Project
Illumina Body Map 2: averaged replicates
Navigation
Downloads
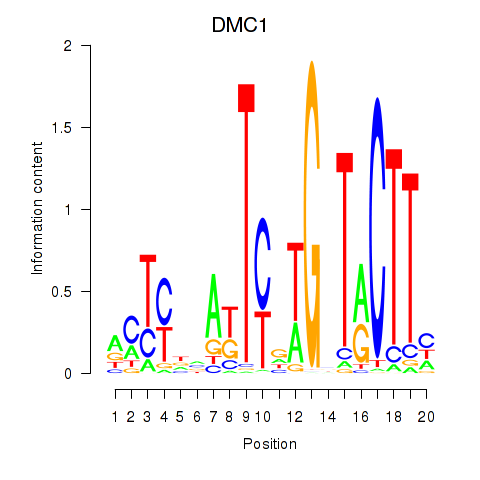
Results for DMC1
Z-value: 1.10
Transcription factors associated with DMC1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
DMC1
|
ENSG00000100206.5 | DNA meiotic recombinase 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| DMC1 | hg19_v2_chr22_-_38966123_38966163 | 0.17 | 3.6e-01 | Click! |
Activity profile of DMC1 motif
Sorted Z-values of DMC1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_+_69865866 | 3.94 |
ENST00000354393.2
|
MYPN
|
myopalladin |
| chr5_+_94727048 | 2.43 |
ENST00000283357.5
|
FAM81B
|
family with sequence similarity 81, member B |
| chr7_-_150884332 | 2.04 |
ENST00000275838.1
ENST00000422024.1 ENST00000434669.1 |
ASB10
|
ankyrin repeat and SOCS box containing 10 |
| chr2_-_75788038 | 1.99 |
ENST00000393913.3
ENST00000410113.1 |
EVA1A
|
eva-1 homolog A (C. elegans) |
| chr20_+_123010 | 1.91 |
ENST00000382398.3
|
DEFB126
|
defensin, beta 126 |
| chr1_+_166958346 | 1.80 |
ENST00000367872.4
|
MAEL
|
maelstrom spermatogenic transposon silencer |
| chr3_+_148457585 | 1.77 |
ENST00000402260.1
|
AGTR1
|
angiotensin II receptor, type 1 |
| chr2_+_171640291 | 1.71 |
ENST00000409885.1
|
ERICH2
|
glutamate-rich 2 |
| chr2_+_120187465 | 1.69 |
ENST00000409826.1
ENST00000417645.1 |
TMEM37
|
transmembrane protein 37 |
| chr1_+_166958497 | 1.69 |
ENST00000367870.2
|
MAEL
|
maelstrom spermatogenic transposon silencer |
| chr6_+_46661575 | 1.67 |
ENST00000450697.1
|
TDRD6
|
tudor domain containing 6 |
| chr12_-_9268707 | 1.56 |
ENST00000318602.7
|
A2M
|
alpha-2-macroglobulin |
| chr1_+_166958504 | 1.54 |
ENST00000447624.1
|
MAEL
|
maelstrom spermatogenic transposon silencer |
| chrX_-_49965009 | 1.51 |
ENST00000437370.1
ENST00000376064.3 ENST00000448865.1 |
AKAP4
|
A kinase (PRKA) anchor protein 4 |
| chr10_-_104597286 | 1.45 |
ENST00000369887.3
|
CYP17A1
|
cytochrome P450, family 17, subfamily A, polypeptide 1 |
| chrX_-_63450480 | 1.43 |
ENST00000362002.2
|
ASB12
|
ankyrin repeat and SOCS box containing 12 |
| chr10_+_17851362 | 1.41 |
ENST00000331429.2
ENST00000457317.1 |
MRC1L1
|
cDNA FLJ56855, highly similar to Macrophage mannose receptor 1 |
| chr17_+_31318886 | 1.34 |
ENST00000269053.3
ENST00000394638.1 |
SPACA3
|
sperm acrosome associated 3 |
| chrX_-_49965663 | 1.34 |
ENST00000376056.2
ENST00000376058.2 ENST00000358526.2 |
AKAP4
|
A kinase (PRKA) anchor protein 4 |
| chr15_-_42448788 | 1.33 |
ENST00000382396.4
ENST00000397272.3 |
PLA2G4F
|
phospholipase A2, group IVF |
| chr11_+_71238313 | 1.26 |
ENST00000398536.4
|
KRTAP5-7
|
keratin associated protein 5-7 |
| chr3_-_108672609 | 1.23 |
ENST00000393963.3
ENST00000471108.1 |
GUCA1C
|
guanylate cyclase activator 1C |
| chr12_-_9268819 | 1.19 |
ENST00000404455.2
|
A2M
|
alpha-2-macroglobulin |
| chr2_+_187454749 | 1.17 |
ENST00000261023.3
ENST00000374907.3 |
ITGAV
|
integrin, alpha V |
| chr3_-_108672742 | 1.11 |
ENST00000261047.3
|
GUCA1C
|
guanylate cyclase activator 1C |
| chr5_+_79615790 | 1.10 |
ENST00000296739.4
|
SPZ1
|
spermatogenic leucine zipper 1 |
| chr7_-_15601595 | 1.10 |
ENST00000342526.3
|
AGMO
|
alkylglycerol monooxygenase |
| chr9_-_34729457 | 1.08 |
ENST00000378788.3
|
FAM205A
|
family with sequence similarity 205, member A |
| chr20_+_2796948 | 1.03 |
ENST00000361033.1
ENST00000380585.1 |
TMEM239
|
transmembrane protein 239 |
| chr5_-_36301984 | 1.03 |
ENST00000502994.1
ENST00000515759.1 ENST00000296604.3 |
RANBP3L
|
RAN binding protein 3-like |
| chr12_-_71031185 | 1.01 |
ENST00000548122.1
ENST00000551525.1 ENST00000550358.1 |
PTPRB
|
protein tyrosine phosphatase, receptor type, B |
| chr15_-_61945400 | 1.00 |
ENST00000560686.1
|
RP11-507B12.1
|
RP11-507B12.1 |
| chr12_-_71031220 | 0.93 |
ENST00000334414.6
|
PTPRB
|
protein tyrosine phosphatase, receptor type, B |
| chr2_-_48982708 | 0.91 |
ENST00000428232.1
ENST00000405626.1 ENST00000294954.7 |
LHCGR
|
luteinizing hormone/choriogonadotropin receptor |
| chrX_-_134305322 | 0.88 |
ENST00000276241.6
ENST00000344129.2 |
CXorf48
|
cancer/testis antigen 55 |
| chr17_+_77018896 | 0.81 |
ENST00000578229.1
|
C1QTNF1
|
C1q and tumor necrosis factor related protein 1 |
| chr7_-_150884873 | 0.79 |
ENST00000377867.3
|
ASB10
|
ankyrin repeat and SOCS box containing 10 |
| chr17_+_77019030 | 0.75 |
ENST00000580454.1
|
C1QTNF1
|
C1q and tumor necrosis factor related protein 1 |
| chr2_+_201173667 | 0.72 |
ENST00000409755.3
|
SPATS2L
|
spermatogenesis associated, serine-rich 2-like |
| chr2_-_48982857 | 0.68 |
ENST00000403273.1
ENST00000401907.1 ENST00000344775.3 |
LHCGR
|
luteinizing hormone/choriogonadotropin receptor |
| chr10_-_135187193 | 0.67 |
ENST00000368547.3
|
ECHS1
|
enoyl CoA hydratase, short chain, 1, mitochondrial |
| chr1_-_160924589 | 0.66 |
ENST00000368029.3
|
ITLN2
|
intelectin 2 |
| chr12_-_120805872 | 0.63 |
ENST00000546985.1
|
MSI1
|
musashi RNA-binding protein 1 |
| chr14_+_74318611 | 0.60 |
ENST00000555976.1
ENST00000267568.4 |
PTGR2
|
prostaglandin reductase 2 |
| chr22_+_21133469 | 0.60 |
ENST00000406799.1
|
SERPIND1
|
serpin peptidase inhibitor, clade D (heparin cofactor), member 1 |
| chr6_+_88054530 | 0.60 |
ENST00000388923.4
|
C6orf163
|
chromosome 6 open reading frame 163 |
| chr14_+_74318513 | 0.58 |
ENST00000555228.1
ENST00000555661.1 |
PTGR2
|
prostaglandin reductase 2 |
| chr2_-_18741882 | 0.56 |
ENST00000381249.3
|
RDH14
|
retinol dehydrogenase 14 (all-trans/9-cis/11-cis) |
| chr3_-_141747950 | 0.54 |
ENST00000497579.1
|
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr3_-_141747459 | 0.52 |
ENST00000477292.1
ENST00000478006.1 ENST00000495310.1 ENST00000486111.1 |
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr10_+_62538248 | 0.47 |
ENST00000448257.2
|
CDK1
|
cyclin-dependent kinase 1 |
| chr10_+_62538089 | 0.47 |
ENST00000519078.2
ENST00000395284.3 ENST00000316629.4 |
CDK1
|
cyclin-dependent kinase 1 |
| chr10_+_74870206 | 0.45 |
ENST00000357321.4
ENST00000349051.5 |
NUDT13
|
nudix (nucleoside diphosphate linked moiety X)-type motif 13 |
| chr2_-_190044480 | 0.43 |
ENST00000374866.3
|
COL5A2
|
collagen, type V, alpha 2 |
| chr10_+_74870253 | 0.42 |
ENST00000544879.1
ENST00000537969.1 ENST00000372997.3 |
NUDT13
|
nudix (nucleoside diphosphate linked moiety X)-type motif 13 |
| chr5_-_82969405 | 0.40 |
ENST00000510978.1
|
HAPLN1
|
hyaluronan and proteoglycan link protein 1 |
| chr3_-_99569821 | 0.38 |
ENST00000487087.1
|
FILIP1L
|
filamin A interacting protein 1-like |
| chr8_+_67104940 | 0.37 |
ENST00000517689.1
|
LINC00967
|
long intergenic non-protein coding RNA 967 |
| chr11_-_73720122 | 0.36 |
ENST00000426995.2
|
UCP3
|
uncoupling protein 3 (mitochondrial, proton carrier) |
| chr19_-_42947121 | 0.35 |
ENST00000601181.1
|
CXCL17
|
chemokine (C-X-C motif) ligand 17 |
| chr1_+_18958008 | 0.34 |
ENST00000420770.2
ENST00000400661.3 |
PAX7
|
paired box 7 |
| chr5_-_82969363 | 0.34 |
ENST00000503117.1
|
HAPLN1
|
hyaluronan and proteoglycan link protein 1 |
| chr20_-_21378666 | 0.33 |
ENST00000351817.4
|
NKX2-4
|
NK2 homeobox 4 |
| chr1_-_247876105 | 0.33 |
ENST00000302084.2
|
OR6F1
|
olfactory receptor, family 6, subfamily F, member 1 |
| chr4_-_76944621 | 0.31 |
ENST00000306602.1
|
CXCL10
|
chemokine (C-X-C motif) ligand 10 |
| chr1_-_47131521 | 0.31 |
ENST00000542495.1
ENST00000532925.1 |
ATPAF1
|
ATP synthase mitochondrial F1 complex assembly factor 1 |
| chr2_+_233497931 | 0.31 |
ENST00000264059.3
|
EFHD1
|
EF-hand domain family, member D1 |
| chr2_-_197675000 | 0.30 |
ENST00000342506.2
|
C2orf66
|
chromosome 2 open reading frame 66 |
| chr11_-_73720276 | 0.28 |
ENST00000348534.4
|
UCP3
|
uncoupling protein 3 (mitochondrial, proton carrier) |
| chr10_-_98031155 | 0.28 |
ENST00000495266.1
|
BLNK
|
B-cell linker |
| chr7_-_6010263 | 0.27 |
ENST00000455618.2
ENST00000405415.1 ENST00000404406.1 ENST00000542644.1 |
RSPH10B
|
radial spoke head 10 homolog B (Chlamydomonas) |
| chr1_-_190446759 | 0.26 |
ENST00000367462.3
|
BRINP3
|
bone morphogenetic protein/retinoic acid inducible neural-specific 3 |
| chr7_+_6793740 | 0.25 |
ENST00000403107.1
ENST00000404077.1 ENST00000435395.1 ENST00000418406.1 |
RSPH10B2
|
radial spoke head 10 homolog B2 (Chlamydomonas) |
| chr21_-_35281399 | 0.25 |
ENST00000418933.1
|
ATP5O
|
ATP synthase, H+ transporting, mitochondrial F1 complex, O subunit |
| chr10_-_98031310 | 0.24 |
ENST00000427367.2
ENST00000413476.2 |
BLNK
|
B-cell linker |
| chr4_+_2800722 | 0.22 |
ENST00000508385.1
|
SH3BP2
|
SH3-domain binding protein 2 |
| chr12_-_53901266 | 0.22 |
ENST00000609999.1
ENST00000267017.3 |
NPFF
|
neuropeptide FF-amide peptide precursor |
| chr10_-_62332357 | 0.21 |
ENST00000503366.1
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G) |
| chr14_+_21525981 | 0.19 |
ENST00000308227.2
|
RNASE8
|
ribonuclease, RNase A family, 8 |
| chr3_+_9944303 | 0.19 |
ENST00000421412.1
ENST00000295980.3 |
IL17RE
|
interleukin 17 receptor E |
| chr14_-_21491477 | 0.19 |
ENST00000298684.5
ENST00000557169.1 ENST00000553563.1 |
NDRG2
|
NDRG family member 2 |
| chr17_-_42875115 | 0.19 |
ENST00000591137.1
|
CTC-296K1.4
|
CTC-296K1.4 |
| chr14_+_21492331 | 0.18 |
ENST00000533984.1
ENST00000532213.2 |
AL161668.5
|
AL161668.5 |
| chr1_+_1222489 | 0.15 |
ENST00000379099.3
|
SCNN1D
|
sodium channel, non-voltage-gated 1, delta subunit |
| chr10_-_98031265 | 0.15 |
ENST00000224337.5
ENST00000371176.2 |
BLNK
|
B-cell linker |
| chr4_+_106473768 | 0.14 |
ENST00000265154.2
ENST00000420470.2 |
ARHGEF38
|
Rho guanine nucleotide exchange factor (GEF) 38 |
| chr17_-_60142609 | 0.12 |
ENST00000397786.2
|
MED13
|
mediator complex subunit 13 |
| chr2_-_61244550 | 0.12 |
ENST00000421319.1
|
PUS10
|
pseudouridylate synthase 10 |
| chr10_-_74856608 | 0.11 |
ENST00000307116.2
ENST00000373008.2 ENST00000412021.2 ENST00000394890.2 ENST00000263556.3 ENST00000440381.1 |
P4HA1
|
prolyl 4-hydroxylase, alpha polypeptide I |
| chr1_-_111061797 | 0.10 |
ENST00000369771.2
|
KCNA10
|
potassium voltage-gated channel, shaker-related subfamily, member 10 |
| chr11_-_85780086 | 0.10 |
ENST00000532317.1
ENST00000528256.1 ENST00000526033.1 |
PICALM
|
phosphatidylinositol binding clathrin assembly protein |
| chr18_+_56892724 | 0.09 |
ENST00000456142.3
ENST00000530323.1 |
GRP
|
gastrin-releasing peptide |
| chr19_+_11466062 | 0.08 |
ENST00000251473.5
ENST00000591329.1 ENST00000586380.1 |
DKFZP761J1410
|
Lipid phosphate phosphatase-related protein type 2 |
| chr20_+_11008408 | 0.07 |
ENST00000378252.1
|
C20orf187
|
chromosome 20 open reading frame 187 |
| chr2_+_61244697 | 0.07 |
ENST00000401576.1
ENST00000295030.5 ENST00000414712.2 |
PEX13
|
peroxisomal biogenesis factor 13 |
| chr2_-_31637560 | 0.06 |
ENST00000379416.3
|
XDH
|
xanthine dehydrogenase |
| chr14_-_21492251 | 0.06 |
ENST00000554398.1
|
NDRG2
|
NDRG family member 2 |
| chr21_+_31802572 | 0.02 |
ENST00000334068.2
|
KRTAP13-4
|
keratin associated protein 13-4 |
| chr7_+_150758642 | 0.01 |
ENST00000488420.1
|
SLC4A2
|
solute carrier family 4 (anion exchanger), member 2 |
| chr7_+_150758304 | 0.01 |
ENST00000482950.1
ENST00000463414.1 ENST00000310317.5 |
SLC4A2
|
solute carrier family 4 (anion exchanger), member 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of DMC1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.8 | GO:0001869 | regulation of complement activation, lectin pathway(GO:0001868) negative regulation of complement activation, lectin pathway(GO:0001869) |
| 0.5 | 1.6 | GO:0042700 | luteinizing hormone signaling pathway(GO:0042700) |
| 0.4 | 1.8 | GO:0086097 | phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) |
| 0.3 | 1.2 | GO:0045715 | negative regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045715) |
| 0.2 | 1.6 | GO:2000857 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.2 | 5.0 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.2 | 1.3 | GO:0009253 | peptidoglycan metabolic process(GO:0000270) peptidoglycan catabolic process(GO:0009253) |
| 0.2 | 3.9 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.1 | 2.3 | GO:0031284 | positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.1 | 0.4 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.1 | 0.9 | GO:0014038 | regulation of Schwann cell differentiation(GO:0014038) |
| 0.1 | 1.0 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.1 | 1.1 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.1 | 0.6 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.1 | 2.6 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.1 | 0.3 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.1 | 1.4 | GO:0006702 | androgen biosynthetic process(GO:0006702) |
| 0.1 | 1.3 | GO:0036148 | phosphatidylglycerol acyl-chain remodeling(GO:0036148) |
| 0.0 | 0.2 | GO:0070253 | somatostatin secretion(GO:0070253) |
| 0.0 | 0.2 | GO:0072660 | maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.0 | 0.3 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) |
| 0.0 | 0.2 | GO:0061760 | antifungal innate immune response(GO:0061760) |
| 0.0 | 0.6 | GO:1990845 | adaptive thermogenesis(GO:1990845) |
| 0.0 | 0.3 | GO:0043950 | positive regulation of cAMP-mediated signaling(GO:0043950) |
| 0.0 | 1.2 | GO:0006692 | prostanoid metabolic process(GO:0006692) prostaglandin metabolic process(GO:0006693) |
| 0.0 | 1.1 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 0.1 | GO:1900738 | positive regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900738) |
| 0.0 | 0.1 | GO:0060152 | peroxisome localization(GO:0060151) microtubule-based peroxisome localization(GO:0060152) |
| 0.0 | 0.1 | GO:0046110 | xanthine metabolic process(GO:0046110) |
| 0.0 | 0.4 | GO:0010759 | positive regulation of macrophage chemotaxis(GO:0010759) |
| 0.0 | 0.9 | GO:0015949 | nucleobase-containing small molecule interconversion(GO:0015949) |
| 0.0 | 0.1 | GO:0097398 | response to interleukin-17(GO:0097396) cellular response to interleukin-17(GO:0097398) |
| 0.0 | 0.6 | GO:0042572 | retinol metabolic process(GO:0042572) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 5.0 | GO:0030849 | autosome(GO:0030849) |
| 0.4 | 1.2 | GO:0034685 | integrin alphav-beta6 complex(GO:0034685) integrin alphav-beta8 complex(GO:0034686) |
| 0.2 | 2.9 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.2 | 0.9 | GO:0097125 | cyclin B1-CDK1 complex(GO:0097125) |
| 0.1 | 0.4 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.1 | 1.7 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.1 | 2.3 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
| 0.0 | 1.3 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.0 | 0.1 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.0 | 2.8 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 4.2 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 1.3 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 1.6 | GO:0005581 | collagen trimer(GO:0005581) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.8 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 0.4 | 1.6 | GO:0004963 | follicle-stimulating hormone receptor activity(GO:0004963) |
| 0.4 | 2.8 | GO:0043120 | tumor necrosis factor binding(GO:0043120) |
| 0.3 | 2.3 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.2 | 1.2 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.2 | 1.2 | GO:0036132 | 13-prostaglandin reductase activity(GO:0036132) 15-oxoprostaglandin 13-oxidase activity(GO:0047522) |
| 0.1 | 1.3 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.1 | 3.9 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.1 | 1.1 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.1 | 0.6 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.1 | 1.3 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.1 | 0.3 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.1 | 1.4 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.0 | 1.9 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.7 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 0.0 | 0.6 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.9 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 2.9 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 0.2 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 0.0 | 0.7 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 1.0 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 0.7 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.1 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) procollagen-proline dioxygenase activity(GO:0019798) |
| 0.0 | 0.1 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 1.6 | GO:0005518 | collagen binding(GO:0005518) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.3 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.1 | 0.9 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 3.4 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 1.2 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 2.8 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 0.7 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 0.7 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.8 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.1 | 1.4 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.1 | 1.6 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.1 | 1.3 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.1 | 0.9 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.0 | 1.2 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.0 | 1.9 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.0 | 0.7 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.7 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 0.3 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |