Project
Illumina Body Map 2: averaged replicates
Navigation
Downloads
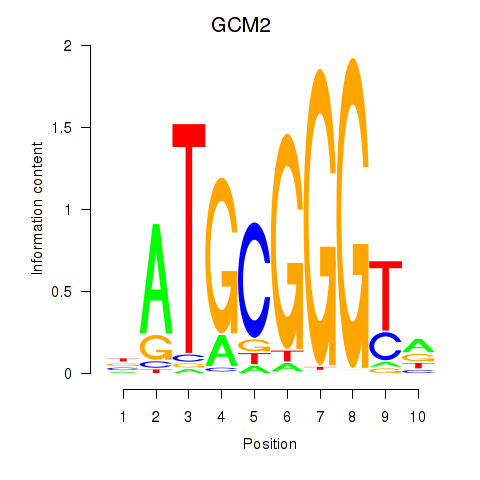
Results for GCM2
Z-value: 1.46
Transcription factors associated with GCM2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
GCM2
|
ENSG00000124827.6 | glial cells missing transcription factor 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| GCM2 | hg19_v2_chr6_-_10882174_10882274 | 0.58 | 4.7e-04 | Click! |
Activity profile of GCM2 motif
Sorted Z-values of GCM2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_44365020 | 6.68 |
ENST00000395747.2
ENST00000347193.4 ENST00000346990.4 ENST00000258682.6 ENST00000353625.4 ENST00000421607.1 ENST00000424197.1 ENST00000502837.2 ENST00000350811.3 ENST00000395749.2 |
CAMK2B
|
calcium/calmodulin-dependent protein kinase II beta |
| chr15_+_78632666 | 3.94 |
ENST00000299529.6
|
CRABP1
|
cellular retinoic acid binding protein 1 |
| chr16_+_71392616 | 3.32 |
ENST00000349553.5
ENST00000302628.4 ENST00000562305.1 |
CALB2
|
calbindin 2 |
| chr7_+_70597109 | 2.73 |
ENST00000333538.5
|
WBSCR17
|
Williams-Beuren syndrome chromosome region 17 |
| chr1_-_205313304 | 2.67 |
ENST00000539253.1
ENST00000607826.1 |
KLHDC8A
|
kelch domain containing 8A |
| chr7_-_44365216 | 2.61 |
ENST00000358707.3
ENST00000457475.1 ENST00000440254.2 |
CAMK2B
|
calcium/calmodulin-dependent protein kinase II beta |
| chr2_-_193059634 | 2.56 |
ENST00000392314.1
|
TMEFF2
|
transmembrane protein with EGF-like and two follistatin-like domains 2 |
| chr3_+_50712672 | 2.53 |
ENST00000266037.9
|
DOCK3
|
dedicator of cytokinesis 3 |
| chr19_-_38878632 | 2.46 |
ENST00000586599.1
ENST00000334928.6 ENST00000587676.1 |
GGN
|
gametogenetin |
| chr14_+_75536280 | 2.35 |
ENST00000238686.8
|
ZC2HC1C
|
zinc finger, C2HC-type containing 1C |
| chr17_-_7197881 | 2.20 |
ENST00000007699.5
|
YBX2
|
Y box binding protein 2 |
| chrX_-_71526813 | 2.18 |
ENST00000246139.5
|
CITED1
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 1 |
| chr11_-_118023490 | 2.18 |
ENST00000324727.4
|
SCN4B
|
sodium channel, voltage-gated, type IV, beta subunit |
| chr3_+_35681728 | 2.11 |
ENST00000421492.1
ENST00000458225.1 ENST00000337271.5 |
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa |
| chr15_-_93632421 | 2.07 |
ENST00000329082.7
|
RGMA
|
repulsive guidance molecule family member a |
| chr7_+_37960163 | 2.01 |
ENST00000199448.4
ENST00000559325.1 ENST00000423717.1 |
EPDR1
|
ependymin related 1 |
| chr1_+_237205476 | 2.00 |
ENST00000366574.2
|
RYR2
|
ryanodine receptor 2 (cardiac) |
| chr19_+_35783028 | 1.95 |
ENST00000600291.1
ENST00000392213.3 |
MAG
|
myelin associated glycoprotein |
| chrX_-_71526999 | 1.94 |
ENST00000453707.2
ENST00000373619.3 |
CITED1
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 1 |
| chr19_+_35783047 | 1.94 |
ENST00000595791.1
ENST00000597035.1 ENST00000537831.2 |
MAG
|
myelin associated glycoprotein |
| chr19_+_35783037 | 1.91 |
ENST00000361922.4
|
MAG
|
myelin associated glycoprotein |
| chr15_+_74466744 | 1.89 |
ENST00000560862.1
ENST00000395118.1 |
ISLR
|
immunoglobulin superfamily containing leucine-rich repeat |
| chr9_+_103340354 | 1.86 |
ENST00000307584.5
|
MURC
|
muscle-related coiled-coil protein |
| chr1_+_20878932 | 1.84 |
ENST00000332947.4
|
FAM43B
|
family with sequence similarity 43, member B |
| chr5_+_137774706 | 1.84 |
ENST00000378339.2
ENST00000254901.5 ENST00000506158.1 |
REEP2
|
receptor accessory protein 2 |
| chr3_-_142682178 | 1.81 |
ENST00000340634.3
|
PAQR9
|
progestin and adipoQ receptor family member IX |
| chr14_+_75536335 | 1.78 |
ENST00000554763.1
ENST00000439583.2 ENST00000526130.1 ENST00000525046.1 |
ZC2HC1C
|
zinc finger, C2HC-type containing 1C |
| chr19_+_51293672 | 1.77 |
ENST00000270593.1
ENST00000270594.3 |
ACPT
|
acid phosphatase, testicular |
| chr2_+_220408724 | 1.77 |
ENST00000421791.1
ENST00000373883.3 ENST00000451952.1 |
TMEM198
|
transmembrane protein 198 |
| chr2_+_220299547 | 1.73 |
ENST00000312358.7
|
SPEG
|
SPEG complex locus |
| chr3_+_42727011 | 1.70 |
ENST00000287777.4
|
KLHL40
|
kelch-like family member 40 |
| chr9_-_130635741 | 1.65 |
ENST00000223836.10
|
AK1
|
adenylate kinase 1 |
| chr11_-_83393429 | 1.64 |
ENST00000426717.2
|
DLG2
|
discs, large homolog 2 (Drosophila) |
| chr19_-_19626351 | 1.63 |
ENST00000585580.3
|
TSSK6
|
testis-specific serine kinase 6 |
| chr14_-_77737543 | 1.62 |
ENST00000298352.4
|
NGB
|
neuroglobin |
| chr22_-_29457832 | 1.61 |
ENST00000216071.4
|
C22orf31
|
chromosome 22 open reading frame 31 |
| chr2_+_16080659 | 1.59 |
ENST00000281043.3
|
MYCN
|
v-myc avian myelocytomatosis viral oncogene neuroblastoma derived homolog |
| chr6_-_110500826 | 1.58 |
ENST00000265601.3
ENST00000447287.1 ENST00000444391.1 |
WASF1
|
WAS protein family, member 1 |
| chr9_-_127269488 | 1.55 |
ENST00000455734.1
|
NR5A1
|
nuclear receptor subfamily 5, group A, member 1 |
| chr9_-_127269661 | 1.52 |
ENST00000373588.4
|
NR5A1
|
nuclear receptor subfamily 5, group A, member 1 |
| chr3_+_35681081 | 1.51 |
ENST00000428373.1
|
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa |
| chr3_+_156008809 | 1.45 |
ENST00000302490.8
|
KCNAB1
|
potassium voltage-gated channel, shaker-related subfamily, beta member 1 |
| chr12_+_5019061 | 1.44 |
ENST00000382545.3
|
KCNA1
|
potassium voltage-gated channel, shaker-related subfamily, member 1 (episodic ataxia with myokymia) |
| chr20_-_44420507 | 1.44 |
ENST00000243938.4
|
WFDC3
|
WAP four-disulfide core domain 3 |
| chr15_+_43809797 | 1.41 |
ENST00000399453.1
ENST00000300231.5 |
MAP1A
|
microtubule-associated protein 1A |
| chr16_+_1031762 | 1.38 |
ENST00000293894.3
|
SOX8
|
SRY (sex determining region Y)-box 8 |
| chr1_-_182642017 | 1.38 |
ENST00000367557.4
ENST00000258302.4 |
RGS8
|
regulator of G-protein signaling 8 |
| chr2_-_220408260 | 1.35 |
ENST00000373891.2
|
CHPF
|
chondroitin polymerizing factor |
| chr17_-_41132088 | 1.34 |
ENST00000591916.1
ENST00000451885.2 ENST00000454303.1 |
PTGES3L
PTGES3L-AARSD1
|
prostaglandin E synthase 3 (cytosolic)-like PTGES3L-AARSD1 readthrough |
| chr14_-_59932044 | 1.33 |
ENST00000395116.1
|
GPR135
|
G protein-coupled receptor 135 |
| chr4_-_153700864 | 1.31 |
ENST00000304337.2
|
TIGD4
|
tigger transposable element derived 4 |
| chr17_+_48911942 | 1.31 |
ENST00000426127.1
|
WFIKKN2
|
WAP, follistatin/kazal, immunoglobulin, kunitz and netrin domain containing 2 |
| chr9_+_72658490 | 1.29 |
ENST00000377182.4
|
MAMDC2
|
MAM domain containing 2 |
| chr9_-_127263265 | 1.28 |
ENST00000373587.3
|
NR5A1
|
nuclear receptor subfamily 5, group A, member 1 |
| chr2_-_19558373 | 1.26 |
ENST00000272223.2
|
OSR1
|
odd-skipped related transciption factor 1 |
| chr6_-_110500905 | 1.26 |
ENST00000392587.2
|
WASF1
|
WAS protein family, member 1 |
| chr19_+_41036371 | 1.24 |
ENST00000392023.1
|
SPTBN4
|
spectrin, beta, non-erythrocytic 4 |
| chr6_-_52859968 | 1.22 |
ENST00000370959.1
|
GSTA4
|
glutathione S-transferase alpha 4 |
| chr14_+_42077552 | 1.21 |
ENST00000554120.1
|
LRFN5
|
leucine rich repeat and fibronectin type III domain containing 5 |
| chrX_+_130192318 | 1.21 |
ENST00000370922.1
|
ARHGAP36
|
Rho GTPase activating protein 36 |
| chr17_+_79989500 | 1.20 |
ENST00000306897.4
|
RAC3
|
ras-related C3 botulinum toxin substrate 3 (rho family, small GTP binding protein Rac3) |
| chr7_-_98467489 | 1.20 |
ENST00000416379.2
|
TMEM130
|
transmembrane protein 130 |
| chr2_-_193059250 | 1.20 |
ENST00000409056.3
|
TMEFF2
|
transmembrane protein with EGF-like and two follistatin-like domains 2 |
| chr14_-_105635090 | 1.16 |
ENST00000331782.3
ENST00000347004.2 |
JAG2
|
jagged 2 |
| chrX_+_46937745 | 1.13 |
ENST00000397180.1
ENST00000457380.1 ENST00000352078.4 |
RGN
|
regucalcin |
| chr1_+_43824669 | 1.13 |
ENST00000372462.1
|
CDC20
|
cell division cycle 20 |
| chrX_+_12156582 | 1.12 |
ENST00000380682.1
|
FRMPD4
|
FERM and PDZ domain containing 4 |
| chr8_-_98290087 | 1.11 |
ENST00000322128.3
|
TSPYL5
|
TSPY-like 5 |
| chr13_-_47471155 | 1.10 |
ENST00000543956.1
ENST00000542664.1 |
HTR2A
|
5-hydroxytryptamine (serotonin) receptor 2A, G protein-coupled |
| chr1_+_43824577 | 1.10 |
ENST00000310955.6
|
CDC20
|
cell division cycle 20 |
| chr17_-_42907564 | 1.08 |
ENST00000592524.1
|
GJC1
|
gap junction protein, gamma 1, 45kDa |
| chr2_-_220435963 | 1.08 |
ENST00000373876.1
ENST00000404537.1 ENST00000603926.1 ENST00000373873.4 ENST00000289656.3 |
OBSL1
|
obscurin-like 1 |
| chr2_+_220309379 | 1.08 |
ENST00000451076.1
|
SPEG
|
SPEG complex locus |
| chr2_-_220252603 | 1.06 |
ENST00000322176.7
ENST00000273075.4 |
DNPEP
|
aspartyl aminopeptidase |
| chr15_+_74421961 | 1.05 |
ENST00000565159.1
ENST00000567206.1 |
ISLR2
|
immunoglobulin superfamily containing leucine-rich repeat 2 |
| chr1_-_67600639 | 1.05 |
ENST00000544837.1
ENST00000603691.1 |
C1orf141
|
chromosome 1 open reading frame 141 |
| chr17_-_8021710 | 1.03 |
ENST00000380149.1
ENST00000448843.2 |
ALOXE3
|
arachidonate lipoxygenase 3 |
| chr20_+_42143053 | 1.02 |
ENST00000373135.3
ENST00000444063.1 |
L3MBTL1
|
l(3)mbt-like 1 (Drosophila) |
| chr2_-_220436248 | 1.01 |
ENST00000265318.4
|
OBSL1
|
obscurin-like 1 |
| chr16_-_25122735 | 0.99 |
ENST00000563176.1
|
RP11-449H11.1
|
RP11-449H11.1 |
| chr4_-_187112626 | 0.98 |
ENST00000596414.1
|
AC110771.1
|
Uncharacterized protein |
| chr17_-_3796334 | 0.98 |
ENST00000381771.2
ENST00000348335.2 ENST00000158166.5 |
CAMKK1
|
calcium/calmodulin-dependent protein kinase kinase 1, alpha |
| chr16_+_22825475 | 0.98 |
ENST00000261374.3
|
HS3ST2
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 2 |
| chr11_-_64410787 | 0.98 |
ENST00000301894.2
|
NRXN2
|
neurexin 2 |
| chr15_-_41624685 | 0.98 |
ENST00000560640.1
ENST00000220514.3 |
OIP5
|
Opa interacting protein 5 |
| chrX_+_150869023 | 0.97 |
ENST00000448324.1
|
PRRG3
|
proline rich Gla (G-carboxyglutamic acid) 3 (transmembrane) |
| chr11_-_118023594 | 0.96 |
ENST00000529878.1
|
SCN4B
|
sodium channel, voltage-gated, type IV, beta subunit |
| chr17_+_53342311 | 0.96 |
ENST00000226067.5
|
HLF
|
hepatic leukemia factor |
| chr3_-_79068138 | 0.96 |
ENST00000495273.1
|
ROBO1
|
roundabout, axon guidance receptor, homolog 1 (Drosophila) |
| chr5_-_160975130 | 0.96 |
ENST00000274547.2
ENST00000520240.1 ENST00000517901.1 ENST00000353437.6 |
GABRB2
|
gamma-aminobutyric acid (GABA) A receptor, beta 2 |
| chr15_+_78556809 | 0.95 |
ENST00000343789.3
ENST00000394852.3 |
DNAJA4
|
DnaJ (Hsp40) homolog, subfamily A, member 4 |
| chr3_-_79068594 | 0.94 |
ENST00000436010.2
|
ROBO1
|
roundabout, axon guidance receptor, homolog 1 (Drosophila) |
| chr11_-_83393457 | 0.94 |
ENST00000404783.3
|
DLG2
|
discs, large homolog 2 (Drosophila) |
| chr3_+_137490748 | 0.93 |
ENST00000478772.1
|
RP11-2A4.3
|
RP11-2A4.3 |
| chr9_-_13279589 | 0.93 |
ENST00000319217.7
|
MPDZ
|
multiple PDZ domain protein |
| chrX_-_27999566 | 0.92 |
ENST00000441525.1
|
DCAF8L1
|
DDB1 and CUL4 associated factor 8-like 1 |
| chr1_+_44444865 | 0.92 |
ENST00000372324.1
|
B4GALT2
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 2 |
| chr2_-_102091144 | 0.92 |
ENST00000428343.1
|
RFX8
|
RFX family member 8, lacking RFX DNA binding domain |
| chr20_+_42143136 | 0.91 |
ENST00000373134.1
|
L3MBTL1
|
l(3)mbt-like 1 (Drosophila) |
| chr1_-_228613026 | 0.91 |
ENST00000366696.1
|
HIST3H3
|
histone cluster 3, H3 |
| chr17_-_71258019 | 0.91 |
ENST00000344935.4
|
CPSF4L
|
cleavage and polyadenylation specific factor 4-like |
| chr7_+_116139821 | 0.91 |
ENST00000393480.2
|
CAV2
|
caveolin 2 |
| chrX_+_122318224 | 0.90 |
ENST00000542149.1
|
GRIA3
|
glutamate receptor, ionotropic, AMPA 3 |
| chr13_-_44735393 | 0.89 |
ENST00000400419.1
|
SMIM2
|
small integral membrane protein 2 |
| chr14_-_70038032 | 0.88 |
ENST00000543986.1
|
CCDC177
|
coiled-coil domain containing 177 |
| chr11_-_10715033 | 0.88 |
ENST00000529547.1
ENST00000558540.1 |
MRVI1
|
murine retrovirus integration site 1 homolog |
| chr19_-_12886327 | 0.88 |
ENST00000397668.3
ENST00000587178.1 ENST00000264827.5 |
HOOK2
|
hook microtubule-tethering protein 2 |
| chr15_-_93199069 | 0.88 |
ENST00000327355.5
|
FAM174B
|
family with sequence similarity 174, member B |
| chr5_-_137878887 | 0.88 |
ENST00000507939.1
ENST00000572514.1 ENST00000499810.2 ENST00000360541.5 |
ETF1
|
eukaryotic translation termination factor 1 |
| chr20_-_982904 | 0.87 |
ENST00000217260.4
ENST00000400634.2 |
RSPO4
|
R-spondin 4 |
| chr8_+_117950422 | 0.86 |
ENST00000378279.3
|
AARD
|
alanine and arginine rich domain containing protein |
| chrX_+_122318318 | 0.85 |
ENST00000371251.1
ENST00000371256.5 |
GRIA3
|
glutamate receptor, ionotropic, AMPA 3 |
| chr8_+_134125727 | 0.83 |
ENST00000521107.1
|
TG
|
thyroglobulin |
| chr15_-_30261066 | 0.83 |
ENST00000558447.1
|
TJP1
|
tight junction protein 1 |
| chr21_+_27011584 | 0.82 |
ENST00000400532.1
ENST00000480456.1 ENST00000312957.5 |
JAM2
|
junctional adhesion molecule 2 |
| chr11_-_10715117 | 0.81 |
ENST00000527509.2
|
MRVI1
|
murine retrovirus integration site 1 homolog |
| chr3_-_50605150 | 0.80 |
ENST00000357203.3
|
C3orf18
|
chromosome 3 open reading frame 18 |
| chr14_+_77647966 | 0.80 |
ENST00000554766.1
|
TMEM63C
|
transmembrane protein 63C |
| chr14_+_77648167 | 0.80 |
ENST00000554346.1
ENST00000298351.4 |
TMEM63C
|
transmembrane protein 63C |
| chr19_-_55954230 | 0.80 |
ENST00000376325.4
|
SHISA7
|
shisa family member 7 |
| chr17_-_7123021 | 0.79 |
ENST00000399510.2
|
DLG4
|
discs, large homolog 4 (Drosophila) |
| chr20_-_60982330 | 0.79 |
ENST00000279101.5
|
CABLES2
|
Cdk5 and Abl enzyme substrate 2 |
| chr2_-_220252530 | 0.78 |
ENST00000521459.1
|
DNPEP
|
aspartyl aminopeptidase |
| chr14_+_105046021 | 0.78 |
ENST00000557649.1
|
C14orf180
|
chromosome 14 open reading frame 180 |
| chr11_-_5345582 | 0.78 |
ENST00000328813.2
|
OR51B2
|
olfactory receptor, family 51, subfamily B, member 2 |
| chr12_+_15125954 | 0.77 |
ENST00000266395.2
|
PDE6H
|
phosphodiesterase 6H, cGMP-specific, cone, gamma |
| chr16_-_3350614 | 0.76 |
ENST00000268674.2
|
TIGD7
|
tigger transposable element derived 7 |
| chr9_-_124976185 | 0.76 |
ENST00000464484.2
|
LHX6
|
LIM homeobox 6 |
| chr15_-_88799384 | 0.75 |
ENST00000540489.2
ENST00000557856.1 ENST00000558676.1 |
NTRK3
|
neurotrophic tyrosine kinase, receptor, type 3 |
| chr7_+_116139744 | 0.74 |
ENST00000343213.2
|
CAV2
|
caveolin 2 |
| chr4_+_95972822 | 0.74 |
ENST00000509540.1
ENST00000440890.2 |
BMPR1B
|
bone morphogenetic protein receptor, type IB |
| chr5_-_87980587 | 0.71 |
ENST00000509783.1
ENST00000509405.1 ENST00000506978.1 ENST00000509265.1 ENST00000513805.1 |
LINC00461
|
long intergenic non-protein coding RNA 461 |
| chr2_+_121493717 | 0.70 |
ENST00000418323.1
|
GLI2
|
GLI family zinc finger 2 |
| chr5_+_157170703 | 0.70 |
ENST00000286307.5
|
LSM11
|
LSM11, U7 small nuclear RNA associated |
| chr12_-_48398104 | 0.69 |
ENST00000337299.6
ENST00000380518.3 |
COL2A1
|
collagen, type II, alpha 1 |
| chr6_-_32077100 | 0.69 |
ENST00000375244.3
ENST00000375247.2 |
TNXB
|
tenascin XB |
| chr3_-_50605077 | 0.69 |
ENST00000426034.1
ENST00000441239.1 |
C3orf18
|
chromosome 3 open reading frame 18 |
| chr2_-_239140276 | 0.69 |
ENST00000334973.4
|
AC016757.3
|
Protein LOC151174 |
| chr6_-_142409936 | 0.69 |
ENST00000258042.1
|
NMBR
|
neuromedin B receptor |
| chr14_+_27342334 | 0.69 |
ENST00000548170.1
ENST00000552926.1 |
RP11-384J4.1
|
RP11-384J4.1 |
| chr11_-_6341724 | 0.67 |
ENST00000530979.1
|
PRKCDBP
|
protein kinase C, delta binding protein |
| chr11_-_62689046 | 0.67 |
ENST00000306960.3
ENST00000543973.1 |
CHRM1
|
cholinergic receptor, muscarinic 1 |
| chr19_-_51530916 | 0.67 |
ENST00000594768.1
|
KLK11
|
kallikrein-related peptidase 11 |
| chr16_-_10276251 | 0.67 |
ENST00000330684.3
|
GRIN2A
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2A |
| chr1_+_15272271 | 0.66 |
ENST00000400797.3
|
KAZN
|
kazrin, periplakin interacting protein |
| chr1_+_77748756 | 0.65 |
ENST00000478407.1
|
AK5
|
adenylate kinase 5 |
| chr16_+_24857309 | 0.65 |
ENST00000565769.1
ENST00000449109.2 ENST00000424767.2 ENST00000545376.1 ENST00000569520.1 |
SLC5A11
|
solute carrier family 5 (sodium/inositol cotransporter), member 11 |
| chr9_-_124976154 | 0.65 |
ENST00000482062.1
|
LHX6
|
LIM homeobox 6 |
| chr2_+_176995011 | 0.64 |
ENST00000548663.1
ENST00000450510.2 |
HOXD8
|
homeobox D8 |
| chr17_-_42908155 | 0.64 |
ENST00000426548.1
ENST00000590758.1 ENST00000591424.1 |
GJC1
|
gap junction protein, gamma 1, 45kDa |
| chr19_-_55953704 | 0.63 |
ENST00000416792.1
|
SHISA7
|
shisa family member 7 |
| chr2_+_179059365 | 0.63 |
ENST00000190611.4
|
OSBPL6
|
oxysterol binding protein-like 6 |
| chr14_+_105046094 | 0.62 |
ENST00000331952.2
|
C14orf180
|
chromosome 14 open reading frame 180 |
| chr2_-_220408430 | 0.61 |
ENST00000243776.6
|
CHPF
|
chondroitin polymerizing factor |
| chr15_-_48470544 | 0.59 |
ENST00000267836.6
|
MYEF2
|
myelin expression factor 2 |
| chr11_+_123886282 | 0.59 |
ENST00000320891.4
|
OR10G4
|
olfactory receptor, family 10, subfamily G, member 4 |
| chr22_-_25335314 | 0.57 |
ENST00000382744.1
|
TMEM211
|
transmembrane protein 211 |
| chr12_-_16758873 | 0.57 |
ENST00000535535.1
|
LMO3
|
LIM domain only 3 (rhombotin-like 2) |
| chr3_-_99245957 | 0.57 |
ENST00000484675.1
|
RP11-779P15.2
|
RP11-779P15.2 |
| chr15_-_48470558 | 0.57 |
ENST00000324324.7
|
MYEF2
|
myelin expression factor 2 |
| chr16_+_3194211 | 0.57 |
ENST00000428155.1
|
CASP16
|
caspase 16, apoptosis-related cysteine peptidase (putative) |
| chrX_+_99839799 | 0.56 |
ENST00000373031.4
|
TNMD
|
tenomodulin |
| chr19_+_589893 | 0.56 |
ENST00000251287.2
|
HCN2
|
hyperpolarization activated cyclic nucleotide-gated potassium channel 2 |
| chr11_-_107729287 | 0.56 |
ENST00000375682.4
|
SLC35F2
|
solute carrier family 35, member F2 |
| chr8_-_145559943 | 0.56 |
ENST00000332135.4
|
SCRT1
|
scratch family zinc finger 1 |
| chr2_+_168149569 | 0.55 |
ENST00000442316.1
|
AC074363.1
|
AC074363.1 |
| chr20_+_18269121 | 0.55 |
ENST00000377671.3
ENST00000360010.5 ENST00000396026.3 ENST00000402618.2 ENST00000401790.1 ENST00000434018.1 ENST00000538547.1 ENST00000535822.1 |
ZNF133
|
zinc finger protein 133 |
| chr17_-_41738931 | 0.55 |
ENST00000329168.3
ENST00000549132.1 |
MEOX1
|
mesenchyme homeobox 1 |
| chr11_-_9113137 | 0.54 |
ENST00000520467.1
ENST00000309263.3 ENST00000457346.2 |
SCUBE2
|
signal peptide, CUB domain, EGF-like 2 |
| chr12_-_16758835 | 0.54 |
ENST00000541295.1
|
LMO3
|
LIM domain only 3 (rhombotin-like 2) |
| chr11_-_10715163 | 0.53 |
ENST00000541483.1
|
MRVI1
|
murine retrovirus integration site 1 homolog |
| chr9_-_13279563 | 0.53 |
ENST00000541718.1
|
MPDZ
|
multiple PDZ domain protein |
| chr15_+_68115895 | 0.53 |
ENST00000554240.1
|
SKOR1
|
SKI family transcriptional corepressor 1 |
| chr14_+_21566980 | 0.52 |
ENST00000418511.2
ENST00000554329.2 |
TMEM253
|
transmembrane protein 253 |
| chr7_+_150811705 | 0.52 |
ENST00000335367.3
|
AGAP3
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 3 |
| chr19_+_639895 | 0.51 |
ENST00000586042.2
ENST00000215530.5 |
FGF22
|
fibroblast growth factor 22 |
| chr2_-_73053126 | 0.50 |
ENST00000272427.6
ENST00000410104.1 |
EXOC6B
|
exocyst complex component 6B |
| chr2_+_179059188 | 0.49 |
ENST00000392505.2
ENST00000359685.3 ENST00000357080.4 ENST00000409045.3 |
OSBPL6
|
oxysterol binding protein-like 6 |
| chr17_-_56565736 | 0.49 |
ENST00000323777.3
|
HSF5
|
heat shock transcription factor family member 5 |
| chr1_+_178062855 | 0.49 |
ENST00000448150.3
|
RASAL2
|
RAS protein activator like 2 |
| chr5_+_140743859 | 0.49 |
ENST00000518069.1
|
PCDHGA5
|
protocadherin gamma subfamily A, 5 |
| chr17_-_10017864 | 0.48 |
ENST00000323816.4
|
GAS7
|
growth arrest-specific 7 |
| chr15_+_73344911 | 0.48 |
ENST00000560262.1
ENST00000558964.1 |
NEO1
|
neogenin 1 |
| chr3_-_11762202 | 0.48 |
ENST00000445411.1
ENST00000404339.1 ENST00000273038.3 |
VGLL4
|
vestigial like 4 (Drosophila) |
| chr6_+_46761118 | 0.47 |
ENST00000230588.4
|
MEP1A
|
meprin A, alpha (PABA peptide hydrolase) |
| chr3_+_126707437 | 0.47 |
ENST00000393409.2
ENST00000251772.4 |
PLXNA1
|
plexin A1 |
| chr22_-_25335502 | 0.46 |
ENST00000423535.1
|
TMEM211
|
transmembrane protein 211 |
| chr11_-_10715287 | 0.46 |
ENST00000423302.2
|
MRVI1
|
murine retrovirus integration site 1 homolog |
| chr6_-_31864977 | 0.46 |
ENST00000395728.3
ENST00000375528.4 |
EHMT2
|
euchromatic histone-lysine N-methyltransferase 2 |
| chr19_-_14316980 | 0.46 |
ENST00000361434.3
ENST00000340736.6 |
LPHN1
|
latrophilin 1 |
| chr16_-_755819 | 0.46 |
ENST00000397621.1
|
FBXL16
|
F-box and leucine-rich repeat protein 16 |
| chr7_-_6048702 | 0.44 |
ENST00000265849.7
|
PMS2
|
PMS2 postmeiotic segregation increased 2 (S. cerevisiae) |
| chr19_-_51531272 | 0.44 |
ENST00000319720.7
|
KLK11
|
kallikrein-related peptidase 11 |
| chr19_-_51531210 | 0.44 |
ENST00000391804.3
|
KLK11
|
kallikrein-related peptidase 11 |
| chr19_+_17581253 | 0.43 |
ENST00000252595.7
ENST00000598424.1 |
SLC27A1
|
solute carrier family 27 (fatty acid transporter), member 1 |
| chr5_+_113697983 | 0.43 |
ENST00000264773.3
|
KCNN2
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 2 |
| chr11_-_61197406 | 0.43 |
ENST00000541963.1
ENST00000477890.2 |
CPSF7
|
cleavage and polyadenylation specific factor 7, 59kDa |
| chrX_-_63005405 | 0.43 |
ENST00000374878.1
ENST00000437457.2 |
ARHGEF9
|
Cdc42 guanine nucleotide exchange factor (GEF) 9 |
| chr6_+_110012462 | 0.43 |
ENST00000441478.2
ENST00000230124.3 |
FIG4
|
FIG4 homolog, SAC1 lipid phosphatase domain containing (S. cerevisiae) |
| chrX_+_128674213 | 0.43 |
ENST00000371113.4
ENST00000357121.5 |
OCRL
|
oculocerebrorenal syndrome of Lowe |
Network of associatons between targets according to the STRING database.
First level regulatory network of GCM2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 3.8 | GO:0045720 | negative regulation of integrin biosynthetic process(GO:0045720) |
| 0.9 | 4.4 | GO:0007538 | primary sex determination(GO:0007538) |
| 0.8 | 3.9 | GO:0016103 | diterpenoid catabolic process(GO:0016103) retinoic acid catabolic process(GO:0034653) |
| 0.7 | 1.4 | GO:0072034 | renal vesicle induction(GO:0072034) |
| 0.7 | 2.0 | GO:0003220 | left ventricular cardiac muscle tissue morphogenesis(GO:0003220) |
| 0.6 | 2.2 | GO:0009386 | translational attenuation(GO:0009386) |
| 0.5 | 11.5 | GO:0090129 | positive regulation of synapse maturation(GO:0090129) |
| 0.5 | 4.1 | GO:0071105 | response to interleukin-9(GO:0071104) response to interleukin-11(GO:0071105) |
| 0.5 | 1.5 | GO:1903630 | regulation of aminoacyl-tRNA ligase activity(GO:1903630) |
| 0.4 | 1.9 | GO:0021823 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) postnatal olfactory bulb interneuron migration(GO:0021827) chemorepulsion involved in postnatal olfactory bulb interneuron migration(GO:0021836) |
| 0.4 | 5.8 | GO:0048711 | positive regulation of astrocyte differentiation(GO:0048711) |
| 0.4 | 1.4 | GO:0050976 | detection of mechanical stimulus involved in sensory perception of touch(GO:0050976) |
| 0.3 | 1.7 | GO:0098528 | skeletal muscle fiber differentiation(GO:0098528) |
| 0.3 | 1.0 | GO:0097116 | gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 0.3 | 1.3 | GO:0023016 | osmosensory signaling pathway(GO:0007231) signal transduction by trans-phosphorylation(GO:0023016) kidney smooth muscle tissue development(GO:0072194) pattern specification involved in metanephros development(GO:0072268) |
| 0.3 | 1.8 | GO:0050916 | sensory perception of sweet taste(GO:0050916) |
| 0.3 | 1.7 | GO:0086021 | SA node cell to atrial cardiac muscle cell communication by electrical coupling(GO:0086021) |
| 0.3 | 0.6 | GO:0035990 | tendon cell differentiation(GO:0035990) tendon formation(GO:0035992) |
| 0.2 | 0.7 | GO:0021776 | smoothened signaling pathway involved in ventral spinal cord interneuron specification(GO:0021775) smoothened signaling pathway involved in spinal cord motor neuron cell fate specification(GO:0021776) |
| 0.2 | 0.7 | GO:0033058 | directional locomotion(GO:0033058) |
| 0.2 | 1.1 | GO:0010513 | positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) |
| 0.2 | 2.8 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.2 | 0.7 | GO:0006173 | dADP biosynthetic process(GO:0006173) |
| 0.2 | 0.8 | GO:1901350 | cell-cell signaling involved in cell-cell junction organization(GO:1901350) |
| 0.2 | 1.7 | GO:0019065 | receptor-mediated endocytosis of virus by host cell(GO:0019065) modulation by host of viral release from host cell(GO:0044789) positive regulation by host of viral release from host cell(GO:0044791) endocytosis involved in viral entry into host cell(GO:0075509) |
| 0.2 | 0.8 | GO:0052026 | modulation by virus of host transcription(GO:0019056) positive regulation of sprouting of injured axon(GO:0048687) positive regulation of axon extension involved in regeneration(GO:0048691) modulation by symbiont of host transcription(GO:0052026) |
| 0.2 | 1.1 | GO:0051835 | positive regulation of synapse structural plasticity(GO:0051835) |
| 0.2 | 0.5 | GO:0006481 | C-terminal protein methylation(GO:0006481) |
| 0.2 | 0.7 | GO:0061056 | sclerotome development(GO:0061056) |
| 0.2 | 3.1 | GO:0086027 | AV node cell action potential(GO:0086016) AV node cell to bundle of His cell signaling(GO:0086027) |
| 0.2 | 2.6 | GO:0021894 | cerebral cortex GABAergic interneuron development(GO:0021894) |
| 0.1 | 0.4 | GO:0001579 | medium-chain fatty acid transport(GO:0001579) |
| 0.1 | 1.0 | GO:0051121 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.1 | 0.5 | GO:0006601 | creatine biosynthetic process(GO:0006601) |
| 0.1 | 0.7 | GO:0048165 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.1 | 1.6 | GO:0035092 | sperm chromatin condensation(GO:0035092) |
| 0.1 | 2.1 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.1 | 0.7 | GO:0015798 | myo-inositol transport(GO:0015798) |
| 0.1 | 0.3 | GO:0097155 | fasciculation of sensory neuron axon(GO:0097155) |
| 0.1 | 1.0 | GO:0060120 | auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 0.1 | 1.2 | GO:0010459 | negative regulation of heart rate(GO:0010459) |
| 0.1 | 0.8 | GO:2000821 | regulation of grooming behavior(GO:2000821) |
| 0.1 | 0.4 | GO:0031642 | negative regulation of myelination(GO:0031642) |
| 0.1 | 0.7 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 0.1 | 1.6 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.1 | 2.1 | GO:0048681 | negative regulation of axon regeneration(GO:0048681) |
| 0.1 | 1.1 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.1 | 1.1 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.1 | 0.3 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) |
| 0.1 | 1.4 | GO:0007213 | G-protein coupled acetylcholine receptor signaling pathway(GO:0007213) |
| 0.1 | 1.4 | GO:0060539 | diaphragm development(GO:0060539) negative regulation of delayed rectifier potassium channel activity(GO:1902260) negative regulation of voltage-gated potassium channel activity(GO:1903817) |
| 0.1 | 0.4 | GO:0001575 | globoside metabolic process(GO:0001575) |
| 0.1 | 0.7 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.1 | 1.6 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.1 | 1.0 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.1 | 0.7 | GO:0060174 | limb bud formation(GO:0060174) |
| 0.1 | 1.4 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.1 | 0.8 | GO:0015705 | iodide transport(GO:0015705) |
| 0.1 | 0.9 | GO:0006449 | regulation of translational termination(GO:0006449) |
| 0.1 | 2.6 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.1 | 0.3 | GO:0030037 | actin filament reorganization involved in cell cycle(GO:0030037) |
| 0.1 | 0.3 | GO:0001933 | negative regulation of protein phosphorylation(GO:0001933) negative regulation of phosphorylation(GO:0042326) |
| 0.1 | 0.9 | GO:0035878 | nail development(GO:0035878) |
| 0.0 | 0.6 | GO:0071321 | cellular response to cGMP(GO:0071321) |
| 0.0 | 1.0 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.0 | 1.0 | GO:0031167 | rRNA methylation(GO:0031167) |
| 0.0 | 0.9 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.0 | 0.5 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.0 | 0.4 | GO:0046078 | dUMP metabolic process(GO:0046078) |
| 0.0 | 0.4 | GO:0098914 | membrane repolarization during atrial cardiac muscle cell action potential(GO:0098914) |
| 0.0 | 0.6 | GO:0007351 | tripartite regional subdivision(GO:0007351) anterior/posterior axis specification, embryo(GO:0008595) |
| 0.0 | 1.3 | GO:0021680 | cerebellar Purkinje cell layer development(GO:0021680) |
| 0.0 | 0.7 | GO:0016446 | somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.0 | 1.8 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.0 | 1.2 | GO:1901685 | glutathione derivative metabolic process(GO:1901685) glutathione derivative biosynthetic process(GO:1901687) |
| 0.0 | 0.9 | GO:0051290 | protein heterotetramerization(GO:0051290) |
| 0.0 | 1.6 | GO:0015949 | nucleobase-containing small molecule interconversion(GO:0015949) |
| 0.0 | 0.2 | GO:0033031 | positive regulation of neutrophil apoptotic process(GO:0033031) |
| 0.0 | 0.4 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.0 | 0.1 | GO:0071288 | cellular response to mercury ion(GO:0071288) |
| 0.0 | 1.0 | GO:0061641 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.0 | 0.5 | GO:0006554 | lysine catabolic process(GO:0006554) |
| 0.0 | 0.8 | GO:0007162 | negative regulation of cell adhesion(GO:0007162) |
| 0.0 | 0.1 | GO:0021997 | neural plate axis specification(GO:0021997) |
| 0.0 | 2.0 | GO:0045652 | regulation of megakaryocyte differentiation(GO:0045652) |
| 0.0 | 1.1 | GO:0071480 | cellular response to gamma radiation(GO:0071480) |
| 0.0 | 0.2 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 | 0.2 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.0 | 0.3 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.0 | 1.5 | GO:0048747 | muscle fiber development(GO:0048747) |
| 0.0 | 0.1 | GO:0006288 | base-excision repair, DNA ligation(GO:0006288) regulation of DNA ligase activity(GO:1904875) |
| 0.0 | 0.8 | GO:0045745 | positive regulation of G-protein coupled receptor protein signaling pathway(GO:0045745) |
| 0.0 | 0.1 | GO:0007388 | anterior compartment pattern formation(GO:0007387) posterior compartment specification(GO:0007388) |
| 0.0 | 0.5 | GO:0070734 | histone H3-K27 methylation(GO:0070734) |
| 0.0 | 3.3 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.0 | 0.7 | GO:0007200 | phospholipase C-activating G-protein coupled receptor signaling pathway(GO:0007200) |
| 0.0 | 0.2 | GO:0030422 | production of siRNA involved in RNA interference(GO:0030422) |
| 0.0 | 0.3 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.0 | 0.4 | GO:0002021 | response to dietary excess(GO:0002021) |
| 0.0 | 0.8 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.0 | 0.6 | GO:2001222 | regulation of neuron migration(GO:2001222) |
| 0.0 | 1.0 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.0 | 1.2 | GO:0014902 | myotube differentiation(GO:0014902) |
| 0.0 | 0.4 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 1.8 | GO:0042476 | odontogenesis(GO:0042476) |
| 0.0 | 1.9 | GO:0090263 | positive regulation of canonical Wnt signaling pathway(GO:0090263) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 5.8 | GO:0097453 | mesaxon(GO:0097453) ensheathing process(GO:1990015) |
| 0.3 | 2.1 | GO:1990393 | 3M complex(GO:1990393) |
| 0.2 | 2.3 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.2 | 0.6 | GO:0098855 | HCN channel complex(GO:0098855) |
| 0.2 | 1.4 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.1 | 11.3 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.1 | 5.9 | GO:0005921 | gap junction(GO:0005921) |
| 0.1 | 2.7 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.1 | 5.1 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.1 | 3.1 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.1 | 1.8 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.1 | 1.8 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 0.3 | GO:0002139 | stereocilia coupling link(GO:0002139) |
| 0.1 | 0.7 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.1 | 0.7 | GO:0005683 | U7 snRNP(GO:0005683) |
| 0.1 | 1.5 | GO:0043220 | Schmidt-Lanterman incisure(GO:0043220) |
| 0.1 | 2.2 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.1 | 3.1 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.1 | 0.9 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.1 | 2.1 | GO:0005849 | mRNA cleavage factor complex(GO:0005849) |
| 0.0 | 0.7 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 0.3 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.0 | 1.7 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.0 | 0.3 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.0 | 4.1 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 1.1 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.5 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 0.4 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 1.0 | GO:0010369 | chromocenter(GO:0010369) |
| 0.0 | 1.0 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.0 | 1.4 | GO:0044298 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 0.7 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 2.9 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.3 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 0.5 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.1 | GO:0070522 | ERCC4-ERCC1 complex(GO:0070522) |
| 0.0 | 4.9 | GO:0044798 | nuclear transcription factor complex(GO:0044798) |
| 0.0 | 0.4 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.0 | 0.7 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.4 | GO:0030133 | transport vesicle(GO:0030133) |
| 0.0 | 1.2 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.4 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.0 | 1.3 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.2 | GO:0070578 | RISC-loading complex(GO:0070578) |
| 0.0 | 0.5 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.7 | GO:0097546 | ciliary base(GO:0097546) |
| 0.0 | 0.2 | GO:0005686 | U2 snRNP(GO:0005686) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.9 | GO:0032093 | SAM domain binding(GO:0032093) |
| 0.5 | 2.0 | GO:0048763 | calcium-induced calcium release activity(GO:0048763) |
| 0.4 | 2.0 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.3 | 3.1 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.3 | 5.8 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.3 | 1.7 | GO:0086020 | gap junction channel activity involved in SA node cell-atrial cardiac muscle cell electrical coupling(GO:0086020) |
| 0.3 | 2.2 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.3 | 3.9 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.2 | 1.0 | GO:0051120 | hepoxilin A3 synthase activity(GO:0051120) |
| 0.2 | 0.8 | GO:0031812 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 0.2 | 1.9 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.2 | 0.9 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.2 | 0.7 | GO:0071209 | U7 snRNA binding(GO:0071209) |
| 0.2 | 1.0 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.2 | 0.5 | GO:0003880 | protein C-terminal carboxyl O-methyltransferase activity(GO:0003880) |
| 0.2 | 0.5 | GO:0015068 | amidinotransferase activity(GO:0015067) glycine amidinotransferase activity(GO:0015068) |
| 0.2 | 3.9 | GO:0019841 | retinol binding(GO:0019841) |
| 0.1 | 10.2 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.1 | 0.9 | GO:0043813 | phosphatidylinositol-3,5-bisphosphate 5-phosphatase activity(GO:0043813) |
| 0.1 | 1.0 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) |
| 0.1 | 2.1 | GO:1990459 | transferrin receptor binding(GO:1990459) |
| 0.1 | 0.6 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.1 | 0.3 | GO:0070506 | high-density lipoprotein particle receptor activity(GO:0070506) |
| 0.1 | 1.0 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.1 | 1.8 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.1 | 0.7 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.1 | 0.7 | GO:0032138 | single base insertion or deletion binding(GO:0032138) |
| 0.1 | 1.1 | GO:0051378 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.1 | 0.7 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.1 | 0.7 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.1 | 1.8 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.1 | 1.0 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.1 | 1.6 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.1 | 0.7 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.1 | 0.4 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.1 | 2.2 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.1 | 2.3 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.1 | 2.7 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.1 | 0.5 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.1 | 2.1 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.1 | 0.7 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.1 | 0.9 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.1 | 1.4 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.1 | 0.8 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) neurotrophin receptor activity(GO:0005030) |
| 0.0 | 0.3 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.0 | 2.2 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 1.2 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 1.4 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 1.3 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.4 | GO:1904929 | coreceptor activity involved in Wnt signaling pathway, planar cell polarity pathway(GO:1904929) |
| 0.0 | 0.0 | GO:0031208 | POZ domain binding(GO:0031208) |
| 0.0 | 5.4 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.0 | 1.6 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.0 | 0.5 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.0 | 2.7 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 0.8 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.0 | 1.1 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 0.4 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 0.0 | 0.5 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.7 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 0.2 | GO:0070883 | pre-miRNA binding(GO:0070883) |
| 0.0 | 1.2 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.4 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.4 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.3 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.0 | 0.3 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.0 | 0.2 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.9 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.5 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.2 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.4 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.0 | 1.0 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.0 | 0.7 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.1 | GO:0015168 | glycerol transmembrane transporter activity(GO:0015168) |
| 0.0 | 0.1 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.0 | 0.4 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.0 | 1.2 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.1 | GO:0005497 | androgen binding(GO:0005497) |
| 0.0 | 0.7 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.0 | 0.4 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 1.1 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 1.3 | GO:0032947 | protein complex scaffold(GO:0032947) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 10.1 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 2.8 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 5.8 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 1.4 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 1.0 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 2.0 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 1.8 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.7 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 1.9 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 1.2 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 3.0 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.6 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 1.5 | PID INTEGRIN1 PATHWAY | Beta1 integrin cell surface interactions |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 9.8 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.1 | 5.8 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.1 | 1.8 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.1 | 1.2 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.1 | 1.1 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.1 | 1.7 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.1 | 2.2 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.1 | 1.9 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.1 | 2.7 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.1 | 2.3 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 2.9 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 1.7 | REACTOME PROCESSING OF CAPPED INTRONLESS PRE MRNA | Genes involved in Processing of Capped Intronless Pre-mRNA |
| 0.0 | 1.3 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 1.4 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.8 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 3.6 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 2.3 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 1.2 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 1.0 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 1.2 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.5 | REACTOME FGFR1 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR1 ligand binding and activation |
| 0.0 | 2.2 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.5 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.4 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 1.0 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 0.9 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.7 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.5 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 0.4 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 1.0 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.0 | 0.5 | REACTOME RNA POL I TRANSCRIPTION INITIATION | Genes involved in RNA Polymerase I Transcription Initiation |