Project
Illumina Body Map 2: averaged replicates
Navigation
Downloads
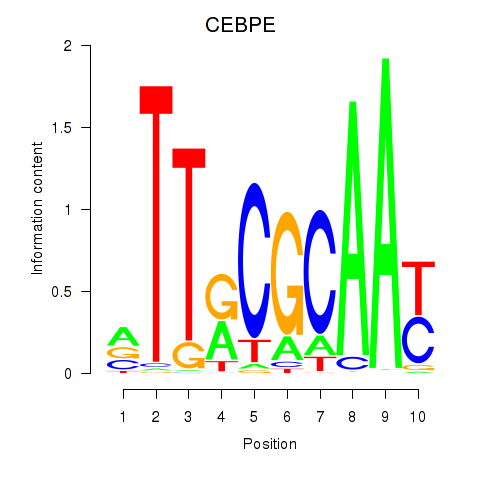
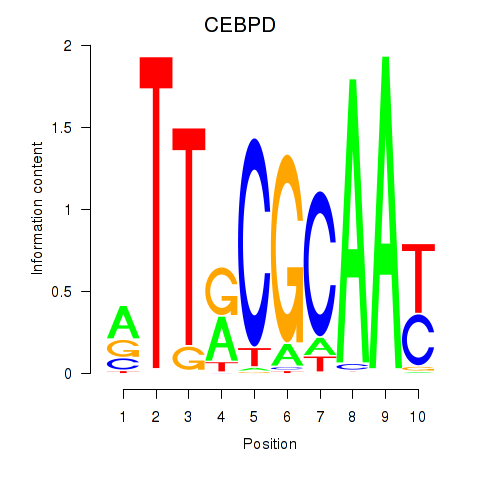
Results for CEBPE_CEBPD
Z-value: 3.43
Transcription factors associated with CEBPE_CEBPD
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
CEBPE
|
ENSG00000092067.5 | CCAAT enhancer binding protein epsilon |
|
CEBPD
|
ENSG00000221869.4 | CCAAT enhancer binding protein delta |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| CEBPE | hg19_v2_chr14_-_23588816_23588836 | 0.20 | 2.8e-01 | Click! |
| CEBPD | hg19_v2_chr8_-_48651648_48651648 | 0.09 | 6.4e-01 | Click! |
Activity profile of CEBPE_CEBPD motif
Sorted Z-values of CEBPE_CEBPD motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_-_133055815 | 24.10 |
ENST00000509351.1
ENST00000417437.2 ENST00000414302.2 ENST00000423615.2 ENST00000427187.2 ENST00000275223.3 ENST00000519686.2 |
VNN3
|
vanin 3 |
| chr6_-_133055896 | 22.87 |
ENST00000367927.5
ENST00000425515.2 ENST00000207771.3 ENST00000392393.3 ENST00000450865.2 ENST00000392394.2 |
VNN3
|
vanin 3 |
| chr12_-_96390108 | 20.71 |
ENST00000538703.1
ENST00000261208.3 |
HAL
|
histidine ammonia-lyase |
| chr12_-_96390063 | 17.91 |
ENST00000541929.1
|
HAL
|
histidine ammonia-lyase |
| chr3_+_186330712 | 16.54 |
ENST00000411641.2
ENST00000273784.5 |
AHSG
|
alpha-2-HS-glycoprotein |
| chr15_+_58430368 | 16.51 |
ENST00000558772.1
ENST00000219919.4 |
AQP9
|
aquaporin 9 |
| chr3_+_186383741 | 15.62 |
ENST00000232003.4
|
HRG
|
histidine-rich glycoprotein |
| chr15_+_58430567 | 15.31 |
ENST00000536493.1
|
AQP9
|
aquaporin 9 |
| chr16_+_72088376 | 13.82 |
ENST00000570083.1
ENST00000355906.5 ENST00000398131.2 ENST00000569639.1 ENST00000564499.1 ENST00000357763.4 ENST00000562526.1 ENST00000565574.1 ENST00000568417.2 ENST00000356967.5 |
HP
HPR
|
haptoglobin haptoglobin-related protein |
| chr11_+_18287801 | 13.62 |
ENST00000532858.1
ENST00000405158.2 |
SAA1
|
serum amyloid A1 |
| chr11_+_18287721 | 13.38 |
ENST00000356524.4
|
SAA1
|
serum amyloid A1 |
| chr10_-_82049424 | 13.12 |
ENST00000372213.3
|
MAT1A
|
methionine adenosyltransferase I, alpha |
| chr11_-_18270182 | 12.72 |
ENST00000528349.1
ENST00000526900.1 ENST00000529528.1 ENST00000414546.2 ENST00000256733.4 |
SAA2
|
serum amyloid A2 |
| chr4_+_104346194 | 12.21 |
ENST00000510200.1
|
RP11-328K4.1
|
RP11-328K4.1 |
| chrX_+_138612889 | 12.04 |
ENST00000218099.2
ENST00000394090.2 |
F9
|
coagulation factor IX |
| chr2_+_113885138 | 9.74 |
ENST00000409930.3
|
IL1RN
|
interleukin 1 receptor antagonist |
| chr3_+_186435137 | 9.74 |
ENST00000447445.1
|
KNG1
|
kininogen 1 |
| chr14_-_70263979 | 9.51 |
ENST00000216540.4
|
SLC10A1
|
solute carrier family 10 (sodium/bile acid cotransporter), member 1 |
| chr3_+_186435065 | 9.03 |
ENST00000287611.2
ENST00000265023.4 |
KNG1
|
kininogen 1 |
| chr1_+_196743912 | 8.84 |
ENST00000367425.4
|
CFHR3
|
complement factor H-related 3 |
| chrX_-_105282712 | 8.72 |
ENST00000372563.1
|
SERPINA7
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 7 |
| chr17_-_7018128 | 8.51 |
ENST00000380952.2
ENST00000254850.7 |
ASGR2
|
asialoglycoprotein receptor 2 |
| chr17_-_7017968 | 8.40 |
ENST00000355035.5
|
ASGR2
|
asialoglycoprotein receptor 2 |
| chr6_+_31895287 | 8.36 |
ENST00000447952.2
|
C2
|
complement component 2 |
| chr5_+_49963239 | 8.31 |
ENST00000505554.1
|
PARP8
|
poly (ADP-ribose) polymerase family, member 8 |
| chr3_-_148939835 | 8.17 |
ENST00000264613.6
|
CP
|
ceruloplasmin (ferroxidase) |
| chr1_+_192127578 | 8.16 |
ENST00000367460.3
|
RGS18
|
regulator of G-protein signaling 18 |
| chr3_-_58200398 | 7.99 |
ENST00000318316.3
ENST00000460422.1 ENST00000483681.1 |
DNASE1L3
|
deoxyribonuclease I-like 3 |
| chr1_+_196743943 | 7.86 |
ENST00000471440.2
ENST00000391985.3 |
CFHR3
|
complement factor H-related 3 |
| chr4_-_155511887 | 7.69 |
ENST00000302053.3
ENST00000403106.3 |
FGA
|
fibrinogen alpha chain |
| chr19_+_6887571 | 7.49 |
ENST00000250572.8
ENST00000381407.5 ENST00000312053.4 ENST00000450315.3 ENST00000381404.4 |
EMR1
|
egf-like module containing, mucin-like, hormone receptor-like 1 |
| chr6_+_31895254 | 7.47 |
ENST00000299367.5
ENST00000442278.2 |
C2
|
complement component 2 |
| chr3_+_46395219 | 7.19 |
ENST00000445132.2
ENST00000292301.4 |
CCR2
|
chemokine (C-C motif) receptor 2 |
| chr21_-_46334186 | 7.01 |
ENST00000522931.1
|
ITGB2
|
integrin, beta 2 (complement component 3 receptor 3 and 4 subunit) |
| chr2_+_219247021 | 6.88 |
ENST00000539932.1
|
SLC11A1
|
solute carrier family 11 (proton-coupled divalent metal ion transporter), member 1 |
| chr3_-_148939598 | 6.76 |
ENST00000455472.3
|
CP
|
ceruloplasmin (ferroxidase) |
| chr20_-_22566089 | 6.71 |
ENST00000377115.4
|
FOXA2
|
forkhead box A2 |
| chr1_+_196857144 | 6.42 |
ENST00000367416.2
ENST00000251424.4 ENST00000367418.2 |
CFHR4
|
complement factor H-related 4 |
| chr13_-_46679144 | 6.42 |
ENST00000181383.4
|
CPB2
|
carboxypeptidase B2 (plasma) |
| chr13_-_46679185 | 6.25 |
ENST00000439329.3
|
CPB2
|
carboxypeptidase B2 (plasma) |
| chr6_+_31895480 | 5.98 |
ENST00000418949.2
ENST00000383177.3 ENST00000477310.1 |
C2
CFB
|
complement component 2 Complement factor B; Uncharacterized protein; cDNA FLJ55673, highly similar to Complement factor B |
| chrX_-_131547596 | 5.93 |
ENST00000538204.1
ENST00000370849.3 |
MBNL3
|
muscleblind-like splicing regulator 3 |
| chr4_+_156775910 | 5.92 |
ENST00000506072.1
ENST00000507590.1 |
TDO2
|
tryptophan 2,3-dioxygenase |
| chr17_-_34417479 | 5.90 |
ENST00000225245.5
|
CCL3
|
chemokine (C-C motif) ligand 3 |
| chr3_+_46395579 | 5.85 |
ENST00000421659.1
|
CCR2
|
chemokine (C-C motif) receptor 2 |
| chrX_-_131547625 | 5.75 |
ENST00000394311.2
|
MBNL3
|
muscleblind-like splicing regulator 3 |
| chr14_-_23284703 | 5.66 |
ENST00000555911.1
|
SLC7A7
|
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7 |
| chr5_-_39364586 | 5.60 |
ENST00000263408.4
|
C9
|
complement component 9 |
| chr4_-_185747188 | 5.40 |
ENST00000507295.1
ENST00000504900.1 ENST00000281455.2 ENST00000454703.2 |
ACSL1
|
acyl-CoA synthetase long-chain family member 1 |
| chr2_-_31637560 | 5.39 |
ENST00000379416.3
|
XDH
|
xanthine dehydrogenase |
| chr6_+_31895467 | 5.36 |
ENST00000556679.1
ENST00000456570.1 |
CFB
CFB
|
complement factor B Complement factor B; Uncharacterized protein; cDNA FLJ55673, highly similar to Complement factor B |
| chr16_-_84651673 | 5.36 |
ENST00000262428.4
|
COTL1
|
coactosin-like 1 (Dictyostelium) |
| chr2_+_219246746 | 5.29 |
ENST00000233202.6
|
SLC11A1
|
solute carrier family 11 (proton-coupled divalent metal ion transporter), member 1 |
| chr13_-_29292956 | 5.24 |
ENST00000266943.6
|
SLC46A3
|
solute carrier family 46, member 3 |
| chr16_-_84651647 | 5.20 |
ENST00000564057.1
|
COTL1
|
coactosin-like 1 (Dictyostelium) |
| chr6_-_24489842 | 5.17 |
ENST00000230036.1
|
GPLD1
|
glycosylphosphatidylinositol specific phospholipase D1 |
| chr1_+_40862501 | 5.17 |
ENST00000539317.1
|
SMAP2
|
small ArfGAP2 |
| chr16_-_30457048 | 5.15 |
ENST00000500504.2
ENST00000542752.1 |
SEPHS2
|
selenophosphate synthetase 2 |
| chr11_-_18258342 | 5.12 |
ENST00000278222.4
|
SAA4
|
serum amyloid A4, constitutive |
| chr1_+_154378049 | 4.97 |
ENST00000512471.1
|
IL6R
|
interleukin 6 receptor |
| chr19_-_41356347 | 4.96 |
ENST00000301141.5
|
CYP2A6
|
cytochrome P450, family 2, subfamily A, polypeptide 6 |
| chr14_+_20937538 | 4.91 |
ENST00000361505.5
ENST00000553591.1 |
PNP
|
purine nucleoside phosphorylase |
| chr19_+_11350278 | 4.85 |
ENST00000252453.8
|
C19orf80
|
chromosome 19 open reading frame 80 |
| chr1_-_1711508 | 4.83 |
ENST00000378625.1
|
NADK
|
NAD kinase |
| chr14_-_23284675 | 4.80 |
ENST00000555959.1
|
SLC7A7
|
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7 |
| chr2_-_61245363 | 4.73 |
ENST00000316752.6
|
PUS10
|
pseudouridylate synthase 10 |
| chr3_-_10547192 | 4.60 |
ENST00000360273.2
ENST00000343816.4 |
ATP2B2
|
ATPase, Ca++ transporting, plasma membrane 2 |
| chr11_+_63057412 | 4.41 |
ENST00000544661.1
|
SLC22A10
|
solute carrier family 22, member 10 |
| chr11_-_116663127 | 4.35 |
ENST00000433069.1
ENST00000542499.1 |
APOA5
|
apolipoprotein A-V |
| chr5_+_49962772 | 4.29 |
ENST00000281631.5
ENST00000513738.1 ENST00000503665.1 ENST00000514067.2 ENST00000503046.1 |
PARP8
|
poly (ADP-ribose) polymerase family, member 8 |
| chr12_-_96389702 | 4.20 |
ENST00000552509.1
|
HAL
|
histidine ammonia-lyase |
| chr5_-_138718973 | 4.14 |
ENST00000353963.3
ENST00000348729.3 |
SLC23A1
|
solute carrier family 23 (ascorbic acid transporter), member 1 |
| chr19_-_40971643 | 4.11 |
ENST00000595483.1
|
BLVRB
|
biliverdin reductase B (flavin reductase (NADPH)) |
| chr19_+_41497178 | 4.09 |
ENST00000324071.4
|
CYP2B6
|
cytochrome P450, family 2, subfamily B, polypeptide 6 |
| chr14_-_23285011 | 4.08 |
ENST00000397532.3
|
SLC7A7
|
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7 |
| chr20_+_56136136 | 4.00 |
ENST00000319441.4
ENST00000543666.1 |
PCK1
|
phosphoenolpyruvate carboxykinase 1 (soluble) |
| chr10_+_30723045 | 3.92 |
ENST00000542547.1
ENST00000415139.1 |
MAP3K8
|
mitogen-activated protein kinase kinase kinase 8 |
| chr8_-_6735451 | 3.91 |
ENST00000297439.3
|
DEFB1
|
defensin, beta 1 |
| chr14_-_23285069 | 3.84 |
ENST00000554758.1
ENST00000397528.4 |
SLC7A7
|
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7 |
| chr1_+_144339738 | 3.83 |
ENST00000538264.1
|
AL592284.1
|
Protein LOC642441 |
| chr1_-_111743285 | 3.75 |
ENST00000357640.4
|
DENND2D
|
DENN/MADD domain containing 2D |
| chr3_-_120400960 | 3.73 |
ENST00000476082.2
|
HGD
|
homogentisate 1,2-dioxygenase |
| chr3_-_190580404 | 3.73 |
ENST00000442080.1
|
GMNC
|
geminin coiled-coil domain containing |
| chr11_-_58980342 | 3.73 |
ENST00000361050.3
|
MPEG1
|
macrophage expressed 1 |
| chr10_-_113943447 | 3.73 |
ENST00000369425.1
ENST00000348367.4 ENST00000423155.1 |
GPAM
|
glycerol-3-phosphate acyltransferase, mitochondrial |
| chr1_-_47697387 | 3.67 |
ENST00000371884.2
|
TAL1
|
T-cell acute lymphocytic leukemia 1 |
| chr15_+_45926919 | 3.65 |
ENST00000561735.1
ENST00000260324.7 |
SQRDL
|
sulfide quinone reductase-like (yeast) |
| chr17_-_77813186 | 3.57 |
ENST00000448310.1
ENST00000269397.4 |
CBX4
|
chromobox homolog 4 |
| chr19_-_41388657 | 3.35 |
ENST00000301146.4
ENST00000291764.3 |
CYP2A7
|
cytochrome P450, family 2, subfamily A, polypeptide 7 |
| chr1_+_207262170 | 3.35 |
ENST00000367078.3
|
C4BPB
|
complement component 4 binding protein, beta |
| chr1_+_209941942 | 3.33 |
ENST00000487271.1
ENST00000477431.1 |
TRAF3IP3
|
TRAF3 interacting protein 3 |
| chr6_-_112194484 | 3.25 |
ENST00000518295.1
ENST00000484067.2 ENST00000229470.5 ENST00000356013.2 ENST00000368678.4 ENST00000523238.1 ENST00000354650.3 |
FYN
|
FYN oncogene related to SRC, FGR, YES |
| chr2_-_169887827 | 3.23 |
ENST00000263817.6
|
ABCB11
|
ATP-binding cassette, sub-family B (MDR/TAP), member 11 |
| chr7_-_36764004 | 3.22 |
ENST00000431169.1
|
AOAH
|
acyloxyacyl hydrolase (neutrophil) |
| chr3_+_16306837 | 3.21 |
ENST00000606098.1
|
OXNAD1
|
oxidoreductase NAD-binding domain containing 1 |
| chr11_-_14993819 | 3.13 |
ENST00000396372.2
ENST00000361010.3 ENST00000359642.3 ENST00000331587.4 |
CALCA
|
calcitonin-related polypeptide alpha |
| chr17_+_7477040 | 3.13 |
ENST00000581384.1
ENST00000577929.1 |
EIF4A1
|
eukaryotic translation initiation factor 4A1 |
| chr11_-_102595681 | 3.12 |
ENST00000236826.3
|
MMP8
|
matrix metallopeptidase 8 (neutrophil collagenase) |
| chr19_-_6670128 | 3.11 |
ENST00000245912.3
|
TNFSF14
|
tumor necrosis factor (ligand) superfamily, member 14 |
| chr1_+_154377669 | 3.10 |
ENST00000368485.3
ENST00000344086.4 |
IL6R
|
interleukin 6 receptor |
| chr3_-_151102529 | 3.04 |
ENST00000302632.3
|
P2RY12
|
purinergic receptor P2Y, G-protein coupled, 12 |
| chr17_+_27369918 | 3.04 |
ENST00000323372.4
|
PIPOX
|
pipecolic acid oxidase |
| chr10_+_30723533 | 3.02 |
ENST00000413724.1
|
MAP3K8
|
mitogen-activated protein kinase kinase kinase 8 |
| chr3_+_133465228 | 2.98 |
ENST00000482271.1
ENST00000264998.3 |
TF
|
transferrin |
| chr14_-_64971288 | 2.95 |
ENST00000394715.1
|
ZBTB25
|
zinc finger and BTB domain containing 25 |
| chr3_+_16306691 | 2.93 |
ENST00000285083.5
ENST00000605932.1 ENST00000435829.2 |
OXNAD1
|
oxidoreductase NAD-binding domain containing 1 |
| chr1_-_204183071 | 2.90 |
ENST00000308302.3
|
GOLT1A
|
golgi transport 1A |
| chr14_+_64971292 | 2.88 |
ENST00000358738.3
ENST00000394712.2 |
ZBTB1
|
zinc finger and BTB domain containing 1 |
| chr3_+_108541608 | 2.80 |
ENST00000426646.1
|
TRAT1
|
T cell receptor associated transmembrane adaptor 1 |
| chr7_+_137761199 | 2.71 |
ENST00000411726.2
|
AKR1D1
|
aldo-keto reductase family 1, member D1 |
| chr14_-_45722605 | 2.71 |
ENST00000310806.4
|
MIS18BP1
|
MIS18 binding protein 1 |
| chr19_-_52148798 | 2.70 |
ENST00000534261.2
|
SIGLEC5
|
sialic acid binding Ig-like lectin 5 |
| chr20_+_54987168 | 2.65 |
ENST00000360314.3
|
CASS4
|
Cas scaffolding protein family member 4 |
| chr7_+_128784712 | 2.65 |
ENST00000289407.4
|
TSPAN33
|
tetraspanin 33 |
| chr16_-_21663919 | 2.59 |
ENST00000569602.1
|
IGSF6
|
immunoglobulin superfamily, member 6 |
| chr19_-_40971667 | 2.58 |
ENST00000263368.4
|
BLVRB
|
biliverdin reductase B (flavin reductase (NADPH)) |
| chr5_-_140013275 | 2.58 |
ENST00000512545.1
ENST00000302014.6 ENST00000401743.2 |
CD14
|
CD14 molecule |
| chr20_+_54987305 | 2.56 |
ENST00000371336.3
ENST00000434344.1 |
CASS4
|
Cas scaffolding protein family member 4 |
| chr16_-_21663950 | 2.54 |
ENST00000268389.4
|
IGSF6
|
immunoglobulin superfamily, member 6 |
| chr1_+_209941827 | 2.54 |
ENST00000367023.1
|
TRAF3IP3
|
TRAF3 interacting protein 3 |
| chr5_-_140013224 | 2.53 |
ENST00000498971.2
|
CD14
|
CD14 molecule |
| chr7_-_87936195 | 2.50 |
ENST00000414498.1
ENST00000301959.5 ENST00000380079.4 |
STEAP4
|
STEAP family member 4 |
| chr9_-_130712995 | 2.50 |
ENST00000373084.4
|
FAM102A
|
family with sequence similarity 102, member A |
| chrY_+_2709527 | 2.44 |
ENST00000250784.8
|
RPS4Y1
|
ribosomal protein S4, Y-linked 1 |
| chr9_-_94186131 | 2.41 |
ENST00000297689.3
|
NFIL3
|
nuclear factor, interleukin 3 regulated |
| chr5_-_140013241 | 2.39 |
ENST00000519715.1
|
CD14
|
CD14 molecule |
| chr14_+_21423611 | 2.36 |
ENST00000304625.2
|
RNASE2
|
ribonuclease, RNase A family, 2 (liver, eosinophil-derived neurotoxin) |
| chr2_-_101925055 | 2.35 |
ENST00000295317.3
|
RNF149
|
ring finger protein 149 |
| chr22_+_40297079 | 2.31 |
ENST00000344138.4
ENST00000543252.1 |
GRAP2
|
GRB2-related adaptor protein 2 |
| chrX_-_73512177 | 2.21 |
ENST00000603672.1
ENST00000418855.1 |
FTX
|
FTX transcript, XIST regulator (non-protein coding) |
| chr22_+_40297105 | 2.18 |
ENST00000540310.1
|
GRAP2
|
GRB2-related adaptor protein 2 |
| chr2_-_209118974 | 2.15 |
ENST00000415913.1
ENST00000415282.1 ENST00000446179.1 |
IDH1
|
isocitrate dehydrogenase 1 (NADP+), soluble |
| chr1_-_67519782 | 2.15 |
ENST00000235345.5
|
SLC35D1
|
solute carrier family 35 (UDP-GlcA/UDP-GalNAc transporter), member D1 |
| chr10_+_30723105 | 2.10 |
ENST00000375322.2
|
MAP3K8
|
mitogen-activated protein kinase kinase kinase 8 |
| chr14_-_106573756 | 2.09 |
ENST00000390601.2
|
IGHV3-11
|
immunoglobulin heavy variable 3-11 (gene/pseudogene) |
| chrX_-_73511908 | 2.06 |
ENST00000455395.1
|
FTX
|
FTX transcript, XIST regulator (non-protein coding) |
| chr20_-_17662878 | 1.96 |
ENST00000377813.1
ENST00000377807.2 ENST00000360807.4 ENST00000398782.2 |
RRBP1
|
ribosome binding protein 1 |
| chr12_-_123201337 | 1.96 |
ENST00000528880.2
|
HCAR3
|
hydroxycarboxylic acid receptor 3 |
| chr14_+_64971438 | 1.95 |
ENST00000555321.1
|
ZBTB1
|
zinc finger and BTB domain containing 1 |
| chr14_-_45722360 | 1.93 |
ENST00000451174.1
|
MIS18BP1
|
MIS18 binding protein 1 |
| chr19_-_6720686 | 1.92 |
ENST00000245907.6
|
C3
|
complement component 3 |
| chr19_+_50922187 | 1.90 |
ENST00000595883.1
ENST00000597855.1 ENST00000596074.1 ENST00000439922.2 ENST00000594685.1 ENST00000270632.7 |
SPIB
|
Spi-B transcription factor (Spi-1/PU.1 related) |
| chr1_-_156721502 | 1.84 |
ENST00000357325.5
|
HDGF
|
hepatoma-derived growth factor |
| chr14_+_52313833 | 1.83 |
ENST00000553560.1
|
GNG2
|
guanine nucleotide binding protein (G protein), gamma 2 |
| chr1_-_151032040 | 1.81 |
ENST00000540998.1
ENST00000357235.5 |
CDC42SE1
|
CDC42 small effector 1 |
| chr4_+_71263599 | 1.78 |
ENST00000399575.2
|
PROL1
|
proline rich, lacrimal 1 |
| chr14_+_22990951 | 1.78 |
ENST00000390515.1
|
TRAJ22
|
T cell receptor alpha joining 22 |
| chr6_-_113754604 | 1.75 |
ENST00000421737.1
|
RP1-124C6.1
|
RP1-124C6.1 |
| chr8_+_8559406 | 1.74 |
ENST00000519106.1
|
CLDN23
|
claudin 23 |
| chr17_-_34524157 | 1.71 |
ENST00000378354.4
ENST00000394484.1 |
CCL3L3
|
chemokine (C-C motif) ligand 3-like 3 |
| chr14_+_22947861 | 1.69 |
ENST00000390482.1
|
TRAJ57
|
T cell receptor alpha joining 57 |
| chr17_-_79817091 | 1.68 |
ENST00000570907.1
|
P4HB
|
prolyl 4-hydroxylase, beta polypeptide |
| chr20_-_1600642 | 1.66 |
ENST00000381603.3
ENST00000381605.4 ENST00000279477.7 ENST00000568365.1 ENST00000564763.1 |
SIRPB1
RP4-576H24.4
|
signal-regulatory protein beta 1 Uncharacterized protein |
| chr1_-_52521831 | 1.66 |
ENST00000371626.4
|
TXNDC12
|
thioredoxin domain containing 12 (endoplasmic reticulum) |
| chr2_-_178128199 | 1.65 |
ENST00000449627.1
|
NFE2L2
|
nuclear factor, erythroid 2-like 2 |
| chr20_-_33732952 | 1.65 |
ENST00000541621.1
|
EDEM2
|
ER degradation enhancer, mannosidase alpha-like 2 |
| chr11_-_64510409 | 1.63 |
ENST00000394429.1
ENST00000394428.1 |
RASGRP2
|
RAS guanyl releasing protein 2 (calcium and DAG-regulated) |
| chr10_-_97416400 | 1.63 |
ENST00000371224.2
ENST00000371221.3 |
ALDH18A1
|
aldehyde dehydrogenase 18 family, member A1 |
| chr17_-_34625719 | 1.58 |
ENST00000422211.2
ENST00000542124.1 |
CCL3L1
|
chemokine (C-C motif) ligand 3-like 1 |
| chr5_+_137722255 | 1.58 |
ENST00000542866.1
|
KDM3B
|
lysine (K)-specific demethylase 3B |
| chr11_-_102595512 | 1.57 |
ENST00000438475.2
|
MMP8
|
matrix metallopeptidase 8 (neutrophil collagenase) |
| chr3_+_88188254 | 1.56 |
ENST00000309495.5
|
ZNF654
|
zinc finger protein 654 |
| chr7_-_121944491 | 1.56 |
ENST00000331178.4
ENST00000427185.2 ENST00000442488.2 |
FEZF1
|
FEZ family zinc finger 1 |
| chr1_+_162531294 | 1.56 |
ENST00000367926.4
ENST00000271469.3 |
UAP1
|
UDP-N-acteylglucosamine pyrophosphorylase 1 |
| chrX_-_40506766 | 1.53 |
ENST00000378421.1
ENST00000440784.2 ENST00000327877.5 ENST00000378426.1 ENST00000378418.2 |
CXorf38
|
chromosome X open reading frame 38 |
| chr5_-_10761206 | 1.53 |
ENST00000432074.2
ENST00000230895.6 |
DAP
|
death-associated protein |
| chr15_+_81293254 | 1.51 |
ENST00000267984.2
|
MESDC1
|
mesoderm development candidate 1 |
| chr4_-_186732241 | 1.46 |
ENST00000421639.1
|
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr1_-_92951607 | 1.45 |
ENST00000427103.1
|
GFI1
|
growth factor independent 1 transcription repressor |
| chr22_-_20731541 | 1.45 |
ENST00000292729.8
|
USP41
|
ubiquitin specific peptidase 41 |
| chr14_-_38028689 | 1.43 |
ENST00000553425.1
|
RP11-356O9.2
|
RP11-356O9.2 |
| chr7_+_150065278 | 1.42 |
ENST00000519397.1
ENST00000479668.1 ENST00000540729.1 |
REPIN1
|
replication initiator 1 |
| chr3_+_136649311 | 1.42 |
ENST00000469404.1
ENST00000467911.1 |
NCK1
|
NCK adaptor protein 1 |
| chr8_+_77318769 | 1.41 |
ENST00000518732.1
|
RP11-706J10.1
|
long intergenic non-protein coding RNA 1111 |
| chr14_-_107199464 | 1.40 |
ENST00000433072.2
|
IGHV3-72
|
immunoglobulin heavy variable 3-72 |
| chr10_+_90750378 | 1.39 |
ENST00000355740.2
ENST00000352159.4 |
FAS
|
Fas cell surface death receptor |
| chr8_+_71485681 | 1.35 |
ENST00000391684.1
|
AC120194.1
|
AC120194.1 |
| chr17_+_7462103 | 1.34 |
ENST00000396545.4
|
TNFSF13
|
tumor necrosis factor (ligand) superfamily, member 13 |
| chr12_-_123187890 | 1.34 |
ENST00000328880.5
|
HCAR2
|
hydroxycarboxylic acid receptor 2 |
| chrX_+_24483338 | 1.30 |
ENST00000379162.4
ENST00000441463.2 |
PDK3
|
pyruvate dehydrogenase kinase, isozyme 3 |
| chr9_+_130860810 | 1.26 |
ENST00000433501.1
|
SLC25A25
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25 |
| chr11_-_77185094 | 1.26 |
ENST00000278568.4
ENST00000356341.3 |
PAK1
|
p21 protein (Cdc42/Rac)-activated kinase 1 |
| chr11_-_18062872 | 1.23 |
ENST00000250018.2
|
TPH1
|
tryptophan hydroxylase 1 |
| chr11_-_104905840 | 1.22 |
ENST00000526568.1
ENST00000393136.4 ENST00000531166.1 ENST00000534497.1 ENST00000527979.1 ENST00000446369.1 ENST00000353247.5 ENST00000528974.1 ENST00000533400.1 ENST00000525825.1 ENST00000436863.3 |
CASP1
|
caspase 1, apoptosis-related cysteine peptidase |
| chr14_-_23292596 | 1.22 |
ENST00000554741.1
|
SLC7A7
|
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7 |
| chr19_+_48949030 | 1.19 |
ENST00000253237.5
|
GRWD1
|
glutamate-rich WD repeat containing 1 |
| chr9_+_130860583 | 1.18 |
ENST00000373064.5
|
SLC25A25
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25 |
| chr5_-_169407744 | 1.17 |
ENST00000377365.3
|
FAM196B
|
family with sequence similarity 196, member B |
| chr9_-_99381660 | 1.15 |
ENST00000375240.3
ENST00000463569.1 |
CDC14B
|
cell division cycle 14B |
| chr2_-_99279928 | 1.14 |
ENST00000414521.2
|
MGAT4A
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme A |
| chr7_-_141401951 | 1.11 |
ENST00000536163.1
|
KIAA1147
|
KIAA1147 |
| chr17_+_7462031 | 1.11 |
ENST00000380535.4
|
TNFSF13
|
tumor necrosis factor (ligand) superfamily, member 13 |
| chr12_-_10605929 | 1.10 |
ENST00000347831.5
ENST00000359151.3 |
KLRC1
|
killer cell lectin-like receptor subfamily C, member 1 |
| chr17_-_60142609 | 1.10 |
ENST00000397786.2
|
MED13
|
mediator complex subunit 13 |
| chr2_-_191878162 | 1.10 |
ENST00000540176.1
|
STAT1
|
signal transducer and activator of transcription 1, 91kDa |
| chr5_+_95997885 | 1.08 |
ENST00000511097.2
|
CAST
|
calpastatin |
| chr11_+_82868030 | 1.07 |
ENST00000298281.4
ENST00000530660.1 |
PCF11
|
PCF11 cleavage and polyadenylation factor subunit |
| chr2_+_64069459 | 1.07 |
ENST00000445915.2
ENST00000475462.1 |
UGP2
|
UDP-glucose pyrophosphorylase 2 |
| chr5_+_156696362 | 1.06 |
ENST00000377576.3
|
CYFIP2
|
cytoplasmic FMR1 interacting protein 2 |
| chr1_-_186649543 | 1.06 |
ENST00000367468.5
|
PTGS2
|
prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase) |
Network of associatons between targets according to the STRING database.
First level regulatory network of CEBPE_CEBPD
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.1 | 42.8 | GO:0019557 | histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 6.1 | 12.2 | GO:0055073 | cadmium ion homeostasis(GO:0055073) |
| 5.8 | 35.0 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 4.6 | 13.8 | GO:2000296 | negative regulation of hydrogen peroxide catabolic process(GO:2000296) |
| 4.3 | 13.0 | GO:2000409 | astrocyte chemotaxis(GO:0035700) positive regulation of immune complex clearance by monocytes and macrophages(GO:0090265) positive regulation of T cell extravasation(GO:2000409) positive regulation of CD8-positive, alpha-beta T cell extravasation(GO:2000451) regulation of astrocyte chemotaxis(GO:2000458) |
| 4.1 | 16.3 | GO:2001027 | negative regulation of endothelial cell chemotaxis(GO:2001027) |
| 3.3 | 13.1 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 3.2 | 12.7 | GO:0003330 | regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) |
| 2.6 | 46.3 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 2.0 | 19.6 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) |
| 1.9 | 5.6 | GO:0052331 | hemolysis by symbiont of host erythrocytes(GO:0019836) hemolysis in other organism(GO:0044179) hemolysis in other organism involved in symbiotic interaction(GO:0052331) |
| 1.7 | 5.2 | GO:0016260 | selenocysteine biosynthetic process(GO:0016260) |
| 1.7 | 6.7 | GO:0045014 | carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) |
| 1.6 | 4.9 | GO:0046495 | nicotinamide riboside catabolic process(GO:0006738) nicotinamide riboside metabolic process(GO:0046495) pyridine nucleoside metabolic process(GO:0070637) pyridine nucleoside catabolic process(GO:0070638) |
| 1.5 | 12.0 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 1.5 | 3.0 | GO:0097460 | ferrous iron import into cell(GO:0097460) ferrous iron import across plasma membrane(GO:0098707) |
| 1.5 | 5.9 | GO:0019442 | tryptophan catabolic process to acetyl-CoA(GO:0019442) |
| 1.4 | 23.7 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 1.4 | 4.1 | GO:0015882 | L-ascorbic acid transport(GO:0015882) transepithelial L-ascorbic acid transport(GO:0070904) |
| 1.4 | 5.5 | GO:0002384 | hepatic immune response(GO:0002384) |
| 1.3 | 5.4 | GO:0046110 | xanthine metabolic process(GO:0046110) |
| 1.3 | 5.2 | GO:0010983 | positive regulation of high-density lipoprotein particle clearance(GO:0010983) |
| 1.2 | 5.0 | GO:0046271 | phenylpropanoid catabolic process(GO:0046271) |
| 1.2 | 3.7 | GO:0045799 | positive regulation of chromatin assembly or disassembly(GO:0045799) |
| 1.2 | 9.7 | GO:2000660 | negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 1.2 | 4.8 | GO:2000176 | regulation of pro-T cell differentiation(GO:2000174) positive regulation of pro-T cell differentiation(GO:2000176) |
| 1.1 | 7.5 | GO:0071727 | toll-like receptor TLR1:TLR2 signaling pathway(GO:0038123) response to triacyl bacterial lipopeptide(GO:0071725) cellular response to triacyl bacterial lipopeptide(GO:0071727) |
| 1.0 | 3.1 | GO:0001978 | regulation of systemic arterial blood pressure by carotid sinus baroreceptor feedback(GO:0001978) baroreceptor response to increased systemic arterial blood pressure(GO:0001983) |
| 1.0 | 3.9 | GO:0006114 | glycerol biosynthetic process(GO:0006114) |
| 0.9 | 4.4 | GO:0010902 | positive regulation of very-low-density lipoprotein particle remodeling(GO:0010902) |
| 0.9 | 26.5 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.8 | 5.9 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.8 | 3.3 | GO:0072308 | negative regulation by virus of viral protein levels in host cell(GO:0046725) negative regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072308) |
| 0.8 | 63.6 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.8 | 3.9 | GO:0060474 | positive regulation of sperm motility involved in capacitation(GO:0060474) |
| 0.7 | 2.2 | GO:0006097 | glyoxylate cycle(GO:0006097) |
| 0.7 | 4.8 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.7 | 8.7 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.6 | 3.1 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.6 | 6.7 | GO:0033015 | porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.5 | 2.7 | GO:0030573 | bile acid catabolic process(GO:0030573) |
| 0.5 | 1.5 | GO:0070105 | positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 0.5 | 2.4 | GO:0097527 | necroptotic signaling pathway(GO:0097527) |
| 0.5 | 1.9 | GO:0050705 | regulation of interleukin-1 alpha secretion(GO:0050705) positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.5 | 1.4 | GO:1903674 | regulation of cap-dependent translational initiation(GO:1903674) positive regulation of cap-dependent translational initiation(GO:1903676) |
| 0.5 | 3.7 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.5 | 15.8 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.4 | 8.0 | GO:0010623 | programmed cell death involved in cell development(GO:0010623) |
| 0.4 | 3.0 | GO:1904139 | microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) |
| 0.4 | 3.0 | GO:0019477 | L-lysine catabolic process to acetyl-CoA(GO:0019474) L-lysine catabolic process(GO:0019477) L-lysine metabolic process(GO:0046440) |
| 0.4 | 4.7 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.4 | 5.4 | GO:0034201 | response to oleic acid(GO:0034201) |
| 0.4 | 3.7 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.4 | 2.4 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.4 | 3.8 | GO:0006065 | UDP-glucuronate biosynthetic process(GO:0006065) |
| 0.3 | 3.4 | GO:0030450 | regulation of complement activation, classical pathway(GO:0030450) negative regulation of complement activation, classical pathway(GO:0045959) |
| 0.3 | 1.7 | GO:0036509 | trimming of terminal mannose on B branch(GO:0036509) trimming of first mannose on A branch(GO:0036511) trimming of second mannose on A branch(GO:0036512) |
| 0.3 | 4.1 | GO:0042738 | exogenous drug catabolic process(GO:0042738) |
| 0.3 | 2.8 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.3 | 7.0 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 0.3 | 0.6 | GO:0060454 | positive regulation of gastric acid secretion(GO:0060454) |
| 0.3 | 11.7 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.3 | 3.1 | GO:0048298 | positive regulation of isotype switching to IgA isotypes(GO:0048298) |
| 0.3 | 5.0 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.3 | 3.3 | GO:0050966 | detection of mechanical stimulus involved in sensory perception of pain(GO:0050966) |
| 0.3 | 1.6 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.3 | 4.4 | GO:0015747 | urate transport(GO:0015747) |
| 0.2 | 10.9 | GO:0050832 | defense response to fungus(GO:0050832) |
| 0.2 | 1.3 | GO:0033031 | positive regulation of neutrophil apoptotic process(GO:0033031) |
| 0.2 | 2.0 | GO:0001649 | osteoblast differentiation(GO:0001649) |
| 0.2 | 0.8 | GO:0009224 | CMP salvage(GO:0006238) CMP biosynthetic process(GO:0009224) CMP metabolic process(GO:0046035) |
| 0.2 | 1.2 | GO:0042427 | serotonin biosynthetic process(GO:0042427) |
| 0.2 | 0.6 | GO:0035750 | protein localization to myelin sheath abaxonal region(GO:0035750) |
| 0.2 | 1.6 | GO:0019240 | citrulline biosynthetic process(GO:0019240) |
| 0.2 | 9.5 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.2 | 3.4 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.2 | 1.2 | GO:0046223 | mycotoxin metabolic process(GO:0043385) mycotoxin catabolic process(GO:0043387) aflatoxin metabolic process(GO:0046222) aflatoxin catabolic process(GO:0046223) organic heteropentacyclic compound metabolic process(GO:1901376) organic heteropentacyclic compound catabolic process(GO:1901377) regulation of glutathione biosynthetic process(GO:1903786) positive regulation of glutathione biosynthetic process(GO:1903788) |
| 0.2 | 16.9 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.2 | 1.3 | GO:0097411 | hypoxia-inducible factor-1alpha signaling pathway(GO:0097411) |
| 0.1 | 1.6 | GO:0072718 | response to cisplatin(GO:0072718) |
| 0.1 | 1.7 | GO:0034378 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) chylomicron assembly(GO:0034378) positive regulation of viral entry into host cell(GO:0046598) |
| 0.1 | 1.3 | GO:0061052 | negative regulation of cell proliferation involved in contact inhibition(GO:0060244) negative regulation of cell growth involved in cardiac muscle cell development(GO:0061052) |
| 0.1 | 0.4 | GO:0016237 | lysosomal microautophagy(GO:0016237) piecemeal microautophagy of nucleus(GO:0034727) suppression by virus of host autophagy(GO:0039521) |
| 0.1 | 1.0 | GO:0071494 | cellular response to UV-C(GO:0071494) |
| 0.1 | 4.6 | GO:0061641 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.1 | 1.6 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.1 | 1.0 | GO:0030643 | cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.1 | 9.0 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.1 | 0.3 | GO:0042214 | terpene metabolic process(GO:0042214) |
| 0.1 | 0.6 | GO:0032468 | Golgi calcium ion homeostasis(GO:0032468) |
| 0.1 | 0.4 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
| 0.1 | 0.3 | GO:0071314 | cellular response to cocaine(GO:0071314) |
| 0.1 | 1.1 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.1 | 2.4 | GO:0002021 | response to dietary excess(GO:0002021) |
| 0.1 | 0.3 | GO:0039534 | negative regulation of MDA-5 signaling pathway(GO:0039534) |
| 0.1 | 0.3 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 0.1 | 0.8 | GO:1902455 | negative regulation of stem cell population maintenance(GO:1902455) |
| 0.1 | 8.3 | GO:0030449 | regulation of complement activation(GO:0030449) regulation of protein activation cascade(GO:2000257) |
| 0.1 | 0.2 | GO:1901340 | negative regulation of store-operated calcium channel activity(GO:1901340) |
| 0.1 | 1.8 | GO:0070933 | histone H4 deacetylation(GO:0070933) |
| 0.1 | 4.7 | GO:0035987 | endodermal cell differentiation(GO:0035987) |
| 0.1 | 1.8 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.1 | 1.2 | GO:0051256 | mitotic spindle midzone assembly(GO:0051256) |
| 0.1 | 2.1 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.1 | 0.5 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.1 | 1.5 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.1 | 0.4 | GO:0008626 | granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 0.1 | 3.7 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.0 | 0.5 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 | 0.5 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.1 | GO:0070510 | regulation of histone H4-K20 methylation(GO:0070510) positive regulation of histone H4-K20 methylation(GO:0070512) |
| 0.0 | 2.4 | GO:0071353 | cellular response to interleukin-4(GO:0071353) |
| 0.0 | 1.7 | GO:1902236 | negative regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway(GO:1902236) |
| 0.0 | 0.2 | GO:0002399 | MHC class II protein complex assembly(GO:0002399) |
| 0.0 | 0.9 | GO:0042448 | progesterone metabolic process(GO:0042448) |
| 0.0 | 8.2 | GO:0008277 | regulation of G-protein coupled receptor protein signaling pathway(GO:0008277) |
| 0.0 | 3.1 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.0 | 1.1 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 1.0 | GO:0019731 | antibacterial humoral response(GO:0019731) |
| 0.0 | 1.6 | GO:0006379 | mRNA cleavage(GO:0006379) |
| 0.0 | 4.5 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.0 | 0.4 | GO:0017182 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.0 | 1.4 | GO:0072376 | protein activation cascade(GO:0072376) |
| 0.0 | 0.1 | GO:2000676 | positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.0 | 1.1 | GO:0055090 | acylglycerol homeostasis(GO:0055090) triglyceride homeostasis(GO:0070328) |
| 0.0 | 0.7 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.0 | 1.4 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.0 | 1.5 | GO:0034198 | cellular response to amino acid starvation(GO:0034198) |
| 0.0 | 0.8 | GO:0006999 | nuclear pore organization(GO:0006999) |
| 0.0 | 0.2 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 0.5 | GO:0043401 | steroid hormone mediated signaling pathway(GO:0043401) |
| 0.0 | 0.7 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 1.4 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 3.6 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.0 | 0.6 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.5 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.0 | 1.3 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.0 | 0.2 | GO:1901725 | regulation of histone deacetylase activity(GO:1901725) |
| 0.0 | 0.1 | GO:0001544 | initiation of primordial ovarian follicle growth(GO:0001544) |
| 0.0 | 0.6 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 2.4 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 | 0.5 | GO:0000717 | nucleotide-excision repair, DNA duplex unwinding(GO:0000717) |
| 0.0 | 0.5 | GO:0090383 | phagosome acidification(GO:0090383) |
| 0.0 | 0.2 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.1 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 1.2 | GO:0002292 | T cell differentiation involved in immune response(GO:0002292) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.5 | 13.8 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 1.2 | 8.1 | GO:0070110 | ciliary neurotrophic factor receptor complex(GO:0070110) |
| 1.0 | 49.7 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.9 | 7.0 | GO:0034688 | integrin alphaM-beta2 complex(GO:0034688) |
| 0.8 | 7.5 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.7 | 6.0 | GO:0044279 | other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.7 | 3.9 | GO:1990742 | microvesicle(GO:1990742) |
| 0.6 | 8.8 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.4 | 15.6 | GO:0036019 | endolysosome(GO:0036019) |
| 0.4 | 3.0 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.4 | 1.7 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.4 | 2.9 | GO:0097179 | protease inhibitor complex(GO:0097179) |
| 0.3 | 80.3 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.3 | 4.6 | GO:0000778 | condensed nuclear chromosome kinetochore(GO:0000778) |
| 0.3 | 3.2 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.2 | 2.6 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.2 | 3.6 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.2 | 39.4 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 14.6 | GO:0101003 | ficolin-1-rich granule membrane(GO:0101003) |
| 0.1 | 60.0 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.1 | 0.7 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.1 | 3.1 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.1 | 0.3 | GO:0097124 | cyclin A2-CDK2 complex(GO:0097124) |
| 0.1 | 0.5 | GO:0071942 | XPC complex(GO:0071942) |
| 0.1 | 0.6 | GO:0043219 | lateral loop(GO:0043219) |
| 0.1 | 2.4 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.1 | 1.4 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.1 | 3.2 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.1 | 8.8 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.1 | 2.8 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.1 | 0.5 | GO:0098536 | deuterosome(GO:0098536) |
| 0.1 | 16.3 | GO:1904813 | ficolin-1-rich granule(GO:0101002) ficolin-1-rich granule lumen(GO:1904813) |
| 0.1 | 0.5 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.1 | 1.4 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.1 | 1.3 | GO:0071437 | invadopodium(GO:0071437) |
| 0.1 | 5.6 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.1 | 5.0 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.2 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
| 0.0 | 0.8 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.0 | 1.6 | GO:0005849 | mRNA cleavage factor complex(GO:0005849) |
| 0.0 | 1.0 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 9.6 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 6.7 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 0.3 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.0 | 0.1 | GO:0031436 | BRCA1-BARD1 complex(GO:0031436) |
| 0.0 | 2.4 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 3.3 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 1.7 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.0 | 0.6 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 3.4 | GO:0044215 | other organism(GO:0044215) other organism cell(GO:0044216) other organism part(GO:0044217) |
| 0.0 | 5.4 | GO:0044438 | microbody part(GO:0044438) peroxisomal part(GO:0044439) |
| 0.0 | 0.9 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 1.8 | GO:0043596 | nuclear replication fork(GO:0043596) |
| 0.0 | 1.0 | GO:0035580 | specific granule lumen(GO:0035580) |
| 0.0 | 0.3 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 10.2 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 2.4 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.0 | 3.9 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.5 | GO:0033178 | proton-transporting two-sector ATPase complex, catalytic domain(GO:0033178) |
| 0.0 | 1.1 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 2.2 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 1.2 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 0.4 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 14.3 | 42.8 | GO:0004397 | histidine ammonia-lyase activity(GO:0004397) |
| 7.8 | 47.0 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 6.4 | 31.8 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 4.1 | 12.2 | GO:0051139 | metal ion:proton antiporter activity(GO:0051139) |
| 3.2 | 9.7 | GO:0005150 | interleukin-1, Type I receptor binding(GO:0005150) |
| 2.7 | 8.1 | GO:0070119 | ciliary neurotrophic factor binding(GO:0070119) |
| 2.6 | 13.0 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 2.3 | 13.8 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 1.9 | 16.9 | GO:0004873 | asialoglycoprotein receptor activity(GO:0004873) |
| 1.7 | 5.2 | GO:0016781 | selenide, water dikinase activity(GO:0004756) phosphotransferase activity, paired acceptors(GO:0016781) |
| 1.7 | 6.7 | GO:0004074 | biliverdin reductase activity(GO:0004074) |
| 1.6 | 4.9 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 1.4 | 4.1 | GO:0070890 | L-ascorbate:sodium symporter activity(GO:0008520) L-ascorbic acid transporter activity(GO:0015229) sodium-dependent L-ascorbate transmembrane transporter activity(GO:0070890) |
| 1.4 | 9.5 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 1.3 | 4.0 | GO:0004611 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 1.3 | 3.9 | GO:0031731 | CCR6 chemokine receptor binding(GO:0031731) |
| 1.2 | 3.7 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 1.1 | 13.3 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 1.1 | 3.2 | GO:0015432 | bile acid-exporting ATPase activity(GO:0015432) |
| 1.1 | 7.5 | GO:0016019 | peptidoglycan receptor activity(GO:0016019) |
| 1.0 | 3.0 | GO:0072510 | ferric iron transmembrane transporter activity(GO:0015091) trivalent inorganic cation transmembrane transporter activity(GO:0072510) |
| 1.0 | 5.9 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 1.0 | 5.9 | GO:0004698 | calcium-dependent protein kinase C activity(GO:0004698) |
| 0.9 | 19.6 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.8 | 3.2 | GO:0050528 | acyloxyacyl hydrolase activity(GO:0050528) |
| 0.8 | 44.8 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.8 | 3.1 | GO:0031716 | calcitonin receptor binding(GO:0031716) |
| 0.7 | 2.2 | GO:0004450 | isocitrate dehydrogenase (NADP+) activity(GO:0004450) |
| 0.6 | 4.4 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.6 | 5.4 | GO:0043546 | molybdopterin cofactor binding(GO:0043546) |
| 0.6 | 7.0 | GO:0030369 | ICAM-3 receptor activity(GO:0030369) |
| 0.5 | 2.7 | GO:0047787 | delta4-3-oxosteroid 5beta-reductase activity(GO:0047787) |
| 0.5 | 12.4 | GO:0008392 | arachidonic acid monooxygenase activity(GO:0008391) arachidonic acid epoxygenase activity(GO:0008392) |
| 0.5 | 3.0 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.5 | 15.6 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.4 | 5.4 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.4 | 3.3 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.4 | 2.5 | GO:0008823 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.3 | 2.4 | GO:0043120 | tumor necrosis factor binding(GO:0043120) |
| 0.3 | 2.4 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 0.3 | 1.6 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.3 | 1.3 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.3 | 1.2 | GO:0004510 | tryptophan 5-monooxygenase activity(GO:0004510) |
| 0.3 | 35.7 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.3 | 9.8 | GO:0001848 | complement binding(GO:0001848) |
| 0.3 | 1.4 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.3 | 4.4 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.2 | 4.7 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.2 | 12.6 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.2 | 1.6 | GO:0051748 | UTP:glucose-1-phosphate uridylyltransferase activity(GO:0003983) UTP-monosaccharide-1-phosphate uridylyltransferase activity(GO:0051748) |
| 0.2 | 1.1 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) arachidonate 15-lipoxygenase activity(GO:0050473) |
| 0.2 | 9.0 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.2 | 5.4 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.2 | 1.1 | GO:0023024 | MHC class I protein complex binding(GO:0023024) |
| 0.2 | 2.1 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.2 | 6.6 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.2 | 1.6 | GO:0070569 | uridylyltransferase activity(GO:0070569) |
| 0.1 | 5.2 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.1 | 3.3 | GO:0015037 | peptide disulfide oxidoreductase activity(GO:0015037) |
| 0.1 | 3.6 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.1 | 1.1 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.1 | 0.6 | GO:0070052 | fibrinogen binding(GO:0070051) collagen V binding(GO:0070052) |
| 0.1 | 4.8 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.1 | 13.1 | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups(GO:0016765) |
| 0.1 | 0.4 | GO:0019778 | Atg12 activating enzyme activity(GO:0019778) Atg8 activating enzyme activity(GO:0019779) |
| 0.1 | 6.3 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.1 | 4.9 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.1 | 1.8 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.1 | 1.7 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.1 | 3.0 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.1 | 2.8 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.1 | 33.3 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 2.1 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.1 | 1.5 | GO:0070513 | death domain binding(GO:0070513) |
| 0.1 | 1.0 | GO:0015293 | symporter activity(GO:0015293) |
| 0.1 | 3.1 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.1 | 2.5 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.1 | 1.0 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.1 | 0.8 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.1 | 1.5 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.1 | 1.8 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.1 | 0.3 | GO:0031208 | POZ domain binding(GO:0031208) |
| 0.1 | 6.3 | GO:0004520 | endodeoxyribonuclease activity(GO:0004520) |
| 0.1 | 9.0 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 0.6 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.1 | 1.3 | GO:0042301 | phosphate ion binding(GO:0042301) |
| 0.1 | 0.3 | GO:0089720 | caspase binding(GO:0089720) |
| 0.1 | 1.6 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.1 | 1.2 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.1 | 1.8 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.1 | 1.1 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.0 | 0.1 | GO:0034353 | RNA pyrophosphohydrolase activity(GO:0034353) |
| 0.0 | 0.5 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 1.4 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 3.7 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 1.6 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 3.1 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 0.5 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 3.7 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.9 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.0 | 0.7 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 2.6 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 5.8 | GO:0005319 | lipid transporter activity(GO:0005319) |
| 0.0 | 1.4 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.2 | GO:0019534 | toxin transporter activity(GO:0019534) |
| 0.0 | 1.9 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 4.6 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 0.7 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 0.5 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.4 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.6 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 1.0 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 9.7 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 1.3 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 0.3 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 1.2 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.8 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.0 | 0.5 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 0.2 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.0 | 0.2 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 3.7 | GO:0030674 | protein binding, bridging(GO:0030674) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 34.5 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.5 | 35.4 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.4 | 13.5 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.4 | 3.3 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.3 | 14.6 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.3 | 5.5 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.2 | 12.1 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.2 | 14.8 | PID BMP PATHWAY | BMP receptor signaling |
| 0.1 | 2.4 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.1 | 10.7 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 3.3 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.1 | 14.3 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.1 | 1.8 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.1 | 5.7 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.1 | 9.8 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 6.9 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 1.1 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.0 | 16.6 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 2.7 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 2.5 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 3.4 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 1.5 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 3.6 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.0 | 0.3 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 1.0 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 2.5 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 1.1 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 0.6 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 1.2 | PID PLK1 PATHWAY | PLK1 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 31.8 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 1.5 | 49.1 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 1.2 | 30.8 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.8 | 27.0 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.7 | 12.7 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.5 | 27.1 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.5 | 10.3 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.5 | 9.1 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.5 | 7.7 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.4 | 8.7 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.4 | 7.5 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.3 | 16.8 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.3 | 14.2 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.3 | 4.0 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.3 | 19.2 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.2 | 5.9 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.2 | 15.5 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.2 | 3.3 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.2 | 6.7 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.2 | 2.7 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.2 | 6.0 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.2 | 5.4 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.2 | 4.5 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.2 | 6.7 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.1 | 3.8 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.1 | 4.9 | REACTOME ADP SIGNALLING THROUGH P2RY12 | Genes involved in ADP signalling through P2Y purinoceptor 12 |
| 0.1 | 37.9 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.1 | 20.2 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.1 | 9.0 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.1 | 4.7 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.1 | 4.6 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.1 | 7.4 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.1 | 1.9 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.1 | 2.8 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.1 | 1.7 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.1 | 0.9 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.1 | 3.7 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.1 | 6.0 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.1 | 2.4 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.1 | 1.3 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.1 | 3.5 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.1 | 8.1 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.1 | 3.1 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.1 | 1.6 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 2.2 | REACTOME PYRUVATE METABOLISM AND CITRIC ACID TCA CYCLE | Genes involved in Pyruvate metabolism and Citric Acid (TCA) cycle |
| 0.0 | 1.7 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 1.7 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 3.1 | REACTOME REGULATION OF MRNA STABILITY BY PROTEINS THAT BIND AU RICH ELEMENTS | Genes involved in Regulation of mRNA Stability by Proteins that Bind AU-rich Elements |
| 0.0 | 4.2 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 7.2 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 3.5 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 1.1 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 2.4 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 1.0 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.8 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 0.3 | REACTOME SCFSKP2 MEDIATED DEGRADATION OF P27 P21 | Genes involved in SCF(Skp2)-mediated degradation of p27/p21 |
| 0.0 | 1.8 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.8 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.0 | 0.5 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 0.5 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.0 | 0.9 | REACTOME DOWNSTREAM SIGNALING EVENTS OF B CELL RECEPTOR BCR | Genes involved in Downstream Signaling Events Of B Cell Receptor (BCR) |
| 0.0 | 0.4 | REACTOME AUTODEGRADATION OF CDH1 BY CDH1 APC C | Genes involved in Autodegradation of Cdh1 by Cdh1:APC/C |
| 0.0 | 0.6 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.2 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |