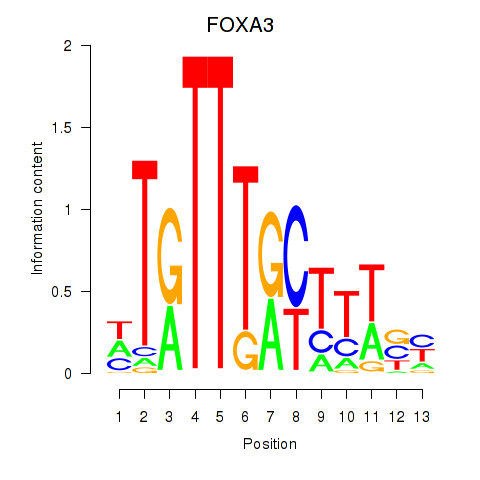
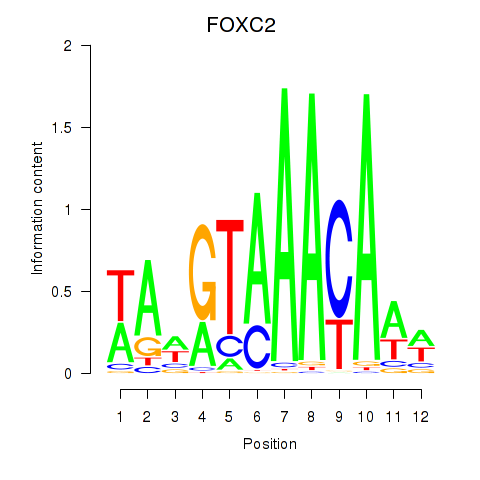
Project
Illumina Body Map 2: averaged replicates
Navigation
Downloads
Results for FOXA3_FOXC2
Z-value: 1.96
Transcription factors associated with FOXA3_FOXC2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
FOXA3
|
ENSG00000170608.2 | forkhead box A3 |
|
FOXC2
|
ENSG00000176692.4 | forkhead box C2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| FOXA3 | hg19_v2_chr19_+_46367518_46367567 | 0.64 | 8.4e-05 | Click! |
| FOXC2 | hg19_v2_chr16_+_86600857_86600921 | 0.13 | 4.7e-01 | Click! |
Activity profile of FOXA3_FOXC2 motif
Sorted Z-values of FOXA3_FOXC2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_+_62186498 | 10.76 |
ENST00000278282.2
|
SCGB1A1
|
secretoglobin, family 1A, member 1 (uteroglobin) |
| chr8_+_120079478 | 8.68 |
ENST00000332843.2
|
COLEC10
|
collectin sub-family member 10 (C-type lectin) |
| chr1_-_57431679 | 8.14 |
ENST00000371237.4
ENST00000535057.1 ENST00000543257.1 |
C8B
|
complement component 8, beta polypeptide |
| chr17_-_26695013 | 7.10 |
ENST00000555059.2
|
CTB-96E2.2
|
Homeobox protein SEBOX |
| chr17_-_26694979 | 7.02 |
ENST00000438614.1
|
VTN
|
vitronectin |
| chr11_-_116708302 | 6.00 |
ENST00000375320.1
ENST00000359492.2 ENST00000375329.2 ENST00000375323.1 |
APOA1
|
apolipoprotein A-I |
| chr6_-_41715128 | 4.69 |
ENST00000356667.4
ENST00000373025.3 ENST00000425343.2 |
PGC
|
progastricsin (pepsinogen C) |
| chr4_+_187187098 | 4.21 |
ENST00000403665.2
ENST00000264692.4 |
F11
|
coagulation factor XI |
| chr2_+_128175997 | 3.87 |
ENST00000234071.3
ENST00000429925.1 ENST00000442644.1 ENST00000453608.2 |
PROC
|
protein C (inactivator of coagulation factors Va and VIIIa) |
| chr13_-_86373536 | 3.74 |
ENST00000400286.2
|
SLITRK6
|
SLIT and NTRK-like family, member 6 |
| chr18_-_25616519 | 3.53 |
ENST00000399380.3
|
CDH2
|
cadherin 2, type 1, N-cadherin (neuronal) |
| chr1_+_207277590 | 3.50 |
ENST00000367070.3
|
C4BPA
|
complement component 4 binding protein, alpha |
| chr20_+_31823792 | 3.48 |
ENST00000375413.4
ENST00000354297.4 ENST00000375422.2 |
BPIFA1
|
BPI fold containing family A, member 1 |
| chr3_+_108855558 | 3.46 |
ENST00000467240.1
ENST00000477643.1 ENST00000479039.1 ENST00000593799.1 |
RP11-59E19.1
|
RP11-59E19.1 |
| chr14_-_70263979 | 3.26 |
ENST00000216540.4
|
SLC10A1
|
solute carrier family 10 (sodium/bile acid cotransporter), member 1 |
| chr1_+_207277632 | 3.22 |
ENST00000421786.1
|
C4BPA
|
complement component 4 binding protein, alpha |
| chr3_+_137717571 | 3.20 |
ENST00000343735.4
|
CLDN18
|
claudin 18 |
| chr5_-_41213123 | 3.18 |
ENST00000417809.1
|
C6
|
complement component 6 |
| chr22_+_21128167 | 3.09 |
ENST00000215727.5
|
SERPIND1
|
serpin peptidase inhibitor, clade D (heparin cofactor), member 1 |
| chr14_-_36988882 | 3.04 |
ENST00000498187.2
|
NKX2-1
|
NK2 homeobox 1 |
| chr16_-_86542652 | 3.02 |
ENST00000599749.1
|
FENDRR
|
FOXF1 adjacent non-coding developmental regulatory RNA |
| chr17_-_19290483 | 2.97 |
ENST00000395592.2
ENST00000299610.4 |
MFAP4
|
microfibrillar-associated protein 4 |
| chr17_-_26697304 | 2.96 |
ENST00000536498.1
|
VTN
|
vitronectin |
| chr10_+_71561649 | 2.88 |
ENST00000398978.3
ENST00000354547.3 ENST00000357811.3 |
COL13A1
|
collagen, type XIII, alpha 1 |
| chr4_+_169013666 | 2.76 |
ENST00000359299.3
|
ANXA10
|
annexin A10 |
| chr2_+_88047606 | 2.57 |
ENST00000359481.4
|
PLGLB2
|
plasminogen-like B2 |
| chr7_-_107443652 | 2.56 |
ENST00000340010.5
ENST00000422236.2 ENST00000453332.1 |
SLC26A3
|
solute carrier family 26 (anion exchanger), member 3 |
| chr2_+_135596180 | 2.55 |
ENST00000283054.4
ENST00000392928.1 |
ACMSD
|
aminocarboxymuconate semialdehyde decarboxylase |
| chr12_+_27849378 | 2.51 |
ENST00000310791.2
|
REP15
|
RAB15 effector protein |
| chr14_-_94789663 | 2.49 |
ENST00000557225.1
ENST00000341584.3 |
SERPINA6
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 6 |
| chr10_+_71561630 | 2.43 |
ENST00000398974.3
ENST00000398971.3 ENST00000398968.3 ENST00000398966.3 ENST00000398964.3 ENST00000398969.3 ENST00000356340.3 ENST00000398972.3 ENST00000398973.3 |
COL13A1
|
collagen, type XIII, alpha 1 |
| chr14_-_38064198 | 2.43 |
ENST00000250448.2
|
FOXA1
|
forkhead box A1 |
| chr17_-_39684550 | 2.40 |
ENST00000455635.1
ENST00000361566.3 |
KRT19
|
keratin 19 |
| chr5_-_41213607 | 2.40 |
ENST00000337836.5
ENST00000433294.1 |
C6
|
complement component 6 |
| chr7_+_142829162 | 2.31 |
ENST00000291009.3
|
PIP
|
prolactin-induced protein |
| chr20_+_31870927 | 2.29 |
ENST00000253354.1
|
BPIFB1
|
BPI fold containing family B, member 1 |
| chr4_-_71532207 | 2.27 |
ENST00000543780.1
ENST00000391614.3 |
IGJ
|
immunoglobulin J polypeptide, linker protein for immunoglobulin alpha and mu polypeptides |
| chr4_+_187187337 | 2.26 |
ENST00000492972.2
|
F11
|
coagulation factor XI |
| chr4_+_74269956 | 2.26 |
ENST00000295897.4
ENST00000415165.2 ENST00000503124.1 ENST00000509063.1 ENST00000401494.3 |
ALB
|
albumin |
| chr4_+_187148556 | 2.24 |
ENST00000264690.6
ENST00000446598.2 ENST00000414291.1 ENST00000513864.1 |
KLKB1
|
kallikrein B, plasma (Fletcher factor) 1 |
| chr3_+_137728842 | 2.23 |
ENST00000183605.5
|
CLDN18
|
claudin 18 |
| chr16_-_86542455 | 2.21 |
ENST00000595886.1
ENST00000597578.1 ENST00000593604.1 |
FENDRR
|
FOXF1 adjacent non-coding developmental regulatory RNA |
| chr2_+_135596106 | 2.20 |
ENST00000356140.5
|
ACMSD
|
aminocarboxymuconate semialdehyde decarboxylase |
| chr12_+_59989918 | 2.19 |
ENST00000547379.1
ENST00000549465.1 |
SLC16A7
|
solute carrier family 16 (monocarboxylate transporter), member 7 |
| chr2_+_113885138 | 2.16 |
ENST00000409930.3
|
IL1RN
|
interleukin 1 receptor antagonist |
| chr4_-_71532668 | 2.11 |
ENST00000510437.1
|
IGJ
|
immunoglobulin J polypeptide, linker protein for immunoglobulin alpha and mu polypeptides |
| chr18_+_47088401 | 2.09 |
ENST00000261292.4
ENST00000427224.2 ENST00000580036.1 |
LIPG
|
lipase, endothelial |
| chr15_+_69857515 | 2.06 |
ENST00000559477.1
|
RP11-279F6.1
|
RP11-279F6.1 |
| chr4_-_71532601 | 2.05 |
ENST00000510614.1
|
IGJ
|
immunoglobulin J polypeptide, linker protein for immunoglobulin alpha and mu polypeptides |
| chr14_+_21156915 | 2.00 |
ENST00000397990.4
ENST00000555597.1 |
ANG
RNASE4
|
angiogenin, ribonuclease, RNase A family, 5 ribonuclease, RNase A family, 4 |
| chr10_+_71561704 | 1.94 |
ENST00000520267.1
|
COL13A1
|
collagen, type XIII, alpha 1 |
| chr12_-_120765565 | 1.93 |
ENST00000423423.3
ENST00000308366.4 |
PLA2G1B
|
phospholipase A2, group IB (pancreas) |
| chr5_+_147258266 | 1.80 |
ENST00000296694.4
|
SCGB3A2
|
secretoglobin, family 3A, member 2 |
| chr8_+_19171128 | 1.79 |
ENST00000265807.3
|
SH2D4A
|
SH2 domain containing 4A |
| chr4_-_71532339 | 1.78 |
ENST00000254801.4
|
IGJ
|
immunoglobulin J polypeptide, linker protein for immunoglobulin alpha and mu polypeptides |
| chr1_+_200011711 | 1.77 |
ENST00000544748.1
|
NR5A2
|
nuclear receptor subfamily 5, group A, member 2 |
| chr10_-_118032697 | 1.76 |
ENST00000439649.3
|
GFRA1
|
GDNF family receptor alpha 1 |
| chrX_-_15402498 | 1.75 |
ENST00000297904.3
|
FIGF
|
c-fos induced growth factor (vascular endothelial growth factor D) |
| chr8_+_19171196 | 1.71 |
ENST00000518040.1
|
SH2D4A
|
SH2 domain containing 4A |
| chr2_-_165424973 | 1.69 |
ENST00000543549.1
|
GRB14
|
growth factor receptor-bound protein 14 |
| chr10_-_118032979 | 1.67 |
ENST00000355422.6
|
GFRA1
|
GDNF family receptor alpha 1 |
| chr14_+_38065052 | 1.67 |
ENST00000556845.1
|
TTC6
|
tetratricopeptide repeat domain 6 |
| chr16_-_48281305 | 1.66 |
ENST00000356608.2
|
ABCC11
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 11 |
| chr9_-_95186739 | 1.63 |
ENST00000375550.4
|
OMD
|
osteomodulin |
| chr2_-_105030466 | 1.61 |
ENST00000449772.1
|
AC068535.3
|
AC068535.3 |
| chr1_-_169555779 | 1.60 |
ENST00000367797.3
ENST00000367796.3 |
F5
|
coagulation factor V (proaccelerin, labile factor) |
| chr12_-_92539614 | 1.59 |
ENST00000256015.3
|
BTG1
|
B-cell translocation gene 1, anti-proliferative |
| chr4_-_72649763 | 1.58 |
ENST00000513476.1
|
GC
|
group-specific component (vitamin D binding protein) |
| chr7_+_134576317 | 1.55 |
ENST00000424922.1
ENST00000495522.1 |
CALD1
|
caldesmon 1 |
| chr2_+_74120094 | 1.54 |
ENST00000409731.3
ENST00000345517.3 ENST00000409918.1 ENST00000442912.1 ENST00000409624.1 |
ACTG2
|
actin, gamma 2, smooth muscle, enteric |
| chr10_-_81708854 | 1.54 |
ENST00000372292.3
|
SFTPD
|
surfactant protein D |
| chr12_-_21487829 | 1.53 |
ENST00000445053.1
ENST00000452078.1 ENST00000458504.1 ENST00000422327.1 ENST00000421294.1 |
SLCO1A2
|
solute carrier organic anion transporter family, member 1A2 |
| chr7_-_81399438 | 1.51 |
ENST00000222390.5
|
HGF
|
hepatocyte growth factor (hepapoietin A; scatter factor) |
| chr15_+_96869165 | 1.49 |
ENST00000421109.2
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr17_-_57229155 | 1.47 |
ENST00000584089.1
|
SKA2
|
spindle and kinetochore associated complex subunit 2 |
| chr14_-_106791536 | 1.46 |
ENST00000390613.2
|
IGHV3-30
|
immunoglobulin heavy variable 3-30 |
| chr21_+_17792672 | 1.46 |
ENST00000602620.1
|
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr8_+_97597148 | 1.45 |
ENST00000521590.1
|
SDC2
|
syndecan 2 |
| chr4_+_15376165 | 1.45 |
ENST00000382383.3
ENST00000429690.1 |
C1QTNF7
|
C1q and tumor necrosis factor related protein 7 |
| chr3_+_119316721 | 1.43 |
ENST00000488919.1
ENST00000495992.1 |
PLA1A
|
phospholipase A1 member A |
| chr12_+_13044787 | 1.43 |
ENST00000534831.1
|
GPRC5A
|
G protein-coupled receptor, family C, group 5, member A |
| chr1_-_146696901 | 1.42 |
ENST00000369272.3
ENST00000441068.2 |
FMO5
|
flavin containing monooxygenase 5 |
| chr18_-_52626622 | 1.40 |
ENST00000591504.1
|
CCDC68
|
coiled-coil domain containing 68 |
| chrX_+_105936982 | 1.40 |
ENST00000418562.1
|
RNF128
|
ring finger protein 128, E3 ubiquitin protein ligase |
| chr6_+_142623758 | 1.38 |
ENST00000541199.1
ENST00000435011.2 |
GPR126
|
G protein-coupled receptor 126 |
| chr2_+_7865923 | 1.38 |
ENST00000417930.1
|
AC092580.4
|
AC092580.4 |
| chr7_-_81399355 | 1.34 |
ENST00000457544.2
|
HGF
|
hepatocyte growth factor (hepapoietin A; scatter factor) |
| chrX_-_70329118 | 1.34 |
ENST00000374188.3
|
IL2RG
|
interleukin 2 receptor, gamma |
| chr20_-_7921090 | 1.32 |
ENST00000378789.3
|
HAO1
|
hydroxyacid oxidase (glycolate oxidase) 1 |
| chr10_+_124320156 | 1.31 |
ENST00000338354.3
ENST00000344338.3 ENST00000330163.4 ENST00000368909.3 ENST00000368955.3 ENST00000368956.2 |
DMBT1
|
deleted in malignant brain tumors 1 |
| chr7_-_81399329 | 1.31 |
ENST00000453411.1
ENST00000444829.2 |
HGF
|
hepatocyte growth factor (hepapoietin A; scatter factor) |
| chr6_+_161123270 | 1.31 |
ENST00000366924.2
ENST00000308192.9 ENST00000418964.1 |
PLG
|
plasminogen |
| chr20_-_7238861 | 1.29 |
ENST00000428954.1
|
RP11-19D2.1
|
RP11-19D2.1 |
| chr13_-_39564993 | 1.29 |
ENST00000423210.1
|
STOML3
|
stomatin (EPB72)-like 3 |
| chr7_-_107643567 | 1.28 |
ENST00000393559.1
ENST00000439976.1 ENST00000393560.1 |
LAMB1
|
laminin, beta 1 |
| chr7_-_81399411 | 1.26 |
ENST00000423064.2
|
HGF
|
hepatocyte growth factor (hepapoietin A; scatter factor) |
| chr3_+_119316689 | 1.26 |
ENST00000273371.4
|
PLA1A
|
phospholipase A1 member A |
| chr17_+_8924837 | 1.25 |
ENST00000173229.2
|
NTN1
|
netrin 1 |
| chr21_+_17791648 | 1.24 |
ENST00000602892.1
ENST00000418813.2 ENST00000435697.1 |
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr18_+_56113488 | 1.24 |
ENST00000590797.1
|
RP11-1151B14.3
|
RP11-1151B14.3 |
| chr6_-_161085291 | 1.24 |
ENST00000316300.5
|
LPA
|
lipoprotein, Lp(a) |
| chr5_+_101569696 | 1.22 |
ENST00000597120.1
|
AC008948.1
|
AC008948.1 |
| chr7_-_107643674 | 1.21 |
ENST00000222399.6
|
LAMB1
|
laminin, beta 1 |
| chr15_-_76352069 | 1.21 |
ENST00000305435.10
ENST00000563910.1 |
NRG4
|
neuregulin 4 |
| chr18_+_68002675 | 1.18 |
ENST00000584919.1
|
RP11-41O4.1
|
Uncharacterized protein |
| chr9_-_4299874 | 1.18 |
ENST00000381971.3
ENST00000477901.1 |
GLIS3
|
GLIS family zinc finger 3 |
| chr1_-_169555709 | 1.17 |
ENST00000546081.1
|
F5
|
coagulation factor V (proaccelerin, labile factor) |
| chr6_+_44310376 | 1.17 |
ENST00000515220.1
ENST00000323108.8 |
SPATS1
|
spermatogenesis associated, serine-rich 1 |
| chr21_+_17791838 | 1.16 |
ENST00000453910.1
|
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr12_-_21927736 | 1.15 |
ENST00000240662.2
|
KCNJ8
|
potassium inwardly-rectifying channel, subfamily J, member 8 |
| chr10_+_71562180 | 1.15 |
ENST00000517713.1
ENST00000522165.1 ENST00000520133.1 |
COL13A1
|
collagen, type XIII, alpha 1 |
| chr3_+_132316081 | 1.14 |
ENST00000249887.2
|
ACKR4
|
atypical chemokine receptor 4 |
| chr14_-_25479811 | 1.14 |
ENST00000550887.1
|
STXBP6
|
syntaxin binding protein 6 (amisyn) |
| chr3_+_114012819 | 1.13 |
ENST00000383671.3
|
TIGIT
|
T cell immunoreceptor with Ig and ITIM domains |
| chr2_+_171640291 | 1.12 |
ENST00000409885.1
|
ERICH2
|
glutamate-rich 2 |
| chr7_+_134464414 | 1.11 |
ENST00000361901.2
|
CALD1
|
caldesmon 1 |
| chr11_-_58378759 | 1.11 |
ENST00000601906.1
|
AP001350.1
|
Uncharacterized protein |
| chr7_-_81399678 | 1.10 |
ENST00000412881.1
|
HGF
|
hepatocyte growth factor (hepapoietin A; scatter factor) |
| chr15_+_58430368 | 1.10 |
ENST00000558772.1
ENST00000219919.4 |
AQP9
|
aquaporin 9 |
| chr14_-_36990354 | 1.09 |
ENST00000518149.1
|
NKX2-1
|
NK2 homeobox 1 |
| chr15_+_58430567 | 1.07 |
ENST00000536493.1
|
AQP9
|
aquaporin 9 |
| chr11_-_108464465 | 1.07 |
ENST00000525344.1
|
EXPH5
|
exophilin 5 |
| chr15_+_69854027 | 1.07 |
ENST00000498938.2
|
RP11-279F6.1
|
RP11-279F6.1 |
| chr7_-_81399287 | 1.06 |
ENST00000354224.6
|
HGF
|
hepatocyte growth factor (hepapoietin A; scatter factor) |
| chr12_-_15038779 | 1.06 |
ENST00000228938.5
ENST00000539261.1 |
MGP
|
matrix Gla protein |
| chr5_-_137674000 | 1.06 |
ENST00000510119.1
ENST00000513970.1 |
CDC25C
|
cell division cycle 25C |
| chr16_-_48281444 | 1.06 |
ENST00000537808.1
ENST00000569991.1 |
ABCC11
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 11 |
| chr14_+_38033252 | 1.06 |
ENST00000554829.1
|
RP11-356O9.1
|
RP11-356O9.1 |
| chr6_+_139135648 | 1.05 |
ENST00000541398.1
|
ECT2L
|
epithelial cell transforming sequence 2 oncogene-like |
| chr2_-_188312971 | 1.05 |
ENST00000410068.1
ENST00000447403.1 ENST00000410102.1 |
CALCRL
|
calcitonin receptor-like |
| chr4_-_103266626 | 1.04 |
ENST00000356736.4
|
SLC39A8
|
solute carrier family 39 (zinc transporter), member 8 |
| chr11_-_108464321 | 1.03 |
ENST00000265843.4
|
EXPH5
|
exophilin 5 |
| chr12_+_59989791 | 1.00 |
ENST00000552432.1
|
SLC16A7
|
solute carrier family 16 (monocarboxylate transporter), member 7 |
| chr20_+_45338126 | 1.00 |
ENST00000359271.2
|
SLC2A10
|
solute carrier family 2 (facilitated glucose transporter), member 10 |
| chr12_-_21928515 | 0.98 |
ENST00000537950.1
|
KCNJ8
|
potassium inwardly-rectifying channel, subfamily J, member 8 |
| chrX_+_135614293 | 0.97 |
ENST00000370634.3
|
VGLL1
|
vestigial like 1 (Drosophila) |
| chr4_-_186696636 | 0.94 |
ENST00000444771.1
|
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr2_-_21266935 | 0.93 |
ENST00000233242.1
|
APOB
|
apolipoprotein B |
| chr4_-_152149033 | 0.93 |
ENST00000514152.1
|
SH3D19
|
SH3 domain containing 19 |
| chr4_-_186696561 | 0.92 |
ENST00000445115.1
ENST00000451701.1 ENST00000457247.1 ENST00000435480.1 ENST00000425679.1 ENST00000457934.1 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chrX_+_135730297 | 0.92 |
ENST00000370629.2
|
CD40LG
|
CD40 ligand |
| chr1_-_177939041 | 0.92 |
ENST00000308284.6
|
SEC16B
|
SEC16 homolog B (S. cerevisiae) |
| chr7_+_134576151 | 0.91 |
ENST00000393118.2
|
CALD1
|
caldesmon 1 |
| chr2_-_87248975 | 0.90 |
ENST00000409310.2
ENST00000355705.3 |
PLGLB1
|
plasminogen-like B1 |
| chr5_-_115872124 | 0.89 |
ENST00000515009.1
|
SEMA6A
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A |
| chr4_-_186696425 | 0.89 |
ENST00000430503.1
ENST00000319454.6 ENST00000450341.1 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr10_+_124320195 | 0.89 |
ENST00000359586.6
|
DMBT1
|
deleted in malignant brain tumors 1 |
| chr11_-_57158109 | 0.88 |
ENST00000525955.1
ENST00000533605.1 ENST00000311862.5 |
PRG2
|
proteoglycan 2, bone marrow (natural killer cell activator, eosinophil granule major basic protein) |
| chr6_+_44310421 | 0.88 |
ENST00000288390.2
|
SPATS1
|
spermatogenesis associated, serine-rich 1 |
| chr3_+_177159695 | 0.87 |
ENST00000442937.1
|
LINC00578
|
long intergenic non-protein coding RNA 578 |
| chr12_-_7596735 | 0.87 |
ENST00000416109.2
ENST00000396630.1 ENST00000313599.3 |
CD163L1
|
CD163 molecule-like 1 |
| chr21_-_43786634 | 0.87 |
ENST00000291527.2
|
TFF1
|
trefoil factor 1 |
| chr6_-_159466136 | 0.87 |
ENST00000367066.3
ENST00000326965.6 |
TAGAP
|
T-cell activation RhoGTPase activating protein |
| chr17_-_66951474 | 0.86 |
ENST00000269080.2
|
ABCA8
|
ATP-binding cassette, sub-family A (ABC1), member 8 |
| chr12_-_102591604 | 0.85 |
ENST00000329406.4
|
PMCH
|
pro-melanin-concentrating hormone |
| chr4_+_15341442 | 0.84 |
ENST00000397700.2
ENST00000295297.4 |
C1QTNF7
|
C1q and tumor necrosis factor related protein 7 |
| chr17_-_38020392 | 0.84 |
ENST00000346872.3
ENST00000439167.2 ENST00000377945.3 ENST00000394189.2 ENST00000377944.3 ENST00000377958.2 ENST00000535189.1 ENST00000377952.2 |
IKZF3
|
IKAROS family zinc finger 3 (Aiolos) |
| chr7_-_81399744 | 0.82 |
ENST00000421558.1
|
HGF
|
hepatocyte growth factor (hepapoietin A; scatter factor) |
| chr1_+_74701062 | 0.81 |
ENST00000326637.3
|
TNNI3K
|
TNNI3 interacting kinase |
| chr7_+_134430212 | 0.81 |
ENST00000436461.2
|
CALD1
|
caldesmon 1 |
| chr2_-_180427304 | 0.79 |
ENST00000336917.5
|
ZNF385B
|
zinc finger protein 385B |
| chr19_-_47157914 | 0.79 |
ENST00000300875.4
|
DACT3
|
dishevelled-binding antagonist of beta-catenin 3 |
| chr7_+_73242490 | 0.78 |
ENST00000431918.1
|
CLDN4
|
claudin 4 |
| chr4_+_75174180 | 0.74 |
ENST00000413830.1
|
EPGN
|
epithelial mitogen |
| chr17_-_77967433 | 0.73 |
ENST00000571872.1
|
TBC1D16
|
TBC1 domain family, member 16 |
| chr2_-_21266816 | 0.73 |
ENST00000399256.4
|
APOB
|
apolipoprotein B |
| chr17_-_38020379 | 0.73 |
ENST00000351680.3
ENST00000346243.3 ENST00000350532.3 ENST00000467757.1 ENST00000439016.2 |
IKZF3
|
IKAROS family zinc finger 3 (Aiolos) |
| chr14_+_37126765 | 0.72 |
ENST00000402703.2
|
PAX9
|
paired box 9 |
| chr11_-_102401469 | 0.71 |
ENST00000260227.4
|
MMP7
|
matrix metallopeptidase 7 (matrilysin, uterine) |
| chr2_-_208030647 | 0.71 |
ENST00000309446.6
|
KLF7
|
Kruppel-like factor 7 (ubiquitous) |
| chr3_+_177159744 | 0.71 |
ENST00000439009.1
|
LINC00578
|
long intergenic non-protein coding RNA 578 |
| chr15_+_75940218 | 0.70 |
ENST00000308527.5
|
SNX33
|
sorting nexin 33 |
| chr16_+_20775358 | 0.69 |
ENST00000440284.2
|
ACSM3
|
acyl-CoA synthetase medium-chain family member 3 |
| chr7_+_134464376 | 0.68 |
ENST00000454108.1
ENST00000361675.2 |
CALD1
|
caldesmon 1 |
| chr1_-_178840157 | 0.68 |
ENST00000367629.1
ENST00000234816.2 |
ANGPTL1
|
angiopoietin-like 1 |
| chr3_+_142315225 | 0.68 |
ENST00000457734.2
ENST00000483373.1 ENST00000475296.1 ENST00000495744.1 ENST00000476044.1 ENST00000461644.1 |
PLS1
|
plastin 1 |
| chr2_+_169659121 | 0.67 |
ENST00000397206.2
ENST00000397209.2 ENST00000421711.2 |
NOSTRIN
|
nitric oxide synthase trafficking |
| chr17_+_7487146 | 0.67 |
ENST00000396501.4
ENST00000584378.1 ENST00000423172.2 ENST00000579445.1 ENST00000585217.1 ENST00000581380.1 |
MPDU1
|
mannose-P-dolichol utilization defect 1 |
| chr14_-_104408032 | 0.65 |
ENST00000455348.2
|
RD3L
|
retinal degeneration 3-like |
| chr4_-_186696515 | 0.65 |
ENST00000456596.1
ENST00000414724.1 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chrX_-_101410762 | 0.64 |
ENST00000543160.1
ENST00000333643.3 |
BEX5
|
brain expressed, X-linked 5 |
| chr8_+_55528627 | 0.64 |
ENST00000220676.1
|
RP1
|
retinitis pigmentosa 1 (autosomal dominant) |
| chr1_+_115642293 | 0.64 |
ENST00000448680.1
|
RP4-666F24.3
|
RP4-666F24.3 |
| chr7_-_117512264 | 0.63 |
ENST00000454375.1
|
CTTNBP2
|
cortactin binding protein 2 |
| chr6_-_112575912 | 0.63 |
ENST00000522006.1
ENST00000230538.7 ENST00000519932.1 |
LAMA4
|
laminin, alpha 4 |
| chr11_-_115375107 | 0.63 |
ENST00000545380.1
ENST00000452722.3 ENST00000537058.1 ENST00000536727.1 ENST00000542447.2 ENST00000331581.6 |
CADM1
|
cell adhesion molecule 1 |
| chr14_+_23299088 | 0.63 |
ENST00000355151.5
ENST00000397496.3 ENST00000555345.1 ENST00000432849.3 ENST00000553711.1 ENST00000556465.1 ENST00000397505.2 ENST00000557221.1 ENST00000311892.6 ENST00000556840.1 ENST00000555536.1 |
MRPL52
|
mitochondrial ribosomal protein L52 |
| chr6_+_122793058 | 0.63 |
ENST00000392491.2
|
PKIB
|
protein kinase (cAMP-dependent, catalytic) inhibitor beta |
| chr6_-_112575838 | 0.62 |
ENST00000455073.1
|
LAMA4
|
laminin, alpha 4 |
| chr6_-_25830785 | 0.61 |
ENST00000468082.1
|
SLC17A1
|
solute carrier family 17 (organic anion transporter), member 1 |
| chr10_+_75668916 | 0.61 |
ENST00000481390.1
|
PLAU
|
plasminogen activator, urokinase |
| chr20_+_9049303 | 0.60 |
ENST00000407043.2
ENST00000441846.1 |
PLCB4
|
phospholipase C, beta 4 |
| chr6_-_112575758 | 0.60 |
ENST00000431543.2
ENST00000453937.2 ENST00000368638.4 ENST00000389463.4 |
LAMA4
|
laminin, alpha 4 |
| chr8_+_75736761 | 0.60 |
ENST00000260113.2
|
PI15
|
peptidase inhibitor 15 |
| chr6_+_21666633 | 0.59 |
ENST00000606851.1
|
CASC15
|
cancer susceptibility candidate 15 (non-protein coding) |
| chr2_+_169658928 | 0.59 |
ENST00000317647.7
ENST00000445023.2 |
NOSTRIN
|
nitric oxide synthase trafficking |
| chr3_-_116163830 | 0.58 |
ENST00000333617.4
|
LSAMP
|
limbic system-associated membrane protein |
| chr1_+_214163033 | 0.58 |
ENST00000607425.1
|
PROX1
|
prospero homeobox 1 |
| chr9_-_70490107 | 0.57 |
ENST00000377395.4
ENST00000429800.2 ENST00000430059.2 ENST00000377384.1 ENST00000382405.3 |
CBWD5
|
COBW domain containing 5 |
| chr6_+_10528560 | 0.57 |
ENST00000379597.3
|
GCNT2
|
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme (I blood group) |
Network of associatons between targets according to the STRING database.
First level regulatory network of FOXA3_FOXC2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 10.0 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 1.8 | 10.8 | GO:0010193 | response to ozone(GO:0010193) |
| 1.5 | 6.0 | GO:0010903 | negative regulation of very-low-density lipoprotein particle remodeling(GO:0010903) |
| 1.1 | 5.6 | GO:0001970 | positive regulation of activation of membrane attack complex(GO:0001970) |
| 1.0 | 3.9 | GO:0044537 | regulation of circulating fibrinogen levels(GO:0044537) |
| 0.9 | 3.5 | GO:1900191 | biofilm formation(GO:0042710) single-species biofilm formation(GO:0044010) single-species biofilm formation in or on host organism(GO:0044407) regulation of single-species biofilm formation(GO:1900190) negative regulation of single-species biofilm formation(GO:1900191) regulation of single-species biofilm formation in or on host organism(GO:1900228) negative regulation of single-species biofilm formation in or on host organism(GO:1900229) |
| 0.8 | 2.4 | GO:0060738 | epithelial-mesenchymal signaling involved in prostate gland development(GO:0060738) |
| 0.8 | 4.7 | GO:0002803 | positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antimicrobial humoral response(GO:0002760) positive regulation of antibacterial peptide production(GO:0002803) |
| 0.8 | 2.3 | GO:0044179 | hemolysis by symbiont of host erythrocytes(GO:0019836) hemolysis in other organism(GO:0044179) hemolysis in other organism involved in symbiotic interaction(GO:0052331) |
| 0.7 | 4.7 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.7 | 6.7 | GO:0030450 | regulation of complement activation, classical pathway(GO:0030450) negative regulation of complement activation, classical pathway(GO:0045959) |
| 0.6 | 4.5 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.6 | 8.4 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.6 | 3.7 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.6 | 10.6 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.5 | 5.4 | GO:0071847 | TNFSF11-mediated signaling pathway(GO:0071847) |
| 0.5 | 8.7 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.5 | 2.1 | GO:0010982 | regulation of high-density lipoprotein particle clearance(GO:0010982) positive regulation of high-density lipoprotein particle clearance(GO:0010983) |
| 0.5 | 2.5 | GO:0021812 | neuronal-glial interaction involved in cerebral cortex radial glia guided migration(GO:0021812) |
| 0.5 | 3.9 | GO:0032431 | activation of phospholipase A2 activity(GO:0032431) |
| 0.5 | 2.8 | GO:0032571 | response to vitamin K(GO:0032571) |
| 0.4 | 8.1 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.4 | 3.2 | GO:1901475 | pyruvate transport(GO:0006848) pyruvate transmembrane transport(GO:1901475) |
| 0.4 | 1.5 | GO:0009956 | radial pattern formation(GO:0009956) |
| 0.4 | 2.2 | GO:0015722 | canalicular bile acid transport(GO:0015722) pyrimidine nucleobase transport(GO:0015855) purine nucleobase transmembrane transport(GO:1904823) |
| 0.3 | 4.1 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.3 | 3.4 | GO:1902897 | regulation of postsynaptic density protein 95 clustering(GO:1902897) |
| 0.3 | 3.7 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.3 | 2.3 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.3 | 1.5 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.2 | 2.2 | GO:2000660 | negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.2 | 1.9 | GO:0009441 | glycolate metabolic process(GO:0009441) |
| 0.2 | 2.3 | GO:0034144 | negative regulation of toll-like receptor 4 signaling pathway(GO:0034144) |
| 0.2 | 0.9 | GO:0002215 | defense response to nematode(GO:0002215) |
| 0.2 | 3.0 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.2 | 1.7 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.2 | 0.6 | GO:0097534 | lymphoid lineage cell migration(GO:0097534) lymphoid lineage cell migration into thymus(GO:0097535) |
| 0.2 | 3.4 | GO:0035860 | glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.2 | 1.3 | GO:0038110 | interleukin-2-mediated signaling pathway(GO:0038110) |
| 0.2 | 2.6 | GO:0045852 | pH elevation(GO:0045852) intracellular pH elevation(GO:0051454) |
| 0.2 | 0.5 | GO:0039007 | pronephric nephron morphogenesis(GO:0039007) pronephric nephron tubule morphogenesis(GO:0039008) pronephric duct development(GO:0039022) pronephric duct morphogenesis(GO:0039023) Kupffer's vesicle development(GO:0070121) |
| 0.2 | 1.3 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.1 | 0.6 | GO:0002305 | gamma-delta intraepithelial T cell differentiation(GO:0002304) CD8-positive, gamma-delta intraepithelial T cell differentiation(GO:0002305) |
| 0.1 | 1.0 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.1 | 0.5 | GO:0050928 | negative regulation of positive chemotaxis(GO:0050928) |
| 0.1 | 0.3 | GO:0032430 | positive regulation of phospholipase A2 activity(GO:0032430) |
| 0.1 | 8.4 | GO:0001958 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.1 | 2.7 | GO:0015865 | purine nucleotide transport(GO:0015865) |
| 0.1 | 0.6 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.1 | 0.9 | GO:1904715 | negative regulation of chaperone-mediated autophagy(GO:1904715) |
| 0.1 | 2.1 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.1 | 1.4 | GO:1904352 | positive regulation of protein catabolic process in the vacuole(GO:1904352) |
| 0.1 | 0.6 | GO:0051958 | methotrexate transport(GO:0051958) |
| 0.1 | 4.9 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.1 | 0.3 | GO:0021934 | medulla oblongata development(GO:0021550) hindbrain tangential cell migration(GO:0021934) lateral line system development(GO:0048925) |
| 0.1 | 1.1 | GO:0034285 | response to sucrose(GO:0009744) response to disaccharide(GO:0034285) |
| 0.1 | 1.2 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 0.1 | 4.2 | GO:0003301 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.1 | 0.5 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.1 | 1.0 | GO:0070574 | cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.1 | 1.1 | GO:0030210 | heparin metabolic process(GO:0030202) heparin biosynthetic process(GO:0030210) |
| 0.1 | 1.6 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.1 | 2.1 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.1 | 1.2 | GO:0034374 | low-density lipoprotein particle remodeling(GO:0034374) |
| 0.1 | 0.2 | GO:0034146 | toll-like receptor 5 signaling pathway(GO:0034146) |
| 0.1 | 1.0 | GO:0038044 | transforming growth factor-beta secretion(GO:0038044) |
| 0.1 | 0.6 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.1 | 2.3 | GO:0070233 | negative regulation of T cell apoptotic process(GO:0070233) |
| 0.1 | 0.5 | GO:0070857 | regulation of bile acid biosynthetic process(GO:0070857) |
| 0.1 | 0.2 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.1 | 1.1 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.1 | 1.6 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.1 | 2.5 | GO:0008211 | glucocorticoid metabolic process(GO:0008211) |
| 0.1 | 1.3 | GO:0070863 | positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.1 | 0.4 | GO:0033512 | L-lysine catabolic process to acetyl-CoA via saccharopine(GO:0033512) |
| 0.1 | 1.6 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.1 | 0.5 | GO:0000480 | endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.1 | 0.5 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.1 | 0.5 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.1 | 1.3 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.1 | 0.5 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.1 | 0.3 | GO:0010966 | regulation of phosphate transport(GO:0010966) |
| 0.1 | 1.6 | GO:0042359 | vitamin D metabolic process(GO:0042359) |
| 0.1 | 0.5 | GO:0003383 | apical constriction(GO:0003383) |
| 0.1 | 0.6 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) retinal cone cell differentiation(GO:0042670) retinal cone cell development(GO:0046549) |
| 0.1 | 0.8 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.1 | 2.5 | GO:0032733 | positive regulation of interleukin-10 production(GO:0032733) |
| 0.1 | 0.5 | GO:1903546 | protein localization to photoreceptor outer segment(GO:1903546) |
| 0.1 | 0.4 | GO:0042536 | negative regulation of tumor necrosis factor biosynthetic process(GO:0042536) |
| 0.0 | 0.8 | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.0 | 0.1 | GO:0060730 | regulation of intestinal epithelial structure maintenance(GO:0060730) |
| 0.0 | 0.4 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 2.4 | GO:0060706 | cell differentiation involved in embryonic placenta development(GO:0060706) |
| 0.0 | 2.5 | GO:0033572 | transferrin transport(GO:0033572) |
| 0.0 | 0.5 | GO:1904153 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.0 | 0.3 | GO:0021993 | initiation of neural tube closure(GO:0021993) |
| 0.0 | 0.6 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.0 | 0.2 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.0 | 1.2 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.2 | GO:0019441 | tryptophan catabolic process to kynurenine(GO:0019441) |
| 0.0 | 0.1 | GO:0032747 | positive regulation of interleukin-23 production(GO:0032747) |
| 0.0 | 1.6 | GO:0045577 | regulation of B cell differentiation(GO:0045577) |
| 0.0 | 0.2 | GO:0036518 | chemorepulsion of dopaminergic neuron axon(GO:0036518) chemorepulsion of axon(GO:0061643) |
| 0.0 | 0.9 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.0 | 1.1 | GO:0001502 | cartilage condensation(GO:0001502) |
| 0.0 | 0.6 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.7 | GO:0044849 | estrous cycle(GO:0044849) |
| 0.0 | 0.1 | GO:0002426 | immunoglobulin production in mucosal tissue(GO:0002426) |
| 0.0 | 0.1 | GO:0061030 | axon target recognition(GO:0007412) epithelial cell differentiation involved in mammary gland alveolus development(GO:0061030) |
| 0.0 | 0.2 | GO:1900147 | Schwann cell migration(GO:0036135) regulation of Schwann cell migration(GO:1900147) |
| 0.0 | 0.6 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 0.0 | 0.9 | GO:0036150 | phosphatidylserine acyl-chain remodeling(GO:0036150) |
| 0.0 | 0.7 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 0.2 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.0 | 0.1 | GO:1903216 | regulation of protein processing involved in protein targeting to mitochondrion(GO:1903216) negative regulation of protein processing involved in protein targeting to mitochondrion(GO:1903217) |
| 0.0 | 1.0 | GO:1904659 | hexose transmembrane transport(GO:0035428) glucose transmembrane transport(GO:1904659) |
| 0.0 | 0.5 | GO:0006012 | galactose metabolic process(GO:0006012) |
| 0.0 | 0.9 | GO:0010039 | response to iron ion(GO:0010039) |
| 0.0 | 2.7 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 0.7 | GO:0042481 | regulation of odontogenesis(GO:0042481) |
| 0.0 | 0.2 | GO:0097647 | calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.0 | 0.3 | GO:0042048 | olfactory behavior(GO:0042048) |
| 0.0 | 0.7 | GO:0007202 | activation of phospholipase C activity(GO:0007202) |
| 0.0 | 0.4 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 0.3 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.0 | 0.5 | GO:1904776 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.0 | 0.1 | GO:0035574 | histone H4-K20 demethylation(GO:0035574) |
| 0.0 | 0.3 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.0 | 1.9 | GO:0006977 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) |
| 0.0 | 0.3 | GO:0060347 | heart trabecula formation(GO:0060347) |
| 0.0 | 0.1 | GO:0006001 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 0.0 | 0.7 | GO:0046627 | negative regulation of insulin receptor signaling pathway(GO:0046627) |
| 0.0 | 1.2 | GO:1901185 | negative regulation of ERBB signaling pathway(GO:1901185) |
| 0.0 | 0.3 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.0 | 0.1 | GO:0003056 | regulation of vascular smooth muscle contraction(GO:0003056) |
| 0.0 | 0.7 | GO:0001919 | regulation of receptor recycling(GO:0001919) |
| 0.0 | 1.5 | GO:0030449 | regulation of complement activation(GO:0030449) |
| 0.0 | 0.2 | GO:0051573 | negative regulation of histone H3-K9 methylation(GO:0051573) |
| 0.0 | 1.5 | GO:0031110 | regulation of microtubule polymerization or depolymerization(GO:0031110) |
| 0.0 | 0.1 | GO:0097106 | postsynaptic density organization(GO:0097106) |
| 0.0 | 0.4 | GO:0070830 | bicellular tight junction assembly(GO:0070830) |
| 0.0 | 0.0 | GO:1903676 | regulation of cap-dependent translational initiation(GO:1903674) positive regulation of cap-dependent translational initiation(GO:1903676) |
| 0.0 | 0.0 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 0.0 | 0.3 | GO:0001893 | maternal placenta development(GO:0001893) |
| 0.0 | 0.4 | GO:0007130 | synaptonemal complex assembly(GO:0007130) |
| 0.0 | 1.1 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.0 | 0.2 | GO:0006275 | regulation of DNA replication(GO:0006275) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 10.0 | GO:0071062 | rough endoplasmic reticulum lumen(GO:0048237) alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 2.1 | 8.4 | GO:0005600 | collagen type XIII trimer(GO:0005600) transmembrane collagen trimer(GO:0030936) |
| 2.0 | 6.0 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 1.4 | 13.7 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.8 | 2.5 | GO:0043257 | laminin-8 complex(GO:0043257) |
| 0.4 | 3.4 | GO:1990357 | terminal web(GO:1990357) |
| 0.3 | 5.1 | GO:0030478 | actin cap(GO:0030478) |
| 0.3 | 1.0 | GO:0034685 | integrin alphav-beta6 complex(GO:0034685) |
| 0.3 | 2.0 | GO:0032311 | angiogenin-PRI complex(GO:0032311) |
| 0.3 | 3.0 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.3 | 2.1 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.2 | 1.7 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.2 | 1.3 | GO:0044279 | other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.2 | 3.5 | GO:0016342 | catenin complex(GO:0016342) |
| 0.1 | 0.9 | GO:0098575 | lumenal side of lysosomal membrane(GO:0098575) |
| 0.1 | 2.6 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.1 | 1.8 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.1 | 13.7 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.1 | 12.0 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 9.7 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.1 | 2.3 | GO:0005605 | basal lamina(GO:0005605) |
| 0.1 | 1.8 | GO:0071682 | endocytic vesicle lumen(GO:0071682) |
| 0.1 | 1.2 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.1 | 2.7 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.0 | 0.5 | GO:0000836 | Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.0 | 6.4 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 9.5 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 0.5 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 1.2 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 1.2 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.2 | GO:0001652 | granular component(GO:0001652) |
| 0.0 | 1.1 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 1.2 | GO:1990777 | plasma lipoprotein particle(GO:0034358) lipoprotein particle(GO:1990777) |
| 0.0 | 0.5 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.5 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.9 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 2.4 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 0.7 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 3.8 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 1.3 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 7.0 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 4.0 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.5 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.0 | 29.6 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 0.5 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.4 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.5 | GO:0043296 | apical junction complex(GO:0043296) |
| 0.0 | 1.4 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 3.0 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.3 | GO:0005922 | connexon complex(GO:0005922) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 6.0 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 1.0 | 10.8 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.8 | 3.2 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.7 | 2.2 | GO:0005150 | interleukin-1, Type I receptor binding(GO:0005150) |
| 0.5 | 2.1 | GO:0017098 | sulfonylurea receptor binding(GO:0017098) |
| 0.5 | 1.6 | GO:1902271 | D3 vitamins binding(GO:1902271) |
| 0.5 | 1.4 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.5 | 3.3 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.4 | 1.3 | GO:0003973 | (S)-2-hydroxy-acid oxidase activity(GO:0003973) very-long-chain-(S)-2-hydroxy-acid oxidase activity(GO:0052852) long-chain-(S)-2-hydroxy-long-chain-acid oxidase activity(GO:0052853) medium-chain-(S)-2-hydroxy-acid oxidase activity(GO:0052854) |
| 0.4 | 1.3 | GO:1904854 | proteasome core complex binding(GO:1904854) |
| 0.4 | 2.2 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 0.4 | 8.7 | GO:0005537 | mannose binding(GO:0005537) |
| 0.4 | 2.7 | GO:0052739 | phosphatidylserine 1-acylhydrolase activity(GO:0052739) |
| 0.4 | 3.4 | GO:0016167 | glial cell-derived neurotrophic factor receptor activity(GO:0016167) |
| 0.3 | 1.3 | GO:0004913 | interleukin-4 receptor activity(GO:0004913) interleukin-7 receptor activity(GO:0004917) |
| 0.3 | 2.0 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 0.3 | 1.1 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.2 | 7.1 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.2 | 2.1 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.2 | 7.4 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.2 | 1.7 | GO:0035473 | lipase binding(GO:0035473) |
| 0.2 | 7.0 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.2 | 0.5 | GO:0033858 | N-acetylgalactosamine kinase activity(GO:0033858) |
| 0.2 | 3.7 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) intronic transcription regulatory region DNA binding(GO:0044213) |
| 0.2 | 0.8 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.2 | 1.9 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.2 | 1.4 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.2 | 2.9 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.1 | 2.7 | GO:0043225 | anion transmembrane-transporting ATPase activity(GO:0043225) |
| 0.1 | 4.7 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.1 | 3.5 | GO:0045294 | alpha-catenin binding(GO:0045294) gamma-catenin binding(GO:0045295) |
| 0.1 | 7.2 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.1 | 1.2 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.1 | 0.3 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 0.1 | 0.8 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.1 | 1.1 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.1 | 2.3 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.1 | 0.5 | GO:0003963 | RNA-3'-phosphate cyclase activity(GO:0003963) |
| 0.1 | 0.6 | GO:0015350 | methotrexate transporter activity(GO:0015350) |
| 0.1 | 4.2 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.1 | 1.4 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.1 | 0.7 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.1 | 0.9 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 0.3 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.1 | 0.6 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.1 | 4.4 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.1 | 14.8 | GO:0008201 | heparin binding(GO:0008201) |
| 0.1 | 2.2 | GO:0008329 | signaling pattern recognition receptor activity(GO:0008329) pattern recognition receptor activity(GO:0038187) |
| 0.1 | 0.7 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.1 | 0.2 | GO:0033754 | indoleamine 2,3-dioxygenase activity(GO:0033754) |
| 0.0 | 1.0 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.0 | 0.2 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.0 | 0.8 | GO:0070513 | death domain binding(GO:0070513) |
| 0.0 | 0.2 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.0 | 1.1 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 5.1 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.6 | GO:0015321 | sodium-dependent phosphate transmembrane transporter activity(GO:0015321) |
| 0.0 | 0.5 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.0 | 1.5 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 2.8 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.0 | 2.1 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 1.3 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 0.6 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.5 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.6 | GO:0030414 | peptidase inhibitor activity(GO:0030414) |
| 0.0 | 8.2 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.2 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.0 | 0.2 | GO:1904929 | coreceptor activity involved in Wnt signaling pathway, planar cell polarity pathway(GO:1904929) |
| 0.0 | 0.4 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.6 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 2.0 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.1 | GO:0047708 | biotinidase activity(GO:0047708) |
| 0.0 | 1.7 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 0.1 | GO:0035575 | histone demethylase activity (H4-K20 specific)(GO:0035575) |
| 0.0 | 3.6 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.0 | 0.3 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 1.8 | GO:0098531 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.0 | 3.6 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 2.1 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.4 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.5 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.0 | 1.7 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.2 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.0 | 0.4 | GO:0016646 | oxidoreductase activity, acting on the CH-NH group of donors, NAD or NADP as acceptor(GO:0016646) |
| 0.0 | 2.5 | GO:0019902 | phosphatase binding(GO:0019902) |
| 0.0 | 0.1 | GO:0004969 | histamine receptor activity(GO:0004969) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 11.6 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.2 | 17.5 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.2 | 18.2 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 12.4 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 7.1 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.1 | 6.0 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.1 | 1.7 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.1 | 3.9 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.1 | 5.6 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 13.7 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 2.5 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.0 | 0.7 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 2.5 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 2.5 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 2.2 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 3.1 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.0 | 1.2 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 1.6 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 1.8 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 1.5 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 1.1 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 1.2 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.0 | 1.4 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 6.2 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 1.1 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 0.5 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 0.2 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 6.9 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.4 | 8.7 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.3 | 6.6 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.3 | 20.4 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.3 | 9.7 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.3 | 7.7 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.2 | 5.0 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.2 | 3.2 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 2.2 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.1 | 6.6 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.1 | 1.9 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.1 | 8.4 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 2.1 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.1 | 1.2 | REACTOME LIPOPROTEIN METABOLISM | Genes involved in Lipoprotein metabolism |
| 0.1 | 13.3 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.1 | 5.2 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 1.7 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.1 | 1.6 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.1 | 3.9 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.1 | 1.3 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.1 | 3.4 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 4.0 | REACTOME CELL CELL JUNCTION ORGANIZATION | Genes involved in Cell-cell junction organization |
| 0.1 | 0.8 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.1 | 2.5 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.1 | 0.5 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.0 | 2.0 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.8 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 1.1 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 1.3 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 1.3 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.0 | 1.2 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.0 | 2.1 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.0 | 1.0 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 1.7 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.0 | 2.1 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 0.6 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 1.4 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.0 | 1.8 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 3.5 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 1.4 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.9 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.1 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.0 | 1.0 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 0.3 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |