Project
Illumina Body Map 2: averaged replicates
Navigation
Downloads
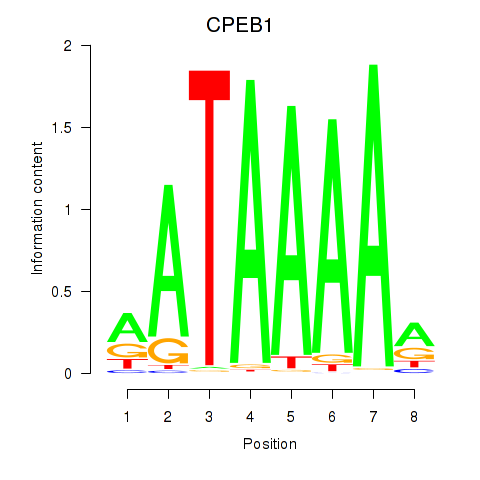
Results for CPEB1
Z-value: 1.66
Transcription factors associated with CPEB1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
CPEB1
|
ENSG00000214575.5 | cytoplasmic polyadenylation element binding protein 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| CPEB1 | hg19_v2_chr15_-_83316254_83316368 | 0.68 | 1.7e-05 | Click! |
Activity profile of CPEB1 motif
Sorted Z-values of CPEB1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_166428839 | 3.34 |
ENST00000342316.4
|
CSRNP3
|
cysteine-serine-rich nuclear protein 3 |
| chr8_+_104831472 | 2.88 |
ENST00000262231.10
ENST00000507740.1 |
RIMS2
|
regulating synaptic membrane exocytosis 2 |
| chr12_+_41221794 | 2.87 |
ENST00000547849.1
|
CNTN1
|
contactin 1 |
| chr8_+_77593474 | 2.72 |
ENST00000455469.2
ENST00000050961.6 |
ZFHX4
|
zinc finger homeobox 4 |
| chr3_-_73483055 | 2.68 |
ENST00000479530.1
|
PDZRN3
|
PDZ domain containing ring finger 3 |
| chr1_+_161228517 | 2.46 |
ENST00000504449.1
|
PCP4L1
|
Purkinje cell protein 4 like 1 |
| chr8_+_77593448 | 2.43 |
ENST00000521891.2
|
ZFHX4
|
zinc finger homeobox 4 |
| chr6_+_153071925 | 2.30 |
ENST00000367244.3
ENST00000367243.3 |
VIP
|
vasoactive intestinal peptide |
| chr2_-_50574856 | 2.30 |
ENST00000342183.5
|
NRXN1
|
neurexin 1 |
| chr2_-_2334888 | 2.20 |
ENST00000428368.2
ENST00000399161.2 |
MYT1L
|
myelin transcription factor 1-like |
| chr1_+_163039143 | 2.18 |
ENST00000531057.1
ENST00000527809.1 ENST00000367908.4 |
RGS4
|
regulator of G-protein signaling 4 |
| chrX_+_117957741 | 2.14 |
ENST00000310164.2
|
ZCCHC12
|
zinc finger, CCHC domain containing 12 |
| chr4_+_76995855 | 2.02 |
ENST00000355810.4
ENST00000349321.3 |
ART3
|
ADP-ribosyltransferase 3 |
| chr7_+_24323782 | 2.01 |
ENST00000242152.2
ENST00000407573.1 |
NPY
|
neuropeptide Y |
| chr14_+_79745746 | 2.00 |
ENST00000281127.7
|
NRXN3
|
neurexin 3 |
| chr1_+_239882842 | 1.93 |
ENST00000448020.1
|
CHRM3
|
cholinergic receptor, muscarinic 3 |
| chr22_+_29876197 | 1.89 |
ENST00000310624.6
|
NEFH
|
neurofilament, heavy polypeptide |
| chr6_+_69942298 | 1.88 |
ENST00000238918.8
|
BAI3
|
brain-specific angiogenesis inhibitor 3 |
| chr14_+_79745682 | 1.88 |
ENST00000557594.1
|
NRXN3
|
neurexin 3 |
| chr9_-_122131696 | 1.82 |
ENST00000373964.2
ENST00000265922.3 |
BRINP1
|
bone morphogenetic protein/retinoic acid inducible neural-specific 1 |
| chr17_-_78450398 | 1.72 |
ENST00000306773.4
|
NPTX1
|
neuronal pentraxin I |
| chr3_+_148545586 | 1.72 |
ENST00000282957.4
ENST00000468341.1 |
CPB1
|
carboxypeptidase B1 (tissue) |
| chr1_-_156675368 | 1.67 |
ENST00000368222.3
|
CRABP2
|
cellular retinoic acid binding protein 2 |
| chr3_-_117716418 | 1.63 |
ENST00000484092.1
|
RP11-384F7.2
|
RP11-384F7.2 |
| chr10_+_102106829 | 1.61 |
ENST00000370355.2
|
SCD
|
stearoyl-CoA desaturase (delta-9-desaturase) |
| chr5_+_161275320 | 1.60 |
ENST00000437025.2
|
GABRA1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1 |
| chr3_-_93747425 | 1.57 |
ENST00000315099.2
|
STX19
|
syntaxin 19 |
| chr11_-_5248294 | 1.52 |
ENST00000335295.4
|
HBB
|
hemoglobin, beta |
| chr8_+_85095497 | 1.52 |
ENST00000522455.1
ENST00000521695.1 |
RALYL
|
RALY RNA binding protein-like |
| chr7_+_35756186 | 1.50 |
ENST00000430518.1
|
AC018647.3
|
AC018647.3 |
| chr18_+_29171689 | 1.48 |
ENST00000237014.3
|
TTR
|
transthyretin |
| chr10_-_69455873 | 1.48 |
ENST00000433211.2
|
CTNNA3
|
catenin (cadherin-associated protein), alpha 3 |
| chr19_-_58485895 | 1.47 |
ENST00000314391.3
|
C19orf18
|
chromosome 19 open reading frame 18 |
| chr1_-_156675535 | 1.46 |
ENST00000368221.1
|
CRABP2
|
cellular retinoic acid binding protein 2 |
| chr5_+_161274685 | 1.45 |
ENST00000428797.2
|
GABRA1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1 |
| chr9_-_23825956 | 1.44 |
ENST00000397312.2
|
ELAVL2
|
ELAV like neuron-specific RNA binding protein 2 |
| chr17_-_66951382 | 1.44 |
ENST00000586539.1
|
ABCA8
|
ATP-binding cassette, sub-family A (ABC1), member 8 |
| chr16_+_56226405 | 1.42 |
ENST00000565363.1
|
GNAO1
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide O |
| chr18_-_35145728 | 1.42 |
ENST00000361795.5
ENST00000603232.1 |
CELF4
|
CUGBP, Elav-like family member 4 |
| chr12_-_81763184 | 1.41 |
ENST00000548670.1
ENST00000541570.2 ENST00000553058.1 |
PPFIA2
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2 |
| chr3_+_158991025 | 1.39 |
ENST00000337808.6
|
IQCJ-SCHIP1
|
IQCJ-SCHIP1 readthrough |
| chr6_+_31683117 | 1.39 |
ENST00000375825.3
ENST00000375824.1 |
LY6G6D
|
lymphocyte antigen 6 complex, locus G6D |
| chr12_-_81763127 | 1.37 |
ENST00000541017.1
|
PPFIA2
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2 |
| chr2_-_51259292 | 1.37 |
ENST00000401669.2
|
NRXN1
|
neurexin 1 |
| chr19_+_35773242 | 1.36 |
ENST00000222304.3
|
HAMP
|
hepcidin antimicrobial peptide |
| chrX_+_135570046 | 1.34 |
ENST00000370648.3
|
BRS3
|
bombesin-like receptor 3 |
| chr18_+_55024383 | 1.33 |
ENST00000586360.1
|
ST8SIA3
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 3 |
| chr17_-_39041479 | 1.33 |
ENST00000167588.3
|
KRT20
|
keratin 20 |
| chr2_-_51259229 | 1.31 |
ENST00000405472.3
|
NRXN1
|
neurexin 1 |
| chr12_+_52203789 | 1.30 |
ENST00000599343.1
|
AC068987.1
|
HCG1997999; cDNA FLJ33996 fis, clone DFNES2008881 |
| chr5_+_161274940 | 1.30 |
ENST00000393943.4
|
GABRA1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1 |
| chr5_+_36608422 | 1.27 |
ENST00000381918.3
|
SLC1A3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3 |
| chr3_-_10547192 | 1.27 |
ENST00000360273.2
ENST00000343816.4 |
ATP2B2
|
ATPase, Ca++ transporting, plasma membrane 2 |
| chr8_+_104831554 | 1.27 |
ENST00000408894.2
|
RIMS2
|
regulating synaptic membrane exocytosis 2 |
| chr3_+_35680339 | 1.26 |
ENST00000450234.1
|
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa |
| chr7_-_16921601 | 1.25 |
ENST00000402239.3
ENST00000310398.2 ENST00000414935.1 |
AGR3
|
anterior gradient 3 |
| chr11_+_34642656 | 1.24 |
ENST00000257831.3
ENST00000450654.2 |
EHF
|
ets homologous factor |
| chr5_-_73937244 | 1.22 |
ENST00000302351.4
ENST00000510316.1 ENST00000508331.1 |
ENC1
|
ectodermal-neural cortex 1 (with BTB domain) |
| chr12_-_81992111 | 1.22 |
ENST00000443686.3
ENST00000407050.4 |
PPFIA2
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2 |
| chr12_+_79258444 | 1.22 |
ENST00000261205.4
|
SYT1
|
synaptotagmin I |
| chrX_+_92929192 | 1.21 |
ENST00000332647.4
|
FAM133A
|
family with sequence similarity 133, member A |
| chr14_-_24047965 | 1.20 |
ENST00000397118.3
ENST00000356300.4 |
JPH4
|
junctophilin 4 |
| chr3_+_160394940 | 1.20 |
ENST00000320767.2
|
ARL14
|
ADP-ribosylation factor-like 14 |
| chr17_-_46806540 | 1.20 |
ENST00000290295.7
|
HOXB13
|
homeobox B13 |
| chrX_+_144908928 | 1.19 |
ENST00000408967.2
|
TMEM257
|
transmembrane protein 257 |
| chr1_-_165414414 | 1.19 |
ENST00000359842.5
|
RXRG
|
retinoid X receptor, gamma |
| chr8_-_120651020 | 1.17 |
ENST00000522826.1
ENST00000520066.1 ENST00000259486.6 ENST00000075322.6 |
ENPP2
|
ectonucleotide pyrophosphatase/phosphodiesterase 2 |
| chr2_-_77749474 | 1.16 |
ENST00000409093.1
ENST00000409088.3 |
LRRTM4
|
leucine rich repeat transmembrane neuronal 4 |
| chr11_-_126810521 | 1.16 |
ENST00000530572.1
|
RP11-688I9.4
|
RP11-688I9.4 |
| chr2_-_183291741 | 1.15 |
ENST00000351439.5
ENST00000409365.1 |
PDE1A
|
phosphodiesterase 1A, calmodulin-dependent |
| chr10_+_119184702 | 1.15 |
ENST00000549104.1
|
CTA-109P11.4
|
CTA-109P11.4 |
| chr16_+_76311169 | 1.14 |
ENST00000307431.8
ENST00000377504.4 |
CNTNAP4
|
contactin associated protein-like 4 |
| chr11_+_112832090 | 1.13 |
ENST00000533760.1
|
NCAM1
|
neural cell adhesion molecule 1 |
| chr9_-_23779367 | 1.11 |
ENST00000440102.1
|
ELAVL2
|
ELAV like neuron-specific RNA binding protein 2 |
| chr9_-_23826298 | 1.11 |
ENST00000380117.1
|
ELAVL2
|
ELAV like neuron-specific RNA binding protein 2 |
| chr10_-_97200772 | 1.09 |
ENST00000371241.1
ENST00000354106.3 ENST00000371239.1 ENST00000361941.3 ENST00000277982.5 ENST00000371245.3 |
SORBS1
|
sorbin and SH3 domain containing 1 |
| chr2_-_166930131 | 1.09 |
ENST00000303395.4
ENST00000409050.1 ENST00000423058.2 ENST00000375405.3 |
SCN1A
|
sodium channel, voltage-gated, type I, alpha subunit |
| chr10_+_121578211 | 1.09 |
ENST00000369080.3
|
INPP5F
|
inositol polyphosphate-5-phosphatase F |
| chr1_+_177140633 | 1.08 |
ENST00000361539.4
|
BRINP2
|
bone morphogenetic protein/retinoic acid inducible neural-specific 2 |
| chr1_-_116311402 | 1.08 |
ENST00000261448.5
|
CASQ2
|
calsequestrin 2 (cardiac muscle) |
| chr12_-_71148413 | 1.07 |
ENST00000440835.2
ENST00000549308.1 ENST00000550661.1 |
PTPRR
|
protein tyrosine phosphatase, receptor type, R |
| chr11_-_8832182 | 1.06 |
ENST00000527510.1
ENST00000528527.1 ENST00000528523.1 ENST00000313726.6 |
ST5
|
suppression of tumorigenicity 5 |
| chr8_+_104831440 | 1.05 |
ENST00000515551.1
|
RIMS2
|
regulating synaptic membrane exocytosis 2 |
| chr6_-_127780510 | 1.05 |
ENST00000487331.2
ENST00000483725.3 |
KIAA0408
|
KIAA0408 |
| chr11_-_18765389 | 1.04 |
ENST00000477854.1
|
PTPN5
|
protein tyrosine phosphatase, non-receptor type 5 (striatum-enriched) |
| chr2_-_80531399 | 1.04 |
ENST00000409148.1
ENST00000415098.1 ENST00000452811.1 |
LRRTM1
|
leucine rich repeat transmembrane neuronal 1 |
| chr12_+_79258547 | 1.04 |
ENST00000457153.2
|
SYT1
|
synaptotagmin I |
| chr7_+_35756092 | 1.02 |
ENST00000458087.3
|
AC018647.3
|
AC018647.3 |
| chr11_-_101778665 | 1.01 |
ENST00000534527.1
|
ANGPTL5
|
angiopoietin-like 5 |
| chr7_-_83824449 | 1.01 |
ENST00000420047.1
|
SEMA3A
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chr20_+_58251716 | 1.00 |
ENST00000355648.4
|
PHACTR3
|
phosphatase and actin regulator 3 |
| chr18_-_35145689 | 1.00 |
ENST00000591287.1
ENST00000601019.1 ENST00000601392.1 |
CELF4
|
CUGBP, Elav-like family member 4 |
| chr11_-_93583697 | 1.00 |
ENST00000409977.1
|
VSTM5
|
V-set and transmembrane domain containing 5 |
| chr3_-_10547333 | 0.99 |
ENST00000383800.4
|
ATP2B2
|
ATPase, Ca++ transporting, plasma membrane 2 |
| chr7_-_14029515 | 0.99 |
ENST00000430479.1
ENST00000405218.2 ENST00000343495.5 |
ETV1
|
ets variant 1 |
| chr8_-_95274536 | 0.99 |
ENST00000297596.2
ENST00000396194.2 |
GEM
|
GTP binding protein overexpressed in skeletal muscle |
| chr10_+_18240834 | 0.99 |
ENST00000377371.3
ENST00000539911.1 |
SLC39A12
|
solute carrier family 39 (zinc transporter), member 12 |
| chr1_+_52682052 | 0.99 |
ENST00000371591.1
|
ZFYVE9
|
zinc finger, FYVE domain containing 9 |
| chr18_-_64271363 | 0.98 |
ENST00000262150.2
|
CDH19
|
cadherin 19, type 2 |
| chr8_+_30496078 | 0.98 |
ENST00000517349.1
|
SMIM18
|
small integral membrane protein 18 |
| chr3_-_164796269 | 0.98 |
ENST00000264382.3
|
SI
|
sucrase-isomaltase (alpha-glucosidase) |
| chrX_+_107683096 | 0.97 |
ENST00000328300.6
ENST00000361603.2 |
COL4A5
|
collagen, type IV, alpha 5 |
| chr2_-_198540751 | 0.97 |
ENST00000429081.1
|
RFTN2
|
raftlin family member 2 |
| chr11_+_112832202 | 0.97 |
ENST00000534015.1
|
NCAM1
|
neural cell adhesion molecule 1 |
| chr9_-_95186739 | 0.95 |
ENST00000375550.4
|
OMD
|
osteomodulin |
| chr2_-_152830479 | 0.95 |
ENST00000360283.6
|
CACNB4
|
calcium channel, voltage-dependent, beta 4 subunit |
| chr8_+_85095769 | 0.95 |
ENST00000518566.1
|
RALYL
|
RALY RNA binding protein-like |
| chr3_-_148939598 | 0.94 |
ENST00000455472.3
|
CP
|
ceruloplasmin (ferroxidase) |
| chr1_+_65730385 | 0.94 |
ENST00000263441.7
ENST00000395325.3 |
DNAJC6
|
DnaJ (Hsp40) homolog, subfamily C, member 6 |
| chr5_+_149546334 | 0.94 |
ENST00000231656.8
|
CDX1
|
caudal type homeobox 1 |
| chr2_-_175711133 | 0.92 |
ENST00000409597.1
ENST00000413882.1 |
CHN1
|
chimerin 1 |
| chr1_+_164528866 | 0.92 |
ENST00000420696.2
|
PBX1
|
pre-B-cell leukemia homeobox 1 |
| chr11_+_112832133 | 0.92 |
ENST00000524665.1
|
NCAM1
|
neural cell adhesion molecule 1 |
| chr2_+_210444748 | 0.92 |
ENST00000392194.1
|
MAP2
|
microtubule-associated protein 2 |
| chr10_+_68685764 | 0.91 |
ENST00000361320.4
|
LRRTM3
|
leucine rich repeat transmembrane neuronal 3 |
| chr3_+_63428982 | 0.91 |
ENST00000479198.1
ENST00000460711.1 ENST00000465156.1 |
SYNPR
|
synaptoporin |
| chr2_-_30144432 | 0.91 |
ENST00000389048.3
|
ALK
|
anaplastic lymphoma receptor tyrosine kinase |
| chr17_+_32582293 | 0.91 |
ENST00000580907.1
ENST00000225831.4 |
CCL2
|
chemokine (C-C motif) ligand 2 |
| chr4_+_95972822 | 0.90 |
ENST00000509540.1
ENST00000440890.2 |
BMPR1B
|
bone morphogenetic protein receptor, type IB |
| chr8_-_93029865 | 0.89 |
ENST00000422361.2
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr17_-_53800217 | 0.89 |
ENST00000424486.2
|
TMEM100
|
transmembrane protein 100 |
| chr3_+_159570722 | 0.89 |
ENST00000482804.1
|
SCHIP1
|
schwannomin interacting protein 1 |
| chr6_+_108487245 | 0.88 |
ENST00000368986.4
|
NR2E1
|
nuclear receptor subfamily 2, group E, member 1 |
| chr10_+_18240753 | 0.88 |
ENST00000377369.2
|
SLC39A12
|
solute carrier family 39 (zinc transporter), member 12 |
| chr12_+_58003935 | 0.88 |
ENST00000333972.7
|
ARHGEF25
|
Rho guanine nucleotide exchange factor (GEF) 25 |
| chr12_-_16759711 | 0.88 |
ENST00000447609.1
|
LMO3
|
LIM domain only 3 (rhombotin-like 2) |
| chr12_-_52715179 | 0.87 |
ENST00000293670.3
|
KRT83
|
keratin 83 |
| chr6_-_46293378 | 0.86 |
ENST00000330430.6
|
RCAN2
|
regulator of calcineurin 2 |
| chr14_-_57277178 | 0.86 |
ENST00000339475.5
ENST00000554559.1 ENST00000555804.1 |
OTX2
|
orthodenticle homeobox 2 |
| chr8_+_85095553 | 0.86 |
ENST00000521268.1
|
RALYL
|
RALY RNA binding protein-like |
| chr10_+_18240814 | 0.85 |
ENST00000377374.4
|
SLC39A12
|
solute carrier family 39 (zinc transporter), member 12 |
| chr15_+_48051920 | 0.85 |
ENST00000559196.1
|
SEMA6D
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D |
| chr2_-_77749446 | 0.84 |
ENST00000409911.1
|
LRRTM4
|
leucine rich repeat transmembrane neuronal 4 |
| chr12_-_16760021 | 0.84 |
ENST00000540445.1
|
LMO3
|
LIM domain only 3 (rhombotin-like 2) |
| chr3_+_43020988 | 0.83 |
ENST00000430121.2
|
FAM198A
|
family with sequence similarity 198, member A |
| chrX_-_117119243 | 0.82 |
ENST00000539496.1
ENST00000469946.1 |
KLHL13
|
kelch-like family member 13 |
| chr11_+_12766583 | 0.82 |
ENST00000361985.2
|
TEAD1
|
TEA domain family member 1 (SV40 transcriptional enhancer factor) |
| chr7_-_27219849 | 0.82 |
ENST00000396344.4
|
HOXA10
|
homeobox A10 |
| chr3_+_158787041 | 0.81 |
ENST00000471575.1
ENST00000476809.1 ENST00000485419.1 |
IQCJ-SCHIP1
|
IQCJ-SCHIP1 readthrough |
| chr1_+_164528616 | 0.80 |
ENST00000340699.3
|
PBX1
|
pre-B-cell leukemia homeobox 1 |
| chr11_-_9025541 | 0.80 |
ENST00000525100.1
ENST00000309166.3 ENST00000531090.1 |
NRIP3
|
nuclear receptor interacting protein 3 |
| chr8_+_104892639 | 0.80 |
ENST00000436393.2
|
RIMS2
|
regulating synaptic membrane exocytosis 2 |
| chr18_-_31802056 | 0.80 |
ENST00000538587.1
|
NOL4
|
nucleolar protein 4 |
| chr12_-_24103841 | 0.80 |
ENST00000541847.1
|
SOX5
|
SRY (sex determining region Y)-box 5 |
| chr7_-_84122033 | 0.80 |
ENST00000424555.1
|
SEMA3A
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chr7_-_83824169 | 0.80 |
ENST00000265362.4
|
SEMA3A
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chr11_-_27723158 | 0.79 |
ENST00000395980.2
|
BDNF
|
brain-derived neurotrophic factor |
| chr12_-_88974236 | 0.78 |
ENST00000228280.5
ENST00000552044.1 ENST00000357116.4 |
KITLG
|
KIT ligand |
| chr18_-_31802282 | 0.77 |
ENST00000535475.1
|
NOL4
|
nucleolar protein 4 |
| chr13_-_84456527 | 0.77 |
ENST00000377084.2
|
SLITRK1
|
SLIT and NTRK-like family, member 1 |
| chr1_-_116311323 | 0.77 |
ENST00000456138.2
|
CASQ2
|
calsequestrin 2 (cardiac muscle) |
| chr21_+_34398153 | 0.76 |
ENST00000382357.3
ENST00000430860.1 ENST00000333337.3 |
OLIG2
|
oligodendrocyte lineage transcription factor 2 |
| chr12_+_113229962 | 0.76 |
ENST00000553114.1
ENST00000548866.1 |
RPH3A
|
rabphilin 3A homolog (mouse) |
| chr13_-_67802549 | 0.75 |
ENST00000328454.5
ENST00000377865.2 |
PCDH9
|
protocadherin 9 |
| chr2_+_166152283 | 0.75 |
ENST00000375427.2
|
SCN2A
|
sodium channel, voltage-gated, type II, alpha subunit |
| chr4_-_174451370 | 0.75 |
ENST00000359562.4
|
HAND2
|
heart and neural crest derivatives expressed 2 |
| chr18_-_35145593 | 0.74 |
ENST00000334919.5
ENST00000591282.1 ENST00000588597.1 |
CELF4
|
CUGBP, Elav-like family member 4 |
| chr13_-_28545276 | 0.74 |
ENST00000381020.7
|
CDX2
|
caudal type homeobox 2 |
| chrX_-_55057403 | 0.74 |
ENST00000396198.3
ENST00000335854.4 ENST00000455688.1 ENST00000330807.5 |
ALAS2
|
aminolevulinate, delta-, synthase 2 |
| chr15_-_76304731 | 0.73 |
ENST00000394907.3
|
NRG4
|
neuregulin 4 |
| chr5_-_146258291 | 0.72 |
ENST00000394411.4
ENST00000453001.1 |
PPP2R2B
|
protein phosphatase 2, regulatory subunit B, beta |
| chrX_-_33146477 | 0.72 |
ENST00000378677.2
|
DMD
|
dystrophin |
| chr18_-_60505948 | 0.72 |
ENST00000593319.1
|
AC015989.2
|
AC015989.2 |
| chr15_+_90319557 | 0.72 |
ENST00000341735.3
|
MESP2
|
mesoderm posterior 2 homolog (mouse) |
| chr2_+_162272605 | 0.71 |
ENST00000389554.3
|
TBR1
|
T-box, brain, 1 |
| chr14_-_64846033 | 0.71 |
ENST00000556556.1
|
CTD-2555O16.1
|
CTD-2555O16.1 |
| chr12_-_16760195 | 0.71 |
ENST00000546281.1
ENST00000537757.1 |
LMO3
|
LIM domain only 3 (rhombotin-like 2) |
| chrX_+_100805496 | 0.71 |
ENST00000372829.3
|
ARMCX1
|
armadillo repeat containing, X-linked 1 |
| chr18_-_25616519 | 0.71 |
ENST00000399380.3
|
CDH2
|
cadherin 2, type 1, N-cadherin (neuronal) |
| chr1_-_112106578 | 0.70 |
ENST00000369717.4
|
ADORA3
|
adenosine A3 receptor |
| chr3_+_43020773 | 0.70 |
ENST00000488863.1
|
FAM198A
|
family with sequence similarity 198, member A |
| chrX_+_14547632 | 0.70 |
ENST00000218075.4
|
GLRA2
|
glycine receptor, alpha 2 |
| chr12_-_91572278 | 0.70 |
ENST00000425043.1
ENST00000420120.2 ENST00000441303.2 ENST00000456569.2 |
DCN
|
decorin |
| chr5_+_139505520 | 0.70 |
ENST00000333305.3
|
IGIP
|
IgA-inducing protein |
| chr3_-_148939835 | 0.70 |
ENST00000264613.6
|
CP
|
ceruloplasmin (ferroxidase) |
| chr7_+_129906660 | 0.69 |
ENST00000222481.4
|
CPA2
|
carboxypeptidase A2 (pancreatic) |
| chr4_-_41750922 | 0.69 |
ENST00000226382.2
|
PHOX2B
|
paired-like homeobox 2b |
| chrX_-_151619746 | 0.69 |
ENST00000370314.4
|
GABRA3
|
gamma-aminobutyric acid (GABA) A receptor, alpha 3 |
| chr5_+_131993856 | 0.69 |
ENST00000304506.3
|
IL13
|
interleukin 13 |
| chr5_+_140800638 | 0.69 |
ENST00000398587.2
ENST00000518882.1 |
PCDHGA11
|
protocadherin gamma subfamily A, 11 |
| chr15_-_37390482 | 0.68 |
ENST00000559085.1
ENST00000397624.3 |
MEIS2
|
Meis homeobox 2 |
| chr11_-_4719072 | 0.68 |
ENST00000396950.3
ENST00000532598.1 |
OR51E2
|
olfactory receptor, family 51, subfamily E, member 2 |
| chr12_-_48111295 | 0.67 |
ENST00000542202.1
|
ENDOU
|
endonuclease, polyU-specific |
| chr12_-_16759440 | 0.67 |
ENST00000537304.1
|
LMO3
|
LIM domain only 3 (rhombotin-like 2) |
| chr2_-_152830441 | 0.67 |
ENST00000534999.1
ENST00000397327.2 |
CACNB4
|
calcium channel, voltage-dependent, beta 4 subunit |
| chr4_-_107957454 | 0.67 |
ENST00000285311.3
|
DKK2
|
dickkopf WNT signaling pathway inhibitor 2 |
| chr5_+_140723601 | 0.67 |
ENST00000253812.6
|
PCDHGA3
|
protocadherin gamma subfamily A, 3 |
| chrX_-_62974941 | 0.66 |
ENST00000374872.1
ENST00000253401.6 ENST00000374870.4 |
ARHGEF9
|
Cdc42 guanine nucleotide exchange factor (GEF) 9 |
| chr5_-_115910630 | 0.66 |
ENST00000343348.6
|
SEMA6A
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A |
| chr9_-_133814455 | 0.66 |
ENST00000448616.1
|
FIBCD1
|
fibrinogen C domain containing 1 |
| chr18_+_34124507 | 0.66 |
ENST00000591635.1
|
FHOD3
|
formin homology 2 domain containing 3 |
| chr12_-_16761007 | 0.66 |
ENST00000354662.1
ENST00000441439.2 |
LMO3
|
LIM domain only 3 (rhombotin-like 2) |
| chr8_-_95220775 | 0.66 |
ENST00000441892.2
ENST00000521491.1 ENST00000027335.3 |
CDH17
|
cadherin 17, LI cadherin (liver-intestine) |
| chr5_-_160973649 | 0.64 |
ENST00000393959.1
ENST00000517547.1 |
GABRB2
|
gamma-aminobutyric acid (GABA) A receptor, beta 2 |
| chr17_-_37308824 | 0.64 |
ENST00000415163.1
ENST00000441877.1 ENST00000444911.2 |
PLXDC1
|
plexin domain containing 1 |
| chr9_-_124990680 | 0.64 |
ENST00000541397.2
ENST00000560485.1 |
LHX6
|
LIM homeobox 6 |
| chr9_-_13165457 | 0.64 |
ENST00000542239.1
ENST00000538841.1 ENST00000433359.2 |
MPDZ
|
multiple PDZ domain protein |
| chr10_-_21806759 | 0.63 |
ENST00000444772.3
|
SKIDA1
|
SKI/DACH domain containing 1 |
| chr12_-_16761117 | 0.63 |
ENST00000538051.1
|
LMO3
|
LIM domain only 3 (rhombotin-like 2) |
| chrX_-_114252193 | 0.63 |
ENST00000243213.1
|
IL13RA2
|
interleukin 13 receptor, alpha 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of CPEB1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.2 | GO:1903045 | neural crest cell migration involved in sympathetic nervous system development(GO:1903045) |
| 0.6 | 1.9 | GO:0033693 | neurofilament bundle assembly(GO:0033693) |
| 0.6 | 5.0 | GO:0097116 | gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 0.5 | 1.6 | GO:1903966 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.5 | 2.1 | GO:0030185 | nitric oxide transport(GO:0030185) |
| 0.5 | 6.0 | GO:0098814 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.5 | 1.4 | GO:1903413 | cellular response to bile acid(GO:1903413) response to iron ion starvation(GO:1990641) |
| 0.4 | 2.3 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 0.3 | 0.7 | GO:0021550 | medulla oblongata development(GO:0021550) |
| 0.3 | 4.4 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.3 | 0.9 | GO:2000502 | negative regulation of natural killer cell chemotaxis(GO:2000502) |
| 0.3 | 3.6 | GO:1902866 | regulation of retina development in camera-type eye(GO:1902866) |
| 0.3 | 1.2 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.3 | 0.9 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 0.3 | 4.5 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.3 | 0.8 | GO:0033024 | mast cell homeostasis(GO:0033023) mast cell apoptotic process(GO:0033024) regulation of mast cell apoptotic process(GO:0033025) regulation of mast cell proliferation(GO:0070666) positive regulation of mast cell proliferation(GO:0070668) |
| 0.3 | 3.9 | GO:0090129 | positive regulation of synapse maturation(GO:0090129) |
| 0.3 | 0.8 | GO:0021529 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.2 | 1.0 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.2 | 1.9 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 0.2 | 3.3 | GO:0008343 | adult feeding behavior(GO:0008343) |
| 0.2 | 0.9 | GO:0035262 | gonad morphogenesis(GO:0035262) |
| 0.2 | 0.7 | GO:1903465 | vacuolar phosphate transport(GO:0007037) positive regulation of mitotic cell cycle DNA replication(GO:1903465) positive regulation of parathyroid hormone secretion(GO:2000830) |
| 0.2 | 0.7 | GO:0033861 | negative regulation of NAD(P)H oxidase activity(GO:0033861) |
| 0.2 | 0.9 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.2 | 5.0 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.2 | 0.8 | GO:1905205 | positive regulation of connective tissue replacement(GO:1905205) |
| 0.2 | 0.7 | GO:0003219 | cardiac right ventricle formation(GO:0003219) |
| 0.2 | 0.5 | GO:0018969 | thiocyanate metabolic process(GO:0018969) |
| 0.2 | 2.3 | GO:0010578 | regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010578) positive regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010579) |
| 0.2 | 1.9 | GO:0070777 | gamma-aminobutyric acid biosynthetic process(GO:0009449) D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.2 | 0.7 | GO:1900133 | regulation of renin secretion into blood stream(GO:1900133) |
| 0.2 | 1.7 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 0.2 | 0.5 | GO:1990709 | presynaptic active zone organization(GO:1990709) |
| 0.2 | 1.3 | GO:2000543 | positive regulation of gastrulation(GO:2000543) |
| 0.2 | 0.6 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.2 | 1.7 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 0.2 | 1.1 | GO:1903385 | regulation of homophilic cell adhesion(GO:1903385) |
| 0.1 | 1.0 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.1 | 3.1 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.1 | 0.4 | GO:0006427 | histidyl-tRNA aminoacylation(GO:0006427) |
| 0.1 | 1.4 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.1 | 0.4 | GO:0090265 | response to high density lipoprotein particle(GO:0055099) positive regulation of immune complex clearance by monocytes and macrophages(GO:0090265) |
| 0.1 | 1.9 | GO:0032596 | protein transport into membrane raft(GO:0032596) |
| 0.1 | 0.4 | GO:0021779 | oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.1 | 1.8 | GO:0086029 | Purkinje myocyte to ventricular cardiac muscle cell signaling(GO:0086029) Purkinje myocyte to ventricular cardiac muscle cell communication(GO:0086068) |
| 0.1 | 0.7 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 0.1 | 0.9 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.1 | 1.1 | GO:2000821 | regulation of grooming behavior(GO:2000821) |
| 0.1 | 0.5 | GO:0097156 | fasciculation of motor neuron axon(GO:0097156) |
| 0.1 | 0.4 | GO:0010621 | negative regulation of transcription by transcription factor localization(GO:0010621) |
| 0.1 | 0.7 | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 0.1 | 1.5 | GO:0086073 | bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 0.1 | 0.3 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.1 | 0.6 | GO:0021764 | amygdala development(GO:0021764) |
| 0.1 | 0.3 | GO:0070781 | arginine biosynthetic process via ornithine(GO:0042450) response to biotin(GO:0070781) |
| 0.1 | 0.4 | GO:0036091 | positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 0.1 | 1.5 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.1 | 0.3 | GO:0061394 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) |
| 0.1 | 1.2 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.1 | 1.4 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 0.1 | 1.1 | GO:0050966 | detection of mechanical stimulus involved in sensory perception of pain(GO:0050966) |
| 0.1 | 0.4 | GO:1903860 | negative regulation of dendrite extension(GO:1903860) |
| 0.1 | 1.0 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.1 | 0.9 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.1 | 0.5 | GO:0071306 | cellular response to vitamin E(GO:0071306) |
| 0.1 | 0.7 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.1 | 1.2 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.1 | 0.9 | GO:0060842 | arterial endothelial cell differentiation(GO:0060842) |
| 0.1 | 0.3 | GO:0014859 | negative regulation of skeletal muscle cell proliferation(GO:0014859) negative regulation of skeletal muscle satellite cell proliferation(GO:1902723) |
| 0.1 | 1.0 | GO:0002091 | negative regulation of receptor internalization(GO:0002091) |
| 0.1 | 0.6 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.1 | 0.6 | GO:1903286 | regulation of potassium ion import(GO:1903286) |
| 0.1 | 1.8 | GO:0060746 | short-term memory(GO:0007614) maternal behavior(GO:0042711) parental behavior(GO:0060746) |
| 0.1 | 0.9 | GO:0050961 | detection of temperature stimulus involved in sensory perception(GO:0050961) detection of temperature stimulus involved in sensory perception of pain(GO:0050965) |
| 0.1 | 0.9 | GO:0021800 | cerebral cortex tangential migration(GO:0021800) |
| 0.1 | 1.6 | GO:0031629 | synaptic vesicle fusion to presynaptic active zone membrane(GO:0031629) vesicle fusion to plasma membrane(GO:0099500) |
| 0.1 | 0.3 | GO:0080154 | regulation of fertilization(GO:0080154) |
| 0.1 | 0.7 | GO:0060012 | synaptic transmission, glycinergic(GO:0060012) |
| 0.1 | 4.7 | GO:0010765 | positive regulation of sodium ion transport(GO:0010765) |
| 0.1 | 1.3 | GO:0001973 | adenosine receptor signaling pathway(GO:0001973) |
| 0.1 | 0.6 | GO:0043305 | negative regulation of mast cell degranulation(GO:0043305) |
| 0.1 | 1.2 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.1 | 0.8 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.1 | 0.5 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.1 | 0.4 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.1 | 1.4 | GO:1905145 | acetylcholine receptor signaling pathway(GO:0095500) postsynaptic signal transduction(GO:0098926) signal transduction involved in cellular response to ammonium ion(GO:1903831) response to acetylcholine(GO:1905144) cellular response to acetylcholine(GO:1905145) |
| 0.1 | 2.2 | GO:0034391 | smooth muscle cell apoptotic process(GO:0034390) regulation of smooth muscle cell apoptotic process(GO:0034391) |
| 0.1 | 0.2 | GO:0060399 | positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.1 | 1.1 | GO:0042532 | negative regulation of tyrosine phosphorylation of STAT protein(GO:0042532) |
| 0.1 | 4.0 | GO:0014047 | glutamate secretion(GO:0014047) |
| 0.1 | 1.4 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.1 | 0.7 | GO:0032364 | oxygen homeostasis(GO:0032364) |
| 0.1 | 0.3 | GO:0050747 | positive regulation of lipoprotein metabolic process(GO:0050747) |
| 0.1 | 1.3 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.1 | 1.4 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.1 | 0.5 | GO:0097084 | vascular smooth muscle cell development(GO:0097084) |
| 0.1 | 4.6 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.1 | 2.1 | GO:0035162 | embryonic hemopoiesis(GO:0035162) |
| 0.1 | 1.0 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.1 | 0.6 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.1 | 0.9 | GO:1902004 | positive regulation of beta-amyloid formation(GO:1902004) |
| 0.1 | 0.1 | GO:0005989 | lactose metabolic process(GO:0005988) lactose biosynthetic process(GO:0005989) |
| 0.0 | 1.2 | GO:0007039 | protein catabolic process in the vacuole(GO:0007039) |
| 0.0 | 2.2 | GO:0045744 | negative regulation of G-protein coupled receptor protein signaling pathway(GO:0045744) |
| 0.0 | 0.4 | GO:2000172 | regulation of branching morphogenesis of a nerve(GO:2000172) |
| 0.0 | 1.6 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.0 | 0.4 | GO:0035989 | tendon development(GO:0035989) |
| 0.0 | 1.8 | GO:0048665 | neuron fate specification(GO:0048665) |
| 0.0 | 0.8 | GO:0008627 | intrinsic apoptotic signaling pathway in response to osmotic stress(GO:0008627) |
| 0.0 | 0.4 | GO:0045653 | negative regulation of megakaryocyte differentiation(GO:0045653) |
| 0.0 | 0.2 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.0 | 0.2 | GO:0038031 | non-canonical Wnt signaling pathway via JNK cascade(GO:0038031) |
| 0.0 | 0.9 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.0 | 0.3 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) |
| 0.0 | 0.5 | GO:2000623 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 | 2.0 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.0 | 1.0 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.0 | 0.8 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.0 | 0.4 | GO:1903025 | regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903025) |
| 0.0 | 0.2 | GO:1903935 | response to sodium arsenite(GO:1903935) cellular response to sodium arsenite(GO:1903936) |
| 0.0 | 0.2 | GO:0097319 | fructose import(GO:0032445) carbohydrate import into cell(GO:0097319) carbohydrate import across plasma membrane(GO:0098704) fructose import across plasma membrane(GO:1990539) |
| 0.0 | 0.2 | GO:0040034 | regulation of development, heterochronic(GO:0040034) regulation of timing of cell differentiation(GO:0048505) |
| 0.0 | 0.2 | GO:0060686 | negative regulation of prostatic bud formation(GO:0060686) |
| 0.0 | 0.2 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.0 | 0.2 | GO:0043163 | cell envelope organization(GO:0043163) external encapsulating structure organization(GO:0045229) |
| 0.0 | 0.1 | GO:0070092 | regulation of glucagon secretion(GO:0070092) |
| 0.0 | 0.3 | GO:0035494 | SNARE complex disassembly(GO:0035494) |
| 0.0 | 0.9 | GO:0042994 | cytoplasmic sequestering of transcription factor(GO:0042994) |
| 0.0 | 0.4 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.1 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.0 | 0.7 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.9 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 3.4 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 1.4 | GO:0043278 | response to isoquinoline alkaloid(GO:0014072) response to morphine(GO:0043278) |
| 0.0 | 0.8 | GO:0060065 | uterus development(GO:0060065) |
| 0.0 | 1.0 | GO:0007184 | SMAD protein import into nucleus(GO:0007184) |
| 0.0 | 0.1 | GO:0072014 | proximal tubule development(GO:0072014) |
| 0.0 | 0.2 | GO:0033138 | positive regulation of peptidyl-serine phosphorylation(GO:0033138) |
| 0.0 | 1.6 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 1.6 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.9 | GO:0070884 | regulation of calcineurin-NFAT signaling cascade(GO:0070884) |
| 0.0 | 0.6 | GO:0050974 | detection of mechanical stimulus involved in sensory perception(GO:0050974) |
| 0.0 | 0.3 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 0.0 | 0.2 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.0 | 0.3 | GO:0035092 | sperm chromatin condensation(GO:0035092) |
| 0.0 | 0.5 | GO:0021535 | cell migration in hindbrain(GO:0021535) |
| 0.0 | 0.2 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.0 | 0.8 | GO:0014912 | negative regulation of smooth muscle cell migration(GO:0014912) |
| 0.0 | 0.7 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 0.3 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.0 | 0.2 | GO:0046325 | negative regulation of glucose import(GO:0046325) |
| 0.0 | 3.9 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.2 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.4 | GO:0042448 | progesterone metabolic process(GO:0042448) |
| 0.0 | 0.1 | GO:1904888 | cranial skeletal system development(GO:1904888) |
| 0.0 | 0.2 | GO:0034625 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.3 | GO:0072189 | ureter development(GO:0072189) |
| 0.0 | 0.8 | GO:0043268 | positive regulation of potassium ion transport(GO:0043268) |
| 0.0 | 1.8 | GO:1903779 | regulation of cardiac conduction(GO:1903779) |
| 0.0 | 0.1 | GO:0003051 | angiotensin-mediated drinking behavior(GO:0003051) positive regulation of gap junction assembly(GO:1903598) |
| 0.0 | 0.7 | GO:0090004 | positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.0 | 2.6 | GO:0060333 | interferon-gamma-mediated signaling pathway(GO:0060333) |
| 0.0 | 0.5 | GO:0071732 | cellular response to nitric oxide(GO:0071732) |
| 0.0 | 0.2 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.0 | 0.0 | GO:0072365 | regulation of cellular ketone metabolic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072365) |
| 0.0 | 0.3 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 0.5 | GO:1901522 | positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus(GO:1901522) |
| 0.0 | 0.2 | GO:0048245 | eosinophil chemotaxis(GO:0048245) |
| 0.0 | 0.4 | GO:0030500 | regulation of bone mineralization(GO:0030500) |
| 0.0 | 0.4 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.0 | 0.8 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.2 | GO:0007398 | ectoderm development(GO:0007398) |
| 0.0 | 0.4 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.4 | GO:0090263 | positive regulation of canonical Wnt signaling pathway(GO:0090263) |
| 0.0 | 0.3 | GO:1902166 | negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:1902166) |
| 0.0 | 0.3 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 0.5 | GO:0051602 | response to electrical stimulus(GO:0051602) |
| 0.0 | 0.1 | GO:0030728 | ovulation(GO:0030728) |
| 0.0 | 0.4 | GO:0003341 | cilium movement(GO:0003341) |
| 0.0 | 0.3 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 0.2 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 0.7 | GO:0008542 | visual learning(GO:0008542) |
| 0.0 | 0.3 | GO:0045104 | intermediate filament cytoskeleton organization(GO:0045104) |
| 0.0 | 0.4 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.0 | GO:1902102 | metaphase/anaphase transition of meiotic cell cycle(GO:0044785) regulation of metaphase/anaphase transition of meiotic cell cycle(GO:1902102) negative regulation of metaphase/anaphase transition of meiotic cell cycle(GO:1902103) regulation of meiotic chromosome separation(GO:1905132) negative regulation of meiotic chromosome separation(GO:1905133) |
| 0.0 | 0.0 | GO:0030222 | eosinophil differentiation(GO:0030222) |
| 0.0 | 0.6 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.0 | 0.1 | GO:2000609 | regulation of thyroid hormone generation(GO:2000609) |
| 0.0 | 0.3 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.2 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.0 | 0.3 | GO:0043968 | histone H2A acetylation(GO:0043968) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.9 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.3 | 2.3 | GO:0060201 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.2 | 3.1 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.2 | 0.8 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 0.2 | 0.8 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.2 | 1.1 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.2 | 0.6 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.1 | 10.0 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.1 | 5.7 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.1 | 0.8 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 0.1 | 0.4 | GO:0044299 | C-fiber(GO:0044299) |
| 0.1 | 1.9 | GO:0005883 | neurofilament(GO:0005883) |
| 0.1 | 0.3 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.1 | 0.9 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.1 | 0.3 | GO:0005584 | collagen type I trimer(GO:0005584) |
| 0.1 | 0.9 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.1 | 0.2 | GO:0033150 | cytoskeletal calyx(GO:0033150) |
| 0.1 | 1.8 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.1 | 0.8 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.1 | 1.4 | GO:0045179 | apical cortex(GO:0045179) |
| 0.1 | 1.6 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.1 | 1.0 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.1 | 0.7 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.1 | 0.9 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.1 | 6.6 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.1 | 0.3 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.1 | 0.9 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.5 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 11.4 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.3 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.4 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 0.6 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.5 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.0 | 0.5 | GO:0031906 | late endosome lumen(GO:0031906) |
| 0.0 | 0.6 | GO:0097025 | lateral loop(GO:0043219) MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 0.5 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 0.2 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.0 | 0.3 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.0 | 0.5 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.3 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.0 | 0.2 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.0 | 0.1 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.0 | 4.7 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 1.6 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 1.0 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 1.6 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 0.4 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.0 | 0.1 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 2.1 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 0.1 | GO:0005846 | nuclear cap binding complex(GO:0005846) |
| 0.0 | 1.9 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 1.4 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.5 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.3 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.0 | 1.0 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 1.3 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 1.8 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.0 | 1.7 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 0.0 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.0 | 1.2 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.4 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.0 | 1.3 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.0 | 0.5 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.3 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 2.8 | GO:0098793 | presynapse(GO:0098793) |
| 0.0 | 1.2 | GO:0014069 | postsynaptic density(GO:0014069) postsynaptic specialization(GO:0099572) |
| 0.0 | 0.4 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 8.2 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.0 | 3.4 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 0.2 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.2 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.0 | 2.8 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 0.6 | GO:0032420 | stereocilium(GO:0032420) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 8.9 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.4 | 4.3 | GO:1904315 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.3 | 1.4 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.3 | 2.9 | GO:0042835 | BRE binding(GO:0042835) |
| 0.3 | 1.2 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.3 | 0.8 | GO:0050429 | calcium-dependent phospholipase C activity(GO:0050429) |
| 0.3 | 0.8 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.3 | 1.3 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.2 | 0.7 | GO:0016749 | 5-aminolevulinate synthase activity(GO:0003870) N-succinyltransferase activity(GO:0016749) |
| 0.2 | 1.0 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 0.2 | 1.9 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.2 | 1.2 | GO:0047391 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) |
| 0.2 | 0.9 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.2 | 0.7 | GO:0015322 | proton-dependent oligopeptide secondary active transmembrane transporter activity(GO:0005427) secondary active oligopeptide transmembrane transporter activity(GO:0015322) |
| 0.2 | 2.3 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.2 | 2.0 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.2 | 1.9 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.2 | 1.6 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) |
| 0.2 | 0.5 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.2 | 0.5 | GO:0036393 | thiocyanate peroxidase activity(GO:0036393) |
| 0.2 | 4.1 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.2 | 0.9 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.2 | 1.3 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) |
| 0.2 | 1.2 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.2 | 0.5 | GO:0031531 | thyrotropin-releasing hormone receptor binding(GO:0031531) |
| 0.1 | 3.7 | GO:0019841 | retinol binding(GO:0019841) |
| 0.1 | 1.6 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 0.4 | GO:0004821 | histidine-tRNA ligase activity(GO:0004821) |
| 0.1 | 0.8 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.1 | 0.9 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.1 | 0.6 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.1 | 1.3 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.1 | 1.1 | GO:0052832 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.1 | 0.9 | GO:0015111 | iodide transmembrane transporter activity(GO:0015111) |
| 0.1 | 0.3 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.1 | 1.5 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 0.1 | 1.4 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.1 | 0.4 | GO:0030899 | calcium-dependent ATPase activity(GO:0030899) |
| 0.1 | 1.4 | GO:0051430 | corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.1 | 3.9 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.1 | 0.3 | GO:0004608 | phosphatidyl-N-methylethanolamine N-methyltransferase activity(GO:0000773) phosphatidylethanolamine N-methyltransferase activity(GO:0004608) phosphatidyl-N-dimethylethanolamine N-methyltransferase activity(GO:0080101) |
| 0.1 | 1.5 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.1 | 0.8 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.1 | 3.0 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.1 | 0.9 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.1 | 2.1 | GO:0042301 | phosphate ion binding(GO:0042301) |
| 0.1 | 0.3 | GO:0017098 | sulfonylurea receptor binding(GO:0017098) |
| 0.1 | 0.3 | GO:0061752 | telomeric repeat-containing RNA binding(GO:0061752) |
| 0.1 | 0.5 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 0.1 | 1.3 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.1 | 0.9 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.1 | 0.7 | GO:0000980 | RNA polymerase II distal enhancer sequence-specific DNA binding(GO:0000980) |
| 0.1 | 0.2 | GO:0072591 | citrate-L-glutamate ligase activity(GO:0072591) |
| 0.1 | 3.1 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 2.3 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.1 | 2.2 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.1 | 1.6 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.1 | 0.7 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.1 | 0.5 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.1 | 0.4 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.1 | 0.8 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.1 | 0.5 | GO:0016167 | glial cell-derived neurotrophic factor receptor activity(GO:0016167) |
| 0.1 | 1.8 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.1 | 1.3 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.1 | 2.1 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.2 | GO:0032564 | adenyl deoxyribonucleotide binding(GO:0032558) dATP binding(GO:0032564) |
| 0.0 | 1.0 | GO:0008061 | chitin binding(GO:0008061) |
| 0.0 | 0.7 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.0 | 0.6 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.7 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 1.3 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 1.5 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 1.8 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 0.7 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.0 | 0.2 | GO:0005353 | fructose transmembrane transporter activity(GO:0005353) |
| 0.0 | 0.6 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 0.5 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.0 | 0.4 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.8 | GO:0001134 | transcription factor activity, transcription factor recruiting(GO:0001134) |
| 0.0 | 0.2 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.0 | 1.0 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 1.1 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 1.6 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.2 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.0 | 4.9 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 0.2 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.0 | 0.7 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.2 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.1 | GO:0031728 | CCR3 chemokine receptor binding(GO:0031728) |
| 0.0 | 0.8 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.0 | 1.3 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.0 | 6.0 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 0.4 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.0 | 1.7 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.1 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.0 | 0.5 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.4 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.5 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.6 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.1 | GO:0047288 | monosialoganglioside sialyltransferase activity(GO:0047288) |
| 0.0 | 0.1 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.0 | 0.6 | GO:0030955 | potassium ion binding(GO:0030955) |
| 0.0 | 0.9 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.2 | GO:0102338 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 1.6 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.1 | GO:0004507 | steroid 11-beta-monooxygenase activity(GO:0004507) corticosterone 18-monooxygenase activity(GO:0047783) |
| 0.0 | 0.5 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 2.2 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 2.0 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.0 | 1.0 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 0.4 | GO:0016641 | oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor(GO:0016641) |
| 0.0 | 0.3 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 0.3 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.0 | 0.4 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.0 | 0.3 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.5 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 0.3 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.0 | 0.4 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.1 | GO:0019798 | procollagen-proline 4-dioxygenase activity(GO:0004656) procollagen-proline dioxygenase activity(GO:0019798) |
| 0.0 | 1.5 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.0 | 0.3 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.0 | PID FGF PATHWAY | FGF signaling pathway |
| 0.1 | 1.3 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.1 | 1.4 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 3.0 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.4 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 1.9 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.0 | 1.4 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 1.7 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 1.1 | ST GAQ PATHWAY | G alpha q Pathway |
| 0.0 | 0.7 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 2.5 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 1.5 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.8 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 1.2 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.4 | ST ADRENERGIC | Adrenergic Pathway |
| 0.0 | 1.0 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.0 | 0.7 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.5 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 0.5 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 1.6 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 3.7 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.6 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 0.6 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.5 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.9 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.9 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.4 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.0 | 0.7 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 1.0 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.7 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 0.5 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.7 | PID ATR PATHWAY | ATR signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 6.3 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.1 | 3.2 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.1 | 1.5 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.1 | 1.4 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.1 | 2.3 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.1 | 0.2 | REACTOME ADP SIGNALLING THROUGH P2RY12 | Genes involved in ADP signalling through P2Y purinoceptor 12 |
| 0.1 | 6.7 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 1.4 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.1 | 0.7 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.1 | 2.2 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 1.8 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 1.5 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.9 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.0 | 0.9 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 2.4 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 2.4 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.0 | 1.3 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 1.0 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 1.4 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 1.4 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 0.6 | REACTOME ACTIVATION OF KAINATE RECEPTORS UPON GLUTAMATE BINDING | Genes involved in Activation of Kainate Receptors upon glutamate binding |
| 0.0 | 1.3 | REACTOME REGULATION OF INSULIN SECRETION BY GLUCAGON LIKE PEPTIDE1 | Genes involved in Regulation of Insulin Secretion by Glucagon-like Peptide-1 |
| 0.0 | 1.4 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 1.7 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.7 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.9 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 0.3 | REACTOME NCAM SIGNALING FOR NEURITE OUT GROWTH | Genes involved in NCAM signaling for neurite out-growth |
| 0.0 | 0.5 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.2 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 2.3 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 1.0 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.3 | REACTOME PLC BETA MEDIATED EVENTS | Genes involved in PLC beta mediated events |
| 0.0 | 0.7 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.7 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.0 | 0.3 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.0 | 0.7 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 0.6 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.0 | 0.9 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.1 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.0 | 0.4 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.4 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.2 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.2 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.2 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |