Project
Illumina Body Map 2: averaged replicates
Navigation
Downloads
Results for FOSL2_SMARCC1
Z-value: 3.79
Transcription factors associated with FOSL2_SMARCC1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
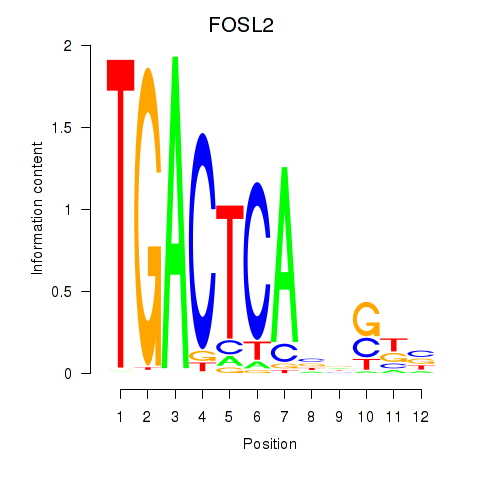
FOSL2
|
ENSG00000075426.7 | FOS like 2, AP-1 transcription factor subunit |
|
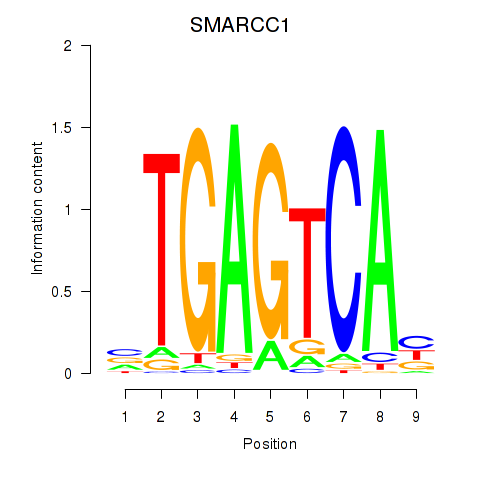
SMARCC1
|
ENSG00000173473.6 | SWI/SNF related, matrix associated, actin dependent regulator of chromatin subfamily c member 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| FOSL2 | hg19_v2_chr2_+_28615669_28615733 | 0.47 | 7.2e-03 | Click! |
| SMARCC1 | hg19_v2_chr3_-_47823298_47823423 | 0.19 | 2.9e-01 | Click! |
Activity profile of FOSL2_SMARCC1 motif
Sorted Z-values of FOSL2_SMARCC1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_89901292 | 23.28 |
ENST00000448155.2
|
IGKV1D-39
|
immunoglobulin kappa variable 1D-39 |
| chr7_+_73245193 | 15.42 |
ENST00000340958.2
|
CLDN4
|
claudin 4 |
| chr2_+_89998789 | 15.33 |
ENST00000453166.2
|
IGKV2D-28
|
immunoglobulin kappa variable 2D-28 |
| chr2_+_90192768 | 15.13 |
ENST00000390275.2
|
IGKV1D-13
|
immunoglobulin kappa variable 1D-13 |
| chr2_-_89521942 | 14.74 |
ENST00000482769.1
|
IGKV2-28
|
immunoglobulin kappa variable 2-28 |
| chr2_-_89417335 | 14.46 |
ENST00000490686.1
|
IGKV1-17
|
immunoglobulin kappa variable 1-17 |
| chr2_+_89999259 | 13.87 |
ENST00000558026.1
|
IGKV2D-28
|
immunoglobulin kappa variable 2D-28 |
| chr22_+_23134974 | 13.45 |
ENST00000390314.2
|
IGLV2-11
|
immunoglobulin lambda variable 2-11 |
| chr2_-_89399845 | 13.24 |
ENST00000479981.1
|
IGKV1-16
|
immunoglobulin kappa variable 1-16 |
| chr22_+_23241661 | 12.14 |
ENST00000390322.2
|
IGLJ2
|
immunoglobulin lambda joining 2 |
| chr22_+_22723969 | 11.44 |
ENST00000390295.2
|
IGLV7-46
|
immunoglobulin lambda variable 7-46 (gene/pseudogene) |
| chr22_+_23029188 | 11.39 |
ENST00000390305.2
|
IGLV3-25
|
immunoglobulin lambda variable 3-25 |
| chr22_+_23077065 | 11.36 |
ENST00000390310.2
|
IGLV2-18
|
immunoglobulin lambda variable 2-18 |
| chr18_+_21452964 | 11.35 |
ENST00000587184.1
|
LAMA3
|
laminin, alpha 3 |
| chr18_+_21452804 | 11.29 |
ENST00000269217.6
|
LAMA3
|
laminin, alpha 3 |
| chr2_+_90139056 | 11.02 |
ENST00000492446.1
|
IGKV1D-16
|
immunoglobulin kappa variable 1D-16 |
| chr2_-_89340242 | 10.98 |
ENST00000480492.1
|
IGKV1-12
|
immunoglobulin kappa variable 1-12 |
| chr2_+_90121477 | 10.78 |
ENST00000483379.1
|
IGKV1D-17
|
immunoglobulin kappa variable 1D-17 |
| chr22_+_23010756 | 10.64 |
ENST00000390304.2
|
IGLV3-27
|
immunoglobulin lambda variable 3-27 |
| chr22_+_22735135 | 10.54 |
ENST00000390297.2
|
IGLV1-44
|
immunoglobulin lambda variable 1-44 |
| chr17_-_39769005 | 9.84 |
ENST00000301653.4
ENST00000593067.1 |
KRT16
|
keratin 16 |
| chr8_-_82395461 | 9.83 |
ENST00000256104.4
|
FABP4
|
fatty acid binding protein 4, adipocyte |
| chr2_+_89952792 | 9.71 |
ENST00000390265.2
|
IGKV1D-33
|
immunoglobulin kappa variable 1D-33 |
| chr2_-_89568263 | 9.65 |
ENST00000473726.1
|
IGKV1-33
|
immunoglobulin kappa variable 1-33 |
| chr7_+_30960915 | 9.31 |
ENST00000441328.2
ENST00000409899.1 ENST00000409611.1 |
AQP1
|
aquaporin 1 (Colton blood group) |
| chr2_-_89459813 | 8.77 |
ENST00000390256.2
|
IGKV6-21
|
immunoglobulin kappa variable 6-21 (non-functional) |
| chr22_+_23089870 | 8.44 |
ENST00000390311.2
|
IGLV3-16
|
immunoglobulin lambda variable 3-16 |
| chr22_+_22569155 | 8.22 |
ENST00000390287.2
|
IGLV10-54
|
immunoglobulin lambda variable 10-54 |
| chr2_-_89619904 | 7.74 |
ENST00000498574.1
|
IGKV1-39
|
immunoglobulin kappa variable 1-39 (gene/pseudogene) |
| chr22_+_23063100 | 7.11 |
ENST00000390309.2
|
IGLV3-19
|
immunoglobulin lambda variable 3-19 |
| chr2_+_89923550 | 7.02 |
ENST00000509129.1
|
IGKV1D-37
|
immunoglobulin kappa variable 1D-37 (non-functional) |
| chr2_+_90198535 | 6.94 |
ENST00000390276.2
|
IGKV1D-12
|
immunoglobulin kappa variable 1D-12 |
| chr22_+_22453093 | 6.66 |
ENST00000390283.2
|
IGLV8-61
|
immunoglobulin lambda variable 8-61 |
| chr11_-_68780824 | 6.12 |
ENST00000441623.1
ENST00000309099.6 |
MRGPRF
|
MAS-related GPR, member F |
| chr22_+_23165153 | 6.05 |
ENST00000390317.2
|
IGLV2-8
|
immunoglobulin lambda variable 2-8 |
| chr22_+_22749343 | 6.00 |
ENST00000390298.2
|
IGLV7-43
|
immunoglobulin lambda variable 7-43 |
| chr1_-_153521714 | 5.87 |
ENST00000368713.3
|
S100A3
|
S100 calcium binding protein A3 |
| chr19_+_46009837 | 5.86 |
ENST00000589627.1
|
VASP
|
vasodilator-stimulated phosphoprotein |
| chr6_+_41604747 | 5.80 |
ENST00000419164.1
ENST00000373051.2 |
MDFI
|
MyoD family inhibitor |
| chr22_+_22730353 | 5.75 |
ENST00000390296.2
|
IGLV5-45
|
immunoglobulin lambda variable 5-45 |
| chr1_-_153521597 | 5.67 |
ENST00000368712.1
|
S100A3
|
S100 calcium binding protein A3 |
| chr22_+_22550113 | 5.53 |
ENST00000390285.3
|
IGLV6-57
|
immunoglobulin lambda variable 6-57 |
| chr2_+_90060377 | 5.45 |
ENST00000436451.2
|
IGKV6D-21
|
immunoglobulin kappa variable 6D-21 (non-functional) |
| chr20_+_44637526 | 5.18 |
ENST00000372330.3
|
MMP9
|
matrix metallopeptidase 9 (gelatinase B, 92kDa gelatinase, 92kDa type IV collagenase) |
| chr22_+_22712087 | 5.14 |
ENST00000390294.2
|
IGLV1-47
|
immunoglobulin lambda variable 1-47 |
| chr22_+_23154239 | 5.03 |
ENST00000390315.2
|
IGLV3-10
|
immunoglobulin lambda variable 3-10 |
| chr1_-_151965048 | 5.01 |
ENST00000368809.1
|
S100A10
|
S100 calcium binding protein A10 |
| chr1_-_223853425 | 4.97 |
ENST00000366873.2
ENST00000419193.2 |
CAPN8
|
calpain 8 |
| chr5_+_150020214 | 4.94 |
ENST00000307662.4
|
SYNPO
|
synaptopodin |
| chr6_+_41604620 | 4.91 |
ENST00000432027.1
|
MDFI
|
MyoD family inhibitor |
| chr1_+_156084461 | 4.82 |
ENST00000347559.2
ENST00000361308.4 ENST00000368300.4 ENST00000368299.3 |
LMNA
|
lamin A/C |
| chr1_-_203198790 | 4.82 |
ENST00000367229.1
ENST00000255427.3 ENST00000535569.1 |
CHIT1
|
chitinase 1 (chitotriosidase) |
| chr22_+_23040274 | 4.75 |
ENST00000390306.2
|
IGLV2-23
|
immunoglobulin lambda variable 2-23 |
| chr4_+_74606223 | 4.74 |
ENST00000307407.3
ENST00000401931.1 |
IL8
|
interleukin 8 |
| chr15_+_41136216 | 4.68 |
ENST00000562057.1
ENST00000344051.4 |
SPINT1
|
serine peptidase inhibitor, Kunitz type 1 |
| chr22_+_22781853 | 4.58 |
ENST00000390300.2
|
IGLV5-37
|
immunoglobulin lambda variable 5-37 |
| chr16_-_30125177 | 4.57 |
ENST00000406256.3
|
GDPD3
|
glycerophosphodiester phosphodiesterase domain containing 3 |
| chr1_-_223853348 | 4.55 |
ENST00000366872.5
|
CAPN8
|
calpain 8 |
| chr2_+_113875466 | 4.52 |
ENST00000361779.3
ENST00000259206.5 ENST00000354115.2 |
IL1RN
|
interleukin 1 receptor antagonist |
| chr15_+_41136586 | 4.46 |
ENST00000431806.1
|
SPINT1
|
serine peptidase inhibitor, Kunitz type 1 |
| chr11_+_393428 | 4.41 |
ENST00000533249.1
ENST00000527442.1 |
PKP3
|
plakophilin 3 |
| chr5_-_141061777 | 4.35 |
ENST00000239440.4
|
ARAP3
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 3 |
| chr13_+_102142296 | 4.32 |
ENST00000376162.3
|
ITGBL1
|
integrin, beta-like 1 (with EGF-like repeat domains) |
| chr2_+_87754989 | 4.30 |
ENST00000409898.2
ENST00000419680.2 ENST00000414584.1 ENST00000455131.1 |
LINC00152
|
long intergenic non-protein coding RNA 152 |
| chr19_-_35992780 | 4.26 |
ENST00000593342.1
ENST00000601650.1 ENST00000408915.2 |
DMKN
|
dermokine |
| chr2_+_87755054 | 4.24 |
ENST00000423846.1
|
LINC00152
|
long intergenic non-protein coding RNA 152 |
| chr10_+_17270214 | 4.19 |
ENST00000544301.1
|
VIM
|
vimentin |
| chr5_-_141061759 | 4.18 |
ENST00000508305.1
|
ARAP3
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 3 |
| chr11_-_102668879 | 4.18 |
ENST00000315274.6
|
MMP1
|
matrix metallopeptidase 1 (interstitial collagenase) |
| chr21_+_10862622 | 4.16 |
ENST00000302092.5
ENST00000559480.1 |
IGHV1OR21-1
|
immunoglobulin heavy variable 1/OR21-1 (non-functional) |
| chr2_+_87754887 | 4.14 |
ENST00000409054.1
ENST00000331944.6 ENST00000409139.1 |
LINC00152
|
long intergenic non-protein coding RNA 152 |
| chr6_-_31704282 | 4.09 |
ENST00000375784.3
ENST00000375779.2 |
CLIC1
|
chloride intracellular channel 1 |
| chr8_+_70476088 | 4.07 |
ENST00000525999.1
|
SULF1
|
sulfatase 1 |
| chr1_+_27189631 | 4.06 |
ENST00000339276.4
|
SFN
|
stratifin |
| chr17_-_38721711 | 4.06 |
ENST00000578085.1
ENST00000246657.2 |
CCR7
|
chemokine (C-C motif) receptor 7 |
| chr12_-_48152611 | 3.98 |
ENST00000389212.3
|
RAPGEF3
|
Rap guanine nucleotide exchange factor (GEF) 3 |
| chr19_-_44174330 | 3.97 |
ENST00000340093.3
|
PLAUR
|
plasminogen activator, urokinase receptor |
| chr16_+_1290694 | 3.91 |
ENST00000338844.3
|
TPSAB1
|
tryptase alpha/beta 1 |
| chr19_-_44174305 | 3.91 |
ENST00000601723.1
ENST00000339082.3 |
PLAUR
|
plasminogen activator, urokinase receptor |
| chr11_+_101983176 | 3.90 |
ENST00000524575.1
|
YAP1
|
Yes-associated protein 1 |
| chr3_+_36421971 | 3.88 |
ENST00000457375.2
ENST00000434649.1 |
STAC
|
SH3 and cysteine rich domain |
| chr12_-_56236734 | 3.84 |
ENST00000548629.1
|
MMP19
|
matrix metallopeptidase 19 |
| chrX_+_99899180 | 3.82 |
ENST00000373004.3
|
SRPX2
|
sushi-repeat containing protein, X-linked 2 |
| chr2_-_89597542 | 3.79 |
ENST00000465170.1
|
IGKV1-37
|
immunoglobulin kappa variable 1-37 (non-functional) |
| chr1_-_156675368 | 3.71 |
ENST00000368222.3
|
CRABP2
|
cellular retinoic acid binding protein 2 |
| chr17_-_39743139 | 3.71 |
ENST00000167586.6
|
KRT14
|
keratin 14 |
| chr2_+_89890533 | 3.70 |
ENST00000429992.2
|
IGKV2D-40
|
immunoglobulin kappa variable 2D-40 |
| chr12_-_56236711 | 3.68 |
ENST00000409200.3
|
MMP19
|
matrix metallopeptidase 19 |
| chr1_+_27669719 | 3.66 |
ENST00000473280.1
|
SYTL1
|
synaptotagmin-like 1 |
| chr7_+_22766766 | 3.63 |
ENST00000426291.1
ENST00000401651.1 ENST00000258743.5 ENST00000420258.2 ENST00000407492.1 ENST00000401630.3 ENST00000406575.1 |
IL6
|
interleukin 6 (interferon, beta 2) |
| chr16_-_122619 | 3.62 |
ENST00000262316.6
|
RHBDF1
|
rhomboid 5 homolog 1 (Drosophila) |
| chr15_+_41136734 | 3.59 |
ENST00000568580.1
|
SPINT1
|
serine peptidase inhibitor, Kunitz type 1 |
| chr12_+_48152774 | 3.58 |
ENST00000549243.1
|
SLC48A1
|
solute carrier family 48 (heme transporter), member 1 |
| chr11_+_394196 | 3.55 |
ENST00000331563.2
ENST00000531857.1 |
PKP3
|
plakophilin 3 |
| chr16_+_1306060 | 3.54 |
ENST00000397534.2
|
TPSD1
|
tryptase delta 1 |
| chrX_-_154563889 | 3.52 |
ENST00000369449.2
ENST00000321926.4 |
CLIC2
|
chloride intracellular channel 2 |
| chr15_+_41136369 | 3.50 |
ENST00000563656.1
|
SPINT1
|
serine peptidase inhibitor, Kunitz type 1 |
| chr2_+_114163945 | 3.50 |
ENST00000453673.3
|
IGKV1OR2-108
|
immunoglobulin kappa variable 1/OR2-108 (non-functional) |
| chr16_+_1290725 | 3.48 |
ENST00000461509.2
|
TPSAB1
|
tryptase alpha/beta 1 |
| chr8_+_105352050 | 3.47 |
ENST00000297581.2
|
DCSTAMP
|
dendrocyte expressed seven transmembrane protein |
| chr12_-_48152853 | 3.47 |
ENST00000171000.4
|
RAPGEF3
|
Rap guanine nucleotide exchange factor (GEF) 3 |
| chr15_+_41136263 | 3.47 |
ENST00000568823.1
|
SPINT1
|
serine peptidase inhibitor, Kunitz type 1 |
| chr10_-_4285835 | 3.46 |
ENST00000454470.1
|
LINC00702
|
long intergenic non-protein coding RNA 702 |
| chr5_-_75919217 | 3.39 |
ENST00000504899.1
|
F2RL2
|
coagulation factor II (thrombin) receptor-like 2 |
| chr7_+_94023873 | 3.30 |
ENST00000297268.6
|
COL1A2
|
collagen, type I, alpha 2 |
| chr1_+_201252580 | 3.23 |
ENST00000367324.3
ENST00000263946.3 |
PKP1
|
plakophilin 1 (ectodermal dysplasia/skin fragility syndrome) |
| chr19_-_44285401 | 3.22 |
ENST00000262888.3
|
KCNN4
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 4 |
| chr12_-_53343602 | 3.20 |
ENST00000546897.1
ENST00000552551.1 |
KRT8
|
keratin 8 |
| chr11_-_6341844 | 3.19 |
ENST00000303927.3
|
PRKCDBP
|
protein kinase C, delta binding protein |
| chr1_-_12679171 | 3.17 |
ENST00000606790.1
|
RP11-474O21.5
|
RP11-474O21.5 |
| chr12_-_56236690 | 3.16 |
ENST00000322569.4
|
MMP19
|
matrix metallopeptidase 19 |
| chr3_+_30648066 | 3.14 |
ENST00000359013.4
|
TGFBR2
|
transforming growth factor, beta receptor II (70/80kDa) |
| chr3_+_30647994 | 3.10 |
ENST00000295754.5
|
TGFBR2
|
transforming growth factor, beta receptor II (70/80kDa) |
| chr16_+_1306093 | 3.08 |
ENST00000211076.3
|
TPSD1
|
tryptase delta 1 |
| chr1_-_43751276 | 3.07 |
ENST00000423420.1
|
C1orf210
|
chromosome 1 open reading frame 210 |
| chr10_-_6019552 | 3.07 |
ENST00000379977.3
ENST00000397251.3 ENST00000397248.2 |
IL15RA
|
interleukin 15 receptor, alpha |
| chr6_+_44194762 | 3.05 |
ENST00000371708.1
|
SLC29A1
|
solute carrier family 29 (equilibrative nucleoside transporter), member 1 |
| chr3_-_87040233 | 3.05 |
ENST00000398399.2
|
VGLL3
|
vestigial like 3 (Drosophila) |
| chr11_-_67141640 | 3.03 |
ENST00000533438.1
|
CLCF1
|
cardiotrophin-like cytokine factor 1 |
| chr15_-_51535208 | 3.02 |
ENST00000405913.3
ENST00000559878.1 |
CYP19A1
|
cytochrome P450, family 19, subfamily A, polypeptide 1 |
| chr2_+_120189422 | 3.02 |
ENST00000306406.4
|
TMEM37
|
transmembrane protein 37 |
| chr13_-_41593425 | 3.01 |
ENST00000239882.3
|
ELF1
|
E74-like factor 1 (ets domain transcription factor) |
| chr3_+_11178779 | 2.98 |
ENST00000438284.2
|
HRH1
|
histamine receptor H1 |
| chr18_+_3450161 | 2.98 |
ENST00000551402.1
ENST00000577543.1 |
TGIF1
|
TGFB-induced factor homeobox 1 |
| chr18_+_61442629 | 2.96 |
ENST00000398019.2
ENST00000540675.1 |
SERPINB7
|
serpin peptidase inhibitor, clade B (ovalbumin), member 7 |
| chr1_+_183155373 | 2.92 |
ENST00000493293.1
ENST00000264144.4 |
LAMC2
|
laminin, gamma 2 |
| chr15_+_89182156 | 2.90 |
ENST00000379224.5
|
ISG20
|
interferon stimulated exonuclease gene 20kDa |
| chr8_+_87111059 | 2.88 |
ENST00000285393.3
|
ATP6V0D2
|
ATPase, H+ transporting, lysosomal 38kDa, V0 subunit d2 |
| chr14_-_24806588 | 2.86 |
ENST00000555591.1
ENST00000554569.1 |
RP11-934B9.3
RIPK3
|
Uncharacterized protein receptor-interacting serine-threonine kinase 3 |
| chr11_-_65667884 | 2.86 |
ENST00000448083.2
ENST00000531493.1 ENST00000532401.1 |
FOSL1
|
FOS-like antigen 1 |
| chr10_-_6019455 | 2.86 |
ENST00000530685.1
ENST00000397255.3 ENST00000379971.1 ENST00000528354.1 ENST00000397250.2 ENST00000429135.2 |
IL15RA
|
interleukin 15 receptor, alpha |
| chr5_-_75919253 | 2.82 |
ENST00000296641.4
|
F2RL2
|
coagulation factor II (thrombin) receptor-like 2 |
| chrX_+_115567767 | 2.78 |
ENST00000371900.4
|
SLC6A14
|
solute carrier family 6 (amino acid transporter), member 14 |
| chr14_-_107078851 | 2.77 |
ENST00000390628.2
|
IGHV1-58
|
immunoglobulin heavy variable 1-58 |
| chr12_+_13349650 | 2.77 |
ENST00000256951.5
ENST00000431267.2 ENST00000542474.1 ENST00000544053.1 |
EMP1
|
epithelial membrane protein 1 |
| chr11_-_62457371 | 2.77 |
ENST00000317449.4
|
LRRN4CL
|
LRRN4 C-terminal like |
| chr1_-_43751230 | 2.76 |
ENST00000523677.1
|
C1orf210
|
chromosome 1 open reading frame 210 |
| chr15_+_89181974 | 2.76 |
ENST00000306072.5
|
ISG20
|
interferon stimulated exonuclease gene 20kDa |
| chr16_-_2908155 | 2.75 |
ENST00000571228.1
ENST00000161006.3 |
PRSS22
|
protease, serine, 22 |
| chr3_-_87039662 | 2.75 |
ENST00000494229.1
|
VGLL3
|
vestigial like 3 (Drosophila) |
| chr2_+_36923830 | 2.75 |
ENST00000379242.3
ENST00000389975.3 |
VIT
|
vitrin |
| chr19_+_39279838 | 2.74 |
ENST00000314980.4
|
LGALS7B
|
lectin, galactoside-binding, soluble, 7B |
| chr1_-_113247543 | 2.72 |
ENST00000414971.1
ENST00000534717.1 |
RHOC
|
ras homolog family member C |
| chr14_-_25103472 | 2.71 |
ENST00000216341.4
ENST00000382542.1 ENST00000382540.1 |
GZMB
|
granzyme B (granzyme 2, cytotoxic T-lymphocyte-associated serine esterase 1) |
| chr15_-_20170354 | 2.71 |
ENST00000338912.5
|
IGHV1OR15-9
|
immunoglobulin heavy variable 1/OR15-9 (non-functional) |
| chr1_-_45272951 | 2.68 |
ENST00000372200.1
|
TCTEX1D4
|
Tctex1 domain containing 4 |
| chr12_-_91573249 | 2.68 |
ENST00000550099.1
ENST00000546391.1 ENST00000551354.1 |
DCN
|
decorin |
| chr19_-_47128294 | 2.68 |
ENST00000596260.1
ENST00000597185.1 ENST00000598865.1 ENST00000594275.1 ENST00000291294.2 |
PTGIR
|
prostaglandin I2 (prostacyclin) receptor (IP) |
| chr17_+_7482785 | 2.67 |
ENST00000250092.6
ENST00000380498.6 ENST00000584502.1 |
CD68
|
CD68 molecule |
| chr11_-_10590118 | 2.66 |
ENST00000529598.1
|
LYVE1
|
lymphatic vessel endothelial hyaluronan receptor 1 |
| chr12_+_13349711 | 2.65 |
ENST00000538364.1
ENST00000396301.3 |
EMP1
|
epithelial membrane protein 1 |
| chr12_-_53343633 | 2.63 |
ENST00000546826.1
|
KRT8
|
keratin 8 |
| chr1_-_153538011 | 2.62 |
ENST00000368707.4
|
S100A2
|
S100 calcium binding protein A2 |
| chr1_+_27719148 | 2.61 |
ENST00000374024.3
|
GPR3
|
G protein-coupled receptor 3 |
| chr10_-_6019984 | 2.59 |
ENST00000525219.2
|
IL15RA
|
interleukin 15 receptor, alpha |
| chr12_-_91573316 | 2.58 |
ENST00000393155.1
|
DCN
|
decorin |
| chr1_+_901847 | 2.57 |
ENST00000379410.3
ENST00000379409.2 ENST00000379407.3 |
PLEKHN1
|
pleckstrin homology domain containing, family N member 1 |
| chr12_-_91573132 | 2.57 |
ENST00000550563.1
ENST00000546370.1 |
DCN
|
decorin |
| chr11_+_128563652 | 2.56 |
ENST00000527786.2
|
FLI1
|
Fli-1 proto-oncogene, ETS transcription factor |
| chr15_+_89182178 | 2.56 |
ENST00000559876.1
|
ISG20
|
interferon stimulated exonuclease gene 20kDa |
| chr16_-_69760409 | 2.55 |
ENST00000561500.1
ENST00000439109.2 ENST00000564043.1 ENST00000379046.2 ENST00000379047.3 |
NQO1
|
NAD(P)H dehydrogenase, quinone 1 |
| chr17_+_59529743 | 2.55 |
ENST00000589003.1
ENST00000393853.4 |
TBX4
|
T-box 4 |
| chr2_+_208104351 | 2.51 |
ENST00000440326.1
|
AC007879.7
|
AC007879.7 |
| chr11_+_35639735 | 2.51 |
ENST00000317811.4
|
FJX1
|
four jointed box 1 (Drosophila) |
| chr11_-_65667997 | 2.51 |
ENST00000312562.2
ENST00000534222.1 |
FOSL1
|
FOS-like antigen 1 |
| chr3_+_191046810 | 2.49 |
ENST00000392455.3
ENST00000392456.3 |
CCDC50
|
coiled-coil domain containing 50 |
| chr14_+_22748980 | 2.48 |
ENST00000390465.2
|
TRAV38-2DV8
|
T cell receptor alpha variable 38-2/delta variable 8 |
| chr22_+_22707260 | 2.48 |
ENST00000390293.1
|
IGLV5-48
|
immunoglobulin lambda variable 5-48 (non-functional) |
| chr1_+_156095951 | 2.45 |
ENST00000448611.2
ENST00000368297.1 |
LMNA
|
lamin A/C |
| chr19_-_40931891 | 2.42 |
ENST00000357949.4
|
SERTAD1
|
SERTA domain containing 1 |
| chr5_-_149535421 | 2.41 |
ENST00000261799.4
|
PDGFRB
|
platelet-derived growth factor receptor, beta polypeptide |
| chr7_-_135433534 | 2.41 |
ENST00000338588.3
|
FAM180A
|
family with sequence similarity 180, member A |
| chrX_-_48901012 | 2.38 |
ENST00000315869.7
|
TFE3
|
transcription factor binding to IGHM enhancer 3 |
| chr3_+_148447887 | 2.37 |
ENST00000475347.1
ENST00000474935.1 ENST00000461609.1 |
AGTR1
|
angiotensin II receptor, type 1 |
| chr17_-_19062424 | 2.36 |
ENST00000399083.1
|
AC007952.6
|
Uncharacterized protein |
| chr2_+_102618428 | 2.36 |
ENST00000457817.1
|
IL1R2
|
interleukin 1 receptor, type II |
| chr13_+_78109884 | 2.35 |
ENST00000377246.3
ENST00000349847.3 |
SCEL
|
sciellin |
| chr19_-_36004543 | 2.33 |
ENST00000339686.3
ENST00000447113.2 ENST00000440396.1 |
DMKN
|
dermokine |
| chr19_+_38794797 | 2.31 |
ENST00000301246.5
ENST00000588605.1 |
C19orf33
|
chromosome 19 open reading frame 33 |
| chr11_+_46402297 | 2.29 |
ENST00000405308.2
|
MDK
|
midkine (neurite growth-promoting factor 2) |
| chr1_-_158656488 | 2.28 |
ENST00000368147.4
|
SPTA1
|
spectrin, alpha, erythrocytic 1 (elliptocytosis 2) |
| chr3_-_149095652 | 2.27 |
ENST00000305366.3
|
TM4SF1
|
transmembrane 4 L six family member 1 |
| chr10_+_75668916 | 2.27 |
ENST00000481390.1
|
PLAU
|
plasminogen activator, urokinase |
| chr8_+_104384616 | 2.26 |
ENST00000520337.1
|
CTHRC1
|
collagen triple helix repeat containing 1 |
| chr11_-_10590238 | 2.26 |
ENST00000256178.3
|
LYVE1
|
lymphatic vessel endothelial hyaluronan receptor 1 |
| chr3_-_194090460 | 2.25 |
ENST00000428839.1
ENST00000347624.3 |
LRRC15
|
leucine rich repeat containing 15 |
| chr16_+_31366536 | 2.24 |
ENST00000562522.1
|
ITGAX
|
integrin, alpha X (complement component 3 receptor 4 subunit) |
| chr3_-_151034734 | 2.23 |
ENST00000260843.4
|
GPR87
|
G protein-coupled receptor 87 |
| chr5_-_141030943 | 2.23 |
ENST00000522783.1
ENST00000519800.1 ENST00000435817.2 |
FCHSD1
|
FCH and double SH3 domains 1 |
| chr2_+_90248739 | 2.18 |
ENST00000468879.1
|
IGKV1D-43
|
immunoglobulin kappa variable 1D-43 |
| chrX_-_129244336 | 2.18 |
ENST00000434609.1
|
ELF4
|
E74-like factor 4 (ets domain transcription factor) |
| chr6_-_149806105 | 2.17 |
ENST00000389942.5
ENST00000416573.2 ENST00000542614.1 ENST00000409806.3 |
ZC3H12D
|
zinc finger CCCH-type containing 12D |
| chr17_-_53800217 | 2.16 |
ENST00000424486.2
|
TMEM100
|
transmembrane protein 100 |
| chr10_+_104155450 | 2.16 |
ENST00000471698.1
ENST00000189444.6 |
NFKB2
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 (p49/p100) |
| chr14_-_25103388 | 2.16 |
ENST00000526004.1
ENST00000415355.3 |
GZMB
|
granzyme B (granzyme 2, cytotoxic T-lymphocyte-associated serine esterase 1) |
| chr13_+_78109804 | 2.15 |
ENST00000535157.1
|
SCEL
|
sciellin |
| chr1_+_26606608 | 2.14 |
ENST00000319041.6
|
SH3BGRL3
|
SH3 domain binding glutamic acid-rich protein like 3 |
| chr14_-_91710852 | 2.14 |
ENST00000535815.1
ENST00000529102.1 |
GPR68
|
G protein-coupled receptor 68 |
| chr11_-_2950642 | 2.13 |
ENST00000314222.4
|
PHLDA2
|
pleckstrin homology-like domain, family A, member 2 |
| chr6_+_74405804 | 2.13 |
ENST00000287097.5
|
CD109
|
CD109 molecule |
Network of associatons between targets according to the STRING database.
First level regulatory network of FOSL2_SMARCC1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.1 | 9.3 | GO:0035378 | carbon dioxide transmembrane transport(GO:0035378) |
| 2.1 | 6.2 | GO:0002663 | B cell tolerance induction(GO:0002514) regulation of B cell tolerance induction(GO:0002661) positive regulation of B cell tolerance induction(GO:0002663) |
| 1.7 | 8.7 | GO:0035105 | sterol regulatory element binding protein import into nucleus(GO:0035105) |
| 1.6 | 8.2 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 1.5 | 7.5 | GO:0030822 | positive regulation of cyclic nucleotide catabolic process(GO:0030807) positive regulation of cAMP catabolic process(GO:0030822) positive regulation of purine nucleotide catabolic process(GO:0033123) |
| 1.4 | 4.3 | GO:0060981 | cell migration involved in coronary angiogenesis(GO:0060981) |
| 1.4 | 237.2 | GO:0030449 | regulation of complement activation(GO:0030449) |
| 1.4 | 4.1 | GO:2000525 | regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) regulation of dendritic cell dendrite assembly(GO:2000547) |
| 1.2 | 3.7 | GO:1902362 | melanocyte apoptotic process(GO:1902362) |
| 1.2 | 10.5 | GO:2000660 | negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 1.2 | 6.9 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 1.1 | 10.2 | GO:0002159 | desmosome assembly(GO:0002159) |
| 1.1 | 5.4 | GO:0090362 | positive regulation of platelet-derived growth factor production(GO:0090362) |
| 1.0 | 7.9 | GO:0038195 | urokinase plasminogen activator signaling pathway(GO:0038195) |
| 1.0 | 2.9 | GO:0042660 | positive regulation of cell fate specification(GO:0042660) |
| 1.0 | 10.7 | GO:0001554 | luteolysis(GO:0001554) |
| 1.0 | 1.0 | GO:0048807 | female genitalia morphogenesis(GO:0048807) |
| 1.0 | 2.9 | GO:0035698 | CD8-positive, alpha-beta cytotoxic T cell extravasation(GO:0035698) positive regulation of necroptotic process(GO:0060545) regulation of CD8-positive, alpha-beta cytotoxic T cell extravasation(GO:2000452) |
| 0.9 | 3.8 | GO:2000097 | regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.9 | 6.6 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.9 | 24.1 | GO:0060670 | branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 0.9 | 7.4 | GO:0072675 | osteoclast fusion(GO:0072675) |
| 0.8 | 30.4 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.8 | 8.4 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.8 | 4.1 | GO:0010482 | epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 0.8 | 4.0 | GO:0021812 | neuronal-glial interaction involved in cerebral cortex radial glia guided migration(GO:0021812) |
| 0.8 | 2.3 | GO:0098886 | modification of dendritic spine(GO:0098886) |
| 0.8 | 3.0 | GO:2000863 | positive regulation of estrogen secretion(GO:2000863) positive regulation of estradiol secretion(GO:2000866) |
| 0.8 | 15.0 | GO:0051546 | keratinocyte migration(GO:0051546) |
| 0.7 | 2.1 | GO:0072573 | regulation of toll-like receptor 5 signaling pathway(GO:0034147) negative regulation of toll-like receptor 5 signaling pathway(GO:0034148) negative regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070429) tolerance induction to lipopolysaccharide(GO:0072573) |
| 0.7 | 4.8 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.7 | 2.7 | GO:0051771 | negative regulation of nitric-oxide synthase biosynthetic process(GO:0051771) |
| 0.7 | 9.5 | GO:0071285 | cellular response to lithium ion(GO:0071285) |
| 0.7 | 5.4 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.7 | 4.0 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.7 | 2.0 | GO:0060734 | regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation(GO:0060734) |
| 0.6 | 14.5 | GO:0060707 | trophoblast giant cell differentiation(GO:0060707) |
| 0.6 | 1.9 | GO:0071613 | granzyme B production(GO:0071613) regulation of granzyme B production(GO:0071661) positive regulation of granzyme B production(GO:0071663) positive regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000566) |
| 0.6 | 1.9 | GO:0045643 | regulation of eosinophil differentiation(GO:0045643) positive regulation of eosinophil differentiation(GO:0045645) |
| 0.6 | 3.1 | GO:0015862 | uridine transport(GO:0015862) |
| 0.6 | 4.9 | GO:0008626 | granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 0.6 | 2.4 | GO:0006218 | uridine catabolic process(GO:0006218) |
| 0.6 | 2.4 | GO:0035425 | autocrine signaling(GO:0035425) |
| 0.6 | 2.4 | GO:0086097 | phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) |
| 0.6 | 7.7 | GO:0046449 | creatinine metabolic process(GO:0046449) |
| 0.6 | 4.1 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.6 | 9.8 | GO:0051599 | response to hydrostatic pressure(GO:0051599) |
| 0.6 | 122.3 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.6 | 2.8 | GO:0014740 | negative regulation of muscle hyperplasia(GO:0014740) |
| 0.5 | 2.2 | GO:0002266 | follicular dendritic cell activation(GO:0002266) follicular dendritic cell differentiation(GO:0002268) |
| 0.5 | 2.1 | GO:0060723 | spongiotrophoblast cell proliferation(GO:0060720) regulation of spongiotrophoblast cell proliferation(GO:0060721) cell proliferation involved in embryonic placenta development(GO:0060722) regulation of cell proliferation involved in embryonic placenta development(GO:0060723) |
| 0.5 | 1.5 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.5 | 18.3 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.5 | 1.5 | GO:0003430 | growth plate cartilage chondrocyte growth(GO:0003430) |
| 0.5 | 5.8 | GO:0001866 | NK T cell proliferation(GO:0001866) |
| 0.5 | 1.4 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.5 | 1.9 | GO:1903238 | positive regulation of leukocyte tethering or rolling(GO:1903238) |
| 0.5 | 2.7 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 0.5 | 2.3 | GO:0046813 | receptor-mediated virion attachment to host cell(GO:0046813) |
| 0.5 | 1.8 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.4 | 1.3 | GO:1901656 | glycoside transport(GO:1901656) |
| 0.4 | 1.3 | GO:0035607 | vacuolar phosphate transport(GO:0007037) fibroblast growth factor receptor signaling pathway involved in orbitofrontal cortex development(GO:0035607) positive regulation of mitotic cell cycle DNA replication(GO:1903465) positive regulation of parathyroid hormone secretion(GO:2000830) |
| 0.4 | 3.1 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 0.4 | 2.2 | GO:0039516 | modulation by virus of host molecular function(GO:0039506) suppression by virus of host molecular function(GO:0039507) suppression by virus of host catalytic activity(GO:0039513) modulation by virus of host catalytic activity(GO:0039516) suppression by virus of host cysteine-type endopeptidase activity involved in apoptotic process(GO:0039650) negative regulation by symbiont of host catalytic activity(GO:0052053) negative regulation by symbiont of host molecular function(GO:0052056) modulation by symbiont of host catalytic activity(GO:0052148) |
| 0.4 | 6.5 | GO:0090385 | phagosome-lysosome fusion(GO:0090385) |
| 0.4 | 0.9 | GO:1903660 | negative regulation of complement-dependent cytotoxicity(GO:1903660) |
| 0.4 | 2.1 | GO:0009608 | response to symbiont(GO:0009608) response to symbiotic bacterium(GO:0009609) |
| 0.4 | 1.3 | GO:0051414 | response to cortisol(GO:0051414) |
| 0.4 | 5.8 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.4 | 2.3 | GO:0030421 | defecation(GO:0030421) |
| 0.4 | 1.1 | GO:0046521 | sphingoid catabolic process(GO:0046521) |
| 0.4 | 1.5 | GO:0071393 | cellular response to progesterone stimulus(GO:0071393) |
| 0.4 | 1.5 | GO:0044028 | DNA hypomethylation(GO:0044028) hypomethylation of CpG island(GO:0044029) |
| 0.4 | 1.4 | GO:0002925 | positive regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002925) |
| 0.4 | 0.4 | GO:0046136 | positive regulation of vitamin metabolic process(GO:0046136) positive regulation of vitamin D biosynthetic process(GO:0060557) positive regulation of calcidiol 1-monooxygenase activity(GO:0060559) |
| 0.4 | 1.1 | GO:0006117 | acetaldehyde metabolic process(GO:0006117) |
| 0.4 | 2.5 | GO:1900623 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.4 | 3.9 | GO:0060449 | bud elongation involved in lung branching(GO:0060449) |
| 0.4 | 1.4 | GO:0044115 | multi-organism catabolic process(GO:0044035) development of symbiont involved in interaction with host(GO:0044115) modulation of development of symbiont involved in interaction with host(GO:0044145) negative regulation of development of symbiont involved in interaction with host(GO:0044147) metabolism of substance in other organism involved in symbiotic interaction(GO:0052214) catabolism of substance in other organism involved in symbiotic interaction(GO:0052227) metabolism of macromolecule in other organism involved in symbiotic interaction(GO:0052229) catabolism by host of symbiont macromolecule(GO:0052360) catabolism by organism of macromolecule in other organism involved in symbiotic interaction(GO:0052361) catabolism by host of symbiont protein(GO:0052362) catabolism by organism of protein in other organism involved in symbiotic interaction(GO:0052363) catabolism by host of substance in symbiont(GO:0052364) metabolism by host of symbiont macromolecule(GO:0052416) metabolism by host of symbiont protein(GO:0052417) metabolism by organism of protein in other organism involved in symbiotic interaction(GO:0052418) metabolism by host of substance in symbiont(GO:0052419) |
| 0.3 | 7.0 | GO:0002138 | retinoic acid biosynthetic process(GO:0002138) diterpenoid biosynthetic process(GO:0016102) |
| 0.3 | 4.2 | GO:0060020 | Bergmann glial cell differentiation(GO:0060020) |
| 0.3 | 2.4 | GO:1903026 | negative regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903026) |
| 0.3 | 4.3 | GO:0043950 | positive regulation of cAMP-mediated signaling(GO:0043950) |
| 0.3 | 2.3 | GO:0043932 | ossification involved in bone remodeling(GO:0043932) |
| 0.3 | 2.2 | GO:0060523 | prostate epithelial cord elongation(GO:0060523) |
| 0.3 | 1.9 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.3 | 4.5 | GO:0031272 | regulation of pseudopodium assembly(GO:0031272) |
| 0.3 | 1.2 | GO:0033625 | positive regulation of integrin activation(GO:0033625) |
| 0.3 | 0.9 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 0.3 | 1.7 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.3 | 7.6 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.3 | 1.4 | GO:0038043 | interleukin-5-mediated signaling pathway(GO:0038043) |
| 0.3 | 1.3 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) |
| 0.3 | 5.6 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.3 | 6.0 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.3 | 1.5 | GO:2000661 | positive regulation of interleukin-1-mediated signaling pathway(GO:2000661) |
| 0.3 | 1.5 | GO:0038112 | interleukin-8-mediated signaling pathway(GO:0038112) |
| 0.3 | 0.3 | GO:1903422 | negative regulation of synaptic vesicle recycling(GO:1903422) |
| 0.2 | 1.9 | GO:1902202 | regulation of hepatocyte growth factor receptor signaling pathway(GO:1902202) |
| 0.2 | 2.1 | GO:2001206 | positive regulation of osteoclast development(GO:2001206) |
| 0.2 | 5.1 | GO:0001765 | membrane raft assembly(GO:0001765) |
| 0.2 | 1.1 | GO:0002415 | immunoglobulin transcytosis in epithelial cells mediated by polymeric immunoglobulin receptor(GO:0002415) |
| 0.2 | 0.9 | GO:2000176 | regulation of pro-T cell differentiation(GO:2000174) positive regulation of pro-T cell differentiation(GO:2000176) |
| 0.2 | 0.7 | GO:1902173 | negative regulation of keratinocyte apoptotic process(GO:1902173) |
| 0.2 | 2.0 | GO:0032962 | positive regulation of inositol trisphosphate biosynthetic process(GO:0032962) |
| 0.2 | 0.7 | GO:0070904 | L-ascorbic acid transport(GO:0015882) transepithelial L-ascorbic acid transport(GO:0070904) |
| 0.2 | 2.9 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.2 | 1.5 | GO:0010286 | heat acclimation(GO:0010286) cellular heat acclimation(GO:0070370) |
| 0.2 | 3.3 | GO:0043589 | skin morphogenesis(GO:0043589) |
| 0.2 | 1.5 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.2 | 2.4 | GO:1900746 | regulation of vascular endothelial growth factor signaling pathway(GO:1900746) regulation of cellular response to vascular endothelial growth factor stimulus(GO:1902547) |
| 0.2 | 3.6 | GO:0015886 | heme transport(GO:0015886) |
| 0.2 | 1.5 | GO:0038183 | bile acid signaling pathway(GO:0038183) |
| 0.2 | 1.6 | GO:0070278 | extracellular matrix constituent secretion(GO:0070278) |
| 0.2 | 1.0 | GO:0034141 | positive regulation of toll-like receptor 3 signaling pathway(GO:0034141) |
| 0.2 | 0.4 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 0.2 | 1.0 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.2 | 3.5 | GO:0060315 | negative regulation of ryanodine-sensitive calcium-release channel activity(GO:0060315) |
| 0.2 | 0.6 | GO:0033590 | response to cobalamin(GO:0033590) |
| 0.2 | 5.4 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.2 | 1.4 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 0.2 | 1.2 | GO:0044800 | fusion of virus membrane with host plasma membrane(GO:0019064) membrane fusion involved in viral entry into host cell(GO:0039663) multi-organism membrane fusion(GO:0044800) |
| 0.2 | 2.2 | GO:2001214 | positive regulation of vasculogenesis(GO:2001214) |
| 0.2 | 0.5 | GO:0097252 | oligodendrocyte apoptotic process(GO:0097252) |
| 0.2 | 2.2 | GO:0046541 | saliva secretion(GO:0046541) |
| 0.2 | 2.3 | GO:0045669 | positive regulation of osteoblast differentiation(GO:0045669) |
| 0.2 | 2.1 | GO:0034242 | negative regulation of syncytium formation by plasma membrane fusion(GO:0034242) negative regulation of myoblast fusion(GO:1901740) regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.2 | 0.7 | GO:0043376 | regulation of CD8-positive, alpha-beta T cell differentiation(GO:0043376) |
| 0.2 | 1.2 | GO:0032455 | nerve growth factor processing(GO:0032455) |
| 0.2 | 1.0 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.2 | 1.1 | GO:0015793 | glycerol transport(GO:0015793) |
| 0.2 | 2.0 | GO:0001542 | ovulation from ovarian follicle(GO:0001542) |
| 0.2 | 1.2 | GO:0098989 | NMDA selective glutamate receptor signaling pathway(GO:0098989) |
| 0.2 | 1.2 | GO:0051607 | defense response to virus(GO:0051607) |
| 0.1 | 4.4 | GO:0032461 | positive regulation of protein oligomerization(GO:0032461) |
| 0.1 | 0.3 | GO:0003099 | positive regulation of the force of heart contraction by chemical signal(GO:0003099) |
| 0.1 | 1.2 | GO:0072734 | response to staurosporine(GO:0072733) cellular response to staurosporine(GO:0072734) |
| 0.1 | 1.4 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.1 | 0.9 | GO:0097211 | response to gonadotropin-releasing hormone(GO:0097210) cellular response to gonadotropin-releasing hormone(GO:0097211) |
| 0.1 | 0.4 | GO:0019243 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.1 | 0.3 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.1 | 0.9 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.1 | 1.9 | GO:0046322 | negative regulation of fatty acid oxidation(GO:0046322) |
| 0.1 | 0.6 | GO:0001845 | phagolysosome assembly(GO:0001845) |
| 0.1 | 0.9 | GO:1903025 | regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903025) |
| 0.1 | 2.7 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.1 | 2.1 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.1 | 3.7 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.1 | 0.8 | GO:0071847 | TNFSF11-mediated signaling pathway(GO:0071847) |
| 0.1 | 0.5 | GO:0010430 | fatty acid omega-oxidation(GO:0010430) |
| 0.1 | 2.2 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.1 | 0.7 | GO:1901222 | regulation of NIK/NF-kappaB signaling(GO:1901222) |
| 0.1 | 5.5 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.1 | 2.0 | GO:0097201 | negative regulation of transcription from RNA polymerase II promoter in response to stress(GO:0097201) |
| 0.1 | 0.7 | GO:0039019 | pronephric nephron development(GO:0039019) |
| 0.1 | 2.0 | GO:2000304 | positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 0.1 | 1.2 | GO:1901164 | negative regulation of trophoblast cell migration(GO:1901164) |
| 0.1 | 2.5 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.1 | 0.2 | GO:0042940 | D-amino acid transport(GO:0042940) |
| 0.1 | 0.6 | GO:0090611 | ubiquitin-independent protein catabolic process via the multivesicular body sorting pathway(GO:0090611) |
| 0.1 | 1.0 | GO:0097070 | ductus arteriosus closure(GO:0097070) |
| 0.1 | 0.7 | GO:0060326 | cell chemotaxis(GO:0060326) |
| 0.1 | 2.0 | GO:0044849 | estrous cycle(GO:0044849) |
| 0.1 | 0.6 | GO:0007406 | negative regulation of neuroblast proliferation(GO:0007406) |
| 0.1 | 0.6 | GO:1903300 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.1 | 0.3 | GO:0033037 | polysaccharide localization(GO:0033037) |
| 0.1 | 0.5 | GO:0009615 | response to virus(GO:0009615) |
| 0.1 | 0.5 | GO:1904994 | regulation of leukocyte tethering or rolling(GO:1903236) regulation of leukocyte adhesion to vascular endothelial cell(GO:1904994) |
| 0.1 | 1.3 | GO:0098734 | macromolecule depalmitoylation(GO:0098734) |
| 0.1 | 0.5 | GO:0070829 | response to vitamin B2(GO:0033274) heterochromatin maintenance(GO:0070829) |
| 0.1 | 0.6 | GO:0048706 | embryonic skeletal system development(GO:0048706) |
| 0.1 | 0.3 | GO:0072139 | glomerular parietal epithelial cell differentiation(GO:0072139) |
| 0.1 | 0.5 | GO:2000286 | receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 0.1 | 0.8 | GO:0060732 | positive regulation of inositol phosphate biosynthetic process(GO:0060732) |
| 0.1 | 0.8 | GO:0002036 | regulation of L-glutamate transport(GO:0002036) |
| 0.1 | 2.9 | GO:0007035 | vacuolar acidification(GO:0007035) |
| 0.1 | 0.5 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.1 | 0.3 | GO:0015680 | intracellular copper ion transport(GO:0015680) |
| 0.1 | 0.7 | GO:0002934 | desmosome organization(GO:0002934) |
| 0.1 | 0.9 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.1 | 1.1 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.1 | 0.6 | GO:0032485 | Ral protein signal transduction(GO:0032484) regulation of Ral protein signal transduction(GO:0032485) |
| 0.1 | 0.5 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 0.1 | 0.3 | GO:0071348 | cellular response to interleukin-11(GO:0071348) |
| 0.1 | 1.3 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.1 | 1.3 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.1 | 0.1 | GO:0035938 | estrogen secretion(GO:0035937) estradiol secretion(GO:0035938) regulation of estrogen secretion(GO:2000861) regulation of estradiol secretion(GO:2000864) |
| 0.1 | 0.3 | GO:0009257 | 10-formyltetrahydrofolate biosynthetic process(GO:0009257) |
| 0.1 | 1.4 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.1 | 0.3 | GO:0001923 | B-1 B cell differentiation(GO:0001923) |
| 0.1 | 0.7 | GO:0043306 | positive regulation of mast cell activation involved in immune response(GO:0033008) positive regulation of mast cell degranulation(GO:0043306) |
| 0.1 | 0.3 | GO:1903596 | regulation of gap junction assembly(GO:1903596) negative regulation of gap junction assembly(GO:1903597) |
| 0.1 | 1.3 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.1 | 0.2 | GO:0048210 | Golgi vesicle fusion to target membrane(GO:0048210) |
| 0.1 | 2.4 | GO:0042481 | regulation of odontogenesis(GO:0042481) |
| 0.1 | 0.1 | GO:1904017 | cellular response to Thyroglobulin triiodothyronine(GO:1904017) |
| 0.1 | 0.9 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.1 | 0.4 | GO:2000825 | positive regulation of androgen receptor activity(GO:2000825) |
| 0.1 | 12.3 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
| 0.1 | 8.9 | GO:0022617 | extracellular matrix disassembly(GO:0022617) |
| 0.1 | 0.6 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.1 | 0.6 | GO:0036149 | phosphatidylinositol acyl-chain remodeling(GO:0036149) |
| 0.1 | 5.1 | GO:0030834 | regulation of actin filament depolymerization(GO:0030834) |
| 0.1 | 1.0 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.1 | 12.5 | GO:0007586 | digestion(GO:0007586) |
| 0.1 | 3.0 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.1 | 0.5 | GO:0001766 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.1 | 0.5 | GO:0007320 | insemination(GO:0007320) |
| 0.1 | 0.4 | GO:0044805 | late nucleophagy(GO:0044805) |
| 0.1 | 0.4 | GO:1900194 | negative regulation of oocyte maturation(GO:1900194) |
| 0.1 | 0.7 | GO:0051443 | positive regulation of ubiquitin-protein transferase activity(GO:0051443) |
| 0.1 | 0.1 | GO:0097029 | mature conventional dendritic cell differentiation(GO:0097029) |
| 0.1 | 1.6 | GO:0007202 | activation of phospholipase C activity(GO:0007202) |
| 0.1 | 5.3 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.1 | 0.5 | GO:0051673 | membrane disruption in other organism(GO:0051673) |
| 0.1 | 0.3 | GO:0052565 | response to defense-related nitric oxide production by other organism involved in symbiotic interaction(GO:0052551) response to defense-related host nitric oxide production(GO:0052565) |
| 0.1 | 0.7 | GO:0045987 | positive regulation of smooth muscle contraction(GO:0045987) |
| 0.1 | 0.5 | GO:0048245 | eosinophil chemotaxis(GO:0048245) |
| 0.1 | 1.5 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.1 | 1.2 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.0 | 0.5 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.0 | 0.4 | GO:0045078 | positive regulation of interferon-gamma biosynthetic process(GO:0045078) |
| 0.0 | 1.6 | GO:0072348 | sulfur compound transport(GO:0072348) |
| 0.0 | 0.4 | GO:0043418 | homocysteine catabolic process(GO:0043418) |
| 0.0 | 1.2 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.0 | 1.3 | GO:0002360 | T cell lineage commitment(GO:0002360) |
| 0.0 | 1.7 | GO:0050819 | negative regulation of coagulation(GO:0050819) |
| 0.0 | 1.2 | GO:0071397 | cellular response to cholesterol(GO:0071397) |
| 0.0 | 1.4 | GO:0072673 | lamellipodium morphogenesis(GO:0072673) |
| 0.0 | 1.0 | GO:0035988 | chondrocyte proliferation(GO:0035988) |
| 0.0 | 0.3 | GO:0045651 | positive regulation of macrophage differentiation(GO:0045651) |
| 0.0 | 0.5 | GO:0046598 | positive regulation of viral entry into host cell(GO:0046598) |
| 0.0 | 0.4 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.0 | 4.7 | GO:0030838 | positive regulation of actin filament polymerization(GO:0030838) |
| 0.0 | 1.1 | GO:0051281 | positive regulation of release of sequestered calcium ion into cytosol(GO:0051281) |
| 0.0 | 1.6 | GO:0019430 | removal of superoxide radicals(GO:0019430) |
| 0.0 | 0.6 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 | 1.2 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.0 | 1.5 | GO:0040020 | regulation of meiotic nuclear division(GO:0040020) |
| 0.0 | 0.9 | GO:0034142 | toll-like receptor 4 signaling pathway(GO:0034142) |
| 0.0 | 0.4 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.4 | GO:0051106 | positive regulation of DNA ligation(GO:0051106) |
| 0.0 | 0.3 | GO:1902748 | melanocyte migration(GO:0097324) positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.0 | 0.3 | GO:1902031 | regulation of NADP metabolic process(GO:1902031) |
| 0.0 | 1.7 | GO:0007176 | regulation of epidermal growth factor-activated receptor activity(GO:0007176) |
| 0.0 | 2.8 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.0 | 0.9 | GO:0031664 | regulation of lipopolysaccharide-mediated signaling pathway(GO:0031664) |
| 0.0 | 0.6 | GO:1904776 | protein localization to cell cortex(GO:0072697) regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.0 | 0.6 | GO:0046677 | response to antibiotic(GO:0046677) |
| 0.0 | 1.8 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.0 | 0.1 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.0 | 0.7 | GO:0051031 | tRNA export from nucleus(GO:0006409) tRNA transport(GO:0051031) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.0 | 1.9 | GO:0032729 | positive regulation of interferon-gamma production(GO:0032729) |
| 0.0 | 1.3 | GO:2000144 | positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.0 | 0.8 | GO:0045730 | respiratory burst(GO:0045730) |
| 0.0 | 0.2 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.0 | 0.2 | GO:0021523 | somatic motor neuron differentiation(GO:0021523) protein localization to ciliary transition zone(GO:1904491) |
| 0.0 | 1.1 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.0 | 1.0 | GO:0007398 | ectoderm development(GO:0007398) |
| 0.0 | 1.2 | GO:0002576 | platelet degranulation(GO:0002576) |
| 0.0 | 1.0 | GO:0036150 | phosphatidylserine acyl-chain remodeling(GO:0036150) |
| 0.0 | 0.8 | GO:0045744 | negative regulation of G-protein coupled receptor protein signaling pathway(GO:0045744) |
| 0.0 | 0.9 | GO:0035987 | endodermal cell differentiation(GO:0035987) |
| 0.0 | 2.1 | GO:0003254 | regulation of membrane depolarization(GO:0003254) |
| 0.0 | 0.2 | GO:0021853 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.0 | 0.6 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.0 | 0.2 | GO:0060356 | leucine import(GO:0060356) |
| 0.0 | 0.5 | GO:0043304 | regulation of mast cell activation involved in immune response(GO:0033006) regulation of mast cell degranulation(GO:0043304) |
| 0.0 | 0.4 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.0 | 0.2 | GO:0050910 | detection of mechanical stimulus involved in sensory perception of sound(GO:0050910) |
| 0.0 | 1.8 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.3 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.0 | 0.6 | GO:0099601 | regulation of neurotransmitter receptor activity(GO:0099601) |
| 0.0 | 2.8 | GO:0006835 | dicarboxylic acid transport(GO:0006835) |
| 0.0 | 0.0 | GO:0002331 | pre-B cell allelic exclusion(GO:0002331) |
| 0.0 | 0.3 | GO:0001973 | adenosine receptor signaling pathway(GO:0001973) |
| 0.0 | 0.4 | GO:0007588 | excretion(GO:0007588) |
| 0.0 | 1.3 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.0 | 0.8 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.0 | 0.1 | GO:0035879 | plasma membrane lactate transport(GO:0035879) |
| 0.0 | 0.3 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.0 | 0.2 | GO:2000404 | regulation of T cell migration(GO:2000404) |
| 0.0 | 0.2 | GO:1902455 | negative regulation of stem cell population maintenance(GO:1902455) |
| 0.0 | 0.6 | GO:0050962 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 | 0.3 | GO:0045124 | regulation of bone resorption(GO:0045124) |
| 0.0 | 1.3 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.0 | 0.2 | GO:0010571 | positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.0 | 0.3 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.0 | 0.5 | GO:0031163 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 0.3 | GO:0008217 | regulation of blood pressure(GO:0008217) |
| 0.0 | 0.6 | GO:0021522 | spinal cord motor neuron differentiation(GO:0021522) |
| 0.0 | 1.2 | GO:0042058 | regulation of epidermal growth factor receptor signaling pathway(GO:0042058) |
| 0.0 | 0.1 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 | 0.3 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.6 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.0 | 1.4 | GO:0009250 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.0 | 2.4 | GO:0008637 | apoptotic mitochondrial changes(GO:0008637) |
| 0.0 | 0.9 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 1.2 | GO:0007044 | cell-substrate junction assembly(GO:0007044) |
| 0.0 | 0.6 | GO:0006359 | regulation of transcription from RNA polymerase III promoter(GO:0006359) |
| 0.0 | 0.2 | GO:0035860 | glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.0 | 0.3 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.0 | 0.5 | GO:2000772 | regulation of cellular senescence(GO:2000772) |
| 0.0 | 0.3 | GO:0043982 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.0 | 0.3 | GO:0030225 | macrophage differentiation(GO:0030225) |
| 0.0 | 0.7 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.0 | 0.8 | GO:0045670 | regulation of osteoclast differentiation(GO:0045670) |
| 0.0 | 0.6 | GO:0006939 | smooth muscle contraction(GO:0006939) |
| 0.0 | 1.9 | GO:0030326 | embryonic limb morphogenesis(GO:0030326) embryonic appendage morphogenesis(GO:0035113) |
| 0.0 | 0.7 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.0 | 0.9 | GO:0071377 | cellular response to glucagon stimulus(GO:0071377) |
| 0.0 | 0.3 | GO:0007601 | visual perception(GO:0007601) sensory perception of light stimulus(GO:0050953) |
| 0.0 | 0.1 | GO:0045229 | cell envelope organization(GO:0043163) external encapsulating structure organization(GO:0045229) |
| 0.0 | 0.9 | GO:0007631 | feeding behavior(GO:0007631) |
| 0.0 | 1.7 | GO:0051225 | spindle assembly(GO:0051225) |
| 0.0 | 1.7 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 | 0.5 | GO:0050954 | sensory perception of sound(GO:0007605) sensory perception of mechanical stimulus(GO:0050954) |
| 0.0 | 0.2 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.1 | 9.3 | GO:0020003 | symbiont-containing vacuole(GO:0020003) symbiont-containing vacuole membrane(GO:0020005) |
| 2.2 | 24.2 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 1.7 | 6.9 | GO:0005607 | laminin-2 complex(GO:0005607) |
| 1.1 | 3.3 | GO:0005584 | collagen type I trimer(GO:0005584) |
| 1.0 | 4.0 | GO:0045160 | myosin I complex(GO:0045160) |
| 0.9 | 2.7 | GO:0097057 | TRAF2-GSTP1 complex(GO:0097057) |
| 0.9 | 8.7 | GO:0005638 | lamin filament(GO:0005638) |
| 0.8 | 3.9 | GO:0071149 | TEAD-2-YAP complex(GO:0071149) |
| 0.8 | 2.3 | GO:0097444 | spine apparatus(GO:0097444) |
| 0.7 | 28.6 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.6 | 11.6 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.6 | 3.0 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) CNTFR-CLCF1 complex(GO:0097059) |
| 0.6 | 6.2 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.5 | 2.2 | GO:0033257 | Bcl3/NF-kappaB2 complex(GO:0033257) |
| 0.5 | 7.8 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.4 | 103.6 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.4 | 1.1 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 0.4 | 1.1 | GO:0035525 | NF-kappaB p50/p65 complex(GO:0035525) |
| 0.3 | 13.3 | GO:0030057 | desmosome(GO:0030057) |
| 0.3 | 2.5 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.3 | 7.1 | GO:0097342 | ripoptosome(GO:0097342) |
| 0.3 | 4.8 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.3 | 1.4 | GO:1990667 | PCSK9-AnxA2 complex(GO:1990667) |
| 0.3 | 3.1 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.3 | 2.2 | GO:0070435 | Shc-EGFR complex(GO:0070435) |
| 0.3 | 1.8 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.2 | 0.9 | GO:0031251 | PAN complex(GO:0031251) |
| 0.2 | 0.6 | GO:0036194 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.2 | 2.7 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.2 | 1.4 | GO:0001652 | granular component(GO:0001652) |
| 0.2 | 0.9 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.2 | 2.9 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.2 | 2.4 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.2 | 1.6 | GO:0030478 | actin cap(GO:0030478) |
| 0.2 | 2.9 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.2 | 2.2 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.2 | 0.9 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.2 | 1.8 | GO:0016942 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) |
| 0.1 | 8.6 | GO:0008305 | integrin complex(GO:0008305) |
| 0.1 | 5.5 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.1 | 10.3 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.1 | 4.3 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.1 | 0.8 | GO:0090498 | extrinsic component of Golgi membrane(GO:0090498) |
| 0.1 | 1.3 | GO:0097413 | Lewy body(GO:0097413) |
| 0.1 | 1.8 | GO:0030658 | transport vesicle membrane(GO:0030658) |
| 0.1 | 0.9 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.1 | 227.3 | GO:0005615 | extracellular space(GO:0005615) |
| 0.1 | 0.8 | GO:0032010 | phagolysosome(GO:0032010) |
| 0.1 | 1.8 | GO:0090543 | Flemming body(GO:0090543) |
| 0.1 | 2.1 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.1 | 2.0 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.1 | 8.2 | GO:0015030 | Cajal body(GO:0015030) |
| 0.1 | 14.8 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.1 | 0.6 | GO:1990462 | omegasome(GO:1990462) |
| 0.1 | 2.0 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.1 | 0.8 | GO:0030905 | retromer, tubulation complex(GO:0030905) |
| 0.1 | 3.6 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.1 | 0.3 | GO:0032449 | CBM complex(GO:0032449) |
| 0.1 | 7.3 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.1 | 2.2 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.1 | 8.2 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.1 | 1.6 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.1 | 0.5 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.1 | 8.2 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 0.3 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.1 | 6.5 | GO:0030315 | T-tubule(GO:0030315) |
| 0.1 | 2.8 | GO:0005901 | caveola(GO:0005901) |
| 0.1 | 3.3 | GO:0002102 | podosome(GO:0002102) |
| 0.1 | 2.0 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.1 | 1.1 | GO:0005861 | troponin complex(GO:0005861) |
| 0.1 | 12.9 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.1 | 1.5 | GO:0010369 | chromocenter(GO:0010369) |
| 0.1 | 2.3 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.1 | 0.6 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.1 | 5.4 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.0 | 0.9 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.0 | 0.6 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.0 | 1.2 | GO:0031094 | platelet dense tubular network(GO:0031094) |
| 0.0 | 3.4 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.0 | 9.1 | GO:0098862 | cluster of actin-based cell projections(GO:0098862) |
| 0.0 | 0.7 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 3.2 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 1.9 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.3 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 5.2 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 0.5 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.5 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 2.1 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 1.7 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 1.0 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.9 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 6.2 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 1.6 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 3.5 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.2 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.0 | 1.4 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 6.3 | GO:0001726 | ruffle(GO:0001726) |
| 0.0 | 124.0 | GO:0005576 | extracellular region(GO:0005576) |
| 0.0 | 4.1 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 0.5 | GO:0098827 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.0 | 0.6 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 1.3 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.3 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.7 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.7 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.7 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.5 | GO:0031301 | integral component of organelle membrane(GO:0031301) |
| 0.0 | 0.8 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 0.8 | GO:0070821 | tertiary granule membrane(GO:0070821) |
| 0.0 | 1.2 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 1.6 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 0.4 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 1.4 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.1 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.1 | 9.3 | GO:0035379 | carbon dioxide transmembrane transporter activity(GO:0035379) |
| 2.7 | 8.2 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 1.6 | 6.2 | GO:0005026 | transforming growth factor beta receptor activity, type II(GO:0005026) |
| 1.5 | 4.5 | GO:0005150 | interleukin-1, Type I receptor binding(GO:0005150) |
| 1.3 | 345.5 | GO:0003823 | antigen binding(GO:0003823) |
| 1.2 | 3.6 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 1.1 | 3.4 | GO:0005146 | leukemia inhibitory factor receptor binding(GO:0005146) |
| 1.1 | 7.9 | GO:1990254 | keratin filament binding(GO:1990254) |
| 1.1 | 7.9 | GO:0030377 | urokinase plasminogen activator receptor activity(GO:0030377) |
| 1.1 | 4.3 | GO:0005019 | platelet-derived growth factor beta-receptor activity(GO:0005019) |
| 1.0 | 3.1 | GO:0015361 | low-affinity sodium:dicarboxylate symporter activity(GO:0015361) |
| 1.0 | 4.1 | GO:0008426 | protein kinase C inhibitor activity(GO:0008426) |
| 0.9 | 7.4 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.9 | 2.6 | GO:0003955 | NAD(P)H dehydrogenase (quinone) activity(GO:0003955) |
| 0.8 | 2.4 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 0.6 | 1.9 | GO:0005171 | hepatocyte growth factor receptor binding(GO:0005171) |
| 0.6 | 1.7 | GO:0047389 | glycerophosphocholine phosphodiesterase activity(GO:0047389) |
| 0.5 | 1.6 | GO:0016649 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.5 | 3.0 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.5 | 2.0 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
| 0.5 | 2.4 | GO:0004850 | uridine phosphorylase activity(GO:0004850) |
| 0.5 | 2.4 | GO:0004910 | interleukin-1, Type II, blocking receptor activity(GO:0004910) |
| 0.5 | 3.3 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.5 | 2.8 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.5 | 2.7 | GO:0070026 | nitric oxide binding(GO:0070026) |
| 0.4 | 3.1 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.4 | 7.5 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.4 | 4.1 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.4 | 2.9 | GO:0042806 | fucose binding(GO:0042806) |
| 0.4 | 4.1 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.4 | 10.5 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.4 | 3.0 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.4 | 1.1 | GO:0004947 | bradykinin receptor activity(GO:0004947) |
| 0.3 | 11.0 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.3 | 3.0 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.3 | 1.7 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.3 | 2.2 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.3 | 0.9 | GO:0004968 | gonadotropin-releasing hormone receptor activity(GO:0004968) |
| 0.3 | 5.2 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.3 | 1.2 | GO:0051431 | corticotropin-releasing hormone receptor 2 binding(GO:0051431) |
| 0.3 | 0.9 | GO:0005427 | proton-dependent oligopeptide secondary active transmembrane transporter activity(GO:0005427) secondary active oligopeptide transmembrane transporter activity(GO:0015322) |
| 0.3 | 1.4 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) interleukin-2 binding(GO:0019976) |
| 0.3 | 1.4 | GO:0004914 | interleukin-5 receptor activity(GO:0004914) |
| 0.3 | 0.8 | GO:0004992 | platelet activating factor receptor activity(GO:0004992) |
| 0.3 | 3.7 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.3 | 5.9 | GO:0005522 | profilin binding(GO:0005522) |
| 0.3 | 2.9 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.3 | 12.3 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.3 | 1.0 | GO:0016404 | 15-hydroxyprostaglandin dehydrogenase (NAD+) activity(GO:0016404) |
| 0.3 | 1.5 | GO:0004918 | interleukin-8 receptor activity(GO:0004918) |
| 0.3 | 1.0 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 0.3 | 9.3 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.2 | 2.2 | GO:0038052 | RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.2 | 1.0 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.2 | 2.2 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.2 | 3.4 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.2 | 1.9 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 0.2 | 4.7 | GO:0019841 | retinol binding(GO:0019841) |
| 0.2 | 1.1 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 0.2 | 0.7 | GO:0008520 | L-ascorbate:sodium symporter activity(GO:0008520) L-ascorbic acid transporter activity(GO:0015229) sodium-dependent L-ascorbate transmembrane transporter activity(GO:0070890) |
| 0.2 | 1.5 | GO:0004909 | interleukin-1, Type I, activating receptor activity(GO:0004909) |
| 0.2 | 0.8 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.2 | 1.5 | GO:0038181 | bile acid receptor activity(GO:0038181) |
| 0.2 | 2.7 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.2 | 1.2 | GO:0047685 | amine sulfotransferase activity(GO:0047685) |
| 0.2 | 35.9 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.2 | 3.6 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.2 | 2.0 | GO:0072542 | protein phosphatase activator activity(GO:0072542) |
| 0.2 | 3.5 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.2 | 2.7 | GO:0004955 | prostaglandin receptor activity(GO:0004955) |
| 0.2 | 8.3 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.2 | 3.3 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.2 | 4.1 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.2 | 1.9 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.2 | 1.3 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.2 | 1.8 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.2 | 5.3 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.2 | 1.1 | GO:0008481 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.2 | 26.0 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.2 | 7.5 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.2 | 0.8 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.2 | 0.8 | GO:0004991 | parathyroid hormone receptor activity(GO:0004991) |
| 0.2 | 4.8 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.1 | 1.8 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 1.4 | GO:0019863 | IgE binding(GO:0019863) |
| 0.1 | 3.1 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 0.7 | GO:0031728 | CCR3 chemokine receptor binding(GO:0031728) |
| 0.1 | 5.4 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.1 | 11.3 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.1 | 2.6 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.1 | 0.4 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.1 | 3.7 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 0.8 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.1 | 1.0 | GO:0019956 | chemokine binding(GO:0019956) |
| 0.1 | 1.5 | GO:0071253 | connexin binding(GO:0071253) |
| 0.1 | 1.1 | GO:0043426 | MRF binding(GO:0043426) |
| 0.1 | 1.0 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.1 | 0.3 | GO:0047225 | acetylgalactosaminyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0047225) |
| 0.1 | 0.9 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.1 | 3.0 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.1 | 1.3 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.1 | 3.3 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.1 | 1.8 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.1 | 0.1 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.1 | 1.9 | GO:0055106 | ligase regulator activity(GO:0055103) ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.1 | 0.3 | GO:0050613 | delta14-sterol reductase activity(GO:0050613) |
| 0.1 | 0.3 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.1 | 24.8 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 1.3 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.1 | 0.5 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 0.1 | 2.0 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.1 | 3.4 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.1 | 3.1 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.1 | 3.3 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.1 | 3.4 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.1 | 3.0 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.1 | 1.5 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.1 | 0.3 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.1 | 0.8 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.1 | 0.3 | GO:0031711 | bradykinin receptor binding(GO:0031711) |
| 0.1 | 1.9 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.1 | 2.4 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.1 | 0.4 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.1 | 0.8 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.1 | 0.2 | GO:0086040 | sodium:proton antiporter activity involved in regulation of cardiac muscle cell membrane potential(GO:0086040) |
| 0.1 | 0.2 | GO:0070984 | SET domain binding(GO:0070984) |
| 0.1 | 0.3 | GO:0016532 | superoxide dismutase copper chaperone activity(GO:0016532) |
| 0.1 | 0.6 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.1 | 2.3 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.1 | 0.3 | GO:0050659 | N-acetylgalactosamine 4-sulfate 6-O-sulfotransferase activity(GO:0050659) |
| 0.1 | 0.9 | GO:0005168 | neurotrophin TRKA receptor binding(GO:0005168) |
| 0.1 | 1.1 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.1 | 1.3 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 2.7 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.1 | 1.8 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.1 | 0.4 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.1 | 2.3 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.1 | 1.6 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.7 | GO:0008526 | phosphatidylcholine transporter activity(GO:0008525) phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 1.2 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 1.1 | GO:0042301 | phosphate ion binding(GO:0042301) |
| 0.0 | 1.8 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.0 | 1.5 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.6 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.0 | 0.3 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.0 | 0.8 | GO:0004875 | complement receptor activity(GO:0004875) |
| 0.0 | 0.6 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.0 | 23.6 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.0 | 6.4 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.5 | GO:0000990 | transcription factor activity, core RNA polymerase binding(GO:0000990) |
| 0.0 | 1.4 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.0 | 4.0 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 1.0 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 42.7 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.0 | 3.2 | GO:0019838 | growth factor binding(GO:0019838) |
| 0.0 | 1.2 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 8.2 | GO:0005539 | glycosaminoglycan binding(GO:0005539) |
| 0.0 | 0.6 | GO:0030548 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.0 | 0.3 | GO:0004473 | malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.0 | 4.2 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 0.1 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.0 | 1.3 | GO:1901505 | carbohydrate derivative transporter activity(GO:1901505) |
| 0.0 | 0.8 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 1.8 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.9 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 1.5 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.4 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.3 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 0.3 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.0 | 0.1 | GO:0030375 | thyroid hormone receptor activator activity(GO:0010861) thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.5 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 0.1 | GO:0070260 | tyrosyl-RNA phosphodiesterase activity(GO:0036317) 5'-tyrosyl-DNA phosphodiesterase activity(GO:0070260) |
| 0.0 | 0.6 | GO:0008329 | signaling pattern recognition receptor activity(GO:0008329) pattern recognition receptor activity(GO:0038187) |
| 0.0 | 1.2 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 4.5 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.0 | 1.5 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 0.9 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 2.7 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.3 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 1.3 | GO:1901682 | sulfur compound transmembrane transporter activity(GO:1901682) |
| 0.0 | 0.4 | GO:0003910 | DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 0.3 | GO:0043996 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.0 | 0.3 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 0.5 | GO:0001163 | RNA polymerase I regulatory region DNA binding(GO:0001013) RNA polymerase I regulatory region sequence-specific DNA binding(GO:0001163) RNA polymerase I CORE element sequence-specific DNA binding(GO:0001164) |
| 0.0 | 0.4 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 3.5 | GO:0008081 | phosphoric diester hydrolase activity(GO:0008081) |
| 0.0 | 0.2 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.0 | 0.2 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.0 | 1.5 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.6 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.0 | 3.0 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 0.5 | GO:0051183 | vitamin transporter activity(GO:0051183) |
| 0.0 | 1.3 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.2 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.2 | GO:0044213 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) intronic transcription regulatory region DNA binding(GO:0044213) |
| 0.0 | 0.1 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 4.0 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.0 | 0.3 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 34.8 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.7 | 52.1 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.3 | 9.5 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.2 | 6.0 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.2 | 8.3 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.2 | 0.6 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.2 | 13.1 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.2 | 5.1 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.2 | 14.8 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.2 | 9.1 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.1 | 12.9 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.1 | 5.3 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.1 | 5.5 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.1 | 6.5 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.1 | 3.1 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.1 | 4.9 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.1 | 8.5 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.1 | 1.4 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.1 | 4.1 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.1 | 8.9 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.1 | 8.5 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.1 | 5.4 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.1 | 2.1 | ST B CELL ANTIGEN RECEPTOR | B Cell Antigen Receptor |
| 0.1 | 2.2 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.1 | 2.4 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.1 | 4.6 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.1 | 2.9 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.1 | 2.4 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.1 | 2.8 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 2.1 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.1 | 1.7 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.1 | 2.3 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.1 | 6.7 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.1 | 22.9 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 1.4 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 2.4 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.0 | 0.9 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.0 | 1.0 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.0 | 1.4 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 1.5 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 15.1 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 3.2 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 1.0 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 1.1 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 1.9 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 1.9 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 1.9 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 1.4 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 4.4 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 1.6 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.3 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.0 | 0.9 | PID INTEGRIN1 PATHWAY | Beta1 integrin cell surface interactions |
| 0.0 | 1.4 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 1.0 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 0.3 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 3.7 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.7 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 1.1 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 0.9 | PID NOTCH PATHWAY | Notch signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 10.5 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.5 | 20.5 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.4 | 8.5 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.4 | 18.3 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.3 | 7.8 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.3 | 36.7 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.3 | 9.8 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.2 | 5.0 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.2 | 2.7 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.2 | 9.6 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.2 | 4.1 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.2 | 6.7 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.2 | 16.0 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.2 | 3.5 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.2 | 3.3 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.2 | 2.5 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.2 | 7.9 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.2 | 5.1 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.2 | 9.4 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.2 | 7.0 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.2 | 7.5 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.2 | 4.8 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.2 | 1.5 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.1 | 1.0 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.1 | 3.1 | REACTOME SHC1 EVENTS IN EGFR SIGNALING | Genes involved in SHC1 events in EGFR signaling |
| 0.1 | 2.4 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.1 | 7.8 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.1 | 5.3 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.1 | 3.0 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.1 | 7.2 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.1 | 2.8 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.1 | 5.4 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.1 | 3.5 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.1 | 1.3 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.1 | 3.2 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.1 | 1.9 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.1 | 1.5 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.1 | 2.1 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.1 | 1.9 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 2.1 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.1 | 1.1 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.1 | 0.4 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.1 | 7.9 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.1 | 4.6 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.1 | 1.1 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.1 | 3.8 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.1 | 0.6 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.1 | 1.9 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.1 | 2.1 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 0.8 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.0 | 3.1 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.3 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.0 | 0.8 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.0 | 4.9 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 2.2 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 0.3 | REACTOME ADP SIGNALLING THROUGH P2RY12 | Genes involved in ADP signalling through P2Y purinoceptor 12 |
| 0.0 | 2.5 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 1.2 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 1.9 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 1.8 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 2.3 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 1.6 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 1.4 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.2 | REACTOME ACTIVATION OF NF KAPPAB IN B CELLS | Genes involved in Activation of NF-kappaB in B Cells |
| 0.0 | 1.8 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 7.5 | REACTOME CLASS A1 RHODOPSIN LIKE RECEPTORS | Genes involved in Class A/1 (Rhodopsin-like receptors) |
| 0.0 | 0.9 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 1.5 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.5 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.8 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.6 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.0 | 1.9 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.5 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.0 | 1.6 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.1 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.3 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 1.3 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 3.5 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.3 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.0 | REACTOME FGFR1 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR1 ligand binding and activation |
| 0.0 | 2.3 | REACTOME SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |
| 0.0 | 0.9 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |