Project
Illumina Body Map 2: averaged replicates
Navigation
Downloads
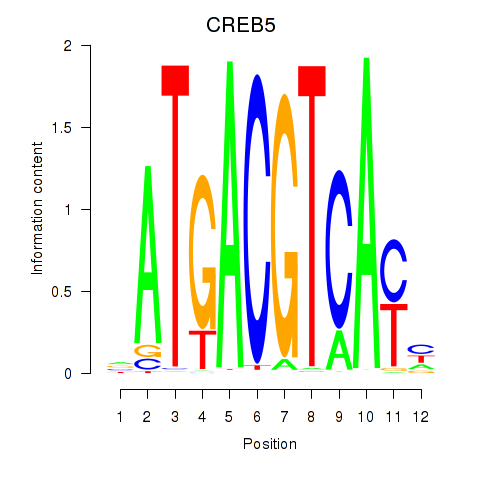
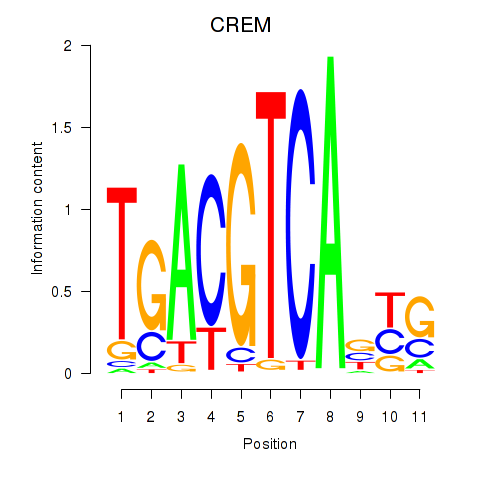
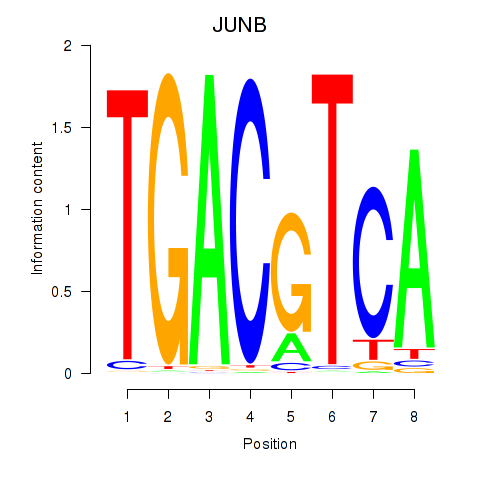
Results for CREB5_CREM_JUNB
Z-value: 2.72
Transcription factors associated with CREB5_CREM_JUNB
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
CREB5
|
ENSG00000146592.12 | cAMP responsive element binding protein 5 |
|
CREM
|
ENSG00000095794.15 | cAMP responsive element modulator |
|
JUNB
|
ENSG00000171223.4 | JunB proto-oncogene, AP-1 transcription factor subunit |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| CREB5 | hg19_v2_chr7_+_28448995_28449011 | 0.60 | 2.7e-04 | Click! |
| JUNB | hg19_v2_chr19_+_12902289_12902310 | -0.44 | 1.1e-02 | Click! |
| CREM | hg19_v2_chr10_+_35484053_35484076 | 0.41 | 1.8e-02 | Click! |
Activity profile of CREB5_CREM_JUNB motif
Sorted Z-values of CREB5_CREM_JUNB motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr22_-_44258360 | 25.18 |
ENST00000330884.4
ENST00000249130.5 |
SULT4A1
|
sulfotransferase family 4A, member 1 |
| chr22_-_44258280 | 21.31 |
ENST00000540422.1
|
SULT4A1
|
sulfotransferase family 4A, member 1 |
| chr20_+_17207636 | 11.37 |
ENST00000262545.2
|
PCSK2
|
proprotein convertase subtilisin/kexin type 2 |
| chr20_+_5892037 | 11.32 |
ENST00000378961.4
|
CHGB
|
chromogranin B (secretogranin 1) |
| chr20_+_33146510 | 11.23 |
ENST00000397709.1
|
MAP1LC3A
|
microtubule-associated protein 1 light chain 3 alpha |
| chr2_-_224467093 | 10.92 |
ENST00000305409.2
|
SCG2
|
secretogranin II |
| chr11_+_66059339 | 10.86 |
ENST00000327259.4
|
TMEM151A
|
transmembrane protein 151A |
| chr11_+_12696102 | 10.68 |
ENST00000527636.1
ENST00000527376.1 |
TEAD1
|
TEA domain family member 1 (SV40 transcriptional enhancer factor) |
| chr20_+_5892147 | 9.24 |
ENST00000455042.1
|
CHGB
|
chromogranin B (secretogranin 1) |
| chr20_+_17207665 | 9.21 |
ENST00000536609.1
|
PCSK2
|
proprotein convertase subtilisin/kexin type 2 |
| chr11_+_12695944 | 9.20 |
ENST00000361905.4
|
TEAD1
|
TEA domain family member 1 (SV40 transcriptional enhancer factor) |
| chr19_+_18723660 | 8.83 |
ENST00000262817.3
|
TMEM59L
|
transmembrane protein 59-like |
| chr16_+_29911864 | 8.36 |
ENST00000308748.5
|
ASPHD1
|
aspartate beta-hydroxylase domain containing 1 |
| chr22_+_31518938 | 8.15 |
ENST00000412985.1
ENST00000331075.5 ENST00000412277.2 ENST00000420017.1 ENST00000400294.2 ENST00000405300.1 ENST00000404390.3 |
INPP5J
|
inositol polyphosphate-5-phosphatase J |
| chr6_-_127780510 | 7.91 |
ENST00000487331.2
ENST00000483725.3 |
KIAA0408
|
KIAA0408 |
| chr4_-_186456652 | 7.78 |
ENST00000284767.5
ENST00000284770.5 |
PDLIM3
|
PDZ and LIM domain 3 |
| chr16_+_29911666 | 7.51 |
ENST00000563177.1
ENST00000483405.1 |
ASPHD1
|
aspartate beta-hydroxylase domain containing 1 |
| chr14_+_93389425 | 7.34 |
ENST00000216492.5
ENST00000334654.4 |
CHGA
|
chromogranin A (parathyroid secretory protein 1) |
| chr4_-_186456766 | 7.30 |
ENST00000284771.6
|
PDLIM3
|
PDZ and LIM domain 3 |
| chr11_-_1593150 | 7.13 |
ENST00000397374.3
|
DUSP8
|
dual specificity phosphatase 8 |
| chr8_-_27115903 | 7.07 |
ENST00000350889.4
ENST00000519997.1 ENST00000519614.1 ENST00000522908.1 ENST00000265770.7 |
STMN4
|
stathmin-like 4 |
| chr17_+_42385927 | 7.06 |
ENST00000426726.3
ENST00000590941.1 ENST00000225441.7 |
RUNDC3A
|
RUN domain containing 3A |
| chr7_+_30174668 | 6.67 |
ENST00000415604.1
|
C7orf41
|
maturin, neural progenitor differentiation regulator homolog (Xenopus) |
| chr8_-_27115931 | 6.47 |
ENST00000523048.1
|
STMN4
|
stathmin-like 4 |
| chr20_+_10199468 | 6.42 |
ENST00000254976.2
ENST00000304886.2 |
SNAP25
|
synaptosomal-associated protein, 25kDa |
| chr2_-_224467002 | 6.41 |
ENST00000421386.1
ENST00000433889.1 |
SCG2
|
secretogranin II |
| chr14_-_103989033 | 6.31 |
ENST00000553878.1
ENST00000557530.1 |
CKB
|
creatine kinase, brain |
| chrX_-_13956737 | 6.29 |
ENST00000454189.2
|
GPM6B
|
glycoprotein M6B |
| chr3_-_187388173 | 6.26 |
ENST00000287641.3
|
SST
|
somatostatin |
| chr20_+_10199566 | 6.10 |
ENST00000430336.1
|
SNAP25
|
synaptosomal-associated protein, 25kDa |
| chr16_+_58535372 | 5.94 |
ENST00000566656.1
ENST00000566618.1 |
NDRG4
|
NDRG family member 4 |
| chr14_+_101292445 | 5.93 |
ENST00000429159.2
ENST00000520714.1 ENST00000522771.2 ENST00000424076.3 ENST00000423456.1 ENST00000521404.1 ENST00000556736.1 ENST00000451743.2 ENST00000398518.2 ENST00000554639.1 ENST00000452120.2 ENST00000519709.1 ENST00000412736.2 |
MEG3
|
maternally expressed 3 (non-protein coding) |
| chr19_-_57352064 | 5.85 |
ENST00000326441.9
ENST00000593695.1 ENST00000599577.1 ENST00000594389.1 ENST00000423103.2 ENST00000598410.1 ENST00000593711.1 ENST00000391708.3 ENST00000221722.5 ENST00000599935.1 |
PEG3
ZIM2
|
paternally expressed 3 zinc finger, imprinted 2 |
| chr6_-_46293378 | 5.83 |
ENST00000330430.6
|
RCAN2
|
regulator of calcineurin 2 |
| chr2_-_241759622 | 5.77 |
ENST00000320389.7
ENST00000498729.2 |
KIF1A
|
kinesin family member 1A |
| chrX_-_13956497 | 5.75 |
ENST00000398361.3
|
GPM6B
|
glycoprotein M6B |
| chr7_+_30174426 | 5.73 |
ENST00000324453.8
|
C7orf41
|
maturin, neural progenitor differentiation regulator homolog (Xenopus) |
| chr6_+_56819773 | 5.72 |
ENST00000370750.2
|
BEND6
|
BEN domain containing 6 |
| chr11_+_109292846 | 5.70 |
ENST00000327419.6
|
C11orf87
|
chromosome 11 open reading frame 87 |
| chr4_+_85504075 | 5.68 |
ENST00000295887.5
|
CDS1
|
CDP-diacylglycerol synthase (phosphatidate cytidylyltransferase) 1 |
| chr1_+_99729813 | 5.38 |
ENST00000457765.1
|
LPPR4
|
Lipid phosphate phosphatase-related protein type 4 |
| chr4_-_156297949 | 5.38 |
ENST00000515654.1
|
MAP9
|
microtubule-associated protein 9 |
| chr6_+_26204825 | 5.37 |
ENST00000360441.4
|
HIST1H4E
|
histone cluster 1, H4e |
| chr7_+_30174574 | 5.24 |
ENST00000409688.1
|
C7orf41
|
maturin, neural progenitor differentiation regulator homolog (Xenopus) |
| chr5_-_138210977 | 5.10 |
ENST00000274711.6
ENST00000521094.2 |
LRRTM2
|
leucine rich repeat transmembrane neuronal 2 |
| chr2_+_149632783 | 4.98 |
ENST00000435030.1
|
KIF5C
|
kinesin family member 5C |
| chrX_+_152953505 | 4.96 |
ENST00000253122.5
|
SLC6A8
|
solute carrier family 6 (neurotransmitter transporter), member 8 |
| chr7_-_45128472 | 4.88 |
ENST00000490531.2
|
NACAD
|
NAC alpha domain containing |
| chr7_-_37956409 | 4.84 |
ENST00000436072.2
|
SFRP4
|
secreted frizzled-related protein 4 |
| chr3_+_158991025 | 4.84 |
ENST00000337808.6
|
IQCJ-SCHIP1
|
IQCJ-SCHIP1 readthrough |
| chr17_-_43210580 | 4.79 |
ENST00000538093.1
ENST00000590644.1 |
PLCD3
|
phospholipase C, delta 3 |
| chr10_-_98116912 | 4.65 |
ENST00000536387.1
|
OPALIN
|
oligodendrocytic myelin paranodal and inner loop protein |
| chr12_+_10365082 | 4.63 |
ENST00000545859.1
|
GABARAPL1
|
GABA(A) receptor-associated protein like 1 |
| chr7_-_31380502 | 4.61 |
ENST00000297142.3
|
NEUROD6
|
neuronal differentiation 6 |
| chr1_+_85527987 | 4.60 |
ENST00000326813.8
ENST00000294664.6 ENST00000528899.1 |
WDR63
|
WD repeat domain 63 |
| chr2_-_216300784 | 4.60 |
ENST00000421182.1
ENST00000432072.2 ENST00000323926.6 ENST00000336916.4 ENST00000357867.4 ENST00000359671.1 ENST00000346544.3 ENST00000345488.5 ENST00000357009.2 ENST00000446046.1 ENST00000356005.4 ENST00000443816.1 ENST00000426059.1 ENST00000354785.4 |
FN1
|
fibronectin 1 |
| chr2_+_11052054 | 4.59 |
ENST00000295082.1
|
KCNF1
|
potassium voltage-gated channel, subfamily F, member 1 |
| chr20_+_58179582 | 4.52 |
ENST00000371015.1
ENST00000395639.4 |
PHACTR3
|
phosphatase and actin regulator 3 |
| chr4_-_156297919 | 4.45 |
ENST00000450097.1
|
MAP9
|
microtubule-associated protein 9 |
| chr9_+_17579084 | 4.43 |
ENST00000380607.4
|
SH3GL2
|
SH3-domain GRB2-like 2 |
| chr6_+_56819895 | 4.42 |
ENST00000370748.3
|
BEND6
|
BEN domain containing 6 |
| chr2_-_220174166 | 4.33 |
ENST00000409251.3
ENST00000451506.1 ENST00000295718.2 ENST00000446182.1 |
PTPRN
|
protein tyrosine phosphatase, receptor type, N |
| chr16_+_56225248 | 4.30 |
ENST00000262493.6
|
GNAO1
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide O |
| chr11_-_119187826 | 4.26 |
ENST00000264036.4
|
MCAM
|
melanoma cell adhesion molecule |
| chr9_+_140032842 | 4.20 |
ENST00000371561.3
ENST00000315048.3 |
GRIN1
|
glutamate receptor, ionotropic, N-methyl D-aspartate 1 |
| chrX_-_47479246 | 4.20 |
ENST00000295987.7
ENST00000340666.4 |
SYN1
|
synapsin I |
| chr2_+_220408724 | 4.18 |
ENST00000421791.1
ENST00000373883.3 ENST00000451952.1 |
TMEM198
|
transmembrane protein 198 |
| chr14_+_67999999 | 4.14 |
ENST00000329153.5
|
PLEKHH1
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 1 |
| chr5_-_138211051 | 4.10 |
ENST00000518785.1
|
LRRTM2
|
leucine rich repeat transmembrane neuronal 2 |
| chr17_-_43209862 | 4.08 |
ENST00000322765.5
|
PLCD3
|
phospholipase C, delta 3 |
| chr6_-_99797522 | 4.05 |
ENST00000389677.5
|
FAXC
|
failed axon connections homolog (Drosophila) |
| chr15_-_30113676 | 4.04 |
ENST00000400011.2
|
TJP1
|
tight junction protein 1 |
| chr17_+_53342311 | 4.02 |
ENST00000226067.5
|
HLF
|
hepatic leukemia factor |
| chr3_+_159570722 | 4.02 |
ENST00000482804.1
|
SCHIP1
|
schwannomin interacting protein 1 |
| chr11_-_107582775 | 3.90 |
ENST00000305991.2
|
SLN
|
sarcolipin |
| chr5_-_132948216 | 3.85 |
ENST00000265342.7
|
FSTL4
|
follistatin-like 4 |
| chr4_+_113739244 | 3.85 |
ENST00000503271.1
ENST00000503423.1 ENST00000506722.1 |
ANK2
|
ankyrin 2, neuronal |
| chr4_-_156298087 | 3.83 |
ENST00000311277.4
|
MAP9
|
microtubule-associated protein 9 |
| chr2_-_220408260 | 3.83 |
ENST00000373891.2
|
CHPF
|
chondroitin polymerizing factor |
| chr1_-_86622421 | 3.73 |
ENST00000370571.2
|
COL24A1
|
collagen, type XXIV, alpha 1 |
| chr4_-_44450814 | 3.70 |
ENST00000360029.3
|
KCTD8
|
potassium channel tetramerization domain containing 8 |
| chr1_+_66999268 | 3.62 |
ENST00000371039.1
ENST00000424320.1 |
SGIP1
|
SH3-domain GRB2-like (endophilin) interacting protein 1 |
| chr3_-_100712352 | 3.61 |
ENST00000471714.1
ENST00000284322.5 |
ABI3BP
|
ABI family, member 3 (NESH) binding protein |
| chr19_-_50979981 | 3.58 |
ENST00000595790.1
ENST00000600100.1 |
FAM71E1
|
family with sequence similarity 71, member E1 |
| chr3_+_99536663 | 3.56 |
ENST00000421999.2
ENST00000463526.1 |
CMSS1
|
cms1 ribosomal small subunit homolog (yeast) |
| chr10_+_123923205 | 3.55 |
ENST00000369004.3
ENST00000260733.3 |
TACC2
|
transforming, acidic coiled-coil containing protein 2 |
| chr13_+_111972980 | 3.54 |
ENST00000283547.1
|
TEX29
|
testis expressed 29 |
| chr3_+_139654018 | 3.51 |
ENST00000458420.3
|
CLSTN2
|
calsyntenin 2 |
| chr10_+_123922941 | 3.48 |
ENST00000360561.3
|
TACC2
|
transforming, acidic coiled-coil containing protein 2 |
| chr16_+_22825475 | 3.47 |
ENST00000261374.3
|
HS3ST2
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 2 |
| chr10_-_90967063 | 3.46 |
ENST00000371852.2
|
CH25H
|
cholesterol 25-hydroxylase |
| chr12_+_12938541 | 3.42 |
ENST00000356591.4
|
APOLD1
|
apolipoprotein L domain containing 1 |
| chr6_-_3157760 | 3.40 |
ENST00000333628.3
|
TUBB2A
|
tubulin, beta 2A class IIa |
| chr10_+_123923105 | 3.39 |
ENST00000368999.1
|
TACC2
|
transforming, acidic coiled-coil containing protein 2 |
| chr12_+_5153085 | 3.38 |
ENST00000252321.3
|
KCNA5
|
potassium voltage-gated channel, shaker-related subfamily, member 5 |
| chr4_-_187647773 | 3.34 |
ENST00000509647.1
|
FAT1
|
FAT atypical cadherin 1 |
| chr8_-_42065075 | 3.31 |
ENST00000429089.2
ENST00000519510.1 ENST00000429710.2 ENST00000524009.1 |
PLAT
|
plasminogen activator, tissue |
| chr12_+_112856690 | 3.30 |
ENST00000392597.1
ENST00000351677.2 |
PTPN11
|
protein tyrosine phosphatase, non-receptor type 11 |
| chr10_+_22634384 | 3.30 |
ENST00000376624.3
ENST00000376603.2 ENST00000376601.1 ENST00000538630.1 ENST00000456231.2 ENST00000313311.6 ENST00000435326.1 |
SPAG6
|
sperm associated antigen 6 |
| chr17_+_41177220 | 3.29 |
ENST00000587250.2
ENST00000544533.1 |
RND2
|
Rho family GTPase 2 |
| chr22_+_41956767 | 3.29 |
ENST00000306149.7
|
CSDC2
|
cold shock domain containing C2, RNA binding |
| chr8_-_42065187 | 3.28 |
ENST00000270189.6
ENST00000352041.3 ENST00000220809.4 |
PLAT
|
plasminogen activator, tissue |
| chrX_+_47863734 | 3.26 |
ENST00000304355.5
|
SPACA5
|
sperm acrosome associated 5 |
| chr11_-_35547151 | 3.25 |
ENST00000378878.3
ENST00000529303.1 ENST00000278360.3 |
PAMR1
|
peptidase domain containing associated with muscle regeneration 1 |
| chr4_-_87281196 | 3.22 |
ENST00000359221.3
|
MAPK10
|
mitogen-activated protein kinase 10 |
| chr20_-_3154162 | 3.21 |
ENST00000360342.3
|
LZTS3
|
Homo sapiens leucine zipper, putative tumor suppressor family member 3 (LZTS3), mRNA. |
| chr1_+_151682909 | 3.18 |
ENST00000326413.3
ENST00000442233.2 |
RIIAD1
AL589765.1
|
regulatory subunit of type II PKA R-subunit (RIIa) domain containing 1 Uncharacterized protein; cDNA FLJ36032 fis, clone TESTI2017069 |
| chr15_-_63674034 | 3.15 |
ENST00000344366.3
ENST00000422263.2 |
CA12
|
carbonic anhydrase XII |
| chr12_-_100660833 | 3.14 |
ENST00000551642.1
ENST00000416321.1 ENST00000550587.1 ENST00000549249.1 |
DEPDC4
|
DEP domain containing 4 |
| chr4_-_156298028 | 3.13 |
ENST00000433024.1
ENST00000379248.2 |
MAP9
|
microtubule-associated protein 9 |
| chr14_+_63671105 | 3.09 |
ENST00000316754.3
|
RHOJ
|
ras homolog family member J |
| chr15_-_63674218 | 3.06 |
ENST00000178638.3
|
CA12
|
carbonic anhydrase XII |
| chr12_+_19389814 | 3.04 |
ENST00000536974.1
|
PLEKHA5
|
pleckstrin homology domain containing, family A member 5 |
| chr11_-_35547277 | 3.02 |
ENST00000527605.1
|
PAMR1
|
peptidase domain containing associated with muscle regeneration 1 |
| chrX_+_10126488 | 3.01 |
ENST00000380829.1
ENST00000421085.2 ENST00000454850.1 |
CLCN4
|
chloride channel, voltage-sensitive 4 |
| chr12_+_10365404 | 2.99 |
ENST00000266458.5
ENST00000421801.2 ENST00000544284.1 ENST00000545047.1 ENST00000543602.1 ENST00000545887.1 |
GABARAPL1
|
GABA(A) receptor-associated protein like 1 |
| chr2_-_183106641 | 2.98 |
ENST00000346717.4
|
PDE1A
|
phosphodiesterase 1A, calmodulin-dependent |
| chr8_-_95274536 | 2.97 |
ENST00000297596.2
ENST00000396194.2 |
GEM
|
GTP binding protein overexpressed in skeletal muscle |
| chr1_+_8378140 | 2.96 |
ENST00000377479.2
|
SLC45A1
|
solute carrier family 45, member 1 |
| chr3_-_100712292 | 2.96 |
ENST00000495063.1
ENST00000530539.1 |
ABI3BP
|
ABI family, member 3 (NESH) binding protein |
| chr11_+_117070037 | 2.95 |
ENST00000392951.4
ENST00000525531.1 ENST00000278968.6 |
TAGLN
|
transgelin |
| chr9_+_134165063 | 2.93 |
ENST00000372264.3
|
PPAPDC3
|
phosphatidic acid phosphatase type 2 domain containing 3 |
| chrX_-_13835461 | 2.92 |
ENST00000316715.4
ENST00000356942.5 |
GPM6B
|
glycoprotein M6B |
| chr13_-_29069232 | 2.91 |
ENST00000282397.4
ENST00000541932.1 ENST00000539099.1 |
FLT1
|
fms-related tyrosine kinase 1 |
| chr16_+_2039946 | 2.88 |
ENST00000248121.2
ENST00000568896.1 |
SYNGR3
|
synaptogyrin 3 |
| chr6_+_43211418 | 2.86 |
ENST00000259750.4
|
TTBK1
|
tau tubulin kinase 1 |
| chr3_-_124653579 | 2.86 |
ENST00000478191.1
ENST00000311075.3 |
MUC13
|
mucin 13, cell surface associated |
| chr3_+_8543533 | 2.85 |
ENST00000454244.1
|
LMCD1
|
LIM and cysteine-rich domains 1 |
| chr11_+_62475130 | 2.83 |
ENST00000294117.5
|
GNG3
|
guanine nucleotide binding protein (G protein), gamma 3 |
| chr19_-_5340730 | 2.83 |
ENST00000372412.4
ENST00000357368.4 ENST00000262963.6 ENST00000348075.2 ENST00000353284.2 |
PTPRS
|
protein tyrosine phosphatase, receptor type, S |
| chr1_+_29563011 | 2.83 |
ENST00000345512.3
ENST00000373779.3 ENST00000356870.3 ENST00000323874.8 ENST00000428026.2 ENST00000460170.2 |
PTPRU
|
protein tyrosine phosphatase, receptor type, U |
| chr4_+_145567297 | 2.82 |
ENST00000434550.2
|
HHIP
|
hedgehog interacting protein |
| chr22_+_38597889 | 2.81 |
ENST00000338483.2
ENST00000538320.1 ENST00000538999.1 ENST00000441709.1 |
MAFF
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog F |
| chr4_-_87281224 | 2.74 |
ENST00000395169.3
ENST00000395161.2 |
MAPK10
|
mitogen-activated protein kinase 10 |
| chr12_+_13044787 | 2.73 |
ENST00000534831.1
|
GPRC5A
|
G protein-coupled receptor, family C, group 5, member A |
| chrY_-_20935572 | 2.72 |
ENST00000382852.1
ENST00000344884.4 ENST00000304790.3 |
HSFY2
|
heat shock transcription factor, Y linked 2 |
| chrX_-_13835398 | 2.71 |
ENST00000475307.1
|
GPM6B
|
glycoprotein M6B |
| chr16_+_30034655 | 2.71 |
ENST00000300575.2
|
C16orf92
|
chromosome 16 open reading frame 92 |
| chr11_-_111782696 | 2.69 |
ENST00000227251.3
ENST00000526180.1 |
CRYAB
|
crystallin, alpha B |
| chr1_+_27719148 | 2.66 |
ENST00000374024.3
|
GPR3
|
G protein-coupled receptor 3 |
| chr7_+_151653464 | 2.66 |
ENST00000431418.2
ENST00000392800.2 |
GALNTL5
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase-like 5 |
| chr2_+_220283091 | 2.65 |
ENST00000373960.3
|
DES
|
desmin |
| chr5_+_71403061 | 2.65 |
ENST00000512974.1
ENST00000296755.7 |
MAP1B
|
microtubule-associated protein 1B |
| chr11_-_119234876 | 2.63 |
ENST00000525735.1
|
USP2
|
ubiquitin specific peptidase 2 |
| chr14_+_104095514 | 2.63 |
ENST00000348520.6
ENST00000380038.3 ENST00000389744.4 ENST00000557575.1 ENST00000553286.1 ENST00000347839.6 ENST00000555836.1 ENST00000334553.6 ENST00000246489.7 ENST00000557450.1 ENST00000452929.2 ENST00000554280.1 ENST00000445352.4 |
KLC1
|
kinesin light chain 1 |
| chr20_-_44420507 | 2.63 |
ENST00000243938.4
|
WFDC3
|
WAP four-disulfide core domain 3 |
| chr9_+_137967268 | 2.59 |
ENST00000371799.4
ENST00000277415.11 |
OLFM1
|
olfactomedin 1 |
| chr11_-_123525289 | 2.55 |
ENST00000392770.2
ENST00000299333.3 ENST00000530277.1 |
SCN3B
|
sodium channel, voltage-gated, type III, beta subunit |
| chr11_-_31839422 | 2.54 |
ENST00000423822.2
|
PAX6
|
paired box 6 |
| chrX_+_152338301 | 2.54 |
ENST00000453825.2
|
PNMA6A
|
paraneoplastic Ma antigen family member 6A |
| chr3_-_134092561 | 2.54 |
ENST00000510560.1
ENST00000504234.1 ENST00000515172.1 |
AMOTL2
|
angiomotin like 2 |
| chr14_+_23025534 | 2.52 |
ENST00000557595.1
|
AE000662.92
|
Uncharacterized protein |
| chrX_+_110339439 | 2.51 |
ENST00000372010.1
ENST00000519681.1 ENST00000372007.5 |
PAK3
|
p21 protein (Cdc42/Rac)-activated kinase 3 |
| chrX_+_152224766 | 2.51 |
ENST00000370265.4
ENST00000447306.1 |
PNMA3
|
paraneoplastic Ma antigen 3 |
| chr12_+_71833550 | 2.49 |
ENST00000266674.5
|
LGR5
|
leucine-rich repeat containing G protein-coupled receptor 5 |
| chr11_+_369804 | 2.48 |
ENST00000329962.6
|
B4GALNT4
|
beta-1,4-N-acetyl-galactosaminyl transferase 4 |
| chr3_+_62304712 | 2.46 |
ENST00000494481.1
|
C3orf14
|
chromosome 3 open reading frame 14 |
| chr11_-_111782484 | 2.46 |
ENST00000533971.1
|
CRYAB
|
crystallin, alpha B |
| chr15_-_83240553 | 2.45 |
ENST00000423133.2
ENST00000398591.2 |
CPEB1
|
cytoplasmic polyadenylation element binding protein 1 |
| chr8_+_98788057 | 2.44 |
ENST00000517924.1
|
LAPTM4B
|
lysosomal protein transmembrane 4 beta |
| chr9_+_111624577 | 2.42 |
ENST00000333999.3
|
ACTL7A
|
actin-like 7A |
| chr3_+_158288999 | 2.42 |
ENST00000482628.1
ENST00000478894.2 ENST00000392822.3 ENST00000466246.1 |
MLF1
|
myeloid leukemia factor 1 |
| chr4_+_145567173 | 2.41 |
ENST00000296575.3
|
HHIP
|
hedgehog interacting protein |
| chrX_-_13835147 | 2.40 |
ENST00000493677.1
ENST00000355135.2 |
GPM6B
|
glycoprotein M6B |
| chr11_-_123525648 | 2.38 |
ENST00000527836.1
|
SCN3B
|
sodium channel, voltage-gated, type III, beta subunit |
| chrX_-_152245978 | 2.37 |
ENST00000538162.2
|
PNMA6D
|
paraneoplastic Ma antigen family member 6D (pseudogene) |
| chr13_-_36429763 | 2.37 |
ENST00000379893.1
|
DCLK1
|
doublecortin-like kinase 1 |
| chr11_-_62457371 | 2.36 |
ENST00000317449.4
|
LRRN4CL
|
LRRN4 C-terminal like |
| chr8_-_67090825 | 2.35 |
ENST00000276571.3
|
CRH
|
corticotropin releasing hormone |
| chr19_-_52227221 | 2.35 |
ENST00000222115.1
ENST00000540069.2 |
HAS1
|
hyaluronan synthase 1 |
| chr7_+_28448995 | 2.35 |
ENST00000424599.1
|
CREB5
|
cAMP responsive element binding protein 5 |
| chr19_+_35849362 | 2.35 |
ENST00000327809.4
|
FFAR3
|
free fatty acid receptor 3 |
| chr2_-_151395525 | 2.35 |
ENST00000439275.1
|
RND3
|
Rho family GTPase 3 |
| chr3_+_158288960 | 2.33 |
ENST00000484955.1
ENST00000359117.5 ENST00000498592.1 ENST00000477042.1 ENST00000471745.1 ENST00000469452.1 |
MLF1
|
myeloid leukemia factor 1 |
| chr10_+_23983671 | 2.31 |
ENST00000376462.1
|
KIAA1217
|
KIAA1217 |
| chr5_+_71403280 | 2.31 |
ENST00000511641.2
|
MAP1B
|
microtubule-associated protein 1B |
| chr18_+_77867177 | 2.30 |
ENST00000560752.1
|
ADNP2
|
ADNP homeobox 2 |
| chr14_+_96671016 | 2.29 |
ENST00000542454.2
ENST00000554311.1 ENST00000306005.3 ENST00000539359.1 ENST00000553811.1 |
BDKRB2
RP11-404P21.8
|
bradykinin receptor B2 Uncharacterized protein |
| chr5_+_71014990 | 2.26 |
ENST00000296777.4
|
CARTPT
|
CART prepropeptide |
| chr1_-_242162375 | 2.23 |
ENST00000357246.3
|
MAP1LC3C
|
microtubule-associated protein 1 light chain 3 gamma |
| chr5_-_57756087 | 2.22 |
ENST00000274289.3
|
PLK2
|
polo-like kinase 2 |
| chr11_-_17555421 | 2.19 |
ENST00000526181.1
|
USH1C
|
Usher syndrome 1C (autosomal recessive, severe) |
| chr18_+_31185530 | 2.18 |
ENST00000586327.1
|
ASXL3
|
additional sex combs like 3 (Drosophila) |
| chr15_-_83240507 | 2.18 |
ENST00000564522.1
ENST00000398592.2 |
CPEB1
|
cytoplasmic polyadenylation element binding protein 1 |
| chr19_+_11649532 | 2.17 |
ENST00000252456.2
ENST00000592923.1 ENST00000535659.2 |
CNN1
|
calponin 1, basic, smooth muscle |
| chrX_-_31285042 | 2.16 |
ENST00000378680.2
ENST00000378723.3 |
DMD
|
dystrophin |
| chr14_-_93651186 | 2.15 |
ENST00000556883.1
ENST00000298894.4 |
MOAP1
|
modulator of apoptosis 1 |
| chr11_+_18343800 | 2.14 |
ENST00000453096.2
|
GTF2H1
|
general transcription factor IIH, polypeptide 1, 62kDa |
| chr12_+_56477093 | 2.14 |
ENST00000549672.1
ENST00000415288.2 |
ERBB3
|
v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 3 |
| chr6_+_108487245 | 2.14 |
ENST00000368986.4
|
NR2E1
|
nuclear receptor subfamily 2, group E, member 1 |
| chr6_-_56819385 | 2.14 |
ENST00000370754.5
ENST00000449297.2 |
DST
|
dystonin |
| chr3_+_158288942 | 2.10 |
ENST00000491767.1
ENST00000355893.5 |
MLF1
|
myeloid leukemia factor 1 |
| chr19_+_55795493 | 2.09 |
ENST00000309383.1
|
BRSK1
|
BR serine/threonine kinase 1 |
| chr3_+_8543393 | 2.09 |
ENST00000157600.3
ENST00000415597.1 ENST00000535732.1 |
LMCD1
|
LIM and cysteine-rich domains 1 |
| chr5_-_95768973 | 2.09 |
ENST00000311106.3
|
PCSK1
|
proprotein convertase subtilisin/kexin type 1 |
| chr5_-_147286065 | 2.08 |
ENST00000318315.4
ENST00000515291.1 |
C5orf46
|
chromosome 5 open reading frame 46 |
| chr11_-_35441524 | 2.08 |
ENST00000395750.1
ENST00000449068.1 |
SLC1A2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
| chr3_+_50712672 | 2.06 |
ENST00000266037.9
|
DOCK3
|
dedicator of cytokinesis 3 |
| chr12_-_16761007 | 2.06 |
ENST00000354662.1
ENST00000441439.2 |
LMO3
|
LIM domain only 3 (rhombotin-like 2) |
| chr1_-_102462565 | 2.06 |
ENST00000370103.4
|
OLFM3
|
olfactomedin 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of CREB5_CREM_JUNB
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.1 | 20.6 | GO:0030070 | insulin processing(GO:0030070) |
| 3.1 | 12.5 | GO:1990926 | short-term synaptic potentiation(GO:1990926) |
| 2.4 | 7.3 | GO:2000705 | dense core granule biogenesis(GO:0061110) positive regulation of relaxation of cardiac muscle(GO:1901899) regulation of dense core granule biogenesis(GO:2000705) |
| 2.3 | 11.5 | GO:0010040 | response to iron(II) ion(GO:0010040) |
| 2.0 | 2.0 | GO:1902081 | regulation of calcium ion import into sarcoplasmic reticulum(GO:1902080) negative regulation of calcium ion import into sarcoplasmic reticulum(GO:1902081) |
| 2.0 | 3.9 | GO:1901876 | regulation of calcium ion binding(GO:1901876) negative regulation of calcium ion binding(GO:1901877) |
| 1.8 | 20.1 | GO:0051612 | negative regulation of neurotransmitter uptake(GO:0051581) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 1.8 | 9.1 | GO:0008628 | hormone-mediated apoptotic signaling pathway(GO:0008628) |
| 1.7 | 5.0 | GO:1902598 | creatine transport(GO:0015881) creatine transmembrane transport(GO:1902598) |
| 1.6 | 4.9 | GO:1904235 | regulation of substrate-dependent cell migration, cell attachment to substrate(GO:1904235) positive regulation of substrate-dependent cell migration, cell attachment to substrate(GO:1904237) |
| 1.6 | 4.8 | GO:2000118 | regulation of sodium-dependent phosphate transport(GO:2000118) |
| 1.6 | 6.3 | GO:1990502 | dense core granule maturation(GO:1990502) |
| 1.5 | 6.0 | GO:1901350 | cell-cell signaling involved in cell-cell junction organization(GO:1901350) |
| 1.4 | 4.2 | GO:0043449 | cellular alkene metabolic process(GO:0043449) |
| 1.4 | 16.8 | GO:1902412 | regulation of mitotic cytokinesis(GO:1902412) |
| 1.4 | 12.5 | GO:0055064 | chloride ion homeostasis(GO:0055064) |
| 1.3 | 3.8 | GO:0031548 | regulation of brain-derived neurotrophic factor receptor signaling pathway(GO:0031548) |
| 1.1 | 2.3 | GO:0070093 | negative regulation of glucagon secretion(GO:0070093) |
| 1.0 | 17.3 | GO:0048245 | eosinophil chemotaxis(GO:0048245) |
| 1.0 | 46.5 | GO:0034035 | purine ribonucleoside bisphosphate metabolic process(GO:0034035) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) |
| 0.9 | 2.8 | GO:0034164 | negative regulation of toll-like receptor 9 signaling pathway(GO:0034164) |
| 0.9 | 3.6 | GO:0097156 | fasciculation of motor neuron axon(GO:0097156) |
| 0.8 | 3.3 | GO:0061582 | intestinal epithelial cell migration(GO:0061582) |
| 0.8 | 3.8 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.8 | 8.3 | GO:0086018 | SA node cell action potential(GO:0086015) SA node cell to atrial cardiac muscle cell signalling(GO:0086018) |
| 0.7 | 2.1 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 0.7 | 11.1 | GO:0002091 | negative regulation of receptor internalization(GO:0002091) |
| 0.6 | 0.6 | GO:1904668 | positive regulation of ubiquitin protein ligase activity(GO:1904668) |
| 0.6 | 1.9 | GO:0051795 | positive regulation of catagen(GO:0051795) activation of meiosis(GO:0090427) |
| 0.6 | 3.2 | GO:0060127 | subthalamic nucleus development(GO:0021763) prolactin secreting cell differentiation(GO:0060127) superior vena cava morphogenesis(GO:0060578) |
| 0.6 | 12.5 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.6 | 1.8 | GO:1990519 | pyrimidine nucleotide transport(GO:0006864) mitochondrial pyrimidine nucleotide import(GO:1990519) |
| 0.6 | 1.8 | GO:0021530 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.6 | 2.4 | GO:0046379 | extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.6 | 2.3 | GO:1902219 | negative regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902219) |
| 0.6 | 2.3 | GO:0002879 | positive regulation of acute inflammatory response to non-antigenic stimulus(GO:0002879) |
| 0.6 | 4.5 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 0.6 | 1.7 | GO:0003218 | cardiac left ventricle formation(GO:0003218) |
| 0.5 | 2.2 | GO:1904106 | protein localization to microvillus(GO:1904106) |
| 0.5 | 2.6 | GO:0035617 | stress granule disassembly(GO:0035617) |
| 0.5 | 1.5 | GO:1903660 | negative regulation of complement-dependent cytotoxicity(GO:1903660) |
| 0.5 | 2.0 | GO:0072600 | establishment of protein localization to Golgi(GO:0072600) |
| 0.5 | 2.0 | GO:0032223 | negative regulation of synaptic transmission, cholinergic(GO:0032223) neurotransmitter receptor biosynthetic process(GO:0045212) |
| 0.5 | 3.8 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.5 | 1.4 | GO:0061033 | secretion by lung epithelial cell involved in lung growth(GO:0061033) |
| 0.5 | 5.7 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.5 | 3.3 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.5 | 7.1 | GO:0031284 | positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.5 | 5.0 | GO:0070779 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.5 | 7.8 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.5 | 0.9 | GO:0033140 | negative regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033140) |
| 0.4 | 1.8 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) negative regulation of beta-amyloid clearance(GO:1900222) |
| 0.4 | 1.3 | GO:0050894 | determination of affect(GO:0050894) |
| 0.4 | 6.0 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.4 | 5.4 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.4 | 4.5 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.4 | 4.4 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.4 | 2.8 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.4 | 0.8 | GO:0032354 | response to follicle-stimulating hormone(GO:0032354) response to gonadotropin(GO:0034698) |
| 0.4 | 4.4 | GO:0003190 | atrioventricular valve formation(GO:0003190) |
| 0.4 | 2.8 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.4 | 1.4 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.3 | 1.7 | GO:1903644 | regulation of chaperone-mediated protein folding(GO:1903644) |
| 0.3 | 1.7 | GO:0002528 | regulation of vascular permeability involved in acute inflammatory response(GO:0002528) |
| 0.3 | 1.6 | GO:1903347 | negative regulation of bicellular tight junction assembly(GO:1903347) |
| 0.3 | 1.0 | GO:0061394 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) |
| 0.3 | 5.2 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.3 | 8.6 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.3 | 9.1 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.3 | 4.4 | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.3 | 5.9 | GO:0070884 | regulation of calcineurin-NFAT signaling cascade(GO:0070884) |
| 0.3 | 6.2 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.3 | 1.2 | GO:1900738 | positive regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900738) |
| 0.3 | 0.6 | GO:0014022 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.3 | 7.0 | GO:0060973 | cell migration involved in heart development(GO:0060973) |
| 0.3 | 20.4 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.3 | 1.1 | GO:0021538 | epithalamus development(GO:0021538) habenula development(GO:0021986) |
| 0.3 | 4.3 | GO:0061042 | vascular wound healing(GO:0061042) |
| 0.3 | 4.2 | GO:0097091 | synaptic vesicle clustering(GO:0097091) |
| 0.3 | 3.1 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.3 | 8.9 | GO:0060716 | labyrinthine layer blood vessel development(GO:0060716) |
| 0.3 | 0.8 | GO:1904781 | positive regulation of protein localization to centrosome(GO:1904781) |
| 0.3 | 4.3 | GO:0061299 | retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.3 | 1.1 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.3 | 0.8 | GO:0046601 | positive regulation of centriole replication(GO:0046601) |
| 0.3 | 2.6 | GO:0060744 | thelarche(GO:0042695) mammary gland branching involved in thelarche(GO:0060744) |
| 0.3 | 0.8 | GO:0097252 | oligodendrocyte apoptotic process(GO:0097252) |
| 0.3 | 3.5 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.2 | 1.7 | GO:1902410 | mitotic cytokinetic process(GO:1902410) |
| 0.2 | 1.0 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.2 | 5.0 | GO:0061162 | establishment of monopolar cell polarity(GO:0061162) establishment or maintenance of monopolar cell polarity(GO:0061339) |
| 0.2 | 0.2 | GO:0097535 | lymphoid lineage cell migration(GO:0097534) lymphoid lineage cell migration into thymus(GO:0097535) |
| 0.2 | 4.6 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.2 | 11.1 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.2 | 1.8 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.2 | 3.2 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.2 | 3.0 | GO:0035754 | B cell chemotaxis(GO:0035754) |
| 0.2 | 4.3 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.2 | 2.2 | GO:0061000 | negative regulation of dendritic spine development(GO:0061000) |
| 0.2 | 1.1 | GO:0030200 | heparan sulfate proteoglycan catabolic process(GO:0030200) |
| 0.2 | 1.3 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.2 | 0.9 | GO:0048691 | modulation by virus of host transcription(GO:0019056) positive regulation of sprouting of injured axon(GO:0048687) positive regulation of axon extension involved in regeneration(GO:0048691) modulation by symbiont of host transcription(GO:0052026) |
| 0.2 | 0.2 | GO:1904862 | inhibitory synapse assembly(GO:1904862) |
| 0.2 | 1.5 | GO:2000322 | regulation of glucocorticoid receptor signaling pathway(GO:2000322) negative regulation of glucocorticoid receptor signaling pathway(GO:2000323) |
| 0.2 | 0.6 | GO:0060304 | regulation of phosphatidylinositol dephosphorylation(GO:0060304) |
| 0.2 | 6.6 | GO:0031639 | plasminogen activation(GO:0031639) |
| 0.2 | 1.7 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) sphingolipid mediated signaling pathway(GO:0090520) |
| 0.2 | 0.8 | GO:0038169 | somatostatin receptor signaling pathway(GO:0038169) somatostatin signaling pathway(GO:0038170) |
| 0.2 | 3.1 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.2 | 11.2 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.2 | 6.0 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.2 | 2.0 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 0.2 | 1.0 | GO:0051892 | negative regulation of cardioblast differentiation(GO:0051892) regulation of cardiac muscle cell myoblast differentiation(GO:2000690) negative regulation of cardiac muscle cell myoblast differentiation(GO:2000691) |
| 0.2 | 3.3 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.2 | 2.4 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.2 | 1.5 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.2 | 2.2 | GO:0007042 | lysosomal lumen acidification(GO:0007042) |
| 0.2 | 0.5 | GO:0021934 | hindbrain tangential cell migration(GO:0021934) |
| 0.2 | 1.6 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.2 | 1.4 | GO:0098912 | membrane depolarization during atrial cardiac muscle cell action potential(GO:0098912) positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.2 | 5.3 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.2 | 0.5 | GO:0042214 | terpene metabolic process(GO:0042214) phytoalexin metabolic process(GO:0052314) |
| 0.2 | 1.4 | GO:0090238 | positive regulation of arachidonic acid secretion(GO:0090238) |
| 0.2 | 1.6 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.2 | 1.2 | GO:0046985 | negative regulation of megakaryocyte differentiation(GO:0045653) positive regulation of hemoglobin biosynthetic process(GO:0046985) |
| 0.2 | 1.9 | GO:0002329 | pre-B cell differentiation(GO:0002329) |
| 0.2 | 5.8 | GO:0030011 | maintenance of cell polarity(GO:0030011) |
| 0.2 | 1.5 | GO:0002318 | myeloid progenitor cell differentiation(GO:0002318) |
| 0.2 | 1.2 | GO:0045229 | cell envelope organization(GO:0043163) external encapsulating structure organization(GO:0045229) |
| 0.2 | 1.0 | GO:0003418 | growth plate cartilage chondrocyte differentiation(GO:0003418) |
| 0.2 | 4.3 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.2 | 0.9 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.2 | 0.6 | GO:1900619 | acetylcholine metabolic process(GO:0008291) acetate ester metabolic process(GO:1900619) |
| 0.2 | 3.7 | GO:0021542 | dentate gyrus development(GO:0021542) |
| 0.2 | 2.7 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.1 | 3.0 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.1 | 0.9 | GO:0014827 | intestine smooth muscle contraction(GO:0014827) |
| 0.1 | 6.2 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.1 | 0.7 | GO:0009257 | 10-formyltetrahydrofolate biosynthetic process(GO:0009257) |
| 0.1 | 2.5 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.1 | 1.8 | GO:1903546 | protein localization to photoreceptor outer segment(GO:1903546) |
| 0.1 | 1.0 | GO:0097056 | selenocysteinyl-tRNA(Sec) biosynthetic process(GO:0097056) |
| 0.1 | 3.3 | GO:0031573 | intra-S DNA damage checkpoint(GO:0031573) |
| 0.1 | 0.7 | GO:0044856 | plasma membrane raft distribution(GO:0044855) plasma membrane raft localization(GO:0044856) plasma membrane raft polarization(GO:0044858) regulation of plasma membrane raft polarization(GO:1903906) |
| 0.1 | 2.1 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.1 | 0.6 | GO:0060478 | acrosomal vesicle exocytosis(GO:0060478) |
| 0.1 | 0.5 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.1 | 5.2 | GO:0060441 | epithelial tube branching involved in lung morphogenesis(GO:0060441) |
| 0.1 | 0.3 | GO:0006700 | C21-steroid hormone biosynthetic process(GO:0006700) C21-steroid hormone metabolic process(GO:0008207) |
| 0.1 | 1.4 | GO:0009407 | toxin catabolic process(GO:0009407) secondary metabolite catabolic process(GO:0090487) |
| 0.1 | 1.0 | GO:0070317 | negative regulation of G0 to G1 transition(GO:0070317) |
| 0.1 | 2.9 | GO:0030949 | positive regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030949) |
| 0.1 | 2.7 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.1 | 1.8 | GO:0098870 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.1 | 0.6 | GO:0060474 | positive regulation of sperm motility involved in capacitation(GO:0060474) |
| 0.1 | 5.8 | GO:0008089 | anterograde axonal transport(GO:0008089) |
| 0.1 | 1.0 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.1 | 0.8 | GO:1903401 | L-lysine transmembrane transport(GO:1903401) |
| 0.1 | 0.4 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.1 | 0.5 | GO:0070173 | regulation of enamel mineralization(GO:0070173) |
| 0.1 | 0.9 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.1 | 0.3 | GO:0003099 | positive regulation of the force of heart contraction by chemical signal(GO:0003099) pilomotor reflex(GO:0097195) |
| 0.1 | 2.9 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.1 | 1.2 | GO:0045054 | constitutive secretory pathway(GO:0045054) |
| 0.1 | 1.4 | GO:0031268 | pseudopodium organization(GO:0031268) |
| 0.1 | 0.8 | GO:0033601 | positive regulation of mammary gland epithelial cell proliferation(GO:0033601) |
| 0.1 | 1.5 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.1 | 2.2 | GO:1904706 | negative regulation of vascular smooth muscle cell proliferation(GO:1904706) |
| 0.1 | 0.3 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.1 | 0.7 | GO:0006983 | ER overload response(GO:0006983) |
| 0.1 | 3.4 | GO:0006293 | nucleotide-excision repair, preincision complex stabilization(GO:0006293) |
| 0.1 | 1.0 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.1 | 2.8 | GO:0007567 | parturition(GO:0007567) |
| 0.1 | 1.0 | GO:0099624 | membrane repolarization during atrial cardiac muscle cell action potential(GO:0098914) atrial cardiac muscle cell membrane repolarization(GO:0099624) |
| 0.1 | 0.4 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.1 | 0.7 | GO:0015793 | glycerol transport(GO:0015793) |
| 0.1 | 2.3 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.1 | 1.2 | GO:0046855 | inositol phosphate dephosphorylation(GO:0046855) |
| 0.1 | 1.4 | GO:0060856 | establishment of blood-brain barrier(GO:0060856) |
| 0.1 | 0.4 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.1 | 0.3 | GO:0006429 | glutaminyl-tRNA aminoacylation(GO:0006425) leucyl-tRNA aminoacylation(GO:0006429) |
| 0.1 | 1.6 | GO:0050812 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) regulation of acyl-CoA biosynthetic process(GO:0050812) |
| 0.1 | 0.6 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.1 | 2.3 | GO:0061436 | establishment of skin barrier(GO:0061436) |
| 0.1 | 1.0 | GO:1904903 | ESCRT complex disassembly(GO:1904896) ESCRT III complex disassembly(GO:1904903) |
| 0.1 | 4.4 | GO:0043278 | response to isoquinoline alkaloid(GO:0014072) response to morphine(GO:0043278) |
| 0.1 | 0.3 | GO:0048808 | male genitalia morphogenesis(GO:0048808) male anatomical structure morphogenesis(GO:0090598) |
| 0.1 | 0.8 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.1 | 0.5 | GO:0060235 | lens induction in camera-type eye(GO:0060235) |
| 0.1 | 2.5 | GO:0061003 | positive regulation of dendritic spine morphogenesis(GO:0061003) |
| 0.1 | 0.5 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) |
| 0.1 | 0.8 | GO:0018094 | protein polyglycylation(GO:0018094) |
| 0.1 | 0.5 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.1 | 0.4 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.1 | 0.3 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.1 | 0.9 | GO:0016572 | histone phosphorylation(GO:0016572) |
| 0.1 | 1.5 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.1 | 1.0 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.1 | 1.0 | GO:1901096 | regulation of autophagosome maturation(GO:1901096) |
| 0.1 | 0.3 | GO:1902990 | leading strand elongation(GO:0006272) mitotic telomere maintenance via semi-conservative replication(GO:1902990) |
| 0.1 | 1.4 | GO:0030575 | nuclear body organization(GO:0030575) |
| 0.1 | 3.3 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.1 | 0.8 | GO:0070836 | caveola assembly(GO:0070836) |
| 0.1 | 0.1 | GO:0070434 | positive regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070426) positive regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070434) |
| 0.1 | 1.5 | GO:1904152 | regulation of retrograde protein transport, ER to cytosol(GO:1904152) |
| 0.1 | 0.7 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.1 | 9.9 | GO:0021987 | cerebral cortex development(GO:0021987) |
| 0.1 | 0.5 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.1 | 0.5 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.1 | 0.8 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.1 | 2.1 | GO:0090200 | positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.1 | 4.3 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.1 | 0.8 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.1 | 0.7 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.1 | 0.8 | GO:0006451 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.1 | 0.6 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.1 | 0.2 | GO:0010840 | regulation of circadian sleep/wake cycle, wakefulness(GO:0010840) circadian sleep/wake cycle, wakefulness(GO:0042746) |
| 0.1 | 0.6 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.1 | 0.6 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.1 | 1.8 | GO:0050974 | detection of mechanical stimulus involved in sensory perception(GO:0050974) |
| 0.1 | 0.3 | GO:0035751 | regulation of lysosomal lumen pH(GO:0035751) |
| 0.0 | 4.1 | GO:0032411 | positive regulation of transporter activity(GO:0032411) |
| 0.0 | 2.2 | GO:0010765 | positive regulation of sodium ion transport(GO:0010765) |
| 0.0 | 0.6 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.0 | 1.0 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.0 | 0.2 | GO:0018101 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.0 | 1.2 | GO:1904427 | positive regulation of calcium ion transmembrane transport(GO:1904427) |
| 0.0 | 3.0 | GO:0033344 | cholesterol efflux(GO:0033344) |
| 0.0 | 0.2 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.0 | 0.9 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.0 | 1.6 | GO:0007044 | cell-substrate junction assembly(GO:0007044) |
| 0.0 | 0.5 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 | 2.1 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.0 | 0.4 | GO:0001996 | positive regulation of heart rate by epinephrine-norepinephrine(GO:0001996) adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.0 | 2.5 | GO:0071377 | cellular response to glucagon stimulus(GO:0071377) |
| 0.0 | 1.1 | GO:0006206 | pyrimidine nucleobase metabolic process(GO:0006206) |
| 0.0 | 2.5 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 0.6 | GO:1900452 | regulation of long term synaptic depression(GO:1900452) |
| 0.0 | 0.7 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 0.4 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.0 | 0.1 | GO:1990637 | response to prolactin(GO:1990637) |
| 0.0 | 0.4 | GO:0015871 | choline transport(GO:0015871) |
| 0.0 | 0.9 | GO:0032401 | establishment of melanosome localization(GO:0032401) melanosome transport(GO:0032402) |
| 0.0 | 1.2 | GO:0048168 | regulation of neuronal synaptic plasticity(GO:0048168) |
| 0.0 | 0.8 | GO:0032703 | negative regulation of interleukin-2 production(GO:0032703) |
| 0.0 | 1.1 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 0.3 | GO:0045075 | interleukin-12 biosynthetic process(GO:0042090) regulation of interleukin-12 biosynthetic process(GO:0045075) |
| 0.0 | 0.9 | GO:0071480 | cellular response to gamma radiation(GO:0071480) |
| 0.0 | 1.0 | GO:0045744 | negative regulation of G-protein coupled receptor protein signaling pathway(GO:0045744) |
| 0.0 | 0.9 | GO:0000291 | nuclear-transcribed mRNA catabolic process, exonucleolytic(GO:0000291) exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.0 | 0.5 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.0 | 0.5 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.1 | GO:1902617 | response to fluoride(GO:1902617) |
| 0.0 | 0.3 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.0 | 2.6 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 1.7 | GO:0007189 | adenylate cyclase-activating G-protein coupled receptor signaling pathway(GO:0007189) |
| 0.0 | 2.0 | GO:0051298 | centrosome duplication(GO:0051298) |
| 0.0 | 0.4 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.0 | 0.1 | GO:0032747 | positive regulation of interleukin-23 production(GO:0032747) |
| 0.0 | 16.5 | GO:0043687 | post-translational protein modification(GO:0043687) |
| 0.0 | 3.2 | GO:0048813 | dendrite morphogenesis(GO:0048813) |
| 0.0 | 0.4 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.0 | 0.4 | GO:0006105 | succinate metabolic process(GO:0006105) |
| 0.0 | 0.8 | GO:0060706 | cell differentiation involved in embryonic placenta development(GO:0060706) |
| 0.0 | 0.4 | GO:0060965 | negative regulation of gene silencing by miRNA(GO:0060965) |
| 0.0 | 0.7 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.0 | 1.6 | GO:0071260 | cellular response to mechanical stimulus(GO:0071260) |
| 0.0 | 0.4 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.1 | GO:0051414 | response to cortisol(GO:0051414) |
| 0.0 | 0.2 | GO:0044375 | regulation of peroxisome size(GO:0044375) |
| 0.0 | 1.3 | GO:0003073 | regulation of systemic arterial blood pressure(GO:0003073) |
| 0.0 | 2.3 | GO:1903955 | positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.0 | 0.4 | GO:0099500 | synaptic vesicle fusion to presynaptic active zone membrane(GO:0031629) vesicle fusion to plasma membrane(GO:0099500) |
| 0.0 | 0.3 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.0 | 0.4 | GO:0014059 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.0 | 0.3 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 3.3 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 1.1 | GO:0021762 | substantia nigra development(GO:0021762) |
| 0.0 | 2.0 | GO:0010508 | positive regulation of autophagy(GO:0010508) |
| 0.0 | 1.2 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.0 | 0.2 | GO:0036123 | histone H3-K9 dimethylation(GO:0036123) |
| 0.0 | 1.7 | GO:0048010 | vascular endothelial growth factor receptor signaling pathway(GO:0048010) |
| 0.0 | 0.3 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 | 0.2 | GO:0071479 | cellular response to ionizing radiation(GO:0071479) |
| 0.0 | 0.6 | GO:0000187 | activation of MAPK activity(GO:0000187) |
| 0.0 | 0.3 | GO:0050995 | negative regulation of lipid catabolic process(GO:0050995) |
| 0.0 | 0.4 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.0 | 0.5 | GO:0014047 | glutamate secretion(GO:0014047) |
| 0.0 | 0.9 | GO:0030239 | myofibril assembly(GO:0030239) |
| 0.0 | 0.2 | GO:0045956 | positive regulation of calcium ion-dependent exocytosis(GO:0045956) |
| 0.0 | 0.3 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.0 | 0.0 | GO:0007518 | myoblast fate determination(GO:0007518) |
| 0.0 | 0.6 | GO:0042108 | positive regulation of cytokine biosynthetic process(GO:0042108) |
| 0.0 | 0.2 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 1.4 | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.0 | 0.4 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.0 | 0.8 | GO:0035690 | cellular response to drug(GO:0035690) |
| 0.0 | 0.2 | GO:0070987 | error-free translesion synthesis(GO:0070987) |
| 0.0 | 1.7 | GO:0008643 | carbohydrate transport(GO:0008643) |
| 0.0 | 1.3 | GO:0008277 | regulation of G-protein coupled receptor protein signaling pathway(GO:0008277) |
| 0.0 | 0.3 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 0.5 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.0 | 1.3 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.0 | 0.1 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) negative regulation of blood vessel remodeling(GO:0060313) |
| 0.0 | 0.2 | GO:0045821 | positive regulation of glycolytic process(GO:0045821) positive regulation of cofactor metabolic process(GO:0051194) positive regulation of coenzyme metabolic process(GO:0051197) |
| 0.0 | 0.1 | GO:0090141 | positive regulation of mitochondrial fission(GO:0090141) |
| 0.0 | 1.0 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) |
| 0.0 | 0.1 | GO:0050434 | positive regulation of viral transcription(GO:0050434) |
| 0.0 | 0.4 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.3 | 19.9 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 1.6 | 12.5 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 0.9 | 16.0 | GO:0005818 | astral microtubule(GO:0000235) aster(GO:0005818) |
| 0.7 | 2.2 | GO:0033150 | cytoskeletal calyx(GO:0033150) |
| 0.7 | 4.9 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.6 | 11.2 | GO:0044754 | autolysosome(GO:0044754) |
| 0.6 | 17.3 | GO:0031045 | dense core granule(GO:0031045) |
| 0.5 | 1.5 | GO:1990742 | microvesicle(GO:1990742) |
| 0.5 | 5.4 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.4 | 6.1 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.4 | 7.8 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.4 | 7.3 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.4 | 0.9 | GO:0043159 | acrosomal matrix(GO:0043159) |
| 0.4 | 2.1 | GO:0031673 | H zone(GO:0031673) |
| 0.4 | 1.4 | GO:0005873 | plus-end kinesin complex(GO:0005873) |
| 0.3 | 4.9 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.3 | 1.0 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 0.3 | 2.6 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.3 | 5.0 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.3 | 3.4 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.3 | 1.5 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 0.3 | 1.1 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.3 | 8.3 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.2 | 2.2 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.2 | 11.1 | GO:0032590 | dendrite membrane(GO:0032590) |
| 0.2 | 7.0 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.2 | 11.5 | GO:0030673 | axolemma(GO:0030673) |
| 0.2 | 4.2 | GO:0045180 | basal cortex(GO:0045180) |
| 0.2 | 3.8 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.2 | 6.3 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.2 | 1.1 | GO:0044393 | microspike(GO:0044393) |
| 0.2 | 38.5 | GO:0043204 | perikaryon(GO:0043204) |
| 0.2 | 1.7 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.2 | 1.5 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.2 | 4.9 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.2 | 0.9 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.1 | 0.3 | GO:0034272 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.1 | 2.4 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.1 | 6.0 | GO:0005921 | gap junction(GO:0005921) |
| 0.1 | 1.1 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.1 | 6.0 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.1 | 0.6 | GO:0002079 | inner acrosomal membrane(GO:0002079) outer acrosomal membrane(GO:0002081) |
| 0.1 | 4.3 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.1 | 5.9 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.1 | 2.0 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.1 | 1.0 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.1 | 8.7 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 6.0 | GO:0043034 | costamere(GO:0043034) |
| 0.1 | 0.8 | GO:0098536 | deuterosome(GO:0098536) |
| 0.1 | 9.0 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.1 | 1.4 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.1 | 0.4 | GO:0045257 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.1 | 1.6 | GO:0030478 | actin cap(GO:0030478) |
| 0.1 | 1.8 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.1 | 1.5 | GO:0034464 | BBSome(GO:0034464) |
| 0.1 | 3.1 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.1 | 21.1 | GO:0030018 | Z disc(GO:0030018) |
| 0.1 | 3.6 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.1 | 1.3 | GO:0030130 | clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.1 | 2.1 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 4.7 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.1 | 0.3 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 0.1 | 2.6 | GO:0005605 | basal lamina(GO:0005605) |
| 0.1 | 3.8 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.1 | 16.5 | GO:0030426 | growth cone(GO:0030426) |
| 0.1 | 3.7 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 1.4 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.1 | 8.8 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.1 | 0.7 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.1 | 26.5 | GO:0060076 | excitatory synapse(GO:0060076) |
| 0.1 | 0.4 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.1 | 0.3 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.1 | 4.4 | GO:0030669 | clathrin-coated endocytic vesicle membrane(GO:0030669) |
| 0.1 | 0.4 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.1 | 7.8 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.1 | 3.7 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.1 | 2.9 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.1 | 3.6 | GO:0044291 | cell-cell contact zone(GO:0044291) |
| 0.1 | 1.0 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.1 | 1.3 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.1 | 1.0 | GO:0031430 | M band(GO:0031430) |
| 0.1 | 0.4 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.1 | 1.0 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.1 | 6.4 | GO:0005776 | autophagosome(GO:0005776) |
| 0.1 | 1.8 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.1 | 24.7 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.1 | 0.5 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 1.0 | GO:0042627 | chylomicron(GO:0042627) |
| 0.0 | 4.7 | GO:0005930 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.0 | 0.5 | GO:0044354 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.0 | 4.1 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.0 | 4.7 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 3.1 | GO:0030286 | dynein complex(GO:0030286) |
| 0.0 | 5.5 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 2.7 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.0 | 0.8 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.0 | 1.4 | GO:1902711 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.0 | 3.2 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.4 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.0 | 0.4 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.6 | GO:0034709 | methylosome(GO:0034709) |
| 0.0 | 1.9 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.3 | GO:0098560 | cytoplasmic side of late endosome membrane(GO:0098560) |
| 0.0 | 2.5 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 3.9 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 1.4 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 0.1 | GO:0031429 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 0.0 | 0.6 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.0 | 1.4 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 1.1 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 0.8 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.2 | GO:0090543 | Flemming body(GO:0090543) |
| 0.0 | 3.5 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 1.5 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.2 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.9 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 5.3 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 0.2 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 0.4 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 6.3 | GO:0045177 | apical part of cell(GO:0045177) |
| 0.0 | 5.3 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 0.7 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.2 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.0 | 1.2 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.4 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.1 | GO:0032021 | NELF complex(GO:0032021) |
| 0.0 | 2.2 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 0.9 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.4 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.7 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 2.8 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.2 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.3 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 4.1 | GO:0030133 | transport vesicle(GO:0030133) |
| 0.0 | 1.6 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.1 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 0.9 | GO:0005875 | microtubule associated complex(GO:0005875) |
| 0.0 | 1.0 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.2 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.0 | 0.3 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.0 | 0.6 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 0.7 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 2.6 | GO:0045121 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 1.8 | GO:0005874 | microtubule(GO:0005874) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 5.0 | GO:0005308 | creatine transmembrane transporter activity(GO:0005308) |
| 1.6 | 46.5 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 1.4 | 5.7 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) phosphatidate cytidylyltransferase activity(GO:0004605) |
| 1.2 | 3.5 | GO:0004947 | bradykinin receptor activity(GO:0004947) |
| 0.8 | 5.9 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.8 | 7.1 | GO:0030250 | cyclase activator activity(GO:0010853) guanylate cyclase activator activity(GO:0030250) |
| 0.8 | 3.9 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.7 | 20.0 | GO:0001134 | transcription factor activity, transcription factor recruiting(GO:0001134) |
| 0.7 | 9.3 | GO:0052658 | inositol-polyphosphate 5-phosphatase activity(GO:0004445) inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) |
| 0.7 | 5.2 | GO:0097108 | hedgehog family protein binding(GO:0097108) |
| 0.6 | 6.3 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.6 | 10.6 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.6 | 1.8 | GO:0015218 | pyrimidine nucleotide transmembrane transporter activity(GO:0015218) |
| 0.6 | 5.6 | GO:1904315 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.5 | 4.9 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.5 | 1.5 | GO:0019948 | SUMO activating enzyme activity(GO:0019948) |
| 0.5 | 5.0 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.5 | 6.0 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.5 | 1.5 | GO:0045155 | electron transporter, transferring electrons from CoQH2-cytochrome c reductase complex and cytochrome c oxidase complex activity(GO:0045155) |
| 0.5 | 3.4 | GO:0086089 | voltage-gated potassium channel activity involved in atrial cardiac muscle cell action potential repolarization(GO:0086089) |
| 0.5 | 3.8 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) |
| 0.5 | 1.9 | GO:0031812 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 0.5 | 2.4 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.4 | 2.6 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.4 | 1.3 | GO:0031877 | somatostatin receptor binding(GO:0031877) |
| 0.4 | 2.2 | GO:0097677 | STAT family protein binding(GO:0097677) |
| 0.4 | 3.0 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.4 | 3.4 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.4 | 7.6 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.4 | 2.0 | GO:0003990 | acetylcholinesterase activity(GO:0003990) |
| 0.4 | 1.5 | GO:0002134 | pyrimidine nucleoside binding(GO:0001884) UTP binding(GO:0002134) sulfonylurea receptor binding(GO:0017098) pyrimidine ribonucleoside binding(GO:0032551) |
| 0.4 | 5.5 | GO:0051430 | corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.4 | 3.2 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.3 | 6.2 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.3 | 1.6 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.3 | 9.2 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.3 | 18.8 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.3 | 9.6 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.3 | 2.0 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.3 | 4.4 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.3 | 0.8 | GO:0043849 | Ras palmitoyltransferase activity(GO:0043849) |
| 0.3 | 3.6 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.3 | 0.5 | GO:0032558 | adenyl deoxyribonucleotide binding(GO:0032558) dATP binding(GO:0032564) |
| 0.3 | 1.0 | GO:0017129 | triglyceride binding(GO:0017129) |
| 0.2 | 13.8 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.2 | 1.4 | GO:0050610 | oxidoreductase activity, acting on a sulfur group of donors, quinone or similar compound as acceptor(GO:0016672) glutathione dehydrogenase (ascorbate) activity(GO:0045174) methylarsonate reductase activity(GO:0050610) |
| 0.2 | 6.7 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.2 | 3.3 | GO:0031748 | D1 dopamine receptor binding(GO:0031748) |
| 0.2 | 9.2 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.2 | 1.1 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.2 | 3.2 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.2 | 0.9 | GO:0004040 | amidase activity(GO:0004040) |
| 0.2 | 7.3 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.2 | 32.4 | GO:0005179 | hormone activity(GO:0005179) |
| 0.2 | 0.2 | GO:0001003 | polymerase III regulatory region sequence-specific DNA binding(GO:0000992) RNA polymerase III type 1 promoter sequence-specific DNA binding(GO:0001002) RNA polymerase III type 2 promoter sequence-specific DNA binding(GO:0001003) |
| 0.2 | 2.8 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.2 | 4.6 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.2 | 4.8 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.2 | 3.8 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.2 | 1.3 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.2 | 3.3 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.2 | 2.9 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.2 | 1.7 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.2 | 2.8 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.2 | 5.2 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.2 | 1.0 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.2 | 0.8 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.2 | 2.0 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.2 | 10.6 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.2 | 1.4 | GO:0044020 | histone methyltransferase activity (H4-R3 specific)(GO:0044020) |
| 0.2 | 1.3 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.2 | 1.4 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.2 | 0.6 | GO:0033265 | choline binding(GO:0033265) |
| 0.2 | 2.1 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.2 | 10.5 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.2 | 4.0 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.2 | 3.0 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.1 | 0.4 | GO:0008177 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) 3 iron, 4 sulfur cluster binding(GO:0051538) |
| 0.1 | 3.4 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.1 | 0.9 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.1 | 5.3 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.1 | 0.9 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.1 | 0.7 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.1 | 0.7 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 0.1 | 7.4 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.1 | 1.1 | GO:0061731 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.1 | 6.2 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.1 | 0.3 | GO:0010465 | neurotrophin receptor activity(GO:0005030) nerve growth factor receptor activity(GO:0010465) |
| 0.1 | 1.8 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.1 | 2.8 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.1 | 2.3 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.1 | 0.3 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.1 | 0.9 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.1 | 4.2 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.1 | 1.8 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.1 | 13.8 | GO:0005518 | collagen binding(GO:0005518) |
| 0.1 | 0.8 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
| 0.1 | 2.7 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.1 | 0.8 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.1 | 0.5 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.1 | 0.6 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.1 | 0.7 | GO:0070139 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.1 | 1.2 | GO:0043176 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.1 | 5.9 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.1 | 2.1 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.1 | 0.6 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.1 | 14.1 | GO:0051213 | dioxygenase activity(GO:0051213) |
| 0.1 | 0.5 | GO:0009019 | tRNA (guanine-N1-)-methyltransferase activity(GO:0009019) |
| 0.1 | 0.4 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.1 | 2.4 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.1 | 0.2 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.1 | 0.3 | GO:0004823 | glutamine-tRNA ligase activity(GO:0004819) leucine-tRNA ligase activity(GO:0004823) |
| 0.1 | 35.8 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.1 | 3.5 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.1 | 1.5 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 29.6 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 0.3 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.1 | 1.4 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.1 | 0.5 | GO:0034617 | tetrahydrobiopterin binding(GO:0034617) |
| 0.1 | 0.2 | GO:0090541 | MIT domain binding(GO:0090541) |
| 0.1 | 2.2 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.1 | 1.2 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.1 | 2.4 | GO:0046965 | retinoid X receptor binding(GO:0046965) |
| 0.1 | 0.8 | GO:0070735 | protein-glycine ligase activity(GO:0070735) |
| 0.1 | 2.0 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.1 | 0.3 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.1 | 5.9 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.1 | 11.6 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 0.7 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.1 | 1.9 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.1 | 0.7 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.1 | 0.6 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.1 | 7.3 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 2.2 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.1 | 1.5 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.1 | 0.9 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.1 | 1.5 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.1 | 1.6 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.1 | 0.4 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.1 | 1.3 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.1 | 1.1 | GO:0043236 | laminin binding(GO:0043236) |
| 0.1 | 2.6 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.1 | 0.4 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.1 | 0.7 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 1.5 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.8 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.4 | GO:0032138 | single base insertion or deletion binding(GO:0032138) |
| 0.0 | 1.9 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 2.1 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.2 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.0 | 0.2 | GO:0016499 | orexin receptor activity(GO:0016499) |
| 0.0 | 3.8 | GO:0004497 | monooxygenase activity(GO:0004497) |
| 0.0 | 1.5 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 2.5 | GO:0008376 | acetylgalactosaminyltransferase activity(GO:0008376) |
| 0.0 | 0.4 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 4.4 | GO:0015631 | tubulin binding(GO:0015631) |
| 0.0 | 0.2 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.0 | 3.0 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 4.8 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.0 | 4.1 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 1.2 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.2 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 0.0 | 3.5 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.0 | 0.8 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.9 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.0 | 0.7 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.1 | GO:0005260 | channel-conductance-controlling ATPase activity(GO:0005260) |
| 0.0 | 1.3 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.0 | 5.2 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 1.9 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.0 | 0.6 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 1.7 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 0.5 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.2 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) S-adenosyl-L-methionine binding(GO:1904047) |
| 0.0 | 6.7 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 0.7 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.9 | GO:0017022 | myosin binding(GO:0017022) |
| 0.0 | 1.2 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 1.7 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.8 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 0.5 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.5 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 2.0 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 0.4 | GO:0042166 | acetylcholine binding(GO:0042166) |
| 0.0 | 0.2 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 0.4 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.8 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.3 | GO:0043548 | phosphatidylinositol 3-kinase binding(GO:0043548) |
| 0.0 | 0.1 | GO:0034353 | RNA pyrophosphohydrolase activity(GO:0034353) |
| 0.0 | 0.4 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 0.7 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.0 | 0.2 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 12.0 | GO:0004930 | G-protein coupled receptor activity(GO:0004930) |
| 0.0 | 0.1 | GO:0030235 | nitric-oxide synthase regulator activity(GO:0030235) TPR domain binding(GO:0030911) |
| 0.0 | 0.3 | GO:0047617 | very long-chain fatty acid-CoA ligase activity(GO:0031957) acyl-CoA hydrolase activity(GO:0047617) |
| 0.0 | 0.4 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 2.8 | GO:0015293 | symporter activity(GO:0015293) |
| 0.0 | 0.7 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 1.3 | GO:0008013 | beta-catenin binding(GO:0008013) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.3 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.2 | 2.8 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.2 | 11.7 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.2 | 7.9 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.2 | 5.2 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.1 | 2.8 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.1 | 4.9 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.1 | 2.1 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.1 | 3.9 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.1 | 2.4 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.1 | 4.8 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.1 | 3.9 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.1 | 5.1 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.1 | 4.2 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.1 | 6.6 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.1 | 1.8 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.1 | 1.7 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.1 | 3.6 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.1 | 5.8 | SIG CHEMOTAXIS | Genes related to chemotaxis |
| 0.1 | 4.4 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.1 | 2.0 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.1 | 6.5 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.1 | 1.1 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.1 | 4.4 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.1 | 3.8 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.1 | 0.6 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.1 | 4.2 | PID INTEGRIN1 PATHWAY | Beta1 integrin cell surface interactions |
| 0.0 | 2.7 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.6 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 2.7 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.0 | 1.6 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 1.1 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 1.1 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 1.0 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 3.2 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 9.8 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 1.5 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 1.4 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 0.4 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 1.2 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 2.2 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.3 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 1.5 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 0.5 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.5 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 0.5 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
| 0.0 | 4.5 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 3.5 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 2.5 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 46.5 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.9 | 35.2 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.4 | 6.2 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.3 | 18.1 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.3 | 7.7 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.3 | 6.0 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.2 | 5.7 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.2 | 10.7 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.2 | 9.1 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.2 | 3.1 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.2 | 1.6 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.1 | 6.0 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.1 | 8.2 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.1 | 4.6 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.1 | 2.8 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.1 | 8.2 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 3.9 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.1 | 1.8 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.1 | 2.7 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.1 | 0.6 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.1 | 3.7 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.1 | 2.5 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.1 | 3.4 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.1 | 3.0 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS | Genes involved in Synthesis of bile acids and bile salts |
| 0.1 | 2.4 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.1 | 3.2 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.1 | 3.9 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.1 | 6.1 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.1 | 1.6 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.1 | 1.4 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.1 | 5.0 | REACTOME CA DEPENDENT EVENTS | Genes involved in Ca-dependent events |
| 0.1 | 2.2 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.1 | 1.7 | REACTOME SHC MEDIATED SIGNALLING | Genes involved in SHC-mediated signalling |
| 0.1 | 3.3 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.1 | 3.1 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.1 | 1.5 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.1 | 2.2 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.1 | 1.7 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.1 | 1.3 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.1 | 2.1 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.1 | 5.8 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.1 | 0.9 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.1 | 1.0 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.1 | 0.5 | REACTOME GLUCAGON SIGNALING IN METABOLIC REGULATION | Genes involved in Glucagon signaling in metabolic regulation |
| 0.1 | 4.9 | REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | Genes involved in Cell surface interactions at the vascular wall |
| 0.1 | 2.4 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 1.4 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 4.4 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 1.7 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.7 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 2.7 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 1.3 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 1.4 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |
| 0.0 | 2.2 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 1.1 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 1.0 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.9 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 1.8 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 2.5 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 9.8 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 2.4 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.8 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.4 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.6 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.0 | 0.5 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.9 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 1.0 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.0 | 2.6 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.9 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 1.7 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.0 | 0.3 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.0 | 1.0 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 1.1 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.3 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 0.4 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 4.9 | REACTOME G ALPHA Q SIGNALLING EVENTS | Genes involved in G alpha (q) signalling events |
| 0.0 | 0.8 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.0 | 0.4 | REACTOME CTLA4 INHIBITORY SIGNALING | Genes involved in CTLA4 inhibitory signaling |
| 0.0 | 1.1 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.4 | REACTOME REGULATION OF MRNA STABILITY BY PROTEINS THAT BIND AU RICH ELEMENTS | Genes involved in Regulation of mRNA Stability by Proteins that Bind AU-rich Elements |
| 0.0 | 1.8 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.7 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 1.6 | REACTOME SIGNALING BY PDGF | Genes involved in Signaling by PDGF |
| 0.0 | 0.1 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.5 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 0.3 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |