Project
Illumina Body Map 2: averaged replicates
Navigation
Downloads
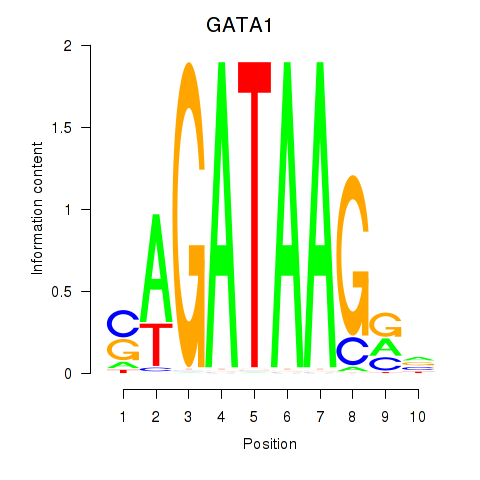
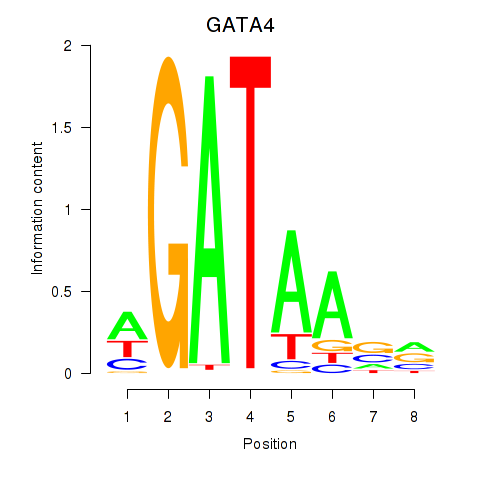
Results for GATA1_GATA4
Z-value: 3.11
Transcription factors associated with GATA1_GATA4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
GATA1
|
ENSG00000102145.9 | GATA binding protein 1 |
|
GATA4
|
ENSG00000136574.13 | GATA binding protein 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| GATA4 | hg19_v2_chr8_+_11561660_11561751 | 0.25 | 1.6e-01 | Click! |
| GATA1 | hg19_v2_chrX_+_48644962_48644983 | -0.01 | 9.6e-01 | Click! |
Activity profile of GATA1_GATA4 motif
Sorted Z-values of GATA1_GATA4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_99531709 | 10.01 |
ENST00000266066.3
|
SFRP5
|
secreted frizzled-related protein 5 |
| chr18_+_19749386 | 8.03 |
ENST00000269216.3
|
GATA6
|
GATA binding protein 6 |
| chr12_-_114841703 | 7.97 |
ENST00000526441.1
|
TBX5
|
T-box 5 |
| chr11_-_116708302 | 6.95 |
ENST00000375320.1
ENST00000359492.2 ENST00000375329.2 ENST00000375323.1 |
APOA1
|
apolipoprotein A-I |
| chr19_-_55669093 | 6.83 |
ENST00000344887.5
|
TNNI3
|
troponin I type 3 (cardiac) |
| chr1_-_27240455 | 6.31 |
ENST00000254227.3
|
NR0B2
|
nuclear receptor subfamily 0, group B, member 2 |
| chr10_-_48416849 | 6.04 |
ENST00000249598.1
|
GDF2
|
growth differentiation factor 2 |
| chr11_-_47374246 | 5.71 |
ENST00000545968.1
ENST00000399249.2 ENST00000256993.4 |
MYBPC3
|
myosin binding protein C, cardiac |
| chr2_-_69098566 | 5.67 |
ENST00000295379.1
|
BMP10
|
bone morphogenetic protein 10 |
| chr7_-_99277610 | 5.65 |
ENST00000343703.5
ENST00000222982.4 ENST00000439761.1 ENST00000339843.2 |
CYP3A5
|
cytochrome P450, family 3, subfamily A, polypeptide 5 |
| chr17_+_4675175 | 5.58 |
ENST00000270560.3
|
TM4SF5
|
transmembrane 4 L six family member 5 |
| chr7_-_99381884 | 5.26 |
ENST00000336411.2
|
CYP3A4
|
cytochrome P450, family 3, subfamily A, polypeptide 4 |
| chr3_+_148583043 | 5.12 |
ENST00000296046.3
|
CPA3
|
carboxypeptidase A3 (mast cell) |
| chr1_-_201368653 | 5.08 |
ENST00000367313.3
|
LAD1
|
ladinin 1 |
| chr3_-_187009646 | 5.01 |
ENST00000296280.6
ENST00000392470.2 ENST00000169293.6 ENST00000439271.1 ENST00000392472.2 ENST00000392475.2 |
MASP1
|
mannan-binding lectin serine peptidase 1 (C4/C2 activating component of Ra-reactive factor) |
| chr4_-_72649763 | 4.95 |
ENST00000513476.1
|
GC
|
group-specific component (vitamin D binding protein) |
| chr1_-_201368707 | 4.89 |
ENST00000391967.2
|
LAD1
|
ladinin 1 |
| chr1_-_209975494 | 4.79 |
ENST00000456314.1
|
IRF6
|
interferon regulatory factor 6 |
| chr3_-_187009798 | 4.60 |
ENST00000337774.5
|
MASP1
|
mannan-binding lectin serine peptidase 1 (C4/C2 activating component of Ra-reactive factor) |
| chr3_+_156807663 | 4.58 |
ENST00000467995.1
ENST00000474477.1 ENST00000471719.1 |
LINC00881
|
long intergenic non-protein coding RNA 881 |
| chr7_-_99332719 | 4.49 |
ENST00000336374.2
|
CYP3A7
|
cytochrome P450, family 3, subfamily A, polypeptide 7 |
| chr11_+_75428857 | 4.29 |
ENST00000198801.5
|
MOGAT2
|
monoacylglycerol O-acyltransferase 2 |
| chr7_-_99381798 | 4.12 |
ENST00000415003.1
ENST00000354593.2 |
CYP3A4
|
cytochrome P450, family 3, subfamily A, polypeptide 4 |
| chr10_-_129691195 | 4.11 |
ENST00000368671.3
|
CLRN3
|
clarin 3 |
| chr17_-_64225508 | 3.85 |
ENST00000205948.6
|
APOH
|
apolipoprotein H (beta-2-glycoprotein I) |
| chr2_+_102928009 | 3.82 |
ENST00000404917.2
ENST00000447231.1 |
IL1RL1
|
interleukin 1 receptor-like 1 |
| chr19_-_39303576 | 3.79 |
ENST00000594209.1
|
LGALS4
|
lectin, galactoside-binding, soluble, 4 |
| chr8_-_86290333 | 3.75 |
ENST00000521846.1
ENST00000523022.1 ENST00000524324.1 ENST00000519991.1 ENST00000520663.1 ENST00000517590.1 ENST00000522579.1 ENST00000522814.1 ENST00000522662.1 ENST00000523858.1 ENST00000519129.1 |
CA1
|
carbonic anhydrase I |
| chr4_-_186697044 | 3.66 |
ENST00000437304.2
|
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr8_+_40010989 | 3.64 |
ENST00000315792.3
|
C8orf4
|
chromosome 8 open reading frame 4 |
| chr3_-_187009468 | 3.59 |
ENST00000425937.1
|
MASP1
|
mannan-binding lectin serine peptidase 1 (C4/C2 activating component of Ra-reactive factor) |
| chr4_-_65275100 | 3.53 |
ENST00000509536.1
|
TECRL
|
trans-2,3-enoyl-CoA reductase-like |
| chr12_-_21928515 | 3.51 |
ENST00000537950.1
|
KCNJ8
|
potassium inwardly-rectifying channel, subfamily J, member 8 |
| chr13_+_24144509 | 3.47 |
ENST00000248484.4
|
TNFRSF19
|
tumor necrosis factor receptor superfamily, member 19 |
| chr20_+_43029911 | 3.47 |
ENST00000443598.2
ENST00000316099.4 ENST00000415691.2 |
HNF4A
|
hepatocyte nuclear factor 4, alpha |
| chr14_-_65409438 | 3.45 |
ENST00000557049.1
|
GPX2
|
glutathione peroxidase 2 (gastrointestinal) |
| chr14_-_65409502 | 3.42 |
ENST00000389614.5
|
GPX2
|
glutathione peroxidase 2 (gastrointestinal) |
| chr4_+_74702214 | 3.39 |
ENST00000226317.5
ENST00000515050.1 |
CXCL6
|
chemokine (C-X-C motif) ligand 6 |
| chr1_-_120311517 | 3.36 |
ENST00000369406.3
ENST00000544913.2 |
HMGCS2
|
3-hydroxy-3-methylglutaryl-CoA synthase 2 (mitochondrial) |
| chr6_-_107235331 | 3.12 |
ENST00000433965.1
ENST00000430094.1 |
RP1-60O19.1
|
RP1-60O19.1 |
| chr2_+_3642545 | 3.10 |
ENST00000382062.2
ENST00000236693.7 ENST00000349077.4 |
COLEC11
|
collectin sub-family member 11 |
| chr16_+_215965 | 3.09 |
ENST00000356815.3
|
HBM
|
hemoglobin, mu |
| chr4_-_186733363 | 3.07 |
ENST00000393523.2
ENST00000393528.3 ENST00000449407.2 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr4_-_185820602 | 3.04 |
ENST00000515864.1
ENST00000507183.1 |
RP11-701P16.4
|
long intergenic non-protein coding RNA 1093 |
| chr3_-_119379427 | 2.92 |
ENST00000264231.3
ENST00000468801.1 ENST00000538678.1 |
POPDC2
|
popeye domain containing 2 |
| chr7_-_100239132 | 2.91 |
ENST00000223051.3
ENST00000431692.1 |
TFR2
|
transferrin receptor 2 |
| chr1_+_199996733 | 2.82 |
ENST00000236914.3
|
NR5A2
|
nuclear receptor subfamily 5, group A, member 2 |
| chr4_-_186733119 | 2.81 |
ENST00000419063.1
|
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr2_+_44066101 | 2.80 |
ENST00000272286.2
|
ABCG8
|
ATP-binding cassette, sub-family G (WHITE), member 8 |
| chrX_+_110754888 | 2.80 |
ENST00000569275.1
ENST00000563467.1 |
LINC00890
|
long intergenic non-protein coding RNA 890 |
| chrX_+_38211777 | 2.77 |
ENST00000039007.4
|
OTC
|
ornithine carbamoyltransferase |
| chr4_-_186696636 | 2.76 |
ENST00000444771.1
|
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr6_-_30128657 | 2.74 |
ENST00000449742.2
ENST00000376704.3 |
TRIM10
|
tripartite motif containing 10 |
| chr1_-_24469602 | 2.74 |
ENST00000270800.1
|
IL22RA1
|
interleukin 22 receptor, alpha 1 |
| chr4_-_186696561 | 2.66 |
ENST00000445115.1
ENST00000451701.1 ENST00000457247.1 ENST00000435480.1 ENST00000425679.1 ENST00000457934.1 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr7_+_80275621 | 2.65 |
ENST00000426978.1
ENST00000432207.1 |
CD36
|
CD36 molecule (thrombospondin receptor) |
| chr3_-_120365866 | 2.64 |
ENST00000475447.2
|
HGD
|
homogentisate 1,2-dioxygenase |
| chr7_-_44180673 | 2.57 |
ENST00000457314.1
ENST00000447951.1 ENST00000431007.1 |
MYL7
|
myosin, light chain 7, regulatory |
| chr1_+_199996702 | 2.57 |
ENST00000367362.3
|
NR5A2
|
nuclear receptor subfamily 5, group A, member 2 |
| chr2_+_234600253 | 2.53 |
ENST00000373424.1
ENST00000441351.1 |
UGT1A6
|
UDP glucuronosyltransferase 1 family, polypeptide A6 |
| chr19_+_11650709 | 2.53 |
ENST00000586059.1
|
CNN1
|
calponin 1, basic, smooth muscle |
| chr3_-_58196688 | 2.52 |
ENST00000486455.1
|
DNASE1L3
|
deoxyribonuclease I-like 3 |
| chr2_-_88427568 | 2.46 |
ENST00000393750.3
ENST00000295834.3 |
FABP1
|
fatty acid binding protein 1, liver |
| chr10_-_21186144 | 2.45 |
ENST00000377119.1
|
NEBL
|
nebulette |
| chr7_-_44180884 | 2.44 |
ENST00000458240.1
ENST00000223364.3 |
MYL7
|
myosin, light chain 7, regulatory |
| chr12_+_109569155 | 2.42 |
ENST00000539864.1
|
ACACB
|
acetyl-CoA carboxylase beta |
| chr21_-_37852359 | 2.42 |
ENST00000399137.1
ENST00000399135.1 |
CLDN14
|
claudin 14 |
| chr12_-_10151773 | 2.42 |
ENST00000298527.6
ENST00000348658.4 |
CLEC1B
|
C-type lectin domain family 1, member B |
| chr13_+_24144796 | 2.42 |
ENST00000403372.2
|
TNFRSF19
|
tumor necrosis factor receptor superfamily, member 19 |
| chr4_-_186732892 | 2.38 |
ENST00000451958.1
ENST00000439914.1 ENST00000428330.1 ENST00000429056.1 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr9_+_33240157 | 2.38 |
ENST00000379721.3
|
SPINK4
|
serine peptidase inhibitor, Kazal type 4 |
| chr12_-_103344615 | 2.37 |
ENST00000546844.1
|
PAH
|
phenylalanine hydroxylase |
| chr2_-_44065946 | 2.37 |
ENST00000260645.1
|
ABCG5
|
ATP-binding cassette, sub-family G (WHITE), member 5 |
| chr4_+_69962185 | 2.29 |
ENST00000305231.7
|
UGT2B7
|
UDP glucuronosyltransferase 2 family, polypeptide B7 |
| chr1_-_238108575 | 2.28 |
ENST00000604646.1
|
MTRNR2L11
|
MT-RNR2-like 11 (pseudogene) |
| chr2_+_102927962 | 2.27 |
ENST00000233954.1
ENST00000393393.3 ENST00000410040.1 |
IL1RL1
IL18R1
|
interleukin 1 receptor-like 1 interleukin 18 receptor 1 |
| chr1_+_81771806 | 2.26 |
ENST00000370721.1
ENST00000370727.1 ENST00000370725.1 ENST00000370723.1 ENST00000370728.1 ENST00000370730.1 |
LPHN2
|
latrophilin 2 |
| chr4_+_169552748 | 2.23 |
ENST00000504519.1
ENST00000512127.1 |
PALLD
|
palladin, cytoskeletal associated protein |
| chr4_+_74269956 | 2.20 |
ENST00000295897.4
ENST00000415165.2 ENST00000503124.1 ENST00000509063.1 ENST00000401494.3 |
ALB
|
albumin |
| chr6_-_46889694 | 2.19 |
ENST00000283296.7
ENST00000362015.4 ENST00000456426.2 |
GPR116
|
G protein-coupled receptor 116 |
| chr4_-_65275162 | 2.16 |
ENST00000381210.3
ENST00000507440.1 |
TECRL
|
trans-2,3-enoyl-CoA reductase-like |
| chr2_-_44065889 | 2.13 |
ENST00000543989.1
ENST00000405322.1 |
ABCG5
|
ATP-binding cassette, sub-family G (WHITE), member 5 |
| chr2_+_234545092 | 2.12 |
ENST00000344644.5
|
UGT1A10
|
UDP glucuronosyltransferase 1 family, polypeptide A10 |
| chr6_-_107235287 | 2.11 |
ENST00000436659.1
ENST00000428750.1 ENST00000427903.1 |
RP1-60O19.1
|
RP1-60O19.1 |
| chr2_+_234627424 | 2.10 |
ENST00000373409.3
|
UGT1A4
|
UDP glucuronosyltransferase 1 family, polypeptide A4 |
| chr11_+_34642656 | 2.08 |
ENST00000257831.3
ENST00000450654.2 |
EHF
|
ets homologous factor |
| chr1_-_155270770 | 2.07 |
ENST00000392414.3
|
PKLR
|
pyruvate kinase, liver and RBC |
| chr4_+_74275057 | 2.07 |
ENST00000511370.1
|
ALB
|
albumin |
| chr2_-_202298268 | 2.06 |
ENST00000440597.1
|
TRAK2
|
trafficking protein, kinesin binding 2 |
| chr10_+_96698406 | 2.06 |
ENST00000260682.6
|
CYP2C9
|
cytochrome P450, family 2, subfamily C, polypeptide 9 |
| chr3_+_151531859 | 2.04 |
ENST00000488869.1
|
AADAC
|
arylacetamide deacetylase |
| chr7_+_80275663 | 2.03 |
ENST00000413265.1
|
CD36
|
CD36 molecule (thrombospondin receptor) |
| chr11_+_10326612 | 2.00 |
ENST00000534464.1
ENST00000530439.1 ENST00000524948.1 ENST00000528655.1 ENST00000526492.1 ENST00000525063.1 |
ADM
|
adrenomedullin |
| chr11_-_5255861 | 1.99 |
ENST00000380299.3
|
HBD
|
hemoglobin, delta |
| chr2_+_234580499 | 1.99 |
ENST00000354728.4
|
UGT1A9
|
UDP glucuronosyltransferase 1 family, polypeptide A9 |
| chr14_+_24584508 | 1.98 |
ENST00000559354.1
ENST00000560459.1 ENST00000559593.1 ENST00000396941.4 ENST00000396936.1 |
DCAF11
|
DDB1 and CUL4 associated factor 11 |
| chr10_+_96443204 | 1.97 |
ENST00000339022.5
|
CYP2C18
|
cytochrome P450, family 2, subfamily C, polypeptide 18 |
| chr12_-_54689532 | 1.97 |
ENST00000540264.2
ENST00000312156.4 |
NFE2
|
nuclear factor, erythroid 2 |
| chrX_+_48380205 | 1.96 |
ENST00000446158.1
ENST00000414061.1 |
EBP
|
emopamil binding protein (sterol isomerase) |
| chr12_+_56477093 | 1.96 |
ENST00000549672.1
ENST00000415288.2 |
ERBB3
|
v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 3 |
| chr2_+_234621551 | 1.93 |
ENST00000608381.1
ENST00000373414.3 |
UGT1A1
UGT1A5
|
UDP glucuronosyltransferase 1 family, polypeptide A8 UDP glucuronosyltransferase 1 family, polypeptide A5 |
| chr21_-_43786634 | 1.93 |
ENST00000291527.2
|
TFF1
|
trefoil factor 1 |
| chr2_-_40739501 | 1.92 |
ENST00000403092.1
ENST00000402441.1 ENST00000448531.1 |
SLC8A1
|
solute carrier family 8 (sodium/calcium exchanger), member 1 |
| chrX_-_55057403 | 1.87 |
ENST00000396198.3
ENST00000335854.4 ENST00000455688.1 ENST00000330807.5 |
ALAS2
|
aminolevulinate, delta-, synthase 2 |
| chr6_-_86099898 | 1.83 |
ENST00000455071.1
|
RP11-30P6.6
|
RP11-30P6.6 |
| chr7_+_80275752 | 1.82 |
ENST00000419819.2
|
CD36
|
CD36 molecule (thrombospondin receptor) |
| chr11_+_58695096 | 1.81 |
ENST00000525608.1
ENST00000526351.1 |
GLYATL1
|
glycine-N-acyltransferase-like 1 |
| chr9_+_90341024 | 1.81 |
ENST00000340342.6
ENST00000342020.5 |
CTSL
|
cathepsin L |
| chr16_+_71560154 | 1.81 |
ENST00000539698.3
|
CHST4
|
carbohydrate (N-acetylglucosamine 6-O) sulfotransferase 4 |
| chr6_-_52628271 | 1.81 |
ENST00000493422.1
|
GSTA2
|
glutathione S-transferase alpha 2 |
| chr3_-_119379719 | 1.80 |
ENST00000493094.1
|
POPDC2
|
popeye domain containing 2 |
| chr7_+_100547156 | 1.76 |
ENST00000379458.4
|
MUC3A
|
Protein LOC100131514 |
| chr18_-_24722995 | 1.76 |
ENST00000581714.1
|
CHST9
|
carbohydrate (N-acetylgalactosamine 4-0) sulfotransferase 9 |
| chr8_-_72268968 | 1.76 |
ENST00000388740.3
|
EYA1
|
eyes absent homolog 1 (Drosophila) |
| chr19_+_7741968 | 1.76 |
ENST00000597445.1
|
C19orf59
|
chromosome 19 open reading frame 59 |
| chr4_-_103266219 | 1.75 |
ENST00000394833.2
|
SLC39A8
|
solute carrier family 39 (zinc transporter), member 8 |
| chr19_-_11494975 | 1.75 |
ENST00000222139.6
ENST00000592375.2 |
EPOR
|
erythropoietin receptor |
| chr12_-_9268707 | 1.74 |
ENST00000318602.7
|
A2M
|
alpha-2-macroglobulin |
| chr13_-_28896641 | 1.73 |
ENST00000543394.1
|
FLT1
|
fms-related tyrosine kinase 1 |
| chr6_-_31846744 | 1.71 |
ENST00000414427.1
ENST00000229729.6 ENST00000375562.4 |
SLC44A4
|
solute carrier family 44, member 4 |
| chr3_+_148447887 | 1.69 |
ENST00000475347.1
ENST00000474935.1 ENST00000461609.1 |
AGTR1
|
angiotensin II receptor, type 1 |
| chr13_+_28527647 | 1.68 |
ENST00000567234.1
|
LINC00543
|
long intergenic non-protein coding RNA 543 |
| chr2_+_234959376 | 1.67 |
ENST00000425558.1
|
SPP2
|
secreted phosphoprotein 2, 24kDa |
| chr6_+_50061315 | 1.66 |
ENST00000415106.1
|
RP11-397G17.1
|
RP11-397G17.1 |
| chr16_+_31539183 | 1.65 |
ENST00000302312.4
|
AHSP
|
alpha hemoglobin stabilizing protein |
| chr16_+_71560023 | 1.65 |
ENST00000572450.1
|
CHST4
|
carbohydrate (N-acetylglucosamine 6-O) sulfotransferase 4 |
| chr1_+_86934526 | 1.65 |
ENST00000394711.1
|
CLCA1
|
chloride channel accessory 1 |
| chr8_-_72268889 | 1.64 |
ENST00000388742.4
|
EYA1
|
eyes absent homolog 1 (Drosophila) |
| chr16_-_67978016 | 1.64 |
ENST00000264005.5
|
LCAT
|
lecithin-cholesterol acyltransferase |
| chr8_-_72268721 | 1.64 |
ENST00000419131.1
ENST00000388743.2 |
EYA1
|
eyes absent homolog 1 (Drosophila) |
| chr11_+_58695174 | 1.62 |
ENST00000317391.4
|
GLYATL1
|
glycine-N-acyltransferase-like 1 |
| chr12_-_120765565 | 1.61 |
ENST00000423423.3
ENST00000308366.4 |
PLA2G1B
|
phospholipase A2, group IB (pancreas) |
| chr16_+_58059470 | 1.61 |
ENST00000219271.3
|
MMP15
|
matrix metallopeptidase 15 (membrane-inserted) |
| chr8_+_76452097 | 1.58 |
ENST00000396423.2
|
HNF4G
|
hepatocyte nuclear factor 4, gamma |
| chr6_+_118869452 | 1.58 |
ENST00000357525.5
|
PLN
|
phospholamban |
| chr2_+_138722028 | 1.57 |
ENST00000280096.5
|
HNMT
|
histamine N-methyltransferase |
| chr2_+_234959323 | 1.57 |
ENST00000373368.1
ENST00000168148.3 |
SPP2
|
secreted phosphoprotein 2, 24kDa |
| chr3_-_178103144 | 1.56 |
ENST00000417383.1
ENST00000418585.1 ENST00000411727.1 ENST00000439810.1 |
RP11-33A14.1
|
RP11-33A14.1 |
| chr4_-_100140331 | 1.55 |
ENST00000407820.2
ENST00000394897.1 ENST00000508558.1 ENST00000394899.2 |
ADH6
|
alcohol dehydrogenase 6 (class V) |
| chr3_-_194188956 | 1.55 |
ENST00000256031.4
ENST00000446356.1 |
ATP13A3
|
ATPase type 13A3 |
| chr14_+_24584372 | 1.55 |
ENST00000559396.1
ENST00000558638.1 ENST00000561041.1 ENST00000559288.1 ENST00000558408.1 |
DCAF11
|
DDB1 and CUL4 associated factor 11 |
| chr20_-_49307897 | 1.54 |
ENST00000535356.1
|
FAM65C
|
family with sequence similarity 65, member C |
| chr1_-_232651312 | 1.53 |
ENST00000262861.4
|
SIPA1L2
|
signal-induced proliferation-associated 1 like 2 |
| chrX_-_65253506 | 1.53 |
ENST00000427538.1
|
VSIG4
|
V-set and immunoglobulin domain containing 4 |
| chr8_+_11534462 | 1.53 |
ENST00000528712.1
ENST00000532977.1 |
GATA4
|
GATA binding protein 4 |
| chr12_-_53297432 | 1.52 |
ENST00000546900.1
|
KRT8
|
keratin 8 |
| chr1_-_155271213 | 1.52 |
ENST00000342741.4
|
PKLR
|
pyruvate kinase, liver and RBC |
| chr17_-_79817091 | 1.52 |
ENST00000570907.1
|
P4HB
|
prolyl 4-hydroxylase, beta polypeptide |
| chr20_-_49308048 | 1.51 |
ENST00000327979.2
|
FAM65C
|
family with sequence similarity 65, member C |
| chr12_-_14996355 | 1.50 |
ENST00000228936.4
|
ART4
|
ADP-ribosyltransferase 4 (Dombrock blood group) |
| chr6_-_56112323 | 1.50 |
ENST00000535941.1
|
COL21A1
|
collagen, type XXI, alpha 1 |
| chr5_+_1201703 | 1.49 |
ENST00000304460.10
|
SLC6A19
|
solute carrier family 6 (neutral amino acid transporter), member 19 |
| chr12_+_6419877 | 1.48 |
ENST00000536531.1
|
PLEKHG6
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 6 |
| chr10_+_70847852 | 1.47 |
ENST00000242465.3
|
SRGN
|
serglycin |
| chrX_-_84634708 | 1.47 |
ENST00000373145.3
|
POF1B
|
premature ovarian failure, 1B |
| chrX_-_15402498 | 1.44 |
ENST00000297904.3
|
FIGF
|
c-fos induced growth factor (vascular endothelial growth factor D) |
| chr19_-_33360647 | 1.44 |
ENST00000590341.1
ENST00000587772.1 ENST00000023064.4 |
SLC7A9
|
solute carrier family 7 (amino acid transporter light chain, bo,+ system), member 9 |
| chr14_-_21566731 | 1.43 |
ENST00000360947.3
|
ZNF219
|
zinc finger protein 219 |
| chr11_+_69831982 | 1.43 |
ENST00000534086.1
|
RP11-626H12.1
|
RP11-626H12.1 |
| chr7_-_107443652 | 1.43 |
ENST00000340010.5
ENST00000422236.2 ENST00000453332.1 |
SLC26A3
|
solute carrier family 26 (anion exchanger), member 3 |
| chr9_+_34652164 | 1.42 |
ENST00000441545.2
ENST00000553620.1 |
IL11RA
|
interleukin 11 receptor, alpha |
| chr19_+_36132631 | 1.42 |
ENST00000379026.2
ENST00000379023.4 ENST00000402764.2 ENST00000479824.1 |
ETV2
|
ets variant 2 |
| chr22_-_43042968 | 1.41 |
ENST00000407623.3
ENST00000396303.3 ENST00000438270.1 |
CYB5R3
|
cytochrome b5 reductase 3 |
| chr8_+_121137333 | 1.38 |
ENST00000309791.4
ENST00000297848.3 ENST00000247781.3 |
COL14A1
|
collagen, type XIV, alpha 1 |
| chr4_-_48683188 | 1.38 |
ENST00000505759.1
|
FRYL
|
FRY-like |
| chr14_+_64680854 | 1.37 |
ENST00000458046.2
|
SYNE2
|
spectrin repeat containing, nuclear envelope 2 |
| chr19_+_4153598 | 1.37 |
ENST00000078445.2
ENST00000252587.3 ENST00000595923.1 ENST00000602257.1 ENST00000602147.1 |
CREB3L3
|
cAMP responsive element binding protein 3-like 3 |
| chr15_+_36887069 | 1.37 |
ENST00000566807.1
ENST00000567389.1 ENST00000562877.1 |
C15orf41
|
chromosome 15 open reading frame 41 |
| chr1_-_109849612 | 1.36 |
ENST00000357155.1
|
MYBPHL
|
myosin binding protein H-like |
| chrX_-_84634737 | 1.35 |
ENST00000262753.4
|
POF1B
|
premature ovarian failure, 1B |
| chr7_+_100318423 | 1.34 |
ENST00000252723.2
|
EPO
|
erythropoietin |
| chr9_-_95244781 | 1.33 |
ENST00000375544.3
ENST00000375543.1 ENST00000395538.3 ENST00000450139.2 |
ASPN
|
asporin |
| chr1_+_206317450 | 1.33 |
ENST00000358184.2
ENST00000361052.3 ENST00000360218.2 |
CTSE
|
cathepsin E |
| chr3_+_195943369 | 1.32 |
ENST00000296327.5
|
SLC51A
|
solute carrier family 51, alpha subunit |
| chr22_-_43042955 | 1.31 |
ENST00000402438.1
|
CYB5R3
|
cytochrome b5 reductase 3 |
| chr4_+_169418195 | 1.31 |
ENST00000261509.6
ENST00000335742.7 |
PALLD
|
palladin, cytoskeletal associated protein |
| chrX_-_100129320 | 1.29 |
ENST00000372966.3
|
NOX1
|
NADPH oxidase 1 |
| chr4_+_169418255 | 1.28 |
ENST00000505667.1
ENST00000511948.1 |
PALLD
|
palladin, cytoskeletal associated protein |
| chr12_-_71031185 | 1.28 |
ENST00000548122.1
ENST00000551525.1 ENST00000550358.1 |
PTPRB
|
protein tyrosine phosphatase, receptor type, B |
| chr16_+_85832146 | 1.27 |
ENST00000565078.1
|
COX4I1
|
cytochrome c oxidase subunit IV isoform 1 |
| chr11_+_10326918 | 1.27 |
ENST00000528544.1
|
ADM
|
adrenomedullin |
| chr7_-_87936195 | 1.27 |
ENST00000414498.1
ENST00000301959.5 ENST00000380079.4 |
STEAP4
|
STEAP family member 4 |
| chr2_+_5832799 | 1.26 |
ENST00000322002.3
|
SOX11
|
SRY (sex determining region Y)-box 11 |
| chr1_-_205912577 | 1.26 |
ENST00000367135.3
ENST00000367134.2 |
SLC26A9
|
solute carrier family 26 (anion exchanger), member 9 |
| chr2_+_201994569 | 1.26 |
ENST00000457277.1
|
CFLAR
|
CASP8 and FADD-like apoptosis regulator |
| chr12_-_96390108 | 1.23 |
ENST00000538703.1
ENST00000261208.3 |
HAL
|
histidine ammonia-lyase |
| chr1_-_120354079 | 1.23 |
ENST00000354219.1
ENST00000369401.4 ENST00000256585.5 |
REG4
|
regenerating islet-derived family, member 4 |
| chr2_+_103035102 | 1.22 |
ENST00000264260.2
|
IL18RAP
|
interleukin 18 receptor accessory protein |
| chr1_+_202431859 | 1.22 |
ENST00000391959.3
ENST00000367270.4 |
PPP1R12B
|
protein phosphatase 1, regulatory subunit 12B |
| chr11_+_34654011 | 1.22 |
ENST00000531794.1
|
EHF
|
ets homologous factor |
| chr12_-_71031220 | 1.22 |
ENST00000334414.6
|
PTPRB
|
protein tyrosine phosphatase, receptor type, B |
| chr16_+_66429358 | 1.22 |
ENST00000539168.1
|
CDH5
|
cadherin 5, type 2 (vascular endothelium) |
| chr4_+_74606223 | 1.22 |
ENST00000307407.3
ENST00000401931.1 |
IL8
|
interleukin 8 |
| chr6_-_107235378 | 1.21 |
ENST00000606430.1
|
RP1-60O19.1
|
RP1-60O19.1 |
| chr1_-_11918988 | 1.20 |
ENST00000376468.3
|
NPPB
|
natriuretic peptide B |
| chr1_+_206317591 | 1.20 |
ENST00000432969.2
|
CTSE
|
cathepsin E |
| chr19_+_16999966 | 1.20 |
ENST00000599210.1
|
F2RL3
|
coagulation factor II (thrombin) receptor-like 3 |
| chr14_+_88490894 | 1.20 |
ENST00000556033.1
ENST00000553929.1 ENST00000555996.1 ENST00000556673.1 ENST00000557339.1 ENST00000556684.1 |
RP11-300J18.3
|
long intergenic non-protein coding RNA 1146 |
| chr5_+_32788945 | 1.19 |
ENST00000326958.1
|
AC026703.1
|
AC026703.1 |
| chr3_-_58196939 | 1.18 |
ENST00000394549.2
ENST00000461914.3 |
DNASE1L3
|
deoxyribonuclease I-like 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of GATA1_GATA4
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.9 | 15.6 | GO:0009822 | alkaloid catabolic process(GO:0009822) |
| 2.7 | 8.0 | GO:0007493 | endodermal cell fate determination(GO:0007493) |
| 2.7 | 8.0 | GO:0003218 | cardiac left ventricle formation(GO:0003218) |
| 2.4 | 7.3 | GO:0010949 | negative regulation of intestinal phytosterol absorption(GO:0010949) negative regulation of intestinal cholesterol absorption(GO:0045796) intestinal phytosterol absorption(GO:0060752) negative regulation of intestinal lipid absorption(GO:1904730) |
| 1.7 | 7.0 | GO:0010903 | negative regulation of very-low-density lipoprotein particle remodeling(GO:0010903) |
| 1.4 | 10.0 | GO:0044333 | Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) |
| 1.4 | 4.3 | GO:0019836 | hemolysis by symbiont of host erythrocytes(GO:0019836) hemolysis in other organism(GO:0044179) hemolysis in other organism involved in symbiotic interaction(GO:0052331) |
| 1.1 | 18.0 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 1.1 | 4.5 | GO:0002933 | lipid hydroxylation(GO:0002933) |
| 1.0 | 3.1 | GO:0038162 | erythropoietin-mediated signaling pathway(GO:0038162) |
| 0.9 | 2.8 | GO:0070781 | arginine biosynthetic process via ornithine(GO:0042450) response to biotin(GO:0070781) |
| 0.9 | 6.4 | GO:0032971 | regulation of muscle filament sliding(GO:0032971) |
| 0.9 | 6.1 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.8 | 10.7 | GO:0052696 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.8 | 4.7 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.7 | 5.0 | GO:0060298 | positive regulation of sarcomere organization(GO:0060298) |
| 0.7 | 4.3 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.7 | 1.4 | GO:0060796 | regulation of transcription involved in primary germ layer cell fate commitment(GO:0060796) |
| 0.7 | 2.1 | GO:0098972 | dendritic transport of mitochondrion(GO:0098939) anterograde dendritic transport of mitochondrion(GO:0098972) |
| 0.7 | 2.7 | GO:0002432 | granuloma formation(GO:0002432) |
| 0.6 | 5.6 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.6 | 2.5 | GO:0045726 | positive regulation of integrin biosynthetic process(GO:0045726) |
| 0.6 | 10.3 | GO:0034197 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.6 | 3.6 | GO:1900020 | regulation of protein kinase C activity(GO:1900019) positive regulation of protein kinase C activity(GO:1900020) |
| 0.6 | 2.4 | GO:0071894 | histone H2B conserved C-terminal lysine ubiquitination(GO:0071894) |
| 0.6 | 1.7 | GO:0048925 | lateral line system development(GO:0048925) |
| 0.6 | 5.6 | GO:0006591 | ornithine metabolic process(GO:0006591) |
| 0.6 | 3.3 | GO:0032430 | positive regulation of phospholipase A2 activity(GO:0032430) |
| 0.5 | 1.1 | GO:1904479 | negative regulation of intestinal absorption(GO:1904479) |
| 0.5 | 1.6 | GO:1901877 | regulation of calcium ion binding(GO:1901876) negative regulation of calcium ion binding(GO:1901877) regulation of calcium ion import into sarcoplasmic reticulum(GO:1902080) negative regulation of calcium ion import into sarcoplasmic reticulum(GO:1902081) |
| 0.5 | 2.1 | GO:0016098 | monoterpenoid metabolic process(GO:0016098) |
| 0.5 | 2.0 | GO:0050928 | negative regulation of positive chemotaxis(GO:0050928) |
| 0.5 | 1.5 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 0.5 | 2.9 | GO:0098707 | ferrous iron import into cell(GO:0097460) ferrous iron import across plasma membrane(GO:0098707) |
| 0.4 | 0.9 | GO:0051552 | flavone metabolic process(GO:0051552) |
| 0.4 | 1.3 | GO:0061386 | closure of optic fissure(GO:0061386) |
| 0.4 | 2.4 | GO:2001295 | malonyl-CoA biosynthetic process(GO:2001295) |
| 0.4 | 0.8 | GO:0032354 | response to follicle-stimulating hormone(GO:0032354) |
| 0.4 | 1.5 | GO:0042418 | epinephrine biosynthetic process(GO:0042418) |
| 0.4 | 1.5 | GO:0003289 | atrial septum primum morphogenesis(GO:0003289) embryonic heart tube anterior/posterior pattern specification(GO:0035054) |
| 0.4 | 0.4 | GO:0060576 | intestinal epithelial cell development(GO:0060576) |
| 0.4 | 3.6 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 0.4 | 7.2 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.4 | 5.7 | GO:0042761 | very long-chain fatty acid biosynthetic process(GO:0042761) |
| 0.4 | 0.4 | GO:0090500 | endocardial cushion to mesenchymal transition(GO:0090500) |
| 0.4 | 17.3 | GO:0003301 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.3 | 1.0 | GO:1903717 | carbamoyl phosphate metabolic process(GO:0070408) carbamoyl phosphate biosynthetic process(GO:0070409) response to ammonia(GO:1903717) cellular response to ammonia(GO:1903718) |
| 0.3 | 1.0 | GO:0060913 | cardiac cell fate determination(GO:0060913) |
| 0.3 | 1.7 | GO:0060741 | prostate gland stromal morphogenesis(GO:0060741) |
| 0.3 | 2.4 | GO:0043606 | histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 0.3 | 1.7 | GO:0015842 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) |
| 0.3 | 1.6 | GO:0090107 | regulation of high-density lipoprotein particle assembly(GO:0090107) |
| 0.3 | 3.3 | GO:0097647 | progesterone biosynthetic process(GO:0006701) calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.3 | 2.0 | GO:0033489 | cholesterol biosynthetic process via desmosterol(GO:0033489) cholesterol biosynthetic process via lathosterol(GO:0033490) |
| 0.3 | 1.9 | GO:0001692 | histamine metabolic process(GO:0001692) |
| 0.3 | 3.8 | GO:1903944 | regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.3 | 0.9 | GO:0061033 | secretion by lung epithelial cell involved in lung growth(GO:0061033) |
| 0.3 | 2.6 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.3 | 0.8 | GO:1902303 | regulation of heart rate by hormone(GO:0003064) negative regulation of potassium ion export(GO:1902303) |
| 0.3 | 1.4 | GO:0033088 | negative regulation of immature T cell proliferation in thymus(GO:0033088) |
| 0.3 | 0.8 | GO:0031247 | actin rod assembly(GO:0031247) |
| 0.3 | 1.3 | GO:0070171 | negative regulation of tooth mineralization(GO:0070171) |
| 0.3 | 3.5 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.3 | 1.0 | GO:0042640 | anagen(GO:0042640) |
| 0.3 | 1.3 | GO:0061107 | seminal vesicle development(GO:0061107) |
| 0.3 | 0.8 | GO:1903465 | vacuolar phosphate transport(GO:0007037) positive regulation of mitotic cell cycle DNA replication(GO:1903465) positive regulation of parathyroid hormone secretion(GO:2000830) |
| 0.2 | 0.5 | GO:1900005 | positive regulation of serine-type endopeptidase activity(GO:1900005) positive regulation of serine-type peptidase activity(GO:1902573) |
| 0.2 | 1.0 | GO:0006480 | N-terminal protein amino acid methylation(GO:0006480) |
| 0.2 | 1.4 | GO:0045852 | pH elevation(GO:0045852) intracellular pH elevation(GO:0051454) |
| 0.2 | 0.7 | GO:2001027 | negative regulation of endothelial cell chemotaxis(GO:2001027) |
| 0.2 | 1.4 | GO:1904274 | tricellular tight junction assembly(GO:1904274) |
| 0.2 | 2.8 | GO:0070574 | cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.2 | 1.6 | GO:0034378 | chylomicron assembly(GO:0034378) |
| 0.2 | 0.9 | GO:0002426 | immunoglobulin production in mucosal tissue(GO:0002426) |
| 0.2 | 4.9 | GO:0002003 | angiotensin maturation(GO:0002003) |
| 0.2 | 7.3 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.2 | 1.1 | GO:0071504 | cellular response to heparin(GO:0071504) |
| 0.2 | 0.2 | GO:0002408 | myeloid dendritic cell chemotaxis(GO:0002408) |
| 0.2 | 1.0 | GO:0090341 | regulation of secretion of lysosomal enzymes(GO:0090182) negative regulation of secretion of lysosomal enzymes(GO:0090341) |
| 0.2 | 0.8 | GO:1902161 | positive regulation of cyclic nucleotide-gated ion channel activity(GO:1902161) |
| 0.2 | 2.4 | GO:0006559 | L-phenylalanine metabolic process(GO:0006558) L-phenylalanine catabolic process(GO:0006559) erythrose 4-phosphate/phosphoenolpyruvate family amino acid metabolic process(GO:1902221) erythrose 4-phosphate/phosphoenolpyruvate family amino acid catabolic process(GO:1902222) |
| 0.2 | 0.6 | GO:0010159 | specification of organ position(GO:0010159) |
| 0.2 | 5.2 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.2 | 2.3 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.2 | 0.6 | GO:0061580 | colon epithelial cell migration(GO:0061580) |
| 0.2 | 0.9 | GO:0030311 | poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.2 | 3.4 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.2 | 3.7 | GO:0010623 | programmed cell death involved in cell development(GO:0010623) |
| 0.2 | 0.4 | GO:0045168 | cell-cell signaling involved in cell fate commitment(GO:0045168) negative regulation of auditory receptor cell differentiation(GO:0045608) |
| 0.2 | 0.6 | GO:1902445 | regulation of mitochondrial membrane permeability involved in programmed necrotic cell death(GO:1902445) |
| 0.2 | 0.7 | GO:0097156 | fasciculation of motor neuron axon(GO:0097156) |
| 0.2 | 4.7 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.2 | 1.4 | GO:0015811 | L-cystine transport(GO:0015811) |
| 0.2 | 0.5 | GO:1904717 | excitatory chemical synaptic transmission(GO:0098976) regulation of AMPA glutamate receptor clustering(GO:1904717) positive regulation of AMPA glutamate receptor clustering(GO:1904719) |
| 0.2 | 0.5 | GO:0032877 | positive regulation of DNA endoreduplication(GO:0032877) |
| 0.2 | 0.9 | GO:0090362 | positive regulation of platelet-derived growth factor production(GO:0090362) |
| 0.2 | 0.5 | GO:1904428 | negative regulation of tubulin deacetylation(GO:1904428) |
| 0.2 | 1.9 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 0.2 | 5.0 | GO:0042359 | vitamin D metabolic process(GO:0042359) |
| 0.2 | 0.7 | GO:0051413 | response to cortisone(GO:0051413) |
| 0.2 | 2.0 | GO:0010898 | positive regulation of triglyceride catabolic process(GO:0010898) |
| 0.2 | 3.5 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.2 | 0.2 | GO:0036482 | neuron death in response to hydrogen peroxide(GO:0036476) neuron intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:0036482) positive regulation of mitochondrial electron transport, NADH to ubiquinone(GO:1902958) regulation of hydrogen peroxide-induced neuron death(GO:1903207) negative regulation of hydrogen peroxide-induced neuron death(GO:1903208) regulation of hydrogen peroxide-induced neuron intrinsic apoptotic signaling pathway(GO:1903383) negative regulation of hydrogen peroxide-induced neuron intrinsic apoptotic signaling pathway(GO:1903384) |
| 0.2 | 1.3 | GO:0097461 | ferric iron import(GO:0033216) ferric iron import into cell(GO:0097461) ferric iron import across plasma membrane(GO:0098706) |
| 0.2 | 0.3 | GO:0043627 | response to estrogen(GO:0043627) |
| 0.2 | 2.5 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.2 | 1.4 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 0.2 | 4.4 | GO:0090023 | positive regulation of neutrophil chemotaxis(GO:0090023) |
| 0.1 | 3.0 | GO:0020027 | hemoglobin metabolic process(GO:0020027) |
| 0.1 | 1.0 | GO:0003199 | endocardial cushion to mesenchymal transition involved in heart valve formation(GO:0003199) |
| 0.1 | 0.4 | GO:2000502 | negative regulation of natural killer cell chemotaxis(GO:2000502) |
| 0.1 | 1.8 | GO:0043950 | positive regulation of cAMP-mediated signaling(GO:0043950) |
| 0.1 | 0.4 | GO:0006097 | glyoxylate cycle(GO:0006097) |
| 0.1 | 0.8 | GO:1904217 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.1 | 0.7 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.1 | 1.4 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.1 | 4.4 | GO:0060644 | mammary gland epithelial cell differentiation(GO:0060644) |
| 0.1 | 0.8 | GO:0060332 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.1 | 1.2 | GO:0055014 | atrial cardiac muscle cell differentiation(GO:0055011) atrial cardiac muscle cell development(GO:0055014) |
| 0.1 | 0.5 | GO:0098582 | innate vocalization behavior(GO:0098582) |
| 0.1 | 1.7 | GO:2000680 | rubidium ion transport(GO:0035826) regulation of rubidium ion transport(GO:2000680) |
| 0.1 | 0.5 | GO:0048596 | embryonic camera-type eye morphogenesis(GO:0048596) |
| 0.1 | 1.3 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.1 | 0.9 | GO:0003228 | atrial cardiac muscle tissue development(GO:0003228) atrial cardiac muscle tissue morphogenesis(GO:0055009) |
| 0.1 | 1.0 | GO:0060372 | regulation of atrial cardiac muscle cell membrane repolarization(GO:0060372) |
| 0.1 | 0.5 | GO:0018874 | benzoate metabolic process(GO:0018874) |
| 0.1 | 1.7 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.1 | 0.6 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.1 | 0.5 | GO:0002215 | defense response to nematode(GO:0002215) |
| 0.1 | 2.5 | GO:0030949 | positive regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030949) |
| 0.1 | 0.4 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.1 | 1.5 | GO:0051599 | response to hydrostatic pressure(GO:0051599) |
| 0.1 | 0.8 | GO:0035931 | mineralocorticoid secretion(GO:0035931) aldosterone secretion(GO:0035932) regulation of mineralocorticoid secretion(GO:2000855) positive regulation of mineralocorticoid secretion(GO:2000857) regulation of aldosterone secretion(GO:2000858) positive regulation of aldosterone secretion(GO:2000860) |
| 0.1 | 0.7 | GO:1990523 | bone regeneration(GO:1990523) |
| 0.1 | 1.1 | GO:0045959 | regulation of complement activation, classical pathway(GO:0030450) negative regulation of complement activation, classical pathway(GO:0045959) |
| 0.1 | 0.3 | GO:0042270 | protection from natural killer cell mediated cytotoxicity(GO:0042270) |
| 0.1 | 1.7 | GO:1990440 | positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990440) |
| 0.1 | 1.8 | GO:0060155 | platelet dense granule organization(GO:0060155) |
| 0.1 | 2.4 | GO:0019852 | L-ascorbic acid metabolic process(GO:0019852) |
| 0.1 | 0.6 | GO:0006045 | N-acetylglucosamine biosynthetic process(GO:0006045) glucosamine-containing compound biosynthetic process(GO:1901073) |
| 0.1 | 0.3 | GO:0051100 | negative regulation of binding(GO:0051100) |
| 0.1 | 1.2 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.1 | 1.0 | GO:0031284 | positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.1 | 2.5 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.1 | 0.6 | GO:1904044 | response to aldosterone(GO:1904044) |
| 0.1 | 0.7 | GO:0046373 | arabinose metabolic process(GO:0019566) L-arabinose metabolic process(GO:0046373) |
| 0.1 | 0.3 | GO:0036451 | cap mRNA methylation(GO:0036451) |
| 0.1 | 0.5 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.1 | 0.4 | GO:0016094 | polyprenol biosynthetic process(GO:0016094) |
| 0.1 | 2.5 | GO:1904706 | negative regulation of vascular smooth muscle cell proliferation(GO:1904706) |
| 0.1 | 0.4 | GO:0044027 | hypermethylation of CpG island(GO:0044027) |
| 0.1 | 0.9 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.1 | 0.5 | GO:0044778 | meiotic DNA integrity checkpoint(GO:0044778) |
| 0.1 | 0.3 | GO:0097065 | anterior head development(GO:0097065) regulation of anterior head development(GO:2000742) positive regulation of anterior head development(GO:2000744) |
| 0.1 | 0.7 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) |
| 0.1 | 1.0 | GO:0034285 | response to sucrose(GO:0009744) response to disaccharide(GO:0034285) |
| 0.1 | 2.1 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.1 | 0.4 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.1 | 2.0 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.1 | 0.2 | GO:0097212 | lysosomal membrane organization(GO:0097212) |
| 0.1 | 2.5 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.1 | 0.4 | GO:0009396 | folic acid-containing compound biosynthetic process(GO:0009396) tetrahydrofolate biosynthetic process(GO:0046654) |
| 0.1 | 1.9 | GO:0045725 | positive regulation of glycogen biosynthetic process(GO:0045725) |
| 0.1 | 2.7 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 0.1 | 1.5 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.1 | 0.4 | GO:0072137 | condensed mesenchymal cell proliferation(GO:0072137) |
| 0.1 | 1.7 | GO:0006525 | arginine metabolic process(GO:0006525) |
| 0.1 | 0.8 | GO:0071847 | TNFSF11-mediated signaling pathway(GO:0071847) |
| 0.1 | 0.7 | GO:0060235 | lens induction in camera-type eye(GO:0060235) |
| 0.1 | 0.4 | GO:2000473 | regulation of hematopoietic stem cell migration(GO:2000471) positive regulation of hematopoietic stem cell migration(GO:2000473) |
| 0.1 | 0.4 | GO:0008050 | female courtship behavior(GO:0008050) |
| 0.1 | 1.9 | GO:0051531 | NFAT protein import into nucleus(GO:0051531) |
| 0.1 | 0.4 | GO:1905167 | positive regulation of lysosomal protein catabolic process(GO:1905167) |
| 0.1 | 1.7 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 0.1 | 0.3 | GO:0021849 | neuroblast division in subventricular zone(GO:0021849) |
| 0.1 | 0.5 | GO:0071461 | cellular response to redox state(GO:0071461) |
| 0.1 | 1.2 | GO:0050900 | leukocyte migration(GO:0050900) |
| 0.1 | 2.4 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.1 | 0.2 | GO:0072684 | mitochondrial tRNA 3'-trailer cleavage, endonucleolytic(GO:0072684) |
| 0.1 | 0.5 | GO:0045602 | negative regulation of endothelial cell differentiation(GO:0045602) |
| 0.1 | 0.4 | GO:0015853 | adenine transport(GO:0015853) |
| 0.1 | 0.4 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.1 | 8.2 | GO:0031100 | organ regeneration(GO:0031100) |
| 0.1 | 1.3 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.1 | 0.3 | GO:1902544 | regulation of DNA N-glycosylase activity(GO:1902544) |
| 0.1 | 1.5 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.1 | 0.3 | GO:0001808 | negative regulation of type IV hypersensitivity(GO:0001808) |
| 0.1 | 1.4 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.1 | 1.0 | GO:0018214 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.1 | 0.6 | GO:0030239 | myofibril assembly(GO:0030239) |
| 0.1 | 0.8 | GO:0044829 | positive regulation by host of viral genome replication(GO:0044829) |
| 0.1 | 0.8 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.1 | 0.4 | GO:0072658 | maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.1 | 0.4 | GO:0071955 | recycling endosome to Golgi transport(GO:0071955) |
| 0.1 | 1.8 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.1 | 0.2 | GO:0071284 | cellular response to lead ion(GO:0071284) |
| 0.1 | 3.8 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.1 | 0.4 | GO:0001880 | Mullerian duct regression(GO:0001880) |
| 0.1 | 0.6 | GO:0097577 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.1 | 1.8 | GO:1901687 | glutathione derivative metabolic process(GO:1901685) glutathione derivative biosynthetic process(GO:1901687) |
| 0.1 | 0.2 | GO:0051342 | regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051342) negative regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051344) |
| 0.1 | 0.4 | GO:0044878 | mitotic cytokinesis checkpoint(GO:0044878) |
| 0.1 | 0.9 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.1 | 1.0 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.1 | 0.2 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.1 | 6.9 | GO:1990748 | cellular oxidant detoxification(GO:0098869) cellular detoxification(GO:1990748) |
| 0.1 | 0.7 | GO:0097475 | motor neuron migration(GO:0097475) |
| 0.1 | 0.3 | GO:0061528 | negative regulation of gamma-aminobutyric acid secretion(GO:0014053) aspartate secretion(GO:0061528) positive regulation of prolactin secretion(GO:1902722) regulation of aspartate secretion(GO:1904448) positive regulation of aspartate secretion(GO:1904450) |
| 0.1 | 0.9 | GO:0016556 | mRNA modification(GO:0016556) |
| 0.1 | 1.6 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.1 | 0.7 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.1 | 0.9 | GO:0010766 | negative regulation of sodium ion transport(GO:0010766) |
| 0.1 | 0.6 | GO:0060056 | mammary gland involution(GO:0060056) |
| 0.1 | 0.7 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.1 | 0.2 | GO:1901350 | cell-cell signaling involved in cell-cell junction organization(GO:1901350) |
| 0.1 | 0.3 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.1 | 0.8 | GO:0045048 | protein insertion into ER membrane(GO:0045048) |
| 0.1 | 0.3 | GO:0045921 | positive regulation of exocytosis(GO:0045921) |
| 0.1 | 0.3 | GO:0035105 | sterol regulatory element binding protein import into nucleus(GO:0035105) |
| 0.1 | 1.0 | GO:0071481 | cellular response to X-ray(GO:0071481) |
| 0.1 | 0.2 | GO:0090383 | phagosome maturation(GO:0090382) phagosome acidification(GO:0090383) |
| 0.0 | 0.5 | GO:0070836 | caveola assembly(GO:0070836) |
| 0.0 | 0.4 | GO:0060318 | regulation of definitive erythrocyte differentiation(GO:0010724) definitive erythrocyte differentiation(GO:0060318) |
| 0.0 | 1.8 | GO:0010039 | response to iron ion(GO:0010039) |
| 0.0 | 1.0 | GO:0051255 | spindle midzone assembly(GO:0051255) |
| 0.0 | 0.6 | GO:0045059 | positive thymic T cell selection(GO:0045059) |
| 0.0 | 1.0 | GO:1903747 | regulation of protein targeting to mitochondrion(GO:1903214) regulation of establishment of protein localization to mitochondrion(GO:1903747) positive regulation of establishment of protein localization to mitochondrion(GO:1903749) positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.0 | 0.5 | GO:0001765 | membrane raft assembly(GO:0001765) |
| 0.0 | 0.1 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.0 | 0.1 | GO:0032707 | negative regulation of interleukin-23 production(GO:0032707) |
| 0.0 | 0.3 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 | 0.6 | GO:0018206 | peptidyl-methionine modification(GO:0018206) |
| 0.0 | 0.7 | GO:0036342 | post-anal tail morphogenesis(GO:0036342) |
| 0.0 | 0.4 | GO:0051873 | killing by host of symbiont cells(GO:0051873) |
| 0.0 | 0.2 | GO:0003366 | cell-matrix adhesion involved in ameboidal cell migration(GO:0003366) |
| 0.0 | 3.1 | GO:0035987 | endodermal cell differentiation(GO:0035987) |
| 0.0 | 0.6 | GO:0030947 | regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030947) |
| 0.0 | 1.8 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.3 | GO:0006041 | glucosamine metabolic process(GO:0006041) |
| 0.0 | 1.2 | GO:0050667 | homocysteine metabolic process(GO:0050667) |
| 0.0 | 0.2 | GO:0030473 | nucleokinesis involved in cell motility in cerebral cortex radial glia guided migration(GO:0021817) nuclear migration along microtubule(GO:0030473) |
| 0.0 | 0.2 | GO:0046498 | S-adenosylhomocysteine metabolic process(GO:0046498) |
| 0.0 | 0.6 | GO:0006924 | activation-induced cell death of T cells(GO:0006924) |
| 0.0 | 4.8 | GO:0002027 | regulation of heart rate(GO:0002027) |
| 0.0 | 5.1 | GO:0001942 | hair follicle development(GO:0001942) |
| 0.0 | 0.7 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.0 | 0.3 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.2 | GO:0038098 | sequestering of BMP from receptor via BMP binding(GO:0038098) |
| 0.0 | 0.4 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 1.0 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 1.6 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.0 | 0.6 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.0 | 0.5 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.0 | 0.1 | GO:0045905 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.0 | 1.3 | GO:0003254 | regulation of membrane depolarization(GO:0003254) |
| 0.0 | 0.7 | GO:0018065 | protein-cofactor linkage(GO:0018065) |
| 0.0 | 0.5 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.0 | 0.1 | GO:0045875 | negative regulation of sister chromatid cohesion(GO:0045875) |
| 0.0 | 1.1 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.0 | 1.8 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.0 | 0.2 | GO:0015793 | glycerol transport(GO:0015793) |
| 0.0 | 0.2 | GO:0050917 | sensory perception of umami taste(GO:0050917) |
| 0.0 | 0.3 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 | 0.2 | GO:2001300 | lipoxin metabolic process(GO:2001300) |
| 0.0 | 0.9 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.0 | 0.3 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.5 | GO:0015886 | heme transport(GO:0015886) |
| 0.0 | 2.7 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 0.0 | 0.2 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.0 | 0.6 | GO:2000144 | positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.0 | 0.7 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 0.5 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.0 | 0.1 | GO:0001777 | T cell homeostatic proliferation(GO:0001777) regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.0 | 0.6 | GO:0046688 | response to copper ion(GO:0046688) |
| 0.0 | 0.1 | GO:0090611 | ubiquitin-independent protein catabolic process via the multivesicular body sorting pathway(GO:0090611) |
| 0.0 | 0.1 | GO:0045663 | positive regulation of myoblast differentiation(GO:0045663) |
| 0.0 | 0.1 | GO:0072262 | metanephric glomerular mesangial cell proliferation involved in metanephros development(GO:0072262) |
| 0.0 | 0.3 | GO:0006739 | NADP metabolic process(GO:0006739) |
| 0.0 | 1.4 | GO:0007566 | embryo implantation(GO:0007566) |
| 0.0 | 0.2 | GO:0006983 | ER overload response(GO:0006983) |
| 0.0 | 1.0 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 | 0.1 | GO:0001812 | positive regulation of type I hypersensitivity(GO:0001812) |
| 0.0 | 0.5 | GO:0060670 | branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 0.0 | 0.7 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.0 | 0.7 | GO:0007202 | activation of phospholipase C activity(GO:0007202) |
| 0.0 | 0.6 | GO:0008535 | respiratory chain complex IV assembly(GO:0008535) |
| 0.0 | 0.4 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.0 | 1.0 | GO:0006882 | cellular zinc ion homeostasis(GO:0006882) |
| 0.0 | 0.1 | GO:0036500 | ATF6-mediated unfolded protein response(GO:0036500) |
| 0.0 | 2.5 | GO:0030521 | androgen receptor signaling pathway(GO:0030521) |
| 0.0 | 0.1 | GO:0051135 | positive regulation of NK T cell activation(GO:0051135) |
| 0.0 | 0.2 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 0.0 | 0.3 | GO:0070933 | histone H4 deacetylation(GO:0070933) |
| 0.0 | 0.1 | GO:0044860 | protein localization to plasma membrane raft(GO:0044860) protein transport into plasma membrane raft(GO:0044861) |
| 0.0 | 0.1 | GO:0034227 | tRNA thio-modification(GO:0034227) |
| 0.0 | 0.3 | GO:1903715 | regulation of aerobic respiration(GO:1903715) |
| 0.0 | 1.2 | GO:0046627 | negative regulation of insulin receptor signaling pathway(GO:0046627) |
| 0.0 | 0.1 | GO:0032627 | interleukin-23 production(GO:0032627) regulation of interleukin-23 production(GO:0032667) positive regulation of interleukin-23 production(GO:0032747) |
| 0.0 | 1.2 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.0 | 0.2 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.0 | 0.3 | GO:0072189 | ureter development(GO:0072189) |
| 0.0 | 0.1 | GO:0033299 | secretion of lysosomal enzymes(GO:0033299) |
| 0.0 | 0.5 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.0 | 1.9 | GO:0046849 | bone remodeling(GO:0046849) |
| 0.0 | 0.1 | GO:0033384 | geranyl diphosphate metabolic process(GO:0033383) geranyl diphosphate biosynthetic process(GO:0033384) farnesyl diphosphate biosynthetic process(GO:0045337) |
| 0.0 | 0.3 | GO:0070884 | regulation of calcineurin-NFAT signaling cascade(GO:0070884) |
| 0.0 | 0.7 | GO:0050732 | negative regulation of peptidyl-tyrosine phosphorylation(GO:0050732) |
| 0.0 | 0.1 | GO:0045872 | positive regulation of rhodopsin gene expression(GO:0045872) |
| 0.0 | 0.3 | GO:0031214 | biomineral tissue development(GO:0031214) |
| 0.0 | 1.3 | GO:0006695 | cholesterol biosynthetic process(GO:0006695) |
| 0.0 | 0.1 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.0 | 0.3 | GO:0001946 | lymphangiogenesis(GO:0001946) |
| 0.0 | 0.2 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.0 | 0.3 | GO:0035162 | embryonic hemopoiesis(GO:0035162) |
| 0.0 | 0.8 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.0 | 0.3 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.2 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.0 | 0.9 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.0 | 1.8 | GO:0030449 | regulation of complement activation(GO:0030449) |
| 0.0 | 0.2 | GO:0050954 | sensory perception of sound(GO:0007605) sensory perception of mechanical stimulus(GO:0050954) |
| 0.0 | 0.4 | GO:0051482 | positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G-protein coupled signaling pathway(GO:0051482) |
| 0.0 | 1.2 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.0 | 1.0 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.0 | 0.6 | GO:0032007 | negative regulation of TOR signaling(GO:0032007) |
| 0.0 | 1.6 | GO:0060333 | interferon-gamma-mediated signaling pathway(GO:0060333) |
| 0.0 | 0.9 | GO:0042493 | response to drug(GO:0042493) |
| 0.0 | 0.1 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.0 | 1.6 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.0 | 0.2 | GO:0050953 | visual perception(GO:0007601) sensory perception of light stimulus(GO:0050953) |
| 0.0 | 0.6 | GO:0007405 | neuroblast proliferation(GO:0007405) |
| 0.0 | 0.3 | GO:0009967 | positive regulation of signal transduction(GO:0009967) |
| 0.0 | 0.5 | GO:0002931 | response to ischemia(GO:0002931) |
| 0.0 | 0.1 | GO:2001206 | positive regulation of osteoclast development(GO:2001206) |
| 0.0 | 0.2 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.1 | GO:0008582 | regulation of synaptic growth at neuromuscular junction(GO:0008582) |
| 0.0 | 0.1 | GO:0039689 | negative stranded viral RNA replication(GO:0039689) multi-organism biosynthetic process(GO:0044034) |
| 0.0 | 0.9 | GO:0070830 | bicellular tight junction assembly(GO:0070830) |
| 0.0 | 0.5 | GO:0042407 | cristae formation(GO:0042407) |
| 0.0 | 0.0 | GO:1990709 | presynaptic active zone organization(GO:1990709) |
| 0.0 | 0.3 | GO:0043403 | skeletal muscle tissue regeneration(GO:0043403) |
| 0.0 | 0.9 | GO:0070301 | cellular response to hydrogen peroxide(GO:0070301) |
| 0.0 | 0.3 | GO:0050850 | positive regulation of calcium-mediated signaling(GO:0050850) |
| 0.0 | 0.3 | GO:0045776 | negative regulation of blood pressure(GO:0045776) |
| 0.0 | 0.6 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 1.8 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.0 | 0.1 | GO:0010891 | negative regulation of sequestering of triglyceride(GO:0010891) |
| 0.0 | 0.3 | GO:0030033 | microvillus assembly(GO:0030033) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 7.0 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 1.7 | 7.0 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 1.3 | 7.5 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.6 | 9.9 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.5 | 1.0 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.4 | 1.8 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.4 | 3.5 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.4 | 1.3 | GO:0036024 | protein C inhibitor-TMPRSS7 complex(GO:0036024) protein C inhibitor-TMPRSS11E complex(GO:0036025) protein C inhibitor-PLAT complex(GO:0036026) protein C inhibitor-PLAU complex(GO:0036027) protein C inhibitor-thrombin complex(GO:0036028) protein C inhibitor-KLK3 complex(GO:0036029) protein C inhibitor-plasma kallikrein complex(GO:0036030) serine protease inhibitor complex(GO:0097180) protein C inhibitor-coagulation factor V complex(GO:0097181) protein C inhibitor-coagulation factor Xa complex(GO:0097182) protein C inhibitor-coagulation factor XI complex(GO:0097183) |
| 0.4 | 7.4 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.4 | 6.9 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.4 | 4.7 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.3 | 0.9 | GO:0030895 | apolipoprotein B mRNA editing enzyme complex(GO:0030895) |
| 0.3 | 1.4 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.3 | 1.9 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.3 | 1.8 | GO:0036021 | endolysosome lumen(GO:0036021) |
| 0.3 | 2.8 | GO:0031090 | organelle membrane(GO:0031090) |
| 0.2 | 1.9 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.2 | 2.9 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.2 | 0.8 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.2 | 3.7 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.2 | 3.8 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) ripoptosome(GO:0097342) |
| 0.2 | 1.4 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.1 | 0.6 | GO:0008043 | intracellular ferritin complex(GO:0008043) ferritin complex(GO:0070288) |
| 0.1 | 3.0 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.1 | 1.5 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.1 | 5.8 | GO:0031672 | A band(GO:0031672) |
| 0.1 | 2.5 | GO:0045179 | apical cortex(GO:0045179) |
| 0.1 | 1.1 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.1 | 3.7 | GO:0043220 | Schmidt-Lanterman incisure(GO:0043220) |
| 0.1 | 0.8 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.1 | 3.1 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.1 | 1.7 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.1 | 5.5 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.1 | 1.0 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.1 | 1.7 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.1 | 1.0 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.1 | 1.9 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.1 | 0.6 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.1 | 28.5 | GO:0030018 | Z disc(GO:0030018) |
| 0.1 | 1.1 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.1 | 0.6 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) |
| 0.1 | 4.2 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.1 | 0.4 | GO:0035370 | UBC13-UEV1A complex(GO:0035370) |
| 0.1 | 1.8 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 0.1 | 0.9 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.1 | 0.7 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.1 | 0.5 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.1 | 0.2 | GO:0097679 | symbiont-containing vacuole(GO:0020003) symbiont-containing vacuole membrane(GO:0020005) other organism cytoplasm(GO:0097679) |
| 0.1 | 9.6 | GO:0005604 | basement membrane(GO:0005604) |
| 0.1 | 1.4 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.1 | 0.5 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.1 | 0.3 | GO:0032144 | 4-aminobutyrate transaminase complex(GO:0032144) |
| 0.1 | 1.6 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.1 | 15.3 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.1 | 0.4 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.1 | 13.8 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.1 | 0.5 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.1 | 0.3 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.1 | 0.5 | GO:0097413 | Lewy body(GO:0097413) |
| 0.0 | 0.5 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.0 | 0.6 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 5.2 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 1.6 | GO:0098533 | ATPase dependent transmembrane transport complex(GO:0098533) |
| 0.0 | 0.2 | GO:0034669 | integrin alpha4-beta7 complex(GO:0034669) |
| 0.0 | 1.7 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.4 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.0 | 0.9 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.9 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 1.3 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.0 | 0.1 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.0 | 2.8 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 0.2 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 1.0 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 2.9 | GO:0031228 | intrinsic component of Golgi membrane(GO:0031228) |
| 0.0 | 0.1 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 0.9 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 6.4 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 0.4 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.0 | 0.4 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 5.6 | GO:0005923 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 0.2 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.0 | 0.3 | GO:0097208 | alveolar lamellar body(GO:0097208) |
| 0.0 | 0.1 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.0 | 0.3 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.6 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 1.4 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 1.1 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 3.9 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 0.2 | GO:0070081 | clathrin-sculpted monoamine transport vesicle(GO:0070081) clathrin-sculpted monoamine transport vesicle membrane(GO:0070083) |
| 0.0 | 0.2 | GO:0005683 | U7 snRNP(GO:0005683) |
| 0.0 | 0.3 | GO:0019774 | proteasome core complex, beta-subunit complex(GO:0019774) |
| 0.0 | 0.7 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 3.2 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 2.6 | GO:0035579 | specific granule membrane(GO:0035579) |
| 0.0 | 32.8 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 1.5 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 1.1 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.1 | GO:0071682 | endocytic vesicle lumen(GO:0071682) |
| 0.0 | 25.6 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 0.3 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.0 | 0.2 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 0.5 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.0 | 0.4 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.0 | 6.3 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 9.0 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 0.4 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.1 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 0.1 | GO:0044352 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.0 | 0.2 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.2 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 0.5 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 0.3 | GO:0042581 | specific granule(GO:0042581) |
| 0.0 | 0.6 | GO:0030863 | cortical cytoskeleton(GO:0030863) |
| 0.0 | 0.2 | GO:0030137 | COPI-coated vesicle(GO:0030137) |
| 0.0 | 0.9 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.0 | 1.6 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 0.2 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 0.4 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 1.0 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.3 | GO:0005801 | cis-Golgi network(GO:0005801) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 9.4 | GO:0030343 | vitamin D3 25-hydroxylase activity(GO:0030343) vitamin D 25-hydroxylase activity(GO:0070643) |
| 2.0 | 6.1 | GO:0002113 | interleukin-33 binding(GO:0002113) |
| 1.7 | 7.0 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 1.7 | 5.0 | GO:1902271 | D3 vitamins binding(GO:1902271) |
| 1.1 | 3.4 | GO:0004421 | hydroxymethylglutaryl-CoA synthase activity(GO:0004421) |
| 0.9 | 3.5 | GO:0017098 | sulfonylurea receptor binding(GO:0017098) |
| 0.9 | 3.4 | GO:0047946 | glutamine N-acyltransferase activity(GO:0047946) |
| 0.9 | 6.8 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.8 | 2.4 | GO:0004397 | histidine ammonia-lyase activity(GO:0004397) |
| 0.7 | 2.9 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 0.7 | 2.1 | GO:0033695 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 0.7 | 4.0 | GO:0004882 | androgen receptor activity(GO:0004882) |
| 0.6 | 4.5 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.6 | 1.9 | GO:0003870 | 5-aminolevulinate synthase activity(GO:0003870) N-succinyltransferase activity(GO:0016749) |
| 0.6 | 3.6 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.6 | 6.5 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.6 | 1.7 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 0.6 | 3.9 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.5 | 4.3 | GO:0003846 | 2-acylglycerol O-acyltransferase activity(GO:0003846) |
| 0.5 | 1.5 | GO:0004603 | phenylethanolamine N-methyltransferase activity(GO:0004603) |
| 0.5 | 3.8 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.5 | 10.6 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.5 | 2.7 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.4 | 1.3 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 0.4 | 3.5 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.4 | 6.2 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.4 | 1.7 | GO:0015222 | serotonin transmembrane transporter activity(GO:0015222) |
| 0.4 | 2.5 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.4 | 2.4 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.4 | 7.2 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.4 | 2.0 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.4 | 1.6 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 0.4 | 2.4 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 0.4 | 17.0 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.4 | 2.5 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.3 | 8.6 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.3 | 3.4 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.3 | 0.9 | GO:0047225 | acetylgalactosaminyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0047225) |
| 0.3 | 1.8 | GO:0030197 | extracellular matrix constituent, lubricant activity(GO:0030197) |
| 0.3 | 5.7 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.3 | 0.8 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.3 | 1.7 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.3 | 1.9 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.3 | 1.6 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.3 | 1.0 | GO:0004087 | carbamoyl-phosphate synthase (ammonia) activity(GO:0004087) carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity(GO:0004088) |
| 0.3 | 1.0 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.3 | 1.8 | GO:0034481 | chondroitin sulfotransferase activity(GO:0034481) chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.2 | 1.7 | GO:0043120 | tumor necrosis factor binding(GO:0043120) |
| 0.2 | 13.0 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.2 | 1.0 | GO:0071885 | N-terminal protein N-methyltransferase activity(GO:0071885) |
| 0.2 | 4.7 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.2 | 1.6 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.2 | 1.3 | GO:0005497 | androgen binding(GO:0005497) |
| 0.2 | 1.7 | GO:0045118 | azole transporter activity(GO:0045118) |
| 0.2 | 7.1 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.2 | 0.8 | GO:0005260 | channel-conductance-controlling ATPase activity(GO:0005260) |
| 0.2 | 1.8 | GO:0019798 | procollagen-proline 4-dioxygenase activity(GO:0004656) procollagen-proline dioxygenase activity(GO:0019798) |
| 0.2 | 4.3 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.2 | 1.3 | GO:0008823 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.2 | 2.7 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.2 | 1.4 | GO:0015184 | L-cystine transmembrane transporter activity(GO:0015184) |
| 0.2 | 5.0 | GO:0017127 | cholesterol transporter activity(GO:0017127) |
| 0.2 | 1.6 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.2 | 3.8 | GO:0005537 | mannose binding(GO:0005537) |
| 0.2 | 0.5 | GO:0050613 | delta14-sterol reductase activity(GO:0050613) |
| 0.2 | 2.0 | GO:0032393 | MHC class I receptor activity(GO:0032393) |
| 0.2 | 0.9 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) |
| 0.2 | 0.6 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.2 | 5.9 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.2 | 1.5 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.1 | 0.6 | GO:0004796 | thromboxane-A synthase activity(GO:0004796) 12-hydroxyheptadecatrienoic acid synthase activity(GO:0036134) |
| 0.1 | 1.5 | GO:1902282 | voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1902282) |
| 0.1 | 0.4 | GO:0004450 | isocitrate dehydrogenase (NADP+) activity(GO:0004450) |
| 0.1 | 0.7 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.1 | 2.7 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.1 | 0.6 | GO:0003943 | N-acetylgalactosamine-4-sulfatase activity(GO:0003943) |
| 0.1 | 2.0 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.1 | 0.6 | GO:0008761 | UDP-N-acetylglucosamine 2-epimerase activity(GO:0008761) N-acylmannosamine kinase activity(GO:0009384) |
| 0.1 | 0.4 | GO:0016964 | alpha-2 macroglobulin receptor activity(GO:0016964) |
| 0.1 | 1.6 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.1 | 3.1 | GO:0042301 | phosphate ion binding(GO:0042301) |
| 0.1 | 2.9 | GO:0043125 | ErbB-3 class receptor binding(GO:0043125) |
| 0.1 | 1.3 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.1 | 1.8 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.1 | 4.0 | GO:0005112 | Notch binding(GO:0005112) |
| 0.1 | 1.2 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.1 | 1.2 | GO:0045029 | UDP-activated nucleotide receptor activity(GO:0045029) |
| 0.1 | 1.1 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.1 | 7.8 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.1 | 3.2 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.1 | 0.4 | GO:0005471 | ATP:ADP antiporter activity(GO:0005471) adenine transmembrane transporter activity(GO:0015207) |
| 0.1 | 1.0 | GO:0032810 | sterol response element binding(GO:0032810) |
| 0.1 | 1.4 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.1 | 0.4 | GO:0004644 | phosphoribosylamine-glycine ligase activity(GO:0004637) phosphoribosylformylglycinamidine cyclo-ligase activity(GO:0004641) phosphoribosylglycinamide formyltransferase activity(GO:0004644) |
| 0.1 | 1.0 | GO:0010853 | cyclase activator activity(GO:0010853) guanylate cyclase activator activity(GO:0030250) |
| 0.1 | 0.7 | GO:0046556 | alpha-L-arabinofuranosidase activity(GO:0046556) |
| 0.1 | 0.4 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.1 | 0.3 | GO:0004483 | mRNA (nucleoside-2'-O-)-methyltransferase activity(GO:0004483) |
| 0.1 | 0.8 | GO:0046790 | virion binding(GO:0046790) |
| 0.1 | 1.7 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.1 | 1.1 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.1 | 10.2 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.1 | 3.8 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.1 | 0.3 | GO:0043739 | G/U mismatch-specific uracil-DNA glycosylase activity(GO:0043739) |
| 0.1 | 0.9 | GO:0017018 | myosin phosphatase activity(GO:0017018) |
| 0.1 | 0.7 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors(GO:0016679) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.1 | 8.8 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.1 | 2.6 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.1 | 3.8 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 0.5 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 1.0 | GO:0086007 | voltage-gated calcium channel activity involved in cardiac muscle cell action potential(GO:0086007) |
| 0.1 | 0.5 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.1 | 2.1 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.1 | 0.8 | GO:0070915 | lysophosphatidic acid binding(GO:0035727) lysophosphatidic acid receptor activity(GO:0070915) |
| 0.1 | 0.5 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.1 | 1.0 | GO:0019958 | C-X-C chemokine binding(GO:0019958) |
| 0.1 | 0.6 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.1 | 0.7 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.1 | 2.9 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.1 | 0.4 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.1 | 1.1 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.1 | 0.3 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.1 | 2.4 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 0.2 | GO:0000773 | phosphatidyl-N-methylethanolamine N-methyltransferase activity(GO:0000773) phosphatidylethanolamine N-methyltransferase activity(GO:0004608) phosphatidyl-N-dimethylethanolamine N-methyltransferase activity(GO:0080101) |
| 0.1 | 2.2 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.1 | 0.2 | GO:1904854 | proteasome core complex binding(GO:1904854) |
| 0.1 | 2.1 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.1 | 0.8 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.1 | 0.7 | GO:0004305 | ethanolamine kinase activity(GO:0004305) |
| 0.1 | 0.2 | GO:0000035 | acyl binding(GO:0000035) |
| 0.1 | 2.4 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.1 | 5.4 | GO:0016627 | oxidoreductase activity, acting on the CH-CH group of donors(GO:0016627) |
| 0.1 | 0.4 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.1 | 4.4 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.1 | 0.4 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.1 | 0.8 | GO:0015194 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.1 | 2.2 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.1 | 0.3 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.1 | 0.3 | GO:0003867 | 4-aminobutyrate transaminase activity(GO:0003867) succinate-semialdehyde dehydrogenase binding(GO:0032145) (S)-3-amino-2-methylpropionate transaminase activity(GO:0047298) |
| 0.1 | 0.4 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.1 | 0.2 | GO:0042781 | 3'-tRNA processing endoribonuclease activity(GO:0042781) |
| 0.1 | 8.2 | GO:0005518 | collagen binding(GO:0005518) |
| 0.1 | 0.7 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.1 | 0.5 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.1 | 0.2 | GO:0005146 | leukemia inhibitory factor receptor binding(GO:0005146) |
| 0.1 | 1.6 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 1.8 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 1.5 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.0 | 0.2 | GO:0052810 | 1-phosphatidylinositol-5-kinase activity(GO:0052810) |
| 0.0 | 1.2 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.0 | 1.4 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.6 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.0 | 0.2 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 0.0 | 1.4 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.4 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.0 | 3.8 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 1.7 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.5 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.0 | 0.7 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.0 | 0.3 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.8 | GO:0004875 | complement receptor activity(GO:0004875) |
| 0.0 | 0.5 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 1.0 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 1.0 | GO:0008242 | omega peptidase activity(GO:0008242) |
| 0.0 | 3.5 | GO:0004520 | endodeoxyribonuclease activity(GO:0004520) |
| 0.0 | 0.8 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.0 | 1.1 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.0 | 0.2 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.0 | 0.4 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.0 | 0.7 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.0 | 1.0 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 1.7 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.0 | 0.5 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.0 | 0.7 | GO:0070513 | death domain binding(GO:0070513) |
| 0.0 | 0.4 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 2.1 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 1.5 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 0.3 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.6 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.0 | 0.4 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.0 | 0.6 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 0.2 | GO:0034617 | nitric-oxide synthase activity(GO:0004517) tetrahydrobiopterin binding(GO:0034617) |
| 0.0 | 0.3 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.1 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.0 | 0.6 | GO:0051213 | dioxygenase activity(GO:0051213) |
| 0.0 | 1.2 | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups(GO:0016765) |
| 0.0 | 0.6 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 1.6 | GO:0008235 | metalloexopeptidase activity(GO:0008235) |
| 0.0 | 0.2 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 0.9 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.0 | 0.8 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.3 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 1.6 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.6 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 0.8 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.1 | GO:0044729 | hemi-methylated DNA-binding(GO:0044729) |
| 0.0 | 0.1 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.0 | 0.2 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.0 | 2.3 | GO:0019213 | deacetylase activity(GO:0019213) |
| 0.0 | 0.2 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 0.0 | 3.1 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.1 | GO:0010348 | lithium:proton antiporter activity(GO:0010348) |
| 0.0 | 0.2 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.0 | 1.5 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.0 | 3.4 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 0.6 | GO:0070063 | RNA polymerase binding(GO:0070063) |
| 0.0 | 11.8 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.2 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.0 | 3.7 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 0.7 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.3 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.2 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.0 | 0.1 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.0 | 0.2 | GO:0008081 | phosphoric diester hydrolase activity(GO:0008081) |
| 0.0 | 0.9 | GO:0016303 | 1-phosphatidylinositol-3-kinase activity(GO:0016303) |
| 0.0 | 0.6 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 2.2 | GO:0008170 | N-methyltransferase activity(GO:0008170) |
| 0.0 | 0.1 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) platelet-derived growth factor beta-receptor activity(GO:0005019) |
| 0.0 | 0.2 | GO:0019864 | IgG binding(GO:0019864) |
| 0.0 | 0.4 | GO:0015464 | acetylcholine receptor activity(GO:0015464) |
| 0.0 | 0.1 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.0 | 0.3 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 7.5 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.7 | GO:0043531 | ADP binding(GO:0043531) |
| 0.0 | 0.1 | GO:0019763 | immunoglobulin receptor activity(GO:0019763) |
| 0.0 | 0.2 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.0 | 0.1 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.0 | 0.9 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 0.3 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.3 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.7 | GO:0046983 | protein dimerization activity(GO:0046983) |
| 0.0 | 0.6 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.0 | 0.1 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.2 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.0 | 0.0 | GO:0045518 | interleukin-22 receptor binding(GO:0045518) |
| 0.0 | 0.9 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 6.3 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.2 | 19.2 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.2 | 3.2 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.1 | 6.5 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.1 | 9.9 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.1 | 0.8 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.1 | 3.8 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.1 | 6.6 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.1 | 1.2 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.1 | 1.8 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.1 | 3.9 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.1 | 1.0 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.1 | 4.3 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.1 | 4.5 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 3.4 | PID EPO PATHWAY | EPO signaling pathway |
| 0.1 | 1.5 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.1 | 4.0 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.1 | 3.2 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.1 | 2.1 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 5.1 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 2.9 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 17.0 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 2.0 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 3.0 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 0.8 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 4.9 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 12.3 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 2.2 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 14.8 | NABA MATRISOME ASSOCIATED | Ensemble of genes encoding ECM-associated proteins including ECM-affilaited proteins, ECM regulators and secreted factors |
| 0.0 | 0.4 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.0 | 2.7 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 0.8 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.8 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 2.0 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.0 | 1.9 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 2.9 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.7 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 1.3 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 0.2 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 0.4 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 2.2 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.8 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 0.2 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 1.0 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.4 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.7 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 0.5 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 1.1 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 0.2 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 1.7 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.1 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.0 | 0.3 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.3 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 0.9 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.0 | 0.4 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 0.4 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 0.5 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 24.1 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.9 | 13.2 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.5 | 14.6 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.3 | 10.8 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.3 | 8.2 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.2 | 3.8 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.2 | 14.2 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.1 | 2.6 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.1 | 3.2 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.1 | 6.3 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.1 | 5.1 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.1 | 3.8 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.1 | 3.9 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 2.9 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.1 | 3.1 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.1 | 1.1 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.1 | 7.0 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.1 | 9.5 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 1.7 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.1 | 2.5 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 13.1 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.1 | 1.6 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.1 | 3.5 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.1 | 1.2 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.1 | 1.5 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.1 | 2.6 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.1 | 1.7 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.1 | 1.6 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.1 | 2.1 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.1 | 0.8 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.1 | 2.9 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.1 | 6.4 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.1 | 11.7 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 4.2 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 1.9 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 2.4 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 1.7 | REACTOME SHC MEDIATED CASCADE | Genes involved in SHC-mediated cascade |
| 0.0 | 1.5 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.0 | 0.6 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 2.4 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.7 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.4 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 2.3 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.0 | 0.8 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.5 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 1.1 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.5 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 10.8 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 1.3 | REACTOME REGULATION OF HYPOXIA INDUCIBLE FACTOR HIF BY OXYGEN | Genes involved in Regulation of Hypoxia-inducible Factor (HIF) by Oxygen |
| 0.0 | 0.8 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.6 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.0 | 1.1 | REACTOME EXTRACELLULAR MATRIX ORGANIZATION | Genes involved in Extracellular matrix organization |
| 0.0 | 0.1 | REACTOME ACTIVATION OF CHAPERONES BY ATF6 ALPHA | Genes involved in Activation of Chaperones by ATF6-alpha |
| 0.0 | 0.6 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 0.2 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.0 | 2.8 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.6 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.6 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.5 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 1.9 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.4 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.6 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 1.3 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.5 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.6 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 1.3 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 1.6 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.6 | REACTOME PYRUVATE METABOLISM AND CITRIC ACID TCA CYCLE | Genes involved in Pyruvate metabolism and Citric Acid (TCA) cycle |
| 0.0 | 0.2 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 0.8 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.2 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 0.2 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.4 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.5 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.0 | 0.5 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |