Project
Illumina Body Map 2: averaged replicates
Navigation
Downloads
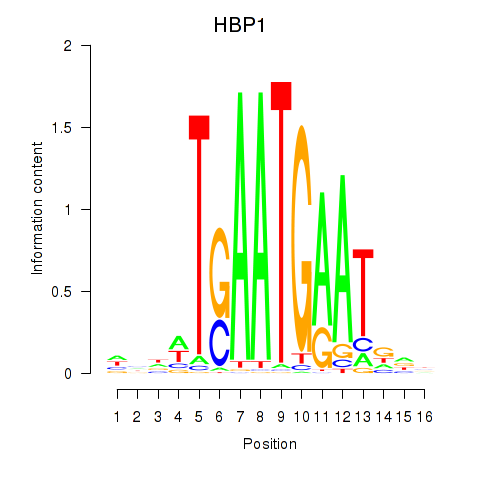
Results for HBP1
Z-value: 1.71
Transcription factors associated with HBP1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HBP1
|
ENSG00000105856.9 | HMG-box transcription factor 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HBP1 | hg19_v2_chr7_+_106809406_106809460 | -0.30 | 9.5e-02 | Click! |
Activity profile of HBP1 motif
Sorted Z-values of HBP1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_+_4198072 | 6.50 |
ENST00000262970.5
|
ANKRD24
|
ankyrin repeat domain 24 |
| chr2_+_231902193 | 3.14 |
ENST00000373640.4
|
C2orf72
|
chromosome 2 open reading frame 72 |
| chr3_+_125985620 | 2.58 |
ENST00000511512.1
ENST00000512435.1 |
RP11-71E19.1
|
RP11-71E19.1 |
| chr1_-_85930246 | 2.24 |
ENST00000426972.3
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1 |
| chr17_+_68071389 | 2.24 |
ENST00000283936.1
ENST00000392671.1 |
KCNJ16
|
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr17_+_68071458 | 2.05 |
ENST00000589377.1
|
KCNJ16
|
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr19_-_10697895 | 2.04 |
ENST00000591240.1
ENST00000589684.1 ENST00000591676.1 ENST00000250244.6 ENST00000590923.1 |
AP1M2
|
adaptor-related protein complex 1, mu 2 subunit |
| chr10_-_134756030 | 2.02 |
ENST00000368586.5
ENST00000368582.2 |
TTC40
|
tetratricopeptide repeat domain 40 |
| chr5_+_159656437 | 2.02 |
ENST00000402432.3
|
FABP6
|
fatty acid binding protein 6, ileal |
| chr5_-_147211226 | 1.99 |
ENST00000296695.5
|
SPINK1
|
serine peptidase inhibitor, Kazal type 1 |
| chr4_+_100495864 | 1.86 |
ENST00000265517.5
ENST00000422897.2 |
MTTP
|
microsomal triglyceride transfer protein |
| chr1_+_85527987 | 1.78 |
ENST00000326813.8
ENST00000294664.6 ENST00000528899.1 |
WDR63
|
WD repeat domain 63 |
| chr17_-_42988004 | 1.73 |
ENST00000586125.1
ENST00000591880.1 |
GFAP
|
glial fibrillary acidic protein |
| chr4_-_186696515 | 1.70 |
ENST00000456596.1
ENST00000414724.1 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr22_-_20367797 | 1.66 |
ENST00000424787.2
|
GGTLC3
|
gamma-glutamyltransferase light chain 3 |
| chr4_-_186696561 | 1.65 |
ENST00000445115.1
ENST00000451701.1 ENST00000457247.1 ENST00000435480.1 ENST00000425679.1 ENST00000457934.1 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr11_+_124789146 | 1.59 |
ENST00000408930.5
|
HEPN1
|
hepatocellular carcinoma, down-regulated 1 |
| chr2_-_161056762 | 1.55 |
ENST00000428609.2
ENST00000409967.2 |
ITGB6
|
integrin, beta 6 |
| chr7_-_100808843 | 1.53 |
ENST00000249330.2
|
VGF
|
VGF nerve growth factor inducible |
| chr22_-_20368028 | 1.49 |
ENST00000404912.1
|
GGTLC3
|
gamma-glutamyltransferase light chain 3 |
| chr18_+_18943554 | 1.39 |
ENST00000580732.2
|
GREB1L
|
growth regulation by estrogen in breast cancer-like |
| chr3_-_165555200 | 1.36 |
ENST00000479451.1
ENST00000540653.1 ENST00000488954.1 ENST00000264381.3 |
BCHE
|
butyrylcholinesterase |
| chr19_+_840963 | 1.36 |
ENST00000234347.5
|
PRTN3
|
proteinase 3 |
| chr9_+_86238016 | 1.34 |
ENST00000530832.1
ENST00000405990.3 ENST00000376417.4 ENST00000376419.4 |
IDNK
|
idnK, gluconokinase homolog (E. coli) |
| chr11_-_66725837 | 1.33 |
ENST00000393958.2
ENST00000393960.1 ENST00000524491.1 ENST00000355677.3 |
PC
|
pyruvate carboxylase |
| chr12_+_100867486 | 1.31 |
ENST00000548884.1
|
NR1H4
|
nuclear receptor subfamily 1, group H, member 4 |
| chr2_-_161056802 | 1.24 |
ENST00000283249.2
ENST00000409872.1 |
ITGB6
|
integrin, beta 6 |
| chr12_+_100867694 | 1.24 |
ENST00000392986.3
ENST00000549996.1 |
NR1H4
|
nuclear receptor subfamily 1, group H, member 4 |
| chr4_+_186347388 | 1.23 |
ENST00000511138.1
ENST00000511581.1 |
C4orf47
|
chromosome 4 open reading frame 47 |
| chr12_+_100867733 | 1.22 |
ENST00000546380.1
|
NR1H4
|
nuclear receptor subfamily 1, group H, member 4 |
| chr1_+_92632542 | 1.21 |
ENST00000409154.4
ENST00000370378.4 |
KIAA1107
|
KIAA1107 |
| chr18_-_68004529 | 1.21 |
ENST00000578633.1
|
RP11-484N16.1
|
RP11-484N16.1 |
| chr4_-_186696425 | 1.17 |
ENST00000430503.1
ENST00000319454.6 ENST00000450341.1 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr11_+_63057412 | 1.17 |
ENST00000544661.1
|
SLC22A10
|
solute carrier family 22, member 10 |
| chrX_+_150151824 | 1.17 |
ENST00000455596.1
ENST00000448905.2 |
HMGB3
|
high mobility group box 3 |
| chr12_-_53320245 | 1.17 |
ENST00000552150.1
|
KRT8
|
keratin 8 |
| chr7_-_99277610 | 1.16 |
ENST00000343703.5
ENST00000222982.4 ENST00000439761.1 ENST00000339843.2 |
CYP3A5
|
cytochrome P450, family 3, subfamily A, polypeptide 5 |
| chr2_-_207630033 | 1.13 |
ENST00000449792.1
|
MDH1B
|
malate dehydrogenase 1B, NAD (soluble) |
| chr12_+_79357815 | 1.13 |
ENST00000547046.1
|
SYT1
|
synaptotagmin I |
| chr4_-_104640973 | 1.11 |
ENST00000304883.2
|
TACR3
|
tachykinin receptor 3 |
| chr15_-_72490114 | 1.11 |
ENST00000309731.7
|
GRAMD2
|
GRAM domain containing 2 |
| chr3_+_140770183 | 1.10 |
ENST00000310546.2
|
SPSB4
|
splA/ryanodine receptor domain and SOCS box containing 4 |
| chr7_+_39125365 | 1.10 |
ENST00000559001.1
ENST00000464276.2 |
POU6F2
|
POU class 6 homeobox 2 |
| chr5_+_175223313 | 1.10 |
ENST00000359546.4
|
CPLX2
|
complexin 2 |
| chr2_-_207629997 | 1.10 |
ENST00000454776.2
|
MDH1B
|
malate dehydrogenase 1B, NAD (soluble) |
| chr4_+_159236462 | 1.08 |
ENST00000460056.2
|
RXFP1
|
relaxin/insulin-like family peptide receptor 1 |
| chr12_-_99548270 | 1.08 |
ENST00000546568.1
ENST00000332712.7 ENST00000546960.1 |
ANKS1B
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr4_-_186697044 | 1.07 |
ENST00000437304.2
|
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr7_+_29874341 | 1.05 |
ENST00000409290.1
ENST00000242140.5 |
WIPF3
|
WAS/WASL interacting protein family, member 3 |
| chr4_-_186696636 | 1.05 |
ENST00000444771.1
|
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr1_+_65730385 | 1.05 |
ENST00000263441.7
ENST00000395325.3 |
DNAJC6
|
DnaJ (Hsp40) homolog, subfamily C, member 6 |
| chr11_+_22689648 | 1.05 |
ENST00000278187.3
|
GAS2
|
growth arrest-specific 2 |
| chr1_+_240255166 | 1.02 |
ENST00000319653.9
|
FMN2
|
formin 2 |
| chr13_-_44735393 | 1.02 |
ENST00000400419.1
|
SMIM2
|
small integral membrane protein 2 |
| chr22_+_41968007 | 1.02 |
ENST00000460790.1
|
CSDC2
|
cold shock domain containing C2, RNA binding |
| chr8_+_107630340 | 1.01 |
ENST00000497705.1
|
OXR1
|
oxidation resistance 1 |
| chrX_+_150151752 | 1.00 |
ENST00000325307.7
|
HMGB3
|
high mobility group box 3 |
| chr14_-_94759361 | 0.94 |
ENST00000393096.1
|
SERPINA10
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 10 |
| chr22_-_30956746 | 0.91 |
ENST00000437282.1
ENST00000447224.1 ENST00000427899.1 ENST00000406955.1 ENST00000452827.1 |
GAL3ST1
|
galactose-3-O-sulfotransferase 1 |
| chr14_-_94759408 | 0.91 |
ENST00000554723.1
|
SERPINA10
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 10 |
| chr14_-_94759595 | 0.91 |
ENST00000261994.4
|
SERPINA10
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 10 |
| chr15_+_72410629 | 0.91 |
ENST00000340912.4
ENST00000544171.1 |
SENP8
|
SUMO/sentrin specific peptidase family member 8 |
| chr11_-_18343669 | 0.90 |
ENST00000396253.3
ENST00000349215.3 ENST00000438420.2 |
HPS5
|
Hermansky-Pudlak syndrome 5 |
| chr10_-_21186144 | 0.89 |
ENST00000377119.1
|
NEBL
|
nebulette |
| chr11_+_66824303 | 0.88 |
ENST00000533360.1
|
RHOD
|
ras homolog family member D |
| chr6_+_149539106 | 0.87 |
ENST00000443992.1
|
RP1-111D6.3
|
RP1-111D6.3 |
| chr5_+_72469014 | 0.87 |
ENST00000296776.5
|
TMEM174
|
transmembrane protein 174 |
| chrX_+_2746850 | 0.87 |
ENST00000381163.3
ENST00000338623.5 ENST00000542787.1 |
GYG2
|
glycogenin 2 |
| chr11_-_7698453 | 0.84 |
ENST00000524608.1
|
CYB5R2
|
cytochrome b5 reductase 2 |
| chrX_+_2746818 | 0.83 |
ENST00000398806.3
|
GYG2
|
glycogenin 2 |
| chr12_+_128399917 | 0.83 |
ENST00000544645.1
|
LINC00507
|
long intergenic non-protein coding RNA 507 |
| chr8_-_27457494 | 0.82 |
ENST00000521770.1
|
CLU
|
clusterin |
| chr20_-_23967432 | 0.82 |
ENST00000286890.4
ENST00000278765.4 |
GGTLC1
|
gamma-glutamyltransferase light chain 1 |
| chr22_+_30477000 | 0.81 |
ENST00000403975.1
|
HORMAD2
|
HORMA domain containing 2 |
| chr4_-_46126093 | 0.81 |
ENST00000295452.4
|
GABRG1
|
gamma-aminobutyric acid (GABA) A receptor, gamma 1 |
| chr5_+_161274940 | 0.81 |
ENST00000393943.4
|
GABRA1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1 |
| chr11_-_18343725 | 0.81 |
ENST00000531848.1
|
HPS5
|
Hermansky-Pudlak syndrome 5 |
| chr20_-_30458432 | 0.79 |
ENST00000375966.4
ENST00000278979.3 |
DUSP15
|
dual specificity phosphatase 15 |
| chr9_-_107361788 | 0.79 |
ENST00000374779.2
|
OR13C5
|
olfactory receptor, family 13, subfamily C, member 5 |
| chr2_-_197226875 | 0.78 |
ENST00000409111.1
|
HECW2
|
HECT, C2 and WW domain containing E3 ubiquitin protein ligase 2 |
| chr1_+_65613217 | 0.78 |
ENST00000545314.1
|
AK4
|
adenylate kinase 4 |
| chr11_-_31014214 | 0.77 |
ENST00000406071.2
ENST00000339794.5 |
DCDC1
|
doublecortin domain containing 1 |
| chr19_+_19144384 | 0.77 |
ENST00000392335.2
ENST00000535612.1 ENST00000537263.1 ENST00000540707.1 ENST00000541725.1 ENST00000269932.6 ENST00000546344.1 ENST00000540792.1 ENST00000536098.1 ENST00000541898.1 ENST00000543877.1 |
ARMC6
|
armadillo repeat containing 6 |
| chr5_+_161274685 | 0.76 |
ENST00000428797.2
|
GABRA1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1 |
| chr20_+_44650348 | 0.75 |
ENST00000454036.2
|
SLC12A5
|
solute carrier family 12 (potassium/chloride transporter), member 5 |
| chr12_+_128399965 | 0.75 |
ENST00000540882.1
ENST00000542089.1 |
LINC00507
|
long intergenic non-protein coding RNA 507 |
| chr7_+_65540780 | 0.74 |
ENST00000304874.9
|
ASL
|
argininosuccinate lyase |
| chr6_+_135818907 | 0.72 |
ENST00000579339.1
ENST00000580741.1 |
LINC00271
|
long intergenic non-protein coding RNA 271 |
| chr6_+_135818979 | 0.71 |
ENST00000421378.2
ENST00000579057.1 ENST00000436554.1 ENST00000438618.2 |
LINC00271
|
long intergenic non-protein coding RNA 271 |
| chr2_-_204399976 | 0.71 |
ENST00000457812.1
|
RAPH1
|
Ras association (RalGDS/AF-6) and pleckstrin homology domains 1 |
| chr7_+_65540853 | 0.71 |
ENST00000380839.4
ENST00000395332.3 ENST00000362000.5 ENST00000395331.3 |
ASL
|
argininosuccinate lyase |
| chr2_+_235887329 | 0.70 |
ENST00000409212.1
ENST00000344528.4 ENST00000444916.1 |
SH3BP4
|
SH3-domain binding protein 4 |
| chr5_+_140514782 | 0.70 |
ENST00000231134.5
|
PCDHB5
|
protocadherin beta 5 |
| chrX_-_131262048 | 0.70 |
ENST00000298542.4
|
FRMD7
|
FERM domain containing 7 |
| chr9_-_123476612 | 0.69 |
ENST00000426959.1
|
MEGF9
|
multiple EGF-like-domains 9 |
| chr4_-_186682716 | 0.68 |
ENST00000445343.1
|
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr16_+_80574854 | 0.68 |
ENST00000305904.6
ENST00000568035.1 |
DYNLRB2
|
dynein, light chain, roadblock-type 2 |
| chr11_+_43702236 | 0.67 |
ENST00000531185.1
ENST00000278353.4 |
HSD17B12
|
hydroxysteroid (17-beta) dehydrogenase 12 |
| chr1_-_24127256 | 0.67 |
ENST00000418277.1
|
GALE
|
UDP-galactose-4-epimerase |
| chr3_+_100428188 | 0.66 |
ENST00000418917.2
ENST00000490574.1 |
TFG
|
TRK-fused gene |
| chr5_-_78809950 | 0.66 |
ENST00000334082.6
|
HOMER1
|
homer homolog 1 (Drosophila) |
| chr11_+_93063137 | 0.65 |
ENST00000534747.1
|
CCDC67
|
coiled-coil domain containing 67 |
| chr7_+_36450169 | 0.65 |
ENST00000428612.1
|
ANLN
|
anillin, actin binding protein |
| chr18_+_52385068 | 0.65 |
ENST00000586570.1
|
RAB27B
|
RAB27B, member RAS oncogene family |
| chr12_+_41221794 | 0.64 |
ENST00000547849.1
|
CNTN1
|
contactin 1 |
| chr2_-_74375081 | 0.63 |
ENST00000327428.5
|
BOLA3
|
bolA family member 3 |
| chr15_-_72410455 | 0.63 |
ENST00000569314.1
|
MYO9A
|
myosin IXA |
| chr11_+_123430948 | 0.63 |
ENST00000529432.1
ENST00000534764.1 |
GRAMD1B
|
GRAM domain containing 1B |
| chr3_+_108308845 | 0.63 |
ENST00000479138.1
|
DZIP3
|
DAZ interacting zinc finger protein 3 |
| chr9_+_86237963 | 0.62 |
ENST00000277124.8
|
IDNK
|
idnK, gluconokinase homolog (E. coli) |
| chr4_+_150999418 | 0.61 |
ENST00000296550.7
|
DCLK2
|
doublecortin-like kinase 2 |
| chr3_+_5163905 | 0.61 |
ENST00000256496.3
ENST00000419534.2 |
ARL8B
|
ADP-ribosylation factor-like 8B |
| chr20_-_30458354 | 0.60 |
ENST00000428829.1
|
DUSP15
|
dual specificity phosphatase 15 |
| chr3_+_46616017 | 0.59 |
ENST00000542931.1
|
TDGF1
|
teratocarcinoma-derived growth factor 1 |
| chr2_-_239197201 | 0.59 |
ENST00000254658.3
|
PER2
|
period circadian clock 2 |
| chr1_+_65613340 | 0.58 |
ENST00000546702.1
|
AK4
|
adenylate kinase 4 |
| chr22_+_22988816 | 0.58 |
ENST00000480559.1
ENST00000448514.1 |
GGTLC2
|
gamma-glutamyltransferase light chain 2 |
| chr20_-_30458491 | 0.58 |
ENST00000339738.5
|
DUSP15
|
dual specificity phosphatase 15 |
| chr8_-_82359662 | 0.58 |
ENST00000519260.1
ENST00000256103.2 |
PMP2
|
peripheral myelin protein 2 |
| chr20_+_25176318 | 0.57 |
ENST00000354989.5
ENST00000360031.2 ENST00000376652.4 ENST00000439162.1 ENST00000433417.1 ENST00000417467.1 ENST00000433259.2 ENST00000427553.1 ENST00000435520.1 ENST00000418890.1 |
ENTPD6
|
ectonucleoside triphosphate diphosphohydrolase 6 (putative) |
| chr11_-_18956556 | 0.57 |
ENST00000302797.3
|
MRGPRX1
|
MAS-related GPR, member X1 |
| chr3_-_42452050 | 0.57 |
ENST00000441172.1
ENST00000287748.3 |
LYZL4
|
lysozyme-like 4 |
| chr12_-_110271178 | 0.57 |
ENST00000261740.2
ENST00000392719.2 ENST00000346520.2 |
TRPV4
|
transient receptor potential cation channel, subfamily V, member 4 |
| chrX_-_23926004 | 0.56 |
ENST00000379226.4
ENST00000379220.3 |
APOO
|
apolipoprotein O |
| chr5_+_125758813 | 0.56 |
ENST00000285689.3
ENST00000515200.1 |
GRAMD3
|
GRAM domain containing 3 |
| chr15_+_34260921 | 0.55 |
ENST00000560035.1
|
CHRM5
|
cholinergic receptor, muscarinic 5 |
| chr3_+_38537960 | 0.55 |
ENST00000453767.1
|
EXOG
|
endo/exonuclease (5'-3'), endonuclease G-like |
| chr11_+_93063878 | 0.54 |
ENST00000298050.3
|
CCDC67
|
coiled-coil domain containing 67 |
| chrX_+_51636629 | 0.54 |
ENST00000375722.1
ENST00000326587.7 ENST00000375695.2 |
MAGED1
|
melanoma antigen family D, 1 |
| chr15_+_34261089 | 0.54 |
ENST00000383263.5
|
CHRM5
|
cholinergic receptor, muscarinic 5 |
| chr2_+_27886330 | 0.53 |
ENST00000326019.6
|
SLC4A1AP
|
solute carrier family 4 (anion exchanger), member 1, adaptor protein |
| chr12_-_100378006 | 0.53 |
ENST00000547776.2
ENST00000329257.7 ENST00000547010.1 |
ANKS1B
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr3_+_100428268 | 0.53 |
ENST00000240851.4
|
TFG
|
TRK-fused gene |
| chr11_+_67183557 | 0.53 |
ENST00000445895.2
|
CARNS1
|
carnosine synthase 1 |
| chr11_+_43702322 | 0.53 |
ENST00000395700.4
|
HSD17B12
|
hydroxysteroid (17-beta) dehydrogenase 12 |
| chr2_-_40739501 | 0.52 |
ENST00000403092.1
ENST00000402441.1 ENST00000448531.1 |
SLC8A1
|
solute carrier family 8 (sodium/calcium exchanger), member 1 |
| chr2_+_98703643 | 0.52 |
ENST00000477737.1
|
VWA3B
|
von Willebrand factor A domain containing 3B |
| chr10_+_81838411 | 0.52 |
ENST00000372281.3
ENST00000372277.3 ENST00000372275.1 ENST00000372274.1 |
TMEM254
|
transmembrane protein 254 |
| chr19_-_19144243 | 0.52 |
ENST00000594445.1
ENST00000452918.2 ENST00000600377.1 ENST00000337018.6 |
SUGP2
|
SURP and G patch domain containing 2 |
| chr7_+_117120017 | 0.51 |
ENST00000003084.6
ENST00000454343.1 |
CFTR
|
cystic fibrosis transmembrane conductance regulator (ATP-binding cassette sub-family C, member 7) |
| chr15_-_52263937 | 0.51 |
ENST00000315141.5
ENST00000299601.5 |
LEO1
|
Leo1, Paf1/RNA polymerase II complex component, homolog (S. cerevisiae) |
| chr12_-_11214893 | 0.50 |
ENST00000533467.1
|
TAS2R46
|
taste receptor, type 2, member 46 |
| chr3_-_197026152 | 0.50 |
ENST00000419354.1
|
DLG1
|
discs, large homolog 1 (Drosophila) |
| chr20_-_33543546 | 0.50 |
ENST00000216951.2
|
GSS
|
glutathione synthetase |
| chr5_+_161494521 | 0.50 |
ENST00000356592.3
|
GABRG2
|
gamma-aminobutyric acid (GABA) A receptor, gamma 2 |
| chr1_+_200860122 | 0.50 |
ENST00000532631.1
ENST00000451872.2 |
C1orf106
|
chromosome 1 open reading frame 106 |
| chr3_+_100428316 | 0.50 |
ENST00000479672.1
ENST00000476228.1 ENST00000463568.1 |
TFG
|
TRK-fused gene |
| chr20_-_39928705 | 0.49 |
ENST00000436099.2
ENST00000309060.3 ENST00000373261.1 ENST00000436440.2 ENST00000540170.1 ENST00000557816.1 ENST00000560361.1 |
ZHX3
|
zinc fingers and homeoboxes 3 |
| chr2_+_242498135 | 0.49 |
ENST00000318407.3
|
BOK
|
BCL2-related ovarian killer |
| chr8_+_69242957 | 0.49 |
ENST00000518698.1
ENST00000539993.1 |
C8orf34
|
chromosome 8 open reading frame 34 |
| chr8_+_104153126 | 0.48 |
ENST00000306391.6
ENST00000330955.5 |
BAALC
|
brain and acute leukemia, cytoplasmic |
| chr20_-_39928756 | 0.47 |
ENST00000432768.2
|
ZHX3
|
zinc fingers and homeoboxes 3 |
| chr6_+_10521574 | 0.47 |
ENST00000495262.1
|
GCNT2
|
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme (I blood group) |
| chr6_-_8102279 | 0.47 |
ENST00000488226.2
|
EEF1E1
|
eukaryotic translation elongation factor 1 epsilon 1 |
| chr20_+_25176450 | 0.47 |
ENST00000425813.1
|
ENTPD6
|
ectonucleoside triphosphate diphosphohydrolase 6 (putative) |
| chr6_+_29555683 | 0.47 |
ENST00000383640.2
|
OR2H2
|
olfactory receptor, family 2, subfamily H, member 2 |
| chr11_+_66824276 | 0.47 |
ENST00000308831.2
|
RHOD
|
ras homolog family member D |
| chr9_-_123476719 | 0.46 |
ENST00000373930.3
|
MEGF9
|
multiple EGF-like-domains 9 |
| chr11_-_76998463 | 0.46 |
ENST00000376217.2
ENST00000315938.4 |
GDPD4
|
glycerophosphodiester phosphodiesterase domain containing 4 |
| chr12_-_11139511 | 0.45 |
ENST00000506868.1
|
TAS2R50
|
taste receptor, type 2, member 50 |
| chr14_-_80677970 | 0.45 |
ENST00000438257.4
|
DIO2
|
deiodinase, iodothyronine, type II |
| chr3_+_159943362 | 0.44 |
ENST00000326474.3
|
C3orf80
|
chromosome 3 open reading frame 80 |
| chr6_-_133035185 | 0.44 |
ENST00000367928.4
|
VNN1
|
vanin 1 |
| chr3_+_185000729 | 0.44 |
ENST00000448876.1
ENST00000446828.1 ENST00000447637.1 ENST00000424227.1 ENST00000454237.1 |
MAP3K13
|
mitogen-activated protein kinase kinase kinase 13 |
| chr4_-_175041663 | 0.44 |
ENST00000503140.1
|
RP11-148L24.1
|
RP11-148L24.1 |
| chr2_-_239197238 | 0.43 |
ENST00000254657.3
|
PER2
|
period circadian clock 2 |
| chr7_-_2354099 | 0.43 |
ENST00000222990.3
|
SNX8
|
sorting nexin 8 |
| chr2_+_208576355 | 0.42 |
ENST00000420822.1
ENST00000295414.3 ENST00000339882.5 |
CCNYL1
|
cyclin Y-like 1 |
| chr19_-_38747172 | 0.42 |
ENST00000347262.4
ENST00000591585.1 ENST00000301242.4 |
PPP1R14A
|
protein phosphatase 1, regulatory (inhibitor) subunit 14A |
| chrX_+_18725758 | 0.42 |
ENST00000472826.1
ENST00000544635.1 ENST00000496075.2 |
PPEF1
|
protein phosphatase, EF-hand calcium binding domain 1 |
| chr14_+_78266436 | 0.41 |
ENST00000557501.1
ENST00000341211.5 |
ADCK1
|
aarF domain containing kinase 1 |
| chr1_-_113392399 | 0.41 |
ENST00000449572.2
ENST00000433505.1 |
RP11-426L16.8
|
RP11-426L16.8 |
| chr3_-_178103144 | 0.41 |
ENST00000417383.1
ENST00000418585.1 ENST00000411727.1 ENST00000439810.1 |
RP11-33A14.1
|
RP11-33A14.1 |
| chr16_+_77756399 | 0.41 |
ENST00000564085.1
ENST00000268533.5 ENST00000568787.1 ENST00000437314.3 ENST00000563839.1 |
NUDT7
|
nudix (nucleoside diphosphate linked moiety X)-type motif 7 |
| chr7_+_128349106 | 0.41 |
ENST00000485070.1
|
FAM71F1
|
family with sequence similarity 71, member F1 |
| chr1_-_25747283 | 0.40 |
ENST00000346452.4
ENST00000340849.4 ENST00000349438.4 ENST00000294413.7 ENST00000413854.1 ENST00000455194.1 ENST00000243186.6 ENST00000425135.1 |
RHCE
|
Rh blood group, CcEe antigens |
| chr3_+_44754126 | 0.40 |
ENST00000449836.1
ENST00000436624.2 ENST00000296091.4 ENST00000411443.1 |
ZNF502
|
zinc finger protein 502 |
| chr7_-_83278322 | 0.40 |
ENST00000307792.3
|
SEMA3E
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3E |
| chr9_-_34665983 | 0.40 |
ENST00000416454.1
ENST00000544078.2 ENST00000421828.2 ENST00000423809.1 |
RP11-195F19.5
|
HCG2040265, isoform CRA_a; Uncharacterized protein; cDNA FLJ50015 |
| chr8_+_104152922 | 0.40 |
ENST00000309982.5
ENST00000438105.2 ENST00000297574.6 |
BAALC
|
brain and acute leukemia, cytoplasmic |
| chr2_+_98703595 | 0.40 |
ENST00000435344.1
|
VWA3B
|
von Willebrand factor A domain containing 3B |
| chr5_-_22853429 | 0.39 |
ENST00000504376.2
|
CDH12
|
cadherin 12, type 2 (N-cadherin 2) |
| chr14_+_78266408 | 0.39 |
ENST00000238561.5
|
ADCK1
|
aarF domain containing kinase 1 |
| chr21_+_45138941 | 0.38 |
ENST00000398081.1
ENST00000468090.1 ENST00000291565.4 |
PDXK
|
pyridoxal (pyridoxine, vitamin B6) kinase |
| chr7_-_120497178 | 0.38 |
ENST00000441017.1
ENST00000424710.1 ENST00000433758.1 |
TSPAN12
|
tetraspanin 12 |
| chr16_-_82203780 | 0.38 |
ENST00000563504.1
ENST00000569021.1 ENST00000258169.4 |
MPHOSPH6
|
M-phase phosphoprotein 6 |
| chr10_+_24528108 | 0.38 |
ENST00000438429.1
|
KIAA1217
|
KIAA1217 |
| chr11_+_18343800 | 0.38 |
ENST00000453096.2
|
GTF2H1
|
general transcription factor IIH, polypeptide 1, 62kDa |
| chr3_+_184032283 | 0.38 |
ENST00000346169.2
ENST00000414031.1 |
EIF4G1
|
eukaryotic translation initiation factor 4 gamma, 1 |
| chr8_+_15397732 | 0.37 |
ENST00000382020.4
ENST00000506802.1 ENST00000509380.1 ENST00000503731.1 |
TUSC3
|
tumor suppressor candidate 3 |
| chr14_+_32963433 | 0.37 |
ENST00000554410.1
|
AKAP6
|
A kinase (PRKA) anchor protein 6 |
| chr5_-_16465901 | 0.37 |
ENST00000308683.2
|
ZNF622
|
zinc finger protein 622 |
| chr21_+_31768348 | 0.36 |
ENST00000355459.2
|
KRTAP13-1
|
keratin associated protein 13-1 |
| chr2_-_27886460 | 0.36 |
ENST00000404798.2
ENST00000405491.1 ENST00000464789.2 ENST00000406540.1 |
SUPT7L
|
suppressor of Ty 7 (S. cerevisiae)-like |
| chr3_-_145878954 | 0.36 |
ENST00000282903.5
ENST00000360060.3 |
PLOD2
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 2 |
| chr6_+_147091575 | 0.36 |
ENST00000326916.8
ENST00000470716.2 ENST00000367488.1 |
ADGB
|
androglobin |
| chr17_-_41856305 | 0.35 |
ENST00000397937.2
ENST00000226004.3 |
DUSP3
|
dual specificity phosphatase 3 |
| chr6_+_140175987 | 0.35 |
ENST00000414038.1
ENST00000431609.1 |
RP5-899B16.1
|
RP5-899B16.1 |
| chr19_-_39390440 | 0.35 |
ENST00000249396.7
ENST00000414941.1 ENST00000392081.2 |
SIRT2
|
sirtuin 2 |
| chr14_-_80678512 | 0.35 |
ENST00000553968.1
|
DIO2
|
deiodinase, iodothyronine, type II |
Network of associatons between targets according to the STRING database.
First level regulatory network of HBP1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 3.8 | GO:2001248 | nitrogen catabolite regulation of transcription from RNA polymerase II promoter(GO:0001079) nitrogen catabolite activation of transcription from RNA polymerase II promoter(GO:0001080) regulation of urea metabolic process(GO:0034255) intracellular bile acid receptor signaling pathway(GO:0038185) interleukin-17 secretion(GO:0072615) nitrogen catabolite regulation of transcription(GO:0090293) nitrogen catabolite activation of transcription(GO:0090294) regulation of nitrogen cycle metabolic process(GO:1903314) positive regulation of glutamate metabolic process(GO:2000213) regulation of ammonia assimilation cycle(GO:2001248) positive regulation of ammonia assimilation cycle(GO:2001250) |
| 0.5 | 2.0 | GO:0019520 | aldonic acid metabolic process(GO:0019520) D-gluconate metabolic process(GO:0019521) |
| 0.5 | 1.4 | GO:0042450 | arginine biosynthetic process via ornithine(GO:0042450) |
| 0.5 | 1.4 | GO:0014016 | neuroblast differentiation(GO:0014016) |
| 0.4 | 2.8 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.4 | 1.1 | GO:0060304 | regulation of phosphatidylinositol dephosphorylation(GO:0060304) |
| 0.3 | 1.0 | GO:0051758 | homologous chromosome movement towards spindle pole involved in homologous chromosome segregation(GO:0051758) |
| 0.3 | 1.3 | GO:0019074 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.3 | 2.0 | GO:0010751 | negative regulation of nitric oxide mediated signal transduction(GO:0010751) |
| 0.3 | 1.2 | GO:0009822 | alkaloid catabolic process(GO:0009822) |
| 0.3 | 0.8 | GO:0070446 | cellular response to caloric restriction(GO:0061433) negative regulation of oligodendrocyte progenitor proliferation(GO:0070446) |
| 0.3 | 4.6 | GO:1901748 | leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.2 | 0.7 | GO:0040040 | thermosensory behavior(GO:0040040) |
| 0.2 | 2.8 | GO:0038044 | transforming growth factor-beta secretion(GO:0038044) |
| 0.2 | 1.6 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.2 | 1.4 | GO:0097029 | mature conventional dendritic cell differentiation(GO:0097029) |
| 0.2 | 1.1 | GO:0048867 | ganglion mother cell fate determination(GO:0007402) stem cell fate determination(GO:0048867) |
| 0.2 | 0.7 | GO:0000921 | septin ring assembly(GO:0000921) septin ring organization(GO:0031106) |
| 0.2 | 0.8 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
| 0.2 | 1.1 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 0.2 | 1.0 | GO:0019249 | lactate biosynthetic process(GO:0019249) regulation of amino acid uptake involved in synaptic transmission(GO:0051941) regulation of glutamate uptake involved in transmission of nerve impulse(GO:0051946) regulation of L-glutamate import(GO:1900920) |
| 0.2 | 0.7 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.1 | 0.6 | GO:1902161 | positive regulation of cyclic nucleotide-gated ion channel activity(GO:1902161) |
| 0.1 | 7.7 | GO:0061049 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.1 | 0.7 | GO:0019242 | methylglyoxal biosynthetic process(GO:0019242) |
| 0.1 | 1.4 | GO:0046940 | nucleoside monophosphate phosphorylation(GO:0046940) |
| 0.1 | 0.4 | GO:0046356 | acetyl-CoA catabolic process(GO:0046356) |
| 0.1 | 2.2 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.1 | 2.2 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.1 | 0.4 | GO:0009443 | pyridoxal 5'-phosphate salvage(GO:0009443) |
| 0.1 | 1.9 | GO:0034378 | chylomicron assembly(GO:0034378) |
| 0.1 | 0.3 | GO:0001798 | positive regulation of type IIa hypersensitivity(GO:0001798) positive regulation of type II hypersensitivity(GO:0002894) |
| 0.1 | 1.1 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.1 | 0.8 | GO:1902998 | regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 0.1 | 0.5 | GO:1901382 | chorionic trophoblast cell proliferation(GO:0097360) regulation of chorionic trophoblast cell proliferation(GO:1901382) |
| 0.1 | 1.0 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.1 | 0.8 | GO:1903764 | regulation of potassium ion export across plasma membrane(GO:1903764) |
| 0.1 | 0.4 | GO:0046946 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.1 | 2.1 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.1 | 1.2 | GO:0051599 | response to hydrostatic pressure(GO:0051599) |
| 0.1 | 0.3 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.1 | 0.8 | GO:0051177 | meiotic sister chromatid cohesion(GO:0051177) |
| 0.1 | 4.3 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.1 | 0.9 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.1 | 1.2 | GO:0015747 | urate transport(GO:0015747) |
| 0.1 | 1.2 | GO:0006451 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.1 | 0.4 | GO:0044789 | modulation by host of viral release from host cell(GO:0044789) positive regulation by host of viral release from host cell(GO:0044791) |
| 0.1 | 0.3 | GO:0031508 | pericentric heterochromatin assembly(GO:0031508) |
| 0.1 | 1.4 | GO:1904714 | regulation of chaperone-mediated autophagy(GO:1904714) |
| 0.1 | 1.0 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.1 | 0.2 | GO:0006480 | N-terminal protein amino acid methylation(GO:0006480) |
| 0.1 | 0.5 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.1 | 1.0 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.1 | 0.5 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 0.1 | 0.7 | GO:2000507 | positive regulation of energy homeostasis(GO:2000507) |
| 0.0 | 1.2 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 0.2 | GO:0002215 | defense response to nematode(GO:0002215) |
| 0.0 | 1.0 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 1.5 | GO:0002021 | response to dietary excess(GO:0002021) |
| 0.0 | 2.2 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.0 | 0.2 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.0 | 1.0 | GO:0034656 | nucleobase-containing small molecule catabolic process(GO:0034656) |
| 0.0 | 0.5 | GO:0052805 | histidine catabolic process(GO:0006548) imidazole-containing compound catabolic process(GO:0052805) |
| 0.0 | 1.1 | GO:0031915 | positive regulation of synaptic plasticity(GO:0031915) |
| 0.0 | 0.6 | GO:0021860 | pyramidal neuron development(GO:0021860) |
| 0.0 | 0.2 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.0 | 1.7 | GO:0005980 | glycogen catabolic process(GO:0005980) |
| 0.0 | 0.9 | GO:0010592 | positive regulation of lamellipodium assembly(GO:0010592) |
| 0.0 | 2.0 | GO:0048713 | regulation of oligodendrocyte differentiation(GO:0048713) |
| 0.0 | 0.6 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.0 | 0.1 | GO:0006868 | glutamine transport(GO:0006868) |
| 0.0 | 0.5 | GO:0032494 | response to peptidoglycan(GO:0032494) |
| 0.0 | 0.3 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.0 | 0.5 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 0.0 | 0.5 | GO:2000786 | positive regulation of autophagosome assembly(GO:2000786) |
| 0.0 | 0.2 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.0 | 0.9 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.0 | 0.6 | GO:0007351 | tripartite regional subdivision(GO:0007351) anterior/posterior axis specification, embryo(GO:0008595) |
| 0.0 | 0.3 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 1.8 | GO:0050690 | regulation of defense response to virus by virus(GO:0050690) |
| 0.0 | 1.8 | GO:2000249 | regulation of actin cytoskeleton reorganization(GO:2000249) |
| 0.0 | 0.4 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.0 | 0.6 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.0 | 0.6 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.0 | 0.4 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.0 | 0.8 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 0.5 | GO:2000774 | positive regulation of cellular senescence(GO:2000774) |
| 0.0 | 1.0 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.3 | GO:0008612 | peptidyl-lysine modification to peptidyl-hypusine(GO:0008612) |
| 0.0 | 0.5 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.0 | 0.4 | GO:0048706 | embryonic skeletal system development(GO:0048706) |
| 0.0 | 1.0 | GO:0010765 | positive regulation of sodium ion transport(GO:0010765) |
| 0.0 | 0.1 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.0 | 0.7 | GO:0043090 | amino acid import(GO:0043090) |
| 0.0 | 0.3 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.0 | 0.5 | GO:0090190 | positive regulation of branching involved in ureteric bud morphogenesis(GO:0090190) |
| 0.0 | 0.3 | GO:2001214 | positive regulation of vasculogenesis(GO:2001214) |
| 0.0 | 0.1 | GO:1900264 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.0 | 0.6 | GO:0022400 | regulation of rhodopsin mediated signaling pathway(GO:0022400) |
| 0.0 | 0.2 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.0 | 0.7 | GO:0042407 | cristae formation(GO:0042407) |
| 0.0 | 1.9 | GO:0032392 | DNA geometric change(GO:0032392) |
| 0.0 | 0.7 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.1 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.0 | 0.4 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 | 0.1 | GO:1903976 | negative regulation of glial cell migration(GO:1903976) |
| 0.0 | 1.0 | GO:0030317 | sperm motility(GO:0030317) |
| 0.0 | 0.2 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.0 | 0.6 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.8 | GO:0034685 | integrin alphav-beta6 complex(GO:0034685) |
| 0.4 | 1.7 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.1 | 1.1 | GO:0060200 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.1 | 1.1 | GO:0070033 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) synaptobrevin 2-SNAP-25-syntaxin-3-complexin complex(GO:0070554) |
| 0.1 | 1.6 | GO:0043219 | lateral loop(GO:0043219) |
| 0.1 | 0.6 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.1 | 1.4 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.1 | 0.7 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.1 | 3.8 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.1 | 0.5 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.1 | 0.2 | GO:0000438 | core TFIIH complex portion of holo TFIIH complex(GO:0000438) |
| 0.1 | 0.6 | GO:0000836 | Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.1 | 1.7 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.1 | 2.1 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.1 | 0.7 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.0 | 0.8 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.0 | 0.4 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.0 | 0.5 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 0.6 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.0 | 0.3 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.4 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.0 | 2.0 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 0.3 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.0 | 0.6 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.0 | 1.8 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 4.3 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 6.7 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.4 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 1.1 | GO:0044298 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 0.7 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.3 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.0 | 1.6 | GO:0030286 | dynein complex(GO:0030286) |
| 0.0 | 0.5 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.0 | 0.4 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.0 | 0.4 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.9 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 1.5 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.2 | GO:0030681 | multimeric ribonuclease P complex(GO:0030681) |
| 0.0 | 2.6 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 0.1 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.0 | 0.4 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 1.7 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.0 | 0.8 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.4 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.0 | 0.5 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 5.6 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.3 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.7 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 1.4 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.0 | 2.0 | GO:0005882 | intermediate filament(GO:0005882) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 3.8 | GO:1902122 | chenodeoxycholic acid binding(GO:1902122) |
| 0.3 | 1.7 | GO:0008466 | glycogenin glucosyltransferase activity(GO:0008466) |
| 0.3 | 2.2 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.3 | 1.4 | GO:0033265 | acetylcholinesterase activity(GO:0003990) choline binding(GO:0033265) |
| 0.3 | 0.8 | GO:0046970 | NAD-dependent histone deacetylase activity (H4-K16 specific)(GO:0046970) |
| 0.3 | 4.6 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.2 | 1.0 | GO:0003978 | UDP-N-acetylglucosamine 4-epimerase activity(GO:0003974) UDP-glucose 4-epimerase activity(GO:0003978) |
| 0.2 | 2.2 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.2 | 1.4 | GO:0046899 | nucleoside triphosphate adenylate kinase activity(GO:0046899) |
| 0.2 | 1.1 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.2 | 1.2 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.2 | 0.6 | GO:0015275 | stretch-activated, cation-selective, calcium channel activity(GO:0015275) |
| 0.2 | 4.3 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.2 | 7.3 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.2 | 1.6 | GO:0099529 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.2 | 1.2 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.1 | 0.6 | GO:0005260 | channel-conductance-controlling ATPase activity(GO:0005260) |
| 0.1 | 1.4 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.1 | 0.7 | GO:0004347 | glucose-6-phosphate isomerase activity(GO:0004347) |
| 0.1 | 1.1 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.1 | 2.0 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.1 | 0.5 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.1 | 0.4 | GO:0008478 | pyridoxal kinase activity(GO:0008478) |
| 0.1 | 1.0 | GO:0045134 | uridine-diphosphatase activity(GO:0045134) |
| 0.1 | 1.3 | GO:0004075 | biotin carboxylase activity(GO:0004075) biotin binding(GO:0009374) |
| 0.1 | 1.1 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.1 | 0.4 | GO:0033823 | procollagen-lysine 5-dioxygenase activity(GO:0008475) procollagen glucosyltransferase activity(GO:0033823) |
| 0.1 | 0.4 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.1 | 1.0 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.1 | 1.8 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.1 | 0.9 | GO:0050694 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.1 | 0.5 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.1 | 0.4 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.1 | 1.2 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.1 | 0.5 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.1 | 2.2 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.1 | 0.5 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.1 | 0.8 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.1 | 0.2 | GO:0071885 | N-terminal protein N-methyltransferase activity(GO:0071885) |
| 0.1 | 0.7 | GO:0022820 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.1 | 0.8 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.1 | 0.5 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 1.0 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.3 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.0 | 1.0 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.0 | 0.3 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 1.3 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 0.1 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.0 | 0.6 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 0.5 | GO:0016594 | glycine binding(GO:0016594) |
| 0.0 | 0.3 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 0.9 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 0.3 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.0 | 0.7 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.0 | 0.6 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 0.7 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.8 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.1 | GO:0052740 | 1-acyl-2-lysophosphatidylserine acylhydrolase activity(GO:0052740) |
| 0.0 | 1.1 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 1.3 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 3.8 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.1 | GO:0030350 | iron-responsive element binding(GO:0030350) |
| 0.0 | 0.6 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.3 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.0 | 0.5 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 0.4 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 2.8 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.0 | 0.2 | GO:0050294 | steroid sulfotransferase activity(GO:0050294) |
| 0.0 | 0.7 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 0.6 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.4 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.2 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.4 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.2 | GO:0005221 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.0 | 0.6 | GO:0008409 | 5'-3' exonuclease activity(GO:0008409) |
| 0.0 | 0.5 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.0 | 0.6 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 0.1 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.0 | 0.4 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.1 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.0 | 0.5 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.0 | 0.1 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.2 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.0 | 1.2 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.0 | 0.3 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.8 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.1 | 3.8 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 1.0 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.8 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.3 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.0 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.1 | 2.0 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.1 | 4.3 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.1 | 1.2 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.1 | 1.9 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.1 | 1.4 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.1 | 2.1 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.1 | 1.7 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 1.1 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.0 | 1.2 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.8 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.5 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 1.2 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 1.0 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 2.3 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 0.7 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 1.0 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 1.1 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 1.0 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 1.3 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.0 | 0.7 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.6 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 1.0 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.6 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 0.5 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.5 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 0.3 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.0 | 0.9 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.4 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.5 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |