Project
Illumina Body Map 2: averaged replicates
Navigation
Downloads
Results for DLX3_EVX1_MEOX1
Z-value: 0.92
Transcription factors associated with DLX3_EVX1_MEOX1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
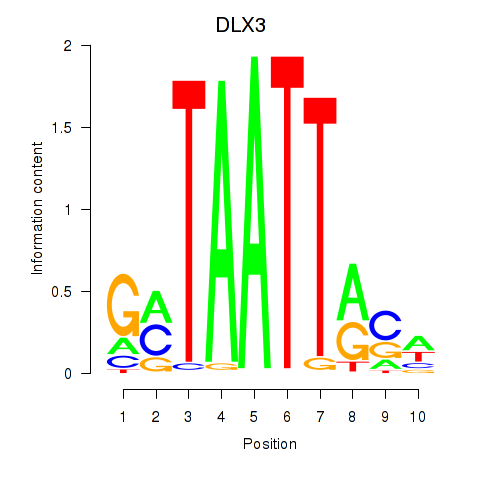
DLX3
|
ENSG00000064195.7 | distal-less homeobox 3 |
|
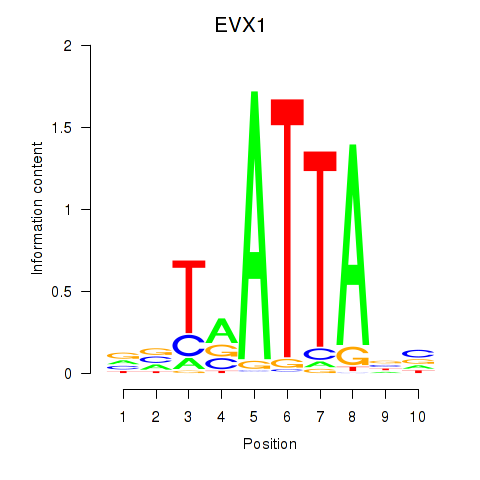
EVX1
|
ENSG00000106038.8 | even-skipped homeobox 1 |
|
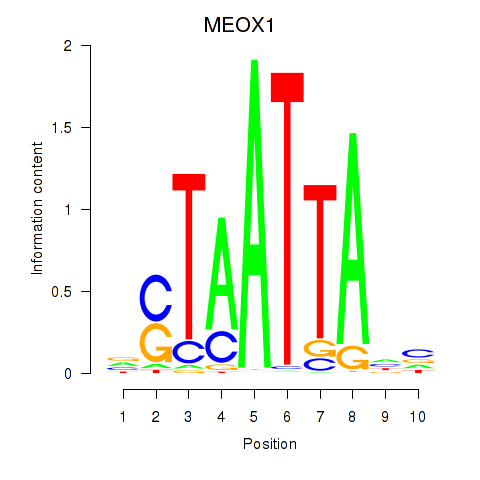
MEOX1
|
ENSG00000005102.8 | mesenchyme homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| MEOX1 | hg19_v2_chr17_-_41738931_41739040 | 0.21 | 2.4e-01 | Click! |
| EVX1 | hg19_v2_chr7_+_27282319_27282394 | -0.18 | 3.1e-01 | Click! |
| DLX3 | hg19_v2_chr17_-_48072574_48072597 | 0.04 | 8.1e-01 | Click! |
Activity profile of DLX3_EVX1_MEOX1 motif
Sorted Z-values of DLX3_EVX1_MEOX1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_6233828 | 1.40 |
ENST00000572068.1
ENST00000261405.5 |
VWF
|
von Willebrand factor |
| chr4_-_36245561 | 1.38 |
ENST00000506189.1
|
ARAP2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
| chr14_+_22748980 | 1.37 |
ENST00000390465.2
|
TRAV38-2DV8
|
T cell receptor alpha variable 38-2/delta variable 8 |
| chr5_-_115890554 | 1.34 |
ENST00000509665.1
|
SEMA6A
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A |
| chr7_-_111424462 | 1.18 |
ENST00000437129.1
|
DOCK4
|
dedicator of cytokinesis 4 |
| chr13_+_58206655 | 1.17 |
ENST00000377918.3
|
PCDH17
|
protocadherin 17 |
| chr6_+_151646800 | 1.14 |
ENST00000354675.6
|
AKAP12
|
A kinase (PRKA) anchor protein 12 |
| chr4_-_138453606 | 1.09 |
ENST00000412923.2
ENST00000344876.4 ENST00000507846.1 ENST00000510305.1 |
PCDH18
|
protocadherin 18 |
| chr7_-_111424506 | 1.07 |
ENST00000450156.1
ENST00000494651.2 |
DOCK4
|
dedicator of cytokinesis 4 |
| chr18_-_53019208 | 1.05 |
ENST00000562607.1
|
TCF4
|
transcription factor 4 |
| chr19_-_47137942 | 0.96 |
ENST00000300873.4
|
GNG8
|
guanine nucleotide binding protein (G protein), gamma 8 |
| chr7_-_99716914 | 0.90 |
ENST00000431404.2
|
TAF6
|
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa |
| chr13_-_46716969 | 0.90 |
ENST00000435666.2
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin) |
| chr4_-_109541610 | 0.89 |
ENST00000510212.1
|
RPL34-AS1
|
RPL34 antisense RNA 1 (head to head) |
| chr7_-_14029283 | 0.86 |
ENST00000433547.1
ENST00000405192.2 |
ETV1
|
ets variant 1 |
| chr15_-_64673630 | 0.85 |
ENST00000558008.1
ENST00000559519.1 ENST00000380258.2 |
KIAA0101
|
KIAA0101 |
| chr18_-_53255379 | 0.84 |
ENST00000565908.2
|
TCF4
|
transcription factor 4 |
| chr10_-_56561022 | 0.81 |
ENST00000373965.2
ENST00000414778.1 ENST00000395438.1 ENST00000409834.1 ENST00000395445.1 ENST00000395446.1 ENST00000395442.1 ENST00000395440.1 ENST00000395432.2 ENST00000361849.3 ENST00000395433.1 ENST00000320301.6 ENST00000395430.1 ENST00000437009.1 |
PCDH15
|
protocadherin-related 15 |
| chr3_+_111718036 | 0.77 |
ENST00000455401.2
|
TAGLN3
|
transgelin 3 |
| chr15_-_55562479 | 0.76 |
ENST00000564609.1
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr3_+_111717600 | 0.74 |
ENST00000273368.4
|
TAGLN3
|
transgelin 3 |
| chr3_+_111717511 | 0.74 |
ENST00000478951.1
ENST00000393917.2 |
TAGLN3
|
transgelin 3 |
| chr14_+_59100774 | 0.72 |
ENST00000556859.1
ENST00000421793.1 |
DACT1
|
dishevelled-binding antagonist of beta-catenin 1 |
| chr7_+_80255472 | 0.71 |
ENST00000428497.1
|
CD36
|
CD36 molecule (thrombospondin receptor) |
| chr13_-_36788718 | 0.70 |
ENST00000317764.6
ENST00000379881.3 |
SOHLH2
|
spermatogenesis and oogenesis specific basic helix-loop-helix 2 |
| chr17_-_46716647 | 0.69 |
ENST00000608940.1
|
RP11-357H14.17
|
RP11-357H14.17 |
| chr15_-_55562582 | 0.69 |
ENST00000396307.2
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chrX_+_99839799 | 0.65 |
ENST00000373031.4
|
TNMD
|
tenomodulin |
| chr8_-_82395461 | 0.65 |
ENST00000256104.4
|
FABP4
|
fatty acid binding protein 4, adipocyte |
| chr3_+_2933893 | 0.65 |
ENST00000397459.2
|
CNTN4
|
contactin 4 |
| chr6_+_130339710 | 0.63 |
ENST00000526087.1
ENST00000533560.1 ENST00000361794.2 |
L3MBTL3
|
l(3)mbt-like 3 (Drosophila) |
| chr3_+_115342349 | 0.61 |
ENST00000393780.3
|
GAP43
|
growth associated protein 43 |
| chrX_-_13835147 | 0.61 |
ENST00000493677.1
ENST00000355135.2 |
GPM6B
|
glycoprotein M6B |
| chr14_+_22739823 | 0.60 |
ENST00000390464.2
|
TRAV38-1
|
T cell receptor alpha variable 38-1 |
| chr15_-_55563072 | 0.60 |
ENST00000567380.1
ENST00000565972.1 ENST00000569493.1 |
RAB27A
|
RAB27A, member RAS oncogene family |
| chr18_-_44181442 | 0.60 |
ENST00000398722.4
|
LOXHD1
|
lipoxygenase homology domains 1 |
| chr22_+_22930626 | 0.58 |
ENST00000390302.2
|
IGLV2-33
|
immunoglobulin lambda variable 2-33 (non-functional) |
| chr17_-_38928414 | 0.58 |
ENST00000335552.4
|
KRT26
|
keratin 26 |
| chr13_+_110958124 | 0.58 |
ENST00000400163.2
|
COL4A2
|
collagen, type IV, alpha 2 |
| chr7_-_14028488 | 0.57 |
ENST00000405358.4
|
ETV1
|
ets variant 1 |
| chr14_-_36988882 | 0.55 |
ENST00000498187.2
|
NKX2-1
|
NK2 homeobox 1 |
| chr1_-_157746909 | 0.55 |
ENST00000392274.3
ENST00000361516.3 ENST00000368181.4 |
FCRL2
|
Fc receptor-like 2 |
| chrX_-_18690210 | 0.54 |
ENST00000379984.3
|
RS1
|
retinoschisin 1 |
| chr7_-_83824449 | 0.53 |
ENST00000420047.1
|
SEMA3A
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chr16_+_14280564 | 0.53 |
ENST00000572567.1
|
MKL2
|
MKL/myocardin-like 2 |
| chr7_+_80253387 | 0.52 |
ENST00000438020.1
|
CD36
|
CD36 molecule (thrombospondin receptor) |
| chr6_+_135502501 | 0.52 |
ENST00000527615.1
ENST00000420123.2 ENST00000525369.1 ENST00000528774.1 ENST00000534121.1 ENST00000534044.1 ENST00000533624.1 |
MYB
|
v-myb avian myeloblastosis viral oncogene homolog |
| chr3_+_111718173 | 0.52 |
ENST00000494932.1
|
TAGLN3
|
transgelin 3 |
| chr4_+_40198527 | 0.52 |
ENST00000381799.5
|
RHOH
|
ras homolog family member H |
| chr15_-_64673665 | 0.52 |
ENST00000300035.4
|
KIAA0101
|
KIAA0101 |
| chr8_-_10512569 | 0.51 |
ENST00000382483.3
|
RP1L1
|
retinitis pigmentosa 1-like 1 |
| chr7_-_83824169 | 0.51 |
ENST00000265362.4
|
SEMA3A
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chr15_-_55562451 | 0.50 |
ENST00000568803.1
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr1_+_202317815 | 0.50 |
ENST00000608999.1
ENST00000336894.4 ENST00000480184.1 |
PPP1R12B
|
protein phosphatase 1, regulatory subunit 12B |
| chr7_-_81399355 | 0.50 |
ENST00000457544.2
|
HGF
|
hepatocyte growth factor (hepapoietin A; scatter factor) |
| chr4_-_138453559 | 0.49 |
ENST00000511115.1
|
PCDH18
|
protocadherin 18 |
| chr6_-_32157947 | 0.49 |
ENST00000375050.4
|
PBX2
|
pre-B-cell leukemia homeobox 2 |
| chr1_-_21625486 | 0.47 |
ENST00000481130.2
|
ECE1
|
endothelin converting enzyme 1 |
| chr15_+_75080883 | 0.47 |
ENST00000567571.1
|
CSK
|
c-src tyrosine kinase |
| chr4_-_109541539 | 0.46 |
ENST00000509984.1
ENST00000507248.1 ENST00000506795.1 |
RPL34-AS1
|
RPL34 antisense RNA 1 (head to head) |
| chr10_+_118187379 | 0.46 |
ENST00000369230.3
|
PNLIPRP3
|
pancreatic lipase-related protein 3 |
| chr12_-_53045948 | 0.46 |
ENST00000309680.3
|
KRT2
|
keratin 2 |
| chr2_-_224467093 | 0.45 |
ENST00000305409.2
|
SCG2
|
secretogranin II |
| chr20_+_45947246 | 0.44 |
ENST00000599904.1
|
AL031666.2
|
HCG2018772; Uncharacterized protein; cDNA FLJ31609 fis, clone NT2RI2002852 |
| chr10_-_73848086 | 0.43 |
ENST00000536168.1
|
SPOCK2
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 2 |
| chr2_+_7865923 | 0.43 |
ENST00000417930.1
|
AC092580.4
|
AC092580.4 |
| chr3_-_3151664 | 0.42 |
ENST00000256452.3
ENST00000311981.8 ENST00000430514.2 ENST00000456302.1 |
IL5RA
|
interleukin 5 receptor, alpha |
| chr12_-_52995291 | 0.41 |
ENST00000293745.2
ENST00000354310.4 |
KRT72
|
keratin 72 |
| chr5_-_141249154 | 0.41 |
ENST00000357517.5
ENST00000536585.1 |
PCDH1
|
protocadherin 1 |
| chr5_+_173930676 | 0.41 |
ENST00000504512.1
|
RP11-267A15.1
|
RP11-267A15.1 |
| chr13_-_28545276 | 0.41 |
ENST00000381020.7
|
CDX2
|
caudal type homeobox 2 |
| chr2_+_169757750 | 0.40 |
ENST00000375363.3
ENST00000429379.2 ENST00000421979.1 |
G6PC2
|
glucose-6-phosphatase, catalytic, 2 |
| chr7_-_27169801 | 0.40 |
ENST00000511914.1
|
HOXA4
|
homeobox A4 |
| chr2_+_196313239 | 0.40 |
ENST00000413290.1
|
AC064834.1
|
AC064834.1 |
| chrX_+_135730297 | 0.40 |
ENST00000370629.2
|
CD40LG
|
CD40 ligand |
| chr1_+_198608146 | 0.38 |
ENST00000367376.2
ENST00000352140.3 ENST00000594404.1 ENST00000598951.1 ENST00000530727.1 ENST00000442510.2 ENST00000367367.4 ENST00000348564.6 ENST00000367364.1 ENST00000413409.2 |
PTPRC
|
protein tyrosine phosphatase, receptor type, C |
| chr1_-_232651312 | 0.38 |
ENST00000262861.4
|
SIPA1L2
|
signal-induced proliferation-associated 1 like 2 |
| chr6_+_127898312 | 0.38 |
ENST00000329722.7
|
C6orf58
|
chromosome 6 open reading frame 58 |
| chr4_-_25865159 | 0.38 |
ENST00000502949.1
ENST00000264868.5 ENST00000513691.1 ENST00000514872.1 |
SEL1L3
|
sel-1 suppressor of lin-12-like 3 (C. elegans) |
| chr11_-_33913708 | 0.37 |
ENST00000257818.2
|
LMO2
|
LIM domain only 2 (rhombotin-like 1) |
| chr1_+_160370344 | 0.36 |
ENST00000368061.2
|
VANGL2
|
VANGL planar cell polarity protein 2 |
| chr2_-_224467002 | 0.36 |
ENST00000421386.1
ENST00000433889.1 |
SCG2
|
secretogranin II |
| chr7_+_129906660 | 0.36 |
ENST00000222481.4
|
CPA2
|
carboxypeptidase A2 (pancreatic) |
| chr11_+_128563652 | 0.35 |
ENST00000527786.2
|
FLI1
|
Fli-1 proto-oncogene, ETS transcription factor |
| chr6_-_116833500 | 0.35 |
ENST00000356128.4
|
TRAPPC3L
|
trafficking protein particle complex 3-like |
| chr6_+_126221034 | 0.35 |
ENST00000433571.1
|
NCOA7
|
nuclear receptor coactivator 7 |
| chr1_-_183622442 | 0.34 |
ENST00000308641.4
|
APOBEC4
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 4 (putative) |
| chr13_-_30683005 | 0.34 |
ENST00000413591.1
ENST00000432770.1 |
LINC00365
|
long intergenic non-protein coding RNA 365 |
| chr7_+_99717230 | 0.34 |
ENST00000262932.3
|
CNPY4
|
canopy FGF signaling regulator 4 |
| chr9_-_75488984 | 0.34 |
ENST00000423171.1
ENST00000449235.1 ENST00000453787.1 |
RP11-151D14.1
|
RP11-151D14.1 |
| chr11_-_40315640 | 0.34 |
ENST00000278198.2
|
LRRC4C
|
leucine rich repeat containing 4C |
| chr3_+_2553281 | 0.33 |
ENST00000434053.1
|
CNTN4
|
contactin 4 |
| chr7_-_81399438 | 0.33 |
ENST00000222390.5
|
HGF
|
hepatocyte growth factor (hepapoietin A; scatter factor) |
| chr7_-_83278322 | 0.32 |
ENST00000307792.3
|
SEMA3E
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3E |
| chr11_-_107729887 | 0.32 |
ENST00000525815.1
|
SLC35F2
|
solute carrier family 35, member F2 |
| chr2_-_145188137 | 0.32 |
ENST00000440875.1
|
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr14_+_97925151 | 0.32 |
ENST00000554862.1
ENST00000554260.1 ENST00000499910.2 |
CTD-2506J14.1
|
CTD-2506J14.1 |
| chr6_-_76203454 | 0.31 |
ENST00000237172.7
|
FILIP1
|
filamin A interacting protein 1 |
| chr3_+_44840679 | 0.31 |
ENST00000425755.1
|
KIF15
|
kinesin family member 15 |
| chr6_-_138866823 | 0.30 |
ENST00000342260.5
|
NHSL1
|
NHS-like 1 |
| chr14_-_95236551 | 0.30 |
ENST00000238558.3
|
GSC
|
goosecoid homeobox |
| chr11_+_115498761 | 0.30 |
ENST00000424313.2
|
AP000997.1
|
AP000997.1 |
| chr6_-_116866773 | 0.29 |
ENST00000368602.3
|
TRAPPC3L
|
trafficking protein particle complex 3-like |
| chrX_-_100129128 | 0.29 |
ENST00000372960.4
ENST00000372964.1 ENST00000217885.5 |
NOX1
|
NADPH oxidase 1 |
| chr12_+_15699286 | 0.29 |
ENST00000442921.2
ENST00000542557.1 ENST00000445537.2 ENST00000544244.1 |
PTPRO
|
protein tyrosine phosphatase, receptor type, O |
| chr6_+_46761118 | 0.28 |
ENST00000230588.4
|
MEP1A
|
meprin A, alpha (PABA peptide hydrolase) |
| chr1_+_198608292 | 0.28 |
ENST00000418674.1
|
PTPRC
|
protein tyrosine phosphatase, receptor type, C |
| chr7_-_81399329 | 0.28 |
ENST00000453411.1
ENST00000444829.2 |
HGF
|
hepatocyte growth factor (hepapoietin A; scatter factor) |
| chr1_+_107683436 | 0.28 |
ENST00000370068.1
|
NTNG1
|
netrin G1 |
| chr11_-_107729287 | 0.28 |
ENST00000375682.4
|
SLC35F2
|
solute carrier family 35, member F2 |
| chr12_-_89746264 | 0.28 |
ENST00000548755.1
|
DUSP6
|
dual specificity phosphatase 6 |
| chr11_-_107729504 | 0.28 |
ENST00000265836.7
|
SLC35F2
|
solute carrier family 35, member F2 |
| chr3_-_123339343 | 0.28 |
ENST00000578202.1
|
MYLK
|
myosin light chain kinase |
| chrX_+_78426469 | 0.27 |
ENST00000276077.1
|
GPR174
|
G protein-coupled receptor 174 |
| chr14_-_92247032 | 0.27 |
ENST00000556661.1
ENST00000553676.1 ENST00000554560.1 |
CATSPERB
|
catsper channel auxiliary subunit beta |
| chr1_+_198607801 | 0.27 |
ENST00000367379.1
|
PTPRC
|
protein tyrosine phosphatase, receptor type, C |
| chr4_-_176812842 | 0.27 |
ENST00000507540.1
|
GPM6A
|
glycoprotein M6A |
| chr15_-_31393910 | 0.27 |
ENST00000397795.2
ENST00000256552.6 ENST00000559179.1 |
TRPM1
|
transient receptor potential cation channel, subfamily M, member 1 |
| chr3_-_108248169 | 0.27 |
ENST00000273353.3
|
MYH15
|
myosin, heavy chain 15 |
| chr6_-_31107127 | 0.26 |
ENST00000259845.4
|
PSORS1C2
|
psoriasis susceptibility 1 candidate 2 |
| chr12_-_68619586 | 0.26 |
ENST00000229134.4
|
IL26
|
interleukin 26 |
| chr2_-_203735484 | 0.26 |
ENST00000420558.1
ENST00000418208.1 |
ICA1L
|
islet cell autoantigen 1,69kDa-like |
| chr11_-_117748138 | 0.26 |
ENST00000527717.1
|
FXYD6
|
FXYD domain containing ion transport regulator 6 |
| chr14_+_62585332 | 0.25 |
ENST00000554895.1
|
LINC00643
|
long intergenic non-protein coding RNA 643 |
| chr12_-_89746173 | 0.25 |
ENST00000308385.6
|
DUSP6
|
dual specificity phosphatase 6 |
| chr7_+_50348268 | 0.25 |
ENST00000438033.1
ENST00000439701.1 |
IKZF1
|
IKAROS family zinc finger 1 (Ikaros) |
| chr11_-_117747607 | 0.25 |
ENST00000540359.1
ENST00000539526.1 |
FXYD6
|
FXYD domain containing ion transport regulator 6 |
| chr11_-_117747434 | 0.24 |
ENST00000529335.2
ENST00000530956.1 ENST00000260282.4 |
FXYD6
|
FXYD domain containing ion transport regulator 6 |
| chr1_+_107683644 | 0.24 |
ENST00000370067.1
|
NTNG1
|
netrin G1 |
| chr7_-_81399411 | 0.23 |
ENST00000423064.2
|
HGF
|
hepatocyte growth factor (hepapoietin A; scatter factor) |
| chr7_-_100860851 | 0.23 |
ENST00000223127.3
|
PLOD3
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 3 |
| chr7_-_14029515 | 0.23 |
ENST00000430479.1
ENST00000405218.2 ENST00000343495.5 |
ETV1
|
ets variant 1 |
| chr12_+_8666126 | 0.23 |
ENST00000299665.2
|
CLEC4D
|
C-type lectin domain family 4, member D |
| chr3_+_77088989 | 0.23 |
ENST00000461745.1
|
ROBO2
|
roundabout, axon guidance receptor, homolog 2 (Drosophila) |
| chr3_+_155755482 | 0.23 |
ENST00000472028.1
|
KCNAB1
|
potassium voltage-gated channel, shaker-related subfamily, beta member 1 |
| chr2_+_158114051 | 0.23 |
ENST00000259056.4
|
GALNT5
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 5 (GalNAc-T5) |
| chr14_+_22180536 | 0.23 |
ENST00000390424.2
|
TRAV2
|
T cell receptor alpha variable 2 |
| chr5_-_34043310 | 0.22 |
ENST00000231338.7
|
C1QTNF3
|
C1q and tumor necrosis factor related protein 3 |
| chr19_-_7968427 | 0.22 |
ENST00000539278.1
|
AC010336.1
|
Uncharacterized protein |
| chr12_-_94673956 | 0.22 |
ENST00000551941.1
|
RP11-1105G2.3
|
Uncharacterized protein |
| chr16_+_14280742 | 0.22 |
ENST00000341243.5
|
MKL2
|
MKL/myocardin-like 2 |
| chr13_-_30160925 | 0.22 |
ENST00000450494.1
|
SLC7A1
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 1 |
| chr10_-_56560939 | 0.22 |
ENST00000373955.1
|
PCDH15
|
protocadherin-related 15 |
| chr19_-_4902877 | 0.21 |
ENST00000381781.2
|
ARRDC5
|
arrestin domain containing 5 |
| chr4_-_120243545 | 0.21 |
ENST00000274024.3
|
FABP2
|
fatty acid binding protein 2, intestinal |
| chr4_-_46126093 | 0.21 |
ENST00000295452.4
|
GABRG1
|
gamma-aminobutyric acid (GABA) A receptor, gamma 1 |
| chr2_-_99279928 | 0.21 |
ENST00000414521.2
|
MGAT4A
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme A |
| chr17_-_54893250 | 0.21 |
ENST00000397862.2
|
C17orf67
|
chromosome 17 open reading frame 67 |
| chr16_-_67517716 | 0.20 |
ENST00000290953.2
|
AGRP
|
agouti related protein homolog (mouse) |
| chr18_-_56985776 | 0.20 |
ENST00000587244.1
|
CPLX4
|
complexin 4 |
| chr4_-_87028478 | 0.20 |
ENST00000515400.1
ENST00000395157.3 |
MAPK10
|
mitogen-activated protein kinase 10 |
| chr14_+_22564294 | 0.19 |
ENST00000390452.2
|
TRDV1
|
T cell receptor delta variable 1 |
| chr17_-_57229155 | 0.19 |
ENST00000584089.1
|
SKA2
|
spindle and kinetochore associated complex subunit 2 |
| chr3_-_195538760 | 0.19 |
ENST00000475231.1
|
MUC4
|
mucin 4, cell surface associated |
| chr1_+_111415757 | 0.19 |
ENST00000429072.2
ENST00000271324.5 |
CD53
|
CD53 molecule |
| chr15_+_80351910 | 0.19 |
ENST00000261749.6
ENST00000561060.1 |
ZFAND6
|
zinc finger, AN1-type domain 6 |
| chr1_+_50575292 | 0.19 |
ENST00000371821.1
ENST00000371819.1 |
ELAVL4
|
ELAV like neuron-specific RNA binding protein 4 |
| chr8_+_70404996 | 0.18 |
ENST00000402687.4
ENST00000419716.3 |
SULF1
|
sulfatase 1 |
| chr12_-_14133053 | 0.18 |
ENST00000609686.1
|
GRIN2B
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2B |
| chr14_-_69261310 | 0.18 |
ENST00000336440.3
|
ZFP36L1
|
ZFP36 ring finger protein-like 1 |
| chr15_-_37393406 | 0.18 |
ENST00000338564.5
ENST00000558313.1 ENST00000340545.5 |
MEIS2
|
Meis homeobox 2 |
| chr4_-_66536057 | 0.18 |
ENST00000273854.3
|
EPHA5
|
EPH receptor A5 |
| chr14_+_101359265 | 0.18 |
ENST00000599197.1
|
AL117190.3
|
Esophagus cancer-related gene-2 interaction susceptibility protein; Uncharacterized protein |
| chr3_-_100565249 | 0.18 |
ENST00000495591.1
ENST00000383691.4 ENST00000466947.1 |
ABI3BP
|
ABI family, member 3 (NESH) binding protein |
| chr19_+_50016610 | 0.18 |
ENST00000596975.1
|
FCGRT
|
Fc fragment of IgG, receptor, transporter, alpha |
| chr4_-_48683188 | 0.18 |
ENST00000505759.1
|
FRYL
|
FRY-like |
| chrX_-_110655306 | 0.18 |
ENST00000371993.2
|
DCX
|
doublecortin |
| chr20_+_3776371 | 0.18 |
ENST00000245960.5
|
CDC25B
|
cell division cycle 25B |
| chr11_-_67220200 | 0.18 |
ENST00000312457.2
|
GPR152
|
G protein-coupled receptor 152 |
| chr6_+_43968306 | 0.17 |
ENST00000442114.2
ENST00000336600.5 ENST00000439969.2 |
C6orf223
|
chromosome 6 open reading frame 223 |
| chr3_-_195538728 | 0.17 |
ENST00000349607.4
ENST00000346145.4 |
MUC4
|
mucin 4, cell surface associated |
| chr11_+_101918153 | 0.17 |
ENST00000434758.2
ENST00000526781.1 ENST00000534360.1 |
C11orf70
|
chromosome 11 open reading frame 70 |
| chrX_-_19689106 | 0.17 |
ENST00000379716.1
|
SH3KBP1
|
SH3-domain kinase binding protein 1 |
| chr9_+_116225999 | 0.17 |
ENST00000317613.6
|
RGS3
|
regulator of G-protein signaling 3 |
| chr3_+_185431080 | 0.17 |
ENST00000296270.1
|
C3orf65
|
chromosome 3 open reading frame 65 |
| chr17_-_39140549 | 0.17 |
ENST00000377755.4
|
KRT40
|
keratin 40 |
| chr7_+_37723336 | 0.17 |
ENST00000450180.1
|
GPR141
|
G protein-coupled receptor 141 |
| chr11_+_67219867 | 0.16 |
ENST00000438189.2
|
CABP4
|
calcium binding protein 4 |
| chr14_-_78083112 | 0.16 |
ENST00000216484.2
|
SPTLC2
|
serine palmitoyltransferase, long chain base subunit 2 |
| chr10_+_114710425 | 0.16 |
ENST00000352065.5
ENST00000369395.1 |
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr20_+_42523336 | 0.16 |
ENST00000428529.1
|
RP5-1030M6.3
|
RP5-1030M6.3 |
| chr14_+_56584414 | 0.16 |
ENST00000559044.1
|
PELI2
|
pellino E3 ubiquitin protein ligase family member 2 |
| chr6_+_167536230 | 0.15 |
ENST00000341935.5
ENST00000349984.4 |
CCR6
|
chemokine (C-C motif) receptor 6 |
| chr1_-_197115818 | 0.15 |
ENST00000367409.4
ENST00000294732.7 |
ASPM
|
asp (abnormal spindle) homolog, microcephaly associated (Drosophila) |
| chr7_+_129984630 | 0.15 |
ENST00000355388.3
ENST00000497503.1 ENST00000463587.1 ENST00000461828.1 ENST00000494311.1 ENST00000466363.2 ENST00000485477.1 ENST00000431780.2 ENST00000474905.1 |
CPA5
|
carboxypeptidase A5 |
| chr10_-_105845674 | 0.15 |
ENST00000353479.5
ENST00000369733.3 |
COL17A1
|
collagen, type XVII, alpha 1 |
| chr10_-_23633720 | 0.15 |
ENST00000323327.4
|
C10orf67
|
chromosome 10 open reading frame 67 |
| chr11_-_118083600 | 0.15 |
ENST00000524477.1
|
AMICA1
|
adhesion molecule, interacts with CXADR antigen 1 |
| chr15_+_96869165 | 0.15 |
ENST00000421109.2
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr5_+_66300446 | 0.15 |
ENST00000261569.7
|
MAST4
|
microtubule associated serine/threonine kinase family member 4 |
| chr8_-_101963482 | 0.15 |
ENST00000419477.2
|
YWHAZ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta |
| chr3_+_152879985 | 0.15 |
ENST00000323534.2
|
RAP2B
|
RAP2B, member of RAS oncogene family |
| chr3_-_143567262 | 0.14 |
ENST00000474151.1
ENST00000316549.6 |
SLC9A9
|
solute carrier family 9, subfamily A (NHE9, cation proton antiporter 9), member 9 |
| chr14_+_100485712 | 0.14 |
ENST00000544450.2
|
EVL
|
Enah/Vasp-like |
| chr2_+_219135115 | 0.14 |
ENST00000248451.3
ENST00000273077.4 |
PNKD
|
paroxysmal nonkinesigenic dyskinesia |
| chr12_-_120241187 | 0.14 |
ENST00000392520.2
|
CIT
|
citron (rho-interacting, serine/threonine kinase 21) |
| chr17_-_48133054 | 0.14 |
ENST00000499842.1
|
RP11-1094H24.4
|
RP11-1094H24.4 |
| chr12_+_41831485 | 0.14 |
ENST00000539469.2
ENST00000298919.7 |
PDZRN4
|
PDZ domain containing ring finger 4 |
| chr2_-_214016314 | 0.14 |
ENST00000434687.1
ENST00000374319.4 |
IKZF2
|
IKAROS family zinc finger 2 (Helios) |
| chr17_+_59489112 | 0.14 |
ENST00000335108.2
|
C17orf82
|
chromosome 17 open reading frame 82 |
Network of associatons between targets according to the STRING database.
First level regulatory network of DLX3_EVX1_MEOX1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | GO:1990709 | presynaptic active zone organization(GO:1990709) |
| 0.4 | 2.5 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 0.3 | 2.3 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.3 | 1.0 | GO:1903045 | neural crest cell migration involved in sympathetic nervous system development(GO:1903045) |
| 0.2 | 0.9 | GO:2000471 | regulation of hematopoietic stem cell migration(GO:2000471) positive regulation of hematopoietic stem cell migration(GO:2000473) |
| 0.2 | 0.5 | GO:1904899 | regulation of hepatic stellate cell proliferation(GO:1904897) positive regulation of hepatic stellate cell proliferation(GO:1904899) hepatic stellate cell proliferation(GO:1990922) |
| 0.1 | 0.5 | GO:0016062 | adaptation of rhodopsin mediated signaling(GO:0016062) light adaption(GO:0036367) |
| 0.1 | 0.4 | GO:0060490 | orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) |
| 0.1 | 1.0 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.1 | 1.5 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.1 | 0.5 | GO:0042663 | regulation of endodermal cell fate specification(GO:0042663) |
| 0.1 | 0.7 | GO:0048619 | embryonic hindgut morphogenesis(GO:0048619) |
| 0.1 | 1.2 | GO:2000334 | blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.1 | 0.3 | GO:0014034 | neural crest cell fate commitment(GO:0014034) |
| 0.1 | 0.3 | GO:0036060 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.1 | 1.0 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.1 | 0.7 | GO:0035992 | tendon cell differentiation(GO:0035990) tendon formation(GO:0035992) |
| 0.1 | 0.4 | GO:0038043 | interleukin-5-mediated signaling pathway(GO:0038043) |
| 0.1 | 1.2 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.1 | 0.5 | GO:0010814 | substance P catabolic process(GO:0010814) calcitonin catabolic process(GO:0010816) endothelin maturation(GO:0034959) |
| 0.1 | 0.3 | GO:0045726 | positive regulation of integrin biosynthetic process(GO:0045726) |
| 0.1 | 0.8 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 0.1 | 0.2 | GO:0002416 | IgG immunoglobulin transcytosis in epithelial cells mediated by FcRn immunoglobulin receptor(GO:0002416) |
| 0.1 | 1.2 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.1 | 0.2 | GO:0046946 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.1 | 0.6 | GO:0051581 | negative regulation of neurotransmitter uptake(GO:0051581) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.1 | 0.2 | GO:0006667 | sphinganine metabolic process(GO:0006667) |
| 0.1 | 0.6 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.1 | 0.5 | GO:0010989 | negative regulation of low-density lipoprotein particle clearance(GO:0010989) |
| 0.1 | 0.2 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 0.0 | 0.1 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.0 | 0.8 | GO:0048245 | eosinophil chemotaxis(GO:0048245) |
| 0.0 | 0.1 | GO:0019243 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.0 | 1.3 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.0 | 0.4 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 0.0 | 1.5 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.0 | 0.6 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.2 | GO:0097403 | cellular response to raffinose(GO:0097403) response to raffinose(GO:1901545) |
| 0.0 | 0.1 | GO:0009956 | radial pattern formation(GO:0009956) |
| 0.0 | 0.9 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.0 | 0.3 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.0 | 0.1 | GO:0032072 | plasmacytoid dendritic cell activation(GO:0002270) regulation of restriction endodeoxyribonuclease activity(GO:0032072) |
| 0.0 | 1.0 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.0 | 0.1 | GO:0060474 | positive regulation of sperm motility involved in capacitation(GO:0060474) |
| 0.0 | 0.1 | GO:0048631 | negative regulation of skeletal muscle cell proliferation(GO:0014859) regulation of skeletal muscle tissue growth(GO:0048631) negative regulation of skeletal muscle satellite cell proliferation(GO:1902723) negative regulation of satellite cell differentiation(GO:1902725) |
| 0.0 | 0.1 | GO:1904116 | response to vasopressin(GO:1904116) cellular response to vasopressin(GO:1904117) |
| 0.0 | 0.2 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.0 | 0.5 | GO:0051546 | keratinocyte migration(GO:0051546) |
| 0.0 | 0.4 | GO:0097475 | motor neuron migration(GO:0097475) |
| 0.0 | 0.2 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.0 | 0.5 | GO:0071285 | cellular response to lithium ion(GO:0071285) |
| 0.0 | 0.1 | GO:1904425 | negative regulation of GTP binding(GO:1904425) |
| 0.0 | 0.2 | GO:0044334 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.0 | 0.2 | GO:1903826 | arginine transmembrane transport(GO:1903826) |
| 0.0 | 0.1 | GO:0090080 | positive regulation of MAPKKK cascade by fibroblast growth factor receptor signaling pathway(GO:0090080) |
| 0.0 | 0.1 | GO:0038033 | positive regulation of endothelial cell chemotaxis by VEGF-activated vascular endothelial growth factor receptor signaling pathway(GO:0038033) |
| 0.0 | 0.2 | GO:0071638 | adiponectin secretion(GO:0070162) regulation of adiponectin secretion(GO:0070163) negative regulation of monocyte chemotactic protein-1 production(GO:0071638) |
| 0.0 | 0.2 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.0 | 0.6 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.0 | 0.1 | GO:1901994 | negative regulation of meiotic cell cycle phase transition(GO:1901994) |
| 0.0 | 0.1 | GO:2000691 | regulation of cardiac muscle cell myoblast differentiation(GO:2000690) negative regulation of cardiac muscle cell myoblast differentiation(GO:2000691) |
| 0.0 | 0.2 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.0 | 0.1 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.0 | 0.2 | GO:1901315 | negative regulation of histone ubiquitination(GO:0033183) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) |
| 0.0 | 0.0 | GO:0021847 | ventricular zone neuroblast division(GO:0021847) |
| 0.0 | 0.3 | GO:0014820 | tonic smooth muscle contraction(GO:0014820) |
| 0.0 | 0.4 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.2 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.2 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.0 | 1.0 | GO:0042461 | photoreceptor cell development(GO:0042461) |
| 0.0 | 0.4 | GO:2000353 | positive regulation of endothelial cell apoptotic process(GO:2000353) |
| 0.0 | 0.3 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.0 | 0.1 | GO:0050653 | chondroitin sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0050653) |
| 0.0 | 0.3 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 0.1 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.0 | 0.4 | GO:0007039 | protein catabolic process in the vacuole(GO:0007039) |
| 0.0 | 0.1 | GO:0060023 | soft palate development(GO:0060023) |
| 0.0 | 0.4 | GO:0042789 | mRNA transcription from RNA polymerase II promoter(GO:0042789) |
| 0.0 | 0.2 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.0 | 0.2 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.0 | 0.4 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.0 | 0.0 | GO:0001994 | norepinephrine-epinephrine vasoconstriction involved in regulation of systemic arterial blood pressure(GO:0001994) |
| 0.0 | 0.1 | GO:1903385 | regulation of homophilic cell adhesion(GO:1903385) |
| 0.0 | 0.4 | GO:0051156 | glucose 6-phosphate metabolic process(GO:0051156) |
| 0.0 | 0.1 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 4.4 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.1 | 0.4 | GO:0060187 | cell pole(GO:0060187) |
| 0.1 | 0.3 | GO:0005873 | plus-end kinesin complex(GO:0005873) |
| 0.1 | 0.3 | GO:0035841 | new growing cell tip(GO:0035841) |
| 0.1 | 0.6 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.1 | 0.6 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.1 | 3.9 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 0.2 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.0 | 1.0 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
| 0.0 | 0.7 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.1 | GO:1990723 | cytoplasmic periphery of the nuclear pore complex(GO:1990723) |
| 0.0 | 1.2 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.2 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.0 | 0.3 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.0 | 0.8 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 0.9 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.6 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 0.3 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.0 | 0.1 | GO:0031265 | death-inducing signaling complex(GO:0031264) CD95 death-inducing signaling complex(GO:0031265) |
| 0.0 | 0.2 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.9 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.2 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.5 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 0.1 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.0 | 0.1 | GO:0097443 | sorting endosome(GO:0097443) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.1 | 1.2 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.1 | 0.4 | GO:0004346 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.1 | 1.9 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.1 | 0.4 | GO:0004914 | interleukin-5 receptor activity(GO:0004914) |
| 0.1 | 2.5 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.1 | 0.7 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.1 | 0.4 | GO:0030197 | extracellular matrix constituent, lubricant activity(GO:0030197) |
| 0.1 | 1.4 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 0.2 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) procollagen glucosyltransferase activity(GO:0033823) procollagen galactosyltransferase activity(GO:0050211) |
| 0.1 | 0.6 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.1 | 1.0 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.3 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.0 | 0.1 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.0 | 2.4 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.0 | 1.6 | GO:0019865 | immunoglobulin binding(GO:0019865) |
| 0.0 | 0.5 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.0 | 1.3 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.6 | GO:0044213 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) intronic transcription regulatory region DNA binding(GO:0044213) |
| 0.0 | 0.5 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.1 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.0 | 0.6 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.3 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.5 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.5 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.3 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.1 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 0.0 | 0.2 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.2 | GO:0015181 | arginine transmembrane transporter activity(GO:0015181) |
| 0.0 | 0.2 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 2.3 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 1.3 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 0.3 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 0.5 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 0.2 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.1 | GO:0008426 | protein kinase C inhibitor activity(GO:0008426) |
| 0.0 | 0.1 | GO:0008955 | peptidoglycan glycosyltransferase activity(GO:0008955) |
| 0.0 | 0.2 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.5 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.0 | 0.2 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.0 | 0.0 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 0.1 | GO:0034040 | lipid-transporting ATPase activity(GO:0034040) |
| 0.0 | 0.4 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.2 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.0 | 0.3 | GO:0045125 | bioactive lipid receptor activity(GO:0045125) |
| 0.0 | 0.1 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.0 | 0.3 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.1 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.1 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 2.8 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 0.4 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 1.1 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.5 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 1.4 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.0 | 0.6 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 1.4 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 1.9 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.3 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 0.6 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 2.2 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.4 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.1 | 2.5 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.1 | 1.2 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.1 | 1.7 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 1.5 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 1.4 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.9 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.0 | 0.5 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.6 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 1.0 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.4 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.0 | 3.1 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 1.5 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.9 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |