Project
Illumina Body Map 2: averaged replicates
Navigation
Downloads
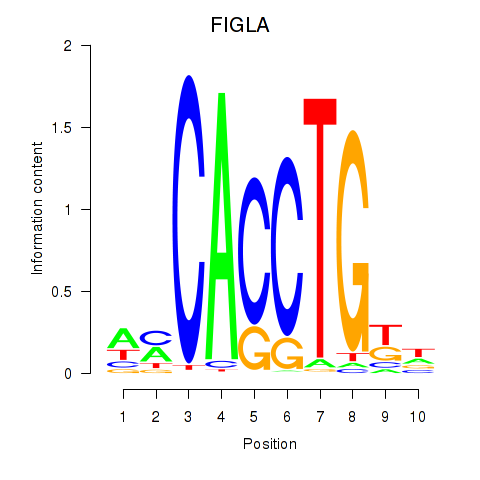
Results for FIGLA
Z-value: 1.59
Transcription factors associated with FIGLA
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
FIGLA
|
ENSG00000183733.6 | folliculogenesis specific bHLH transcription factor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| FIGLA | hg19_v2_chr2_-_71017775_71017775 | 0.44 | 1.2e-02 | Click! |
Activity profile of FIGLA motif
Sorted Z-values of FIGLA motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_-_39258461 | 4.74 |
ENST00000440582.1
|
KRTAP4-16P
|
keratin associated protein 4-16, pseudogene |
| chr2_-_152590946 | 3.14 |
ENST00000172853.10
|
NEB
|
nebulin |
| chr16_-_11375179 | 3.03 |
ENST00000312511.3
|
PRM1
|
protamine 1 |
| chr19_+_50936142 | 2.92 |
ENST00000357701.5
|
MYBPC2
|
myosin binding protein C, fast type |
| chr7_-_27196267 | 2.50 |
ENST00000242159.3
|
HOXA7
|
homeobox A7 |
| chr17_-_39324424 | 2.37 |
ENST00000391356.2
|
KRTAP4-3
|
keratin associated protein 4-3 |
| chr1_-_201391149 | 2.33 |
ENST00000555948.1
ENST00000556362.1 |
TNNI1
|
troponin I type 1 (skeletal, slow) |
| chr17_-_7165662 | 2.20 |
ENST00000571881.2
ENST00000360325.7 |
CLDN7
|
claudin 7 |
| chr17_+_48133459 | 2.15 |
ENST00000320031.8
|
ITGA3
|
integrin, alpha 3 (antigen CD49C, alpha 3 subunit of VLA-3 receptor) |
| chr3_-_50383096 | 2.06 |
ENST00000442887.1
ENST00000360165.3 |
ZMYND10
|
zinc finger, MYND-type containing 10 |
| chr4_-_177116772 | 2.04 |
ENST00000280191.2
|
SPATA4
|
spermatogenesis associated 4 |
| chr2_-_42160486 | 2.03 |
ENST00000427054.1
|
AC104654.2
|
AC104654.2 |
| chr12_+_6419877 | 2.02 |
ENST00000536531.1
|
PLEKHG6
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 6 |
| chr2_-_152590982 | 1.98 |
ENST00000409198.1
ENST00000397345.3 ENST00000427231.2 |
NEB
|
nebulin |
| chr17_+_48133330 | 1.92 |
ENST00000544892.1
|
ITGA3
|
integrin, alpha 3 (antigen CD49C, alpha 3 subunit of VLA-3 receptor) |
| chr6_-_39197226 | 1.91 |
ENST00000359534.3
|
KCNK5
|
potassium channel, subfamily K, member 5 |
| chr3_+_52454971 | 1.87 |
ENST00000465863.1
|
PHF7
|
PHD finger protein 7 |
| chr1_-_183622442 | 1.83 |
ENST00000308641.4
|
APOBEC4
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 4 (putative) |
| chr2_+_176987088 | 1.80 |
ENST00000249499.6
|
HOXD9
|
homeobox D9 |
| chr17_-_46035187 | 1.75 |
ENST00000300557.2
|
PRR15L
|
proline rich 15-like |
| chr6_-_19804973 | 1.74 |
ENST00000457670.1
ENST00000607810.1 ENST00000606628.1 |
RP4-625H18.2
|
RP4-625H18.2 |
| chr1_-_115238207 | 1.59 |
ENST00000520113.2
ENST00000369538.3 ENST00000353928.6 |
AMPD1
|
adenosine monophosphate deaminase 1 |
| chr15_+_45722727 | 1.58 |
ENST00000396650.2
ENST00000558435.1 ENST00000344300.3 |
C15orf48
|
chromosome 15 open reading frame 48 |
| chr12_-_13248562 | 1.56 |
ENST00000457134.2
ENST00000537302.1 |
GSG1
|
germ cell associated 1 |
| chr17_-_39254391 | 1.53 |
ENST00000333822.4
|
KRTAP4-8
|
keratin associated protein 4-8 |
| chrX_-_11445856 | 1.49 |
ENST00000380736.1
|
ARHGAP6
|
Rho GTPase activating protein 6 |
| chr16_+_30383613 | 1.45 |
ENST00000568749.1
|
MYLPF
|
myosin light chain, phosphorylatable, fast skeletal muscle |
| chr2_-_75788038 | 1.45 |
ENST00000393913.3
ENST00000410113.1 |
EVA1A
|
eva-1 homolog A (C. elegans) |
| chr22_+_22988816 | 1.43 |
ENST00000480559.1
ENST00000448514.1 |
GGTLC2
|
gamma-glutamyltransferase light chain 2 |
| chr6_-_100912785 | 1.41 |
ENST00000369208.3
|
SIM1
|
single-minded family bHLH transcription factor 1 |
| chr20_+_30407151 | 1.40 |
ENST00000375985.4
|
MYLK2
|
myosin light chain kinase 2 |
| chr20_+_30407105 | 1.36 |
ENST00000375994.2
|
MYLK2
|
myosin light chain kinase 2 |
| chr11_-_117698787 | 1.36 |
ENST00000260287.2
|
FXYD2
|
FXYD domain containing ion transport regulator 2 |
| chr12_-_13248732 | 1.34 |
ENST00000396302.3
|
GSG1
|
germ cell associated 1 |
| chr7_-_92855762 | 1.31 |
ENST00000453812.2
ENST00000394468.2 |
HEPACAM2
|
HEPACAM family member 2 |
| chr17_+_73521763 | 1.29 |
ENST00000167462.5
ENST00000375227.4 ENST00000392550.3 ENST00000578363.1 ENST00000579392.1 |
LLGL2
|
lethal giant larvae homolog 2 (Drosophila) |
| chr8_-_49834299 | 1.29 |
ENST00000396822.1
|
SNAI2
|
snail family zinc finger 2 |
| chr5_-_54281491 | 1.28 |
ENST00000381405.4
|
ESM1
|
endothelial cell-specific molecule 1 |
| chr4_+_106816592 | 1.26 |
ENST00000379987.2
ENST00000453617.2 ENST00000427316.2 ENST00000514622.1 ENST00000305572.8 |
NPNT
|
nephronectin |
| chr17_-_39280419 | 1.26 |
ENST00000394014.1
|
KRTAP4-12
|
keratin associated protein 4-12 |
| chr17_+_42015654 | 1.26 |
ENST00000565120.1
|
RP11-527L4.2
|
Uncharacterized protein |
| chr8_-_49833978 | 1.23 |
ENST00000020945.1
|
SNAI2
|
snail family zinc finger 2 |
| chr11_+_111789580 | 1.22 |
ENST00000278601.5
|
C11orf52
|
chromosome 11 open reading frame 52 |
| chr1_-_171621815 | 1.20 |
ENST00000037502.6
|
MYOC
|
myocilin, trabecular meshwork inducible glucocorticoid response |
| chr12_-_13248705 | 1.19 |
ENST00000396310.2
|
GSG1
|
germ cell associated 1 |
| chr18_+_77439775 | 1.18 |
ENST00000299543.7
ENST00000075430.7 |
CTDP1
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) phosphatase, subunit 1 |
| chrX_+_13587712 | 1.17 |
ENST00000361306.1
ENST00000380602.3 |
EGFL6
|
EGF-like-domain, multiple 6 |
| chr7_+_130020180 | 1.17 |
ENST00000481342.1
ENST00000011292.3 ENST00000604896.1 |
CPA1
|
carboxypeptidase A1 (pancreatic) |
| chr5_-_54281407 | 1.17 |
ENST00000381403.4
|
ESM1
|
endothelial cell-specific molecule 1 |
| chr12_-_91348949 | 1.16 |
ENST00000358859.2
|
CCER1
|
coiled-coil glutamate-rich protein 1 |
| chr3_+_140396881 | 1.16 |
ENST00000286349.3
|
TRIM42
|
tripartite motif containing 42 |
| chr7_+_140103842 | 1.16 |
ENST00000495590.1
ENST00000275874.5 ENST00000537763.1 |
RAB19
|
RAB19, member RAS oncogene family |
| chr5_+_34656529 | 1.13 |
ENST00000513974.1
ENST00000512629.1 |
RAI14
|
retinoic acid induced 14 |
| chr6_+_22569784 | 1.13 |
ENST00000510882.2
|
HDGFL1
|
hepatoma derived growth factor-like 1 |
| chr11_-_117698765 | 1.12 |
ENST00000532119.1
|
FXYD2
|
FXYD domain containing ion transport regulator 2 |
| chr4_+_106816644 | 1.11 |
ENST00000506666.1
ENST00000503451.1 |
NPNT
|
nephronectin |
| chr2_-_227050079 | 1.10 |
ENST00000423838.1
|
AC068138.1
|
AC068138.1 |
| chr6_-_10419871 | 1.10 |
ENST00000319516.4
|
TFAP2A
|
transcription factor AP-2 alpha (activating enhancer binding protein 2 alpha) |
| chr6_+_44184653 | 1.09 |
ENST00000573382.2
ENST00000576476.1 |
RP1-302G2.5
|
RP1-302G2.5 |
| chr19_+_6464243 | 1.09 |
ENST00000600229.1
ENST00000356762.3 |
CRB3
|
crumbs homolog 3 (Drosophila) |
| chr16_+_610407 | 1.07 |
ENST00000409413.3
|
C16orf11
|
chromosome 16 open reading frame 11 |
| chr5_+_34656569 | 1.07 |
ENST00000428746.2
|
RAI14
|
retinoic acid induced 14 |
| chr2_+_233390890 | 1.07 |
ENST00000258385.3
ENST00000536614.1 ENST00000457943.2 |
CHRND
|
cholinergic receptor, nicotinic, delta (muscle) |
| chr19_-_46272106 | 1.06 |
ENST00000560168.1
|
SIX5
|
SIX homeobox 5 |
| chr17_+_8243154 | 1.06 |
ENST00000328248.2
ENST00000584943.1 |
ODF4
|
outer dense fiber of sperm tails 4 |
| chr8_-_144512576 | 1.06 |
ENST00000333480.2
|
MAFA
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog A |
| chr1_+_183155373 | 1.05 |
ENST00000493293.1
ENST00000264144.4 |
LAMC2
|
laminin, gamma 2 |
| chr2_+_78143006 | 1.05 |
ENST00000443419.1
|
AC073628.1
|
AC073628.1 |
| chr2_+_177001685 | 1.05 |
ENST00000432796.2
|
HOXD3
|
homeobox D3 |
| chr6_-_19804877 | 1.04 |
ENST00000447250.1
|
RP4-625H18.2
|
RP4-625H18.2 |
| chr9_-_116861337 | 1.03 |
ENST00000374118.3
|
KIF12
|
kinesin family member 12 |
| chr12_+_50497784 | 1.01 |
ENST00000548814.1
|
GPD1
|
glycerol-3-phosphate dehydrogenase 1 (soluble) |
| chr17_-_46262541 | 0.98 |
ENST00000579336.1
|
SKAP1
|
src kinase associated phosphoprotein 1 |
| chr3_-_49722523 | 0.97 |
ENST00000448220.1
|
MST1
|
macrophage stimulating 1 (hepatocyte growth factor-like) |
| chr6_+_7107830 | 0.97 |
ENST00000379933.3
|
RREB1
|
ras responsive element binding protein 1 |
| chr17_-_39306054 | 0.96 |
ENST00000343246.4
|
KRTAP4-5
|
keratin associated protein 4-5 |
| chr20_+_44441304 | 0.95 |
ENST00000352551.5
|
UBE2C
|
ubiquitin-conjugating enzyme E2C |
| chr20_-_22566089 | 0.94 |
ENST00000377115.4
|
FOXA2
|
forkhead box A2 |
| chr2_-_85890569 | 0.94 |
ENST00000494165.1
|
SFTPB
|
surfactant protein B |
| chr2_+_46524537 | 0.94 |
ENST00000263734.3
|
EPAS1
|
endothelial PAS domain protein 1 |
| chr5_-_16936340 | 0.94 |
ENST00000507288.1
ENST00000513610.1 |
MYO10
|
myosin X |
| chr6_-_31697255 | 0.93 |
ENST00000436437.1
|
DDAH2
|
dimethylarginine dimethylaminohydrolase 2 |
| chr6_+_22569678 | 0.93 |
ENST00000230012.3
|
HDGFL1
|
hepatoma derived growth factor-like 1 |
| chr19_+_35739280 | 0.92 |
ENST00000602122.1
|
LSR
|
lipolysis stimulated lipoprotein receptor |
| chrX_+_128872998 | 0.92 |
ENST00000371106.3
|
XPNPEP2
|
X-prolyl aminopeptidase (aminopeptidase P) 2, membrane-bound |
| chr17_-_46657473 | 0.92 |
ENST00000332503.5
|
HOXB4
|
homeobox B4 |
| chr16_+_4845379 | 0.91 |
ENST00000588606.1
ENST00000586005.1 |
SMIM22
|
small integral membrane protein 22 |
| chr19_+_35739782 | 0.91 |
ENST00000347609.4
|
LSR
|
lipolysis stimulated lipoprotein receptor |
| chr17_-_39296739 | 0.89 |
ENST00000345847.4
|
KRTAP4-6
|
keratin associated protein 4-6 |
| chr12_-_52585765 | 0.85 |
ENST00000313234.5
ENST00000394815.2 |
KRT80
|
keratin 80 |
| chr6_+_7108210 | 0.85 |
ENST00000467782.1
ENST00000334984.6 ENST00000349384.6 |
RREB1
|
ras responsive element binding protein 1 |
| chr2_-_175629135 | 0.85 |
ENST00000409542.1
ENST00000409219.1 |
CHRNA1
|
cholinergic receptor, nicotinic, alpha 1 (muscle) |
| chr5_+_68788594 | 0.85 |
ENST00000396442.2
ENST00000380766.2 |
OCLN
|
occludin |
| chr2_+_233390863 | 0.84 |
ENST00000449596.1
ENST00000543200.1 |
CHRND
|
cholinergic receptor, nicotinic, delta (muscle) |
| chr20_+_44441215 | 0.84 |
ENST00000356455.4
ENST00000405520.1 |
UBE2C
|
ubiquitin-conjugating enzyme E2C |
| chr16_-_57513657 | 0.83 |
ENST00000566936.1
ENST00000568617.1 ENST00000567276.1 ENST00000569548.1 ENST00000569250.1 ENST00000564378.1 |
DOK4
|
docking protein 4 |
| chr5_-_159739532 | 0.82 |
ENST00000520748.1
ENST00000393977.3 ENST00000257536.7 |
CCNJL
|
cyclin J-like |
| chr19_+_35739631 | 0.81 |
ENST00000602003.1
ENST00000360798.3 ENST00000354900.3 |
LSR
|
lipolysis stimulated lipoprotein receptor |
| chr19_+_35739597 | 0.81 |
ENST00000361790.3
|
LSR
|
lipolysis stimulated lipoprotein receptor |
| chr5_-_159739483 | 0.81 |
ENST00000519673.1
ENST00000541762.1 |
CCNJL
|
cyclin J-like |
| chr18_-_45663666 | 0.80 |
ENST00000535628.2
|
ZBTB7C
|
zinc finger and BTB domain containing 7C |
| chr7_-_11871815 | 0.79 |
ENST00000423059.4
|
THSD7A
|
thrombospondin, type I, domain containing 7A |
| chr15_-_53097139 | 0.79 |
ENST00000560818.1
|
RP11-209K10.2
|
RP11-209K10.2 |
| chr7_+_93535817 | 0.79 |
ENST00000248572.5
|
GNGT1
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 1 |
| chr16_+_4606347 | 0.79 |
ENST00000444310.4
|
C16orf96
|
chromosome 16 open reading frame 96 |
| chr15_+_67547113 | 0.78 |
ENST00000512104.1
ENST00000358767.3 ENST00000546225.1 |
IQCH
|
IQ motif containing H |
| chrX_+_128872918 | 0.78 |
ENST00000371105.3
|
XPNPEP2
|
X-prolyl aminopeptidase (aminopeptidase P) 2, membrane-bound |
| chr19_+_14017116 | 0.77 |
ENST00000589606.1
|
CC2D1A
|
coiled-coil and C2 domain containing 1A |
| chr8_+_22435762 | 0.77 |
ENST00000456545.1
|
PDLIM2
|
PDZ and LIM domain 2 (mystique) |
| chr12_-_53228079 | 0.77 |
ENST00000330553.5
|
KRT79
|
keratin 79 |
| chr14_-_92413353 | 0.77 |
ENST00000556154.1
|
FBLN5
|
fibulin 5 |
| chr11_+_47279155 | 0.75 |
ENST00000444396.1
ENST00000457932.1 ENST00000412937.1 |
NR1H3
|
nuclear receptor subfamily 1, group H, member 3 |
| chr1_+_6615241 | 0.74 |
ENST00000333172.6
ENST00000328191.4 ENST00000351136.3 |
TAS1R1
|
taste receptor, type 1, member 1 |
| chr19_-_4902877 | 0.73 |
ENST00000381781.2
|
ARRDC5
|
arrestin domain containing 5 |
| chr4_-_76555657 | 0.73 |
ENST00000307465.4
|
CDKL2
|
cyclin-dependent kinase-like 2 (CDC2-related kinase) |
| chr6_+_7107999 | 0.72 |
ENST00000491191.1
ENST00000379938.2 ENST00000471433.1 |
RREB1
|
ras responsive element binding protein 1 |
| chr10_+_24755416 | 0.71 |
ENST00000396446.1
ENST00000396445.1 ENST00000376451.2 |
KIAA1217
|
KIAA1217 |
| chrX_-_153775426 | 0.71 |
ENST00000393562.2
|
G6PD
|
glucose-6-phosphate dehydrogenase |
| chr19_+_16187085 | 0.71 |
ENST00000300933.4
|
TPM4
|
tropomyosin 4 |
| chr1_-_156675535 | 0.70 |
ENST00000368221.1
|
CRABP2
|
cellular retinoic acid binding protein 2 |
| chr14_-_37051798 | 0.70 |
ENST00000258829.5
|
NKX2-8
|
NK2 homeobox 8 |
| chr1_+_22328144 | 0.70 |
ENST00000290122.3
ENST00000374663.1 |
CELA3A
|
chymotrypsin-like elastase family, member 3A |
| chr2_-_175629164 | 0.70 |
ENST00000409323.1
ENST00000261007.5 ENST00000348749.5 |
CHRNA1
|
cholinergic receptor, nicotinic, alpha 1 (muscle) |
| chr2_+_137523086 | 0.69 |
ENST00000409968.1
|
THSD7B
|
thrombospondin, type I, domain containing 7B |
| chr10_+_24498060 | 0.69 |
ENST00000376454.3
ENST00000376452.3 |
KIAA1217
|
KIAA1217 |
| chr16_+_67233412 | 0.69 |
ENST00000477898.1
|
ELMO3
|
engulfment and cell motility 3 |
| chr1_-_205313304 | 0.68 |
ENST00000539253.1
ENST00000607826.1 |
KLHDC8A
|
kelch domain containing 8A |
| chr17_-_31204124 | 0.68 |
ENST00000579584.1
ENST00000318217.5 ENST00000583621.1 |
MYO1D
|
myosin ID |
| chr14_-_92413727 | 0.67 |
ENST00000267620.10
|
FBLN5
|
fibulin 5 |
| chr14_-_92414055 | 0.67 |
ENST00000342058.4
|
FBLN5
|
fibulin 5 |
| chr11_+_130318869 | 0.67 |
ENST00000299164.2
|
ADAMTS15
|
ADAM metallopeptidase with thrombospondin type 1 motif, 15 |
| chr12_+_52450298 | 0.66 |
ENST00000550582.2
|
NR4A1
|
nuclear receptor subfamily 4, group A, member 1 |
| chr9_-_22009241 | 0.66 |
ENST00000380142.4
|
CDKN2B
|
cyclin-dependent kinase inhibitor 2B (p15, inhibits CDK4) |
| chr10_+_24497704 | 0.66 |
ENST00000376456.4
ENST00000458595.1 |
KIAA1217
|
KIAA1217 |
| chr11_+_47279248 | 0.65 |
ENST00000449369.1
|
NR1H3
|
nuclear receptor subfamily 1, group H, member 3 |
| chr5_+_157098534 | 0.65 |
ENST00000409999.3
|
C5orf52
|
chromosome 5 open reading frame 52 |
| chr1_-_156675368 | 0.64 |
ENST00000368222.3
|
CRABP2
|
cellular retinoic acid binding protein 2 |
| chr5_+_150406527 | 0.64 |
ENST00000520059.1
|
GPX3
|
glutathione peroxidase 3 (plasma) |
| chr7_+_128784712 | 0.63 |
ENST00000289407.4
|
TSPAN33
|
tetraspanin 33 |
| chr12_-_53242770 | 0.63 |
ENST00000304620.4
ENST00000547110.1 |
KRT78
|
keratin 78 |
| chr7_-_148725733 | 0.63 |
ENST00000286091.4
|
PDIA4
|
protein disulfide isomerase family A, member 4 |
| chr4_+_15376165 | 0.63 |
ENST00000382383.3
ENST00000429690.1 |
C1QTNF7
|
C1q and tumor necrosis factor related protein 7 |
| chr15_+_67547163 | 0.62 |
ENST00000335894.4
|
IQCH
|
IQ motif containing H |
| chr4_-_186456652 | 0.62 |
ENST00000284767.5
ENST00000284770.5 |
PDLIM3
|
PDZ and LIM domain 3 |
| chr13_-_20767037 | 0.62 |
ENST00000382848.4
|
GJB2
|
gap junction protein, beta 2, 26kDa |
| chr8_-_99837856 | 0.61 |
ENST00000518165.1
ENST00000419617.2 |
STK3
|
serine/threonine kinase 3 |
| chr17_+_39261584 | 0.60 |
ENST00000391415.1
|
KRTAP4-9
|
keratin associated protein 4-9 |
| chr9_-_22009297 | 0.60 |
ENST00000276925.6
|
CDKN2B
|
cyclin-dependent kinase inhibitor 2B (p15, inhibits CDK4) |
| chr1_-_150669500 | 0.60 |
ENST00000271732.3
|
GOLPH3L
|
golgi phosphoprotein 3-like |
| chr6_+_109169591 | 0.60 |
ENST00000368972.3
ENST00000392644.4 |
ARMC2
|
armadillo repeat containing 2 |
| chr11_+_71249071 | 0.60 |
ENST00000398534.3
|
KRTAP5-8
|
keratin associated protein 5-8 |
| chr19_-_18709357 | 0.59 |
ENST00000597131.1
|
CRLF1
|
cytokine receptor-like factor 1 |
| chrX_-_30326445 | 0.59 |
ENST00000378963.1
|
NR0B1
|
nuclear receptor subfamily 0, group B, member 1 |
| chr22_-_50970506 | 0.59 |
ENST00000428989.2
ENST00000403326.1 |
ODF3B
|
outer dense fiber of sperm tails 3B |
| chr6_-_133055896 | 0.59 |
ENST00000367927.5
ENST00000425515.2 ENST00000207771.3 ENST00000392393.3 ENST00000450865.2 ENST00000392394.2 |
VNN3
|
vanin 3 |
| chr8_-_103668114 | 0.58 |
ENST00000285407.6
|
KLF10
|
Kruppel-like factor 10 |
| chr11_+_27062272 | 0.58 |
ENST00000529202.1
ENST00000533566.1 |
BBOX1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1 |
| chr7_-_84816122 | 0.57 |
ENST00000444867.1
|
SEMA3D
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3D |
| chr2_-_28113965 | 0.57 |
ENST00000302188.3
|
RBKS
|
ribokinase |
| chr1_-_55352834 | 0.57 |
ENST00000371269.3
|
DHCR24
|
24-dehydrocholesterol reductase |
| chr11_+_93754513 | 0.56 |
ENST00000315765.9
|
HEPHL1
|
hephaestin-like 1 |
| chr4_+_5712898 | 0.56 |
ENST00000264956.6
ENST00000382674.2 |
EVC
|
Ellis van Creveld syndrome |
| chr11_-_124632179 | 0.56 |
ENST00000278927.5
ENST00000442070.2 ENST00000444566.1 ENST00000435477.1 |
ESAM
|
endothelial cell adhesion molecule |
| chr2_+_220436917 | 0.56 |
ENST00000243786.2
|
INHA
|
inhibin, alpha |
| chr11_+_120110863 | 0.55 |
ENST00000543440.2
|
POU2F3
|
POU class 2 homeobox 3 |
| chr10_-_128975273 | 0.55 |
ENST00000424811.2
|
FAM196A
|
family with sequence similarity 196, member A |
| chr19_+_6464502 | 0.54 |
ENST00000308243.7
|
CRB3
|
crumbs homolog 3 (Drosophila) |
| chr8_-_144790224 | 0.54 |
ENST00000533508.1
ENST00000542437.1 |
CCDC166
|
coiled-coil domain containing 166 |
| chr10_-_33625154 | 0.53 |
ENST00000265371.4
|
NRP1
|
neuropilin 1 |
| chr7_+_75028199 | 0.53 |
ENST00000437796.1
|
TRIM73
|
tripartite motif containing 73 |
| chr19_+_14017003 | 0.53 |
ENST00000318003.7
|
CC2D1A
|
coiled-coil and C2 domain containing 1A |
| chr2_-_28113217 | 0.53 |
ENST00000444339.2
|
RBKS
|
ribokinase |
| chr2_+_228029281 | 0.53 |
ENST00000396578.3
|
COL4A3
|
collagen, type IV, alpha 3 (Goodpasture antigen) |
| chr11_+_27062502 | 0.53 |
ENST00000263182.3
|
BBOX1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1 |
| chr4_-_186456766 | 0.52 |
ENST00000284771.6
|
PDLIM3
|
PDZ and LIM domain 3 |
| chr19_+_16186903 | 0.51 |
ENST00000588507.1
|
TPM4
|
tropomyosin 4 |
| chr2_-_220436248 | 0.51 |
ENST00000265318.4
|
OBSL1
|
obscurin-like 1 |
| chr15_-_32162833 | 0.51 |
ENST00000560598.1
|
OTUD7A
|
OTU domain containing 7A |
| chr2_+_121493717 | 0.51 |
ENST00000418323.1
|
GLI2
|
GLI family zinc finger 2 |
| chr11_-_62752455 | 0.51 |
ENST00000360421.4
|
SLC22A6
|
solute carrier family 22 (organic anion transporter), member 6 |
| chr6_-_31697563 | 0.50 |
ENST00000375789.2
ENST00000416410.1 |
DDAH2
|
dimethylarginine dimethylaminohydrolase 2 |
| chr14_-_81916501 | 0.49 |
ENST00000555001.1
|
RP11-299L17.3
|
RP11-299L17.3 |
| chr19_-_15590306 | 0.49 |
ENST00000292609.4
|
PGLYRP2
|
peptidoglycan recognition protein 2 |
| chr2_+_42104692 | 0.49 |
ENST00000398796.2
ENST00000442214.1 |
AC104654.1
|
AC104654.1 |
| chr17_-_39191107 | 0.48 |
ENST00000344363.5
|
KRTAP1-3
|
keratin associated protein 1-3 |
| chrX_+_47863734 | 0.48 |
ENST00000304355.5
|
SPACA5
|
sperm acrosome associated 5 |
| chr1_-_39339777 | 0.48 |
ENST00000397572.2
|
MYCBP
|
MYC binding protein |
| chr3_-_49066811 | 0.48 |
ENST00000442157.1
ENST00000326739.4 |
IMPDH2
|
IMP (inosine 5'-monophosphate) dehydrogenase 2 |
| chr11_-_62752162 | 0.48 |
ENST00000458333.2
ENST00000421062.2 |
SLC22A6
|
solute carrier family 22 (organic anion transporter), member 6 |
| chr5_+_34656331 | 0.47 |
ENST00000265109.3
|
RAI14
|
retinoic acid induced 14 |
| chr4_+_5712942 | 0.47 |
ENST00000509451.1
|
EVC
|
Ellis van Creveld syndrome |
| chr1_+_10509971 | 0.47 |
ENST00000320498.4
|
CORT
|
cortistatin |
| chr17_+_9548845 | 0.47 |
ENST00000570475.1
ENST00000285199.7 |
USP43
|
ubiquitin specific peptidase 43 |
| chr11_-_62752429 | 0.46 |
ENST00000377871.3
|
SLC22A6
|
solute carrier family 22 (organic anion transporter), member 6 |
| chr6_+_18387570 | 0.46 |
ENST00000259939.3
|
RNF144B
|
ring finger protein 144B |
| chr19_-_49118067 | 0.46 |
ENST00000593772.1
|
FAM83E
|
family with sequence similarity 83, member E |
| chr7_+_12726623 | 0.45 |
ENST00000439721.1
|
ARL4A
|
ADP-ribosylation factor-like 4A |
| chr19_-_51456198 | 0.45 |
ENST00000594846.1
|
KLK5
|
kallikrein-related peptidase 5 |
| chr14_+_75746340 | 0.44 |
ENST00000555686.1
ENST00000555672.1 |
FOS
|
FBJ murine osteosarcoma viral oncogene homolog |
| chr11_+_86511569 | 0.44 |
ENST00000441050.1
|
PRSS23
|
protease, serine, 23 |
Network of associatons between targets according to the STRING database.
First level regulatory network of FIGLA
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 5.1 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.8 | 2.5 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 0.6 | 2.5 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.6 | 2.4 | GO:0097195 | pilomotor reflex(GO:0097195) |
| 0.6 | 3.5 | GO:1904274 | tricellular tight junction assembly(GO:1904274) |
| 0.5 | 1.4 | GO:0097254 | renal tubular secretion(GO:0097254) |
| 0.4 | 1.2 | GO:0014734 | skeletal muscle hypertrophy(GO:0014734) |
| 0.4 | 2.8 | GO:0032971 | regulation of muscle filament sliding(GO:0032971) |
| 0.4 | 3.5 | GO:0048630 | skeletal muscle tissue growth(GO:0048630) |
| 0.4 | 1.1 | GO:1902723 | negative regulation of skeletal muscle cell proliferation(GO:0014859) negative regulation of skeletal muscle satellite cell proliferation(GO:1902723) |
| 0.3 | 1.0 | GO:0046168 | glycerol-3-phosphate catabolic process(GO:0046168) |
| 0.3 | 2.5 | GO:1902202 | regulation of hepatocyte growth factor receptor signaling pathway(GO:1902202) |
| 0.3 | 1.4 | GO:0090341 | negative regulation of secretion of lysosomal enzymes(GO:0090341) |
| 0.2 | 0.9 | GO:0045013 | carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) |
| 0.2 | 1.1 | GO:0019303 | D-ribose catabolic process(GO:0019303) |
| 0.2 | 1.1 | GO:0003409 | optic cup structural organization(GO:0003409) |
| 0.2 | 1.8 | GO:0031536 | positive regulation of exit from mitosis(GO:0031536) |
| 0.2 | 1.2 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.2 | 2.5 | GO:0045617 | negative regulation of keratinocyte differentiation(GO:0045617) |
| 0.2 | 0.5 | GO:0021775 | smoothened signaling pathway involved in ventral spinal cord interneuron specification(GO:0021775) smoothened signaling pathway involved in spinal cord motor neuron cell fate specification(GO:0021776) |
| 0.2 | 0.9 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.2 | 0.5 | GO:0032824 | negative regulation of natural killer cell differentiation(GO:0032824) negative regulation of natural killer cell differentiation involved in immune response(GO:0032827) |
| 0.2 | 2.1 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.2 | 3.3 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.1 | 1.6 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.1 | 0.6 | GO:0046882 | negative regulation of B cell differentiation(GO:0045578) negative regulation of follicle-stimulating hormone secretion(GO:0046882) |
| 0.1 | 2.3 | GO:0014883 | transition between fast and slow fiber(GO:0014883) |
| 0.1 | 0.8 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.1 | 0.6 | GO:0002669 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.1 | 0.6 | GO:0044752 | response to human chorionic gonadotropin(GO:0044752) |
| 0.1 | 0.6 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.1 | 0.7 | GO:0009051 | pentose-phosphate shunt, oxidative branch(GO:0009051) |
| 0.1 | 2.5 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.1 | 2.1 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.1 | 4.1 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.1 | 1.8 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.1 | 1.0 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.1 | 0.5 | GO:1902378 | vestibulocochlear nerve structural organization(GO:0021649) positive regulation of cytokine activity(GO:0060301) ganglion morphogenesis(GO:0061552) endothelial tip cell fate specification(GO:0097102) VEGF-activated neuropilin signaling pathway involved in axon guidance(GO:1902378) dorsal root ganglion morphogenesis(GO:1904835) otic placode development(GO:1905040) |
| 0.1 | 0.4 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.1 | 0.7 | GO:0050917 | sensory perception of umami taste(GO:0050917) |
| 0.1 | 0.9 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.1 | 0.9 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.1 | 1.0 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.1 | 0.6 | GO:0033490 | cholesterol biosynthetic process via desmosterol(GO:0033489) cholesterol biosynthetic process via lathosterol(GO:0033490) |
| 0.1 | 0.6 | GO:0006982 | response to lipid hydroperoxide(GO:0006982) |
| 0.1 | 0.3 | GO:1901383 | negative regulation of chorionic trophoblast cell proliferation(GO:1901383) |
| 0.1 | 0.3 | GO:0060488 | orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) |
| 0.1 | 0.7 | GO:0061502 | early endosome to recycling endosome transport(GO:0061502) |
| 0.1 | 1.1 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.1 | 0.2 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.1 | 0.3 | GO:0035854 | regulation of primitive erythrocyte differentiation(GO:0010725) eosinophil fate commitment(GO:0035854) |
| 0.1 | 1.4 | GO:1901750 | leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.1 | 0.4 | GO:0090402 | oncogene-induced cell senescence(GO:0090402) |
| 0.1 | 0.2 | GO:1900369 | transcription, RNA-templated(GO:0001172) negative regulation of RNA interference(GO:1900369) |
| 0.1 | 0.3 | GO:0070426 | positive regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070426) positive regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070434) |
| 0.1 | 0.4 | GO:0002378 | immunoglobulin biosynthetic process(GO:0002378) |
| 0.1 | 0.4 | GO:0002803 | positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antimicrobial humoral response(GO:0002760) positive regulation of antibacterial peptide production(GO:0002803) |
| 0.1 | 1.7 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.1 | 0.5 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.1 | 0.3 | GO:0003335 | corneocyte development(GO:0003335) |
| 0.1 | 1.4 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.1 | 3.0 | GO:0030261 | chromosome condensation(GO:0030261) |
| 0.1 | 0.5 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.1 | 0.2 | GO:0090472 | viral protein processing(GO:0019082) regulation of nerve growth factor production(GO:0032903) negative regulation of nerve growth factor production(GO:0032904) dibasic protein processing(GO:0090472) |
| 0.1 | 2.0 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.1 | 0.8 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.1 | 0.2 | GO:0002752 | cell surface pattern recognition receptor signaling pathway(GO:0002752) |
| 0.1 | 0.4 | GO:0042986 | positive regulation of amyloid precursor protein biosynthetic process(GO:0042986) |
| 0.1 | 2.2 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.1 | 0.4 | GO:0097473 | cellular response to high light intensity(GO:0071486) retinal rod cell apoptotic process(GO:0097473) |
| 0.1 | 0.1 | GO:0001922 | B-1 B cell homeostasis(GO:0001922) |
| 0.1 | 0.2 | GO:0036292 | DNA rewinding(GO:0036292) |
| 0.1 | 1.0 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.1 | 1.3 | GO:0032878 | regulation of establishment or maintenance of cell polarity(GO:0032878) |
| 0.1 | 0.6 | GO:1901314 | negative regulation of histone ubiquitination(GO:0033183) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) |
| 0.1 | 0.7 | GO:0046886 | positive regulation of hormone biosynthetic process(GO:0046886) |
| 0.1 | 0.4 | GO:0003199 | endocardial cushion to mesenchymal transition involved in heart valve formation(GO:0003199) |
| 0.1 | 0.8 | GO:0070673 | response to interleukin-18(GO:0070673) |
| 0.1 | 0.9 | GO:1903025 | regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903025) |
| 0.1 | 0.4 | GO:0045084 | positive regulation of interleukin-12 biosynthetic process(GO:0045084) |
| 0.1 | 0.7 | GO:0061469 | response to corticotropin-releasing hormone(GO:0043435) regulation of type B pancreatic cell proliferation(GO:0061469) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.0 | 0.3 | GO:0045200 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.0 | 0.2 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.0 | 1.5 | GO:0051895 | negative regulation of focal adhesion assembly(GO:0051895) |
| 0.0 | 1.1 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.0 | 0.2 | GO:0015917 | aminophospholipid transport(GO:0015917) |
| 0.0 | 0.3 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.2 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 0.0 | 1.3 | GO:0030511 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.0 | 0.3 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.0 | 0.3 | GO:0075525 | viral translational termination-reinitiation(GO:0075525) |
| 0.0 | 1.0 | GO:0048012 | hepatocyte growth factor receptor signaling pathway(GO:0048012) |
| 0.0 | 0.1 | GO:0006617 | SRP-dependent cotranslational protein targeting to membrane, signal sequence recognition(GO:0006617) |
| 0.0 | 0.2 | GO:0006344 | maintenance of chromatin silencing(GO:0006344) |
| 0.0 | 7.4 | GO:0031424 | keratinization(GO:0031424) |
| 0.0 | 0.5 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.0 | 0.5 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.0 | 0.5 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.0 | 1.0 | GO:0003416 | endochondral bone growth(GO:0003416) |
| 0.0 | 0.5 | GO:0043922 | negative regulation by host of viral transcription(GO:0043922) |
| 0.0 | 0.2 | GO:0035502 | metanephric part of ureteric bud development(GO:0035502) |
| 0.0 | 0.5 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
| 0.0 | 0.1 | GO:0042539 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.0 | 0.4 | GO:0035092 | sperm chromatin condensation(GO:0035092) |
| 0.0 | 0.1 | GO:0045632 | auditory receptor cell fate determination(GO:0042668) negative regulation of mechanoreceptor differentiation(GO:0045632) negative regulation of pro-B cell differentiation(GO:2000974) negative regulation of inner ear receptor cell differentiation(GO:2000981) |
| 0.0 | 0.3 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.0 | 0.8 | GO:0022400 | regulation of rhodopsin mediated signaling pathway(GO:0022400) |
| 0.0 | 1.7 | GO:0070830 | bicellular tight junction assembly(GO:0070830) |
| 0.0 | 0.2 | GO:0048298 | positive regulation of isotype switching to IgA isotypes(GO:0048298) |
| 0.0 | 0.2 | GO:0038129 | ERBB3 signaling pathway(GO:0038129) |
| 0.0 | 0.1 | GO:0032416 | negative regulation of sodium:proton antiporter activity(GO:0032416) |
| 0.0 | 0.6 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.0 | 0.2 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 | 0.1 | GO:0008295 | spermidine biosynthetic process(GO:0008295) |
| 0.0 | 0.1 | GO:1901895 | negative regulation of calcium-transporting ATPase activity(GO:1901895) |
| 0.0 | 0.2 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.3 | GO:0031069 | hair follicle morphogenesis(GO:0031069) |
| 0.0 | 0.1 | GO:0060481 | lobar bronchus epithelium development(GO:0060481) |
| 0.0 | 0.1 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.0 | 0.1 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.0 | 0.0 | GO:2001016 | positive regulation of skeletal muscle cell differentiation(GO:2001016) |
| 0.0 | 0.6 | GO:0045672 | positive regulation of osteoclast differentiation(GO:0045672) |
| 0.0 | 0.1 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.0 | 0.1 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.0 | 0.2 | GO:0015747 | urate transport(GO:0015747) |
| 0.0 | 0.2 | GO:0060965 | negative regulation of gene silencing by miRNA(GO:0060965) |
| 0.0 | 0.1 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.0 | 0.2 | GO:0002158 | osteoclast proliferation(GO:0002158) |
| 0.0 | 1.7 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.0 | 1.4 | GO:0001657 | ureteric bud development(GO:0001657) |
| 0.0 | 0.6 | GO:0090140 | regulation of mitochondrial fission(GO:0090140) |
| 0.0 | 0.1 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.1 | GO:0097398 | response to interleukin-17(GO:0097396) cellular response to interleukin-17(GO:0097398) |
| 0.0 | 0.3 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.0 | 0.2 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.0 | 0.1 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.0 | 0.1 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.0 | 0.1 | GO:0046598 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) positive regulation of viral entry into host cell(GO:0046598) |
| 0.0 | 0.9 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.0 | 0.1 | GO:0051592 | response to calcium ion(GO:0051592) |
| 0.0 | 0.9 | GO:0007585 | respiratory gaseous exchange(GO:0007585) |
| 0.0 | 0.1 | GO:0048850 | hypophysis morphogenesis(GO:0048850) |
| 0.0 | 1.8 | GO:0048706 | embryonic skeletal system development(GO:0048706) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 4.1 | GO:0034667 | integrin alpha3-beta1 complex(GO:0034667) |
| 0.8 | 2.4 | GO:0034678 | integrin alpha8-beta1 complex(GO:0034678) |
| 0.3 | 3.5 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.3 | 1.1 | GO:0005607 | laminin-2 complex(GO:0005607) |
| 0.2 | 2.1 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.2 | 0.6 | GO:0043511 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.2 | 0.5 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 0.1 | 1.0 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.1 | 3.5 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.1 | 0.6 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.1 | 1.0 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.1 | 9.8 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 2.3 | GO:0005861 | troponin complex(GO:0005861) |
| 0.1 | 3.0 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.1 | 2.5 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.1 | 0.2 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.1 | 0.9 | GO:0097486 | alveolar lamellar body(GO:0097208) multivesicular body lumen(GO:0097486) |
| 0.1 | 0.2 | GO:0031379 | RNA-directed RNA polymerase complex(GO:0031379) |
| 0.1 | 0.3 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.1 | 1.1 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.1 | 0.5 | GO:1990393 | 3M complex(GO:1990393) |
| 0.1 | 0.4 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.1 | 0.5 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.1 | 1.2 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.1 | 2.1 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 2.2 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.0 | 0.1 | GO:0034686 | integrin alphav-beta8 complex(GO:0034686) |
| 0.0 | 0.6 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 0.5 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.0 | 0.6 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.6 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.0 | 0.9 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 3.0 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 1.5 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.1 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.0 | 0.6 | GO:0097449 | astrocyte projection(GO:0097449) |
| 0.0 | 0.4 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.0 | 1.2 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.0 | 6.7 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 1.2 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 0.8 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
| 0.0 | 0.2 | GO:0030905 | retromer, tubulation complex(GO:0030905) |
| 0.0 | 0.1 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.0 | 0.3 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.5 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 1.4 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 0.2 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 2.0 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 2.3 | GO:0044853 | plasma membrane raft(GO:0044853) |
| 0.0 | 0.4 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 1.1 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.6 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 2.0 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.1 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 4.0 | GO:0030017 | sarcomere(GO:0030017) |
| 0.0 | 0.3 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 2.0 | GO:0070160 | occluding junction(GO:0070160) |
| 0.0 | 0.5 | GO:0097546 | ciliary base(GO:0097546) |
| 0.0 | 0.1 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.0 | 0.3 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.0 | 0.2 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 1.5 | GO:0005604 | basement membrane(GO:0005604) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.5 | GO:0005171 | hepatocyte growth factor receptor binding(GO:0005171) |
| 0.3 | 1.0 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.3 | 0.9 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 0.3 | 2.8 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.3 | 1.1 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.2 | 1.4 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.2 | 0.7 | GO:0004874 | aryl hydrocarbon receptor activity(GO:0004874) |
| 0.2 | 1.0 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.2 | 1.6 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.2 | 1.4 | GO:0032810 | sterol response element binding(GO:0032810) |
| 0.1 | 1.6 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.1 | 0.7 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.1 | 3.5 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.1 | 1.2 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.1 | 0.5 | GO:0008745 | N-acetylmuramoyl-L-alanine amidase activity(GO:0008745) |
| 0.1 | 3.3 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.1 | 2.5 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.1 | 1.6 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.1 | 0.5 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.1 | 4.1 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.1 | 1.2 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.1 | 0.3 | GO:0038100 | nodal binding(GO:0038100) |
| 0.1 | 0.3 | GO:0034353 | RNA pyrophosphohydrolase activity(GO:0034353) |
| 0.1 | 1.4 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.1 | 0.2 | GO:0003968 | RNA-directed RNA polymerase activity(GO:0003968) |
| 0.1 | 1.1 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.1 | 1.7 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.1 | 0.6 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.1 | 1.8 | GO:0019841 | retinol binding(GO:0019841) |
| 0.1 | 0.6 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.1 | 0.2 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.1 | 1.3 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.1 | 1.7 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 5.5 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 3.9 | GO:0070063 | RNA polymerase binding(GO:0070063) |
| 0.0 | 0.6 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.6 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.0 | 0.2 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.0 | 0.1 | GO:0004766 | spermidine synthase activity(GO:0004766) |
| 0.0 | 0.5 | GO:0038085 | vascular endothelial growth factor binding(GO:0038085) |
| 0.0 | 0.7 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.3 | GO:0004331 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.0 | 0.2 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 4.5 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.0 | 1.6 | GO:0016814 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amidines(GO:0016814) |
| 0.0 | 1.3 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.2 | GO:0008079 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.0 | 0.6 | GO:0008430 | selenium binding(GO:0008430) |
| 0.0 | 0.1 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.0 | 6.5 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.2 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 0.5 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 0.1 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.0 | 0.7 | GO:0008527 | taste receptor activity(GO:0008527) |
| 0.0 | 0.1 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.0 | 0.2 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.0 | 0.6 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.4 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.9 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.1 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.0 | 0.7 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.3 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 1.0 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 1.3 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.0 | 0.3 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.4 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.0 | 0.4 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.3 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 3.9 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 0.8 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.4 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.2 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.0 | 8.0 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) |
| 0.0 | 0.1 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.1 | GO:0004515 | nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 0.0 | 3.7 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 0.1 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.0 | 0.6 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.0 | 0.3 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.1 | GO:0005280 | hydrogen:amino acid symporter activity(GO:0005280) |
| 0.0 | 0.6 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.0 | 0.7 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.1 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.0 | 0.3 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.0 | 1.6 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.5 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.3 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.2 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.1 | 1.0 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 2.9 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 2.5 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 1.0 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 2.1 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 1.7 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 1.7 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 0.6 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.5 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.4 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.0 | 1.0 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 2.2 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.8 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 1.8 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.0 | 2.0 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 1.4 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 0.6 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.8 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 0.2 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.0 | 0.1 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.5 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.1 | 8.4 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 4.1 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.1 | 3.8 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.1 | 1.6 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 1.4 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.0 | 0.4 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 2.7 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 1.5 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |
| 0.0 | 0.6 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.8 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 1.9 | REACTOME REGULATION OF HYPOXIA INDUCIBLE FACTOR HIF BY OXYGEN | Genes involved in Regulation of Hypoxia-inducible Factor (HIF) by Oxygen |
| 0.0 | 0.7 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.8 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 1.2 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.0 | 0.5 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.4 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 1.3 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 0.6 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.0 | 0.8 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 2.7 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.4 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 0.5 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.5 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.6 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.4 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.2 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.9 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.2 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |