 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 0.9 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |