Project
Illumina Body Map 2: averaged replicates
Navigation
Downloads
Results for HES7_HES5
Z-value: 1.25
Transcription factors associated with HES7_HES5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
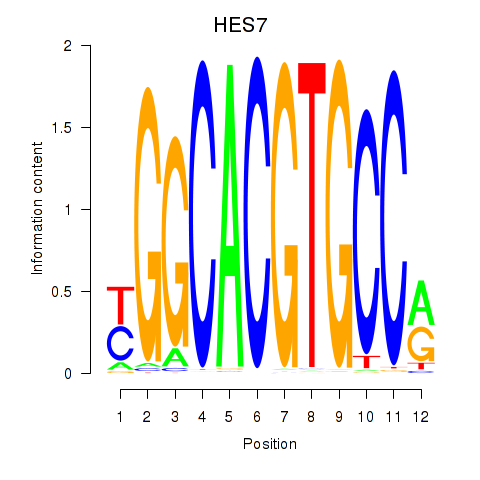
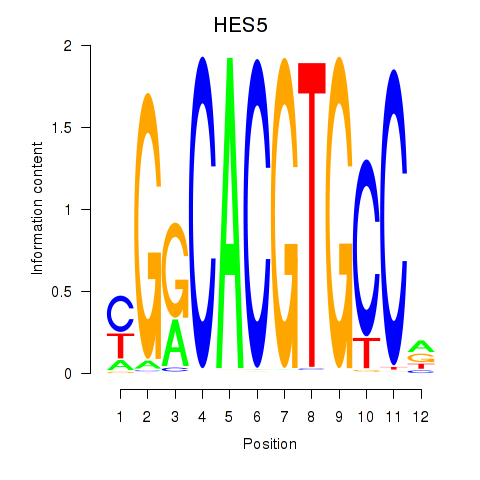
HES7
|
ENSG00000179111.4 | hes family bHLH transcription factor 7 |
|
HES5
|
ENSG00000197921.5 | hes family bHLH transcription factor 5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HES7 | hg19_v2_chr17_-_8027402_8027432 | 0.64 | 7.1e-05 | Click! |
| HES5 | hg19_v2_chr1_-_2461684_2461710 | 0.39 | 2.9e-02 | Click! |
Activity profile of HES7_HES5 motif
Sorted Z-values of HES7_HES5 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_26233423 | 4.01 |
ENST00000357865.2
|
STMN1
|
stathmin 1 |
| chr14_-_77737543 | 3.61 |
ENST00000298352.4
|
NGB
|
neuroglobin |
| chr19_+_4304585 | 3.54 |
ENST00000221856.6
|
FSD1
|
fibronectin type III and SPRY domain containing 1 |
| chr19_+_4304632 | 3.53 |
ENST00000597590.1
|
FSD1
|
fibronectin type III and SPRY domain containing 1 |
| chr1_-_26232951 | 3.51 |
ENST00000426559.2
ENST00000455785.2 |
STMN1
|
stathmin 1 |
| chr1_-_26232522 | 3.35 |
ENST00000399728.1
|
STMN1
|
stathmin 1 |
| chr1_+_16083098 | 2.99 |
ENST00000496928.2
ENST00000508310.1 |
FBLIM1
|
filamin binding LIM protein 1 |
| chr1_+_16083154 | 2.63 |
ENST00000375771.1
|
FBLIM1
|
filamin binding LIM protein 1 |
| chr4_-_6202291 | 2.48 |
ENST00000282924.5
|
JAKMIP1
|
janus kinase and microtubule interacting protein 1 |
| chr1_+_16083123 | 2.41 |
ENST00000510393.1
ENST00000430076.1 |
FBLIM1
|
filamin binding LIM protein 1 |
| chr19_-_19739007 | 2.33 |
ENST00000586703.1
ENST00000591042.1 ENST00000407877.3 |
LPAR2
|
lysophosphatidic acid receptor 2 |
| chr5_+_152870106 | 2.17 |
ENST00000285900.5
|
GRIA1
|
glutamate receptor, ionotropic, AMPA 1 |
| chr2_-_132919528 | 2.11 |
ENST00000409867.1
|
ANKRD30BL
|
ankyrin repeat domain 30B-like |
| chr4_-_6202247 | 2.08 |
ENST00000409021.3
ENST00000409371.3 |
JAKMIP1
|
janus kinase and microtubule interacting protein 1 |
| chr17_-_42907564 | 2.08 |
ENST00000592524.1
|
GJC1
|
gap junction protein, gamma 1, 45kDa |
| chr1_+_210111534 | 2.04 |
ENST00000422431.1
ENST00000534859.1 ENST00000399639.2 ENST00000537238.1 |
SYT14
|
synaptotagmin XIV |
| chr8_+_104383728 | 1.94 |
ENST00000330295.5
|
CTHRC1
|
collagen triple helix repeat containing 1 |
| chr2_-_219925189 | 1.85 |
ENST00000295731.6
|
IHH
|
indian hedgehog |
| chr19_+_4304685 | 1.84 |
ENST00000601006.1
|
FSD1
|
fibronectin type III and SPRY domain containing 1 |
| chr6_+_122931366 | 1.76 |
ENST00000368452.2
ENST00000368448.1 ENST00000392490.1 |
PKIB
|
protein kinase (cAMP-dependent, catalytic) inhibitor beta |
| chr5_+_152870215 | 1.68 |
ENST00000518142.1
|
GRIA1
|
glutamate receptor, ionotropic, AMPA 1 |
| chr12_-_91348949 | 1.61 |
ENST00000358859.2
|
CCER1
|
coiled-coil glutamate-rich protein 1 |
| chr20_-_2821271 | 1.57 |
ENST00000448755.1
ENST00000360652.2 |
PCED1A
|
PC-esterase domain containing 1A |
| chr1_-_109655377 | 1.57 |
ENST00000369948.3
|
C1orf194
|
chromosome 1 open reading frame 194 |
| chr5_-_54529415 | 1.55 |
ENST00000282572.4
|
CCNO
|
cyclin O |
| chr11_+_60197040 | 1.54 |
ENST00000300190.2
|
MS4A5
|
membrane-spanning 4-domains, subfamily A, member 5 |
| chr2_-_27486951 | 1.50 |
ENST00000432351.1
|
SLC30A3
|
solute carrier family 30 (zinc transporter), member 3 |
| chr1_+_28261621 | 1.49 |
ENST00000549094.1
|
SMPDL3B
|
sphingomyelin phosphodiesterase, acid-like 3B |
| chr14_+_65879437 | 1.49 |
ENST00000394585.1
|
FUT8
|
fucosyltransferase 8 (alpha (1,6) fucosyltransferase) |
| chr5_+_152870287 | 1.47 |
ENST00000340592.5
|
GRIA1
|
glutamate receptor, ionotropic, AMPA 1 |
| chr4_-_144940477 | 1.43 |
ENST00000513128.1
ENST00000429670.2 ENST00000502664.1 |
GYPB
|
glycophorin B (MNS blood group) |
| chr20_+_45523227 | 1.37 |
ENST00000327619.5
ENST00000357410.3 |
EYA2
|
eyes absent homolog 2 (Drosophila) |
| chr14_+_104689918 | 1.37 |
ENST00000553757.1
ENST00000556528.1 ENST00000555282.1 |
RP11-260M19.2
|
RP11-260M19.2 |
| chr1_+_28261492 | 1.35 |
ENST00000373894.3
|
SMPDL3B
|
sphingomyelin phosphodiesterase, acid-like 3B |
| chr5_+_143550396 | 1.32 |
ENST00000512467.1
|
KCTD16
|
potassium channel tetramerization domain containing 16 |
| chr19_+_6135646 | 1.32 |
ENST00000588304.1
ENST00000588485.1 ENST00000588722.1 ENST00000591403.1 ENST00000586696.1 ENST00000589401.1 ENST00000252669.5 |
ACSBG2
|
acyl-CoA synthetase bubblegum family member 2 |
| chr3_-_189840223 | 1.27 |
ENST00000427335.2
|
LEPREL1
|
leprecan-like 1 |
| chr1_+_28261533 | 1.26 |
ENST00000411604.1
ENST00000373888.4 |
SMPDL3B
|
sphingomyelin phosphodiesterase, acid-like 3B |
| chrX_+_152224766 | 1.25 |
ENST00000370265.4
ENST00000447306.1 |
PNMA3
|
paraneoplastic Ma antigen 3 |
| chr13_+_35516390 | 1.19 |
ENST00000540320.1
ENST00000400445.3 ENST00000310336.4 |
NBEA
|
neurobeachin |
| chr14_+_65879668 | 1.19 |
ENST00000553924.1
ENST00000358307.2 ENST00000557338.1 ENST00000554610.1 |
FUT8
|
fucosyltransferase 8 (alpha (1,6) fucosyltransferase) |
| chr2_+_27665232 | 1.18 |
ENST00000543753.1
ENST00000288873.3 |
KRTCAP3
|
keratinocyte associated protein 3 |
| chr11_+_60197069 | 1.16 |
ENST00000528905.1
ENST00000528093.1 |
MS4A5
|
membrane-spanning 4-domains, subfamily A, member 5 |
| chr1_+_210111570 | 1.16 |
ENST00000367019.1
ENST00000472886.1 |
SYT14
|
synaptotagmin XIV |
| chr16_+_222846 | 1.15 |
ENST00000251595.6
ENST00000397806.1 |
HBA2
|
hemoglobin, alpha 2 |
| chrX_-_16887963 | 1.14 |
ENST00000380084.4
|
RBBP7
|
retinoblastoma binding protein 7 |
| chr17_-_42908155 | 1.09 |
ENST00000426548.1
ENST00000590758.1 ENST00000591424.1 |
GJC1
|
gap junction protein, gamma 1, 45kDa |
| chr7_-_73184588 | 1.08 |
ENST00000395145.2
|
CLDN3
|
claudin 3 |
| chr11_-_26593649 | 1.07 |
ENST00000455601.2
|
MUC15
|
mucin 15, cell surface associated |
| chr4_-_144826682 | 1.01 |
ENST00000358615.4
ENST00000437468.2 |
GYPE
|
glycophorin E (MNS blood group) |
| chr12_-_123728548 | 1.00 |
ENST00000545406.1
|
MPHOSPH9
|
M-phase phosphoprotein 9 |
| chrX_-_48326764 | 0.98 |
ENST00000413668.1
ENST00000441948.1 |
SLC38A5
|
solute carrier family 38, member 5 |
| chr4_+_76439649 | 0.98 |
ENST00000507557.1
|
THAP6
|
THAP domain containing 6 |
| chr6_+_13182751 | 0.97 |
ENST00000415087.1
|
PHACTR1
|
phosphatase and actin regulator 1 |
| chr9_-_23821842 | 0.97 |
ENST00000544538.1
|
ELAVL2
|
ELAV like neuron-specific RNA binding protein 2 |
| chr16_-_5115913 | 0.95 |
ENST00000474471.3
|
C16orf89
|
chromosome 16 open reading frame 89 |
| chr20_+_44650348 | 0.94 |
ENST00000454036.2
|
SLC12A5
|
solute carrier family 12 (potassium/chloride transporter), member 5 |
| chr1_+_166958504 | 0.93 |
ENST00000447624.1
|
MAEL
|
maelstrom spermatogenic transposon silencer |
| chr3_+_39851094 | 0.93 |
ENST00000302541.6
|
MYRIP
|
myosin VIIA and Rab interacting protein |
| chr1_+_166958497 | 0.91 |
ENST00000367870.2
|
MAEL
|
maelstrom spermatogenic transposon silencer |
| chr12_-_10320256 | 0.90 |
ENST00000538745.1
|
OLR1
|
oxidized low density lipoprotein (lectin-like) receptor 1 |
| chr17_+_30813576 | 0.90 |
ENST00000313401.3
|
CDK5R1
|
cyclin-dependent kinase 5, regulatory subunit 1 (p35) |
| chr22_+_30752963 | 0.89 |
ENST00000445005.1
ENST00000430839.1 |
CCDC157
|
coiled-coil domain containing 157 |
| chr19_-_55458860 | 0.88 |
ENST00000592784.1
ENST00000448121.2 ENST00000340844.2 |
NLRP7
|
NLR family, pyrin domain containing 7 |
| chrX_+_134124968 | 0.86 |
ENST00000330288.4
|
SMIM10
|
small integral membrane protein 10 |
| chr14_-_106967788 | 0.86 |
ENST00000390622.2
|
IGHV1-46
|
immunoglobulin heavy variable 1-46 |
| chr14_+_94385235 | 0.86 |
ENST00000557719.1
ENST00000267594.5 |
FAM181A
|
family with sequence similarity 181, member A |
| chr1_+_44457441 | 0.84 |
ENST00000466180.1
|
CCDC24
|
coiled-coil domain containing 24 |
| chr14_+_24641062 | 0.82 |
ENST00000311457.3
ENST00000557806.1 ENST00000559919.1 |
REC8
|
REC8 meiotic recombination protein |
| chr5_+_78532003 | 0.80 |
ENST00000396137.4
|
JMY
|
junction mediating and regulatory protein, p53 cofactor |
| chr2_+_27665289 | 0.78 |
ENST00000407293.1
|
KRTCAP3
|
keratinocyte associated protein 3 |
| chr4_+_94750014 | 0.77 |
ENST00000306011.3
|
ATOH1
|
atonal homolog 1 (Drosophila) |
| chr1_-_201476220 | 0.76 |
ENST00000526723.1
ENST00000524951.1 |
CSRP1
|
cysteine and glycine-rich protein 1 |
| chr8_+_104383759 | 0.71 |
ENST00000415886.2
|
CTHRC1
|
collagen triple helix repeat containing 1 |
| chr19_-_3700388 | 0.70 |
ENST00000589578.1
ENST00000537021.1 ENST00000539785.1 ENST00000335312.3 |
PIP5K1C
|
phosphatidylinositol-4-phosphate 5-kinase, type I, gamma |
| chr4_+_2965307 | 0.70 |
ENST00000398051.4
ENST00000503518.2 ENST00000398052.4 ENST00000345167.6 ENST00000504933.1 ENST00000442472.2 |
GRK4
|
G protein-coupled receptor kinase 4 |
| chr1_-_166944652 | 0.69 |
ENST00000528703.1
ENST00000525740.1 ENST00000529387.1 ENST00000469934.2 ENST00000529071.1 ENST00000526687.1 |
ILDR2
|
immunoglobulin-like domain containing receptor 2 |
| chr1_-_166944561 | 0.65 |
ENST00000271417.3
|
ILDR2
|
immunoglobulin-like domain containing receptor 2 |
| chr2_-_232328867 | 0.65 |
ENST00000453992.1
ENST00000417652.1 ENST00000454824.1 |
NCL
|
nucleolin |
| chr2_-_175870085 | 0.64 |
ENST00000409156.3
|
CHN1
|
chimerin 1 |
| chr11_-_26593677 | 0.63 |
ENST00000527569.1
|
MUC15
|
mucin 15, cell surface associated |
| chr4_-_145061788 | 0.63 |
ENST00000512064.1
ENST00000512789.1 ENST00000504786.1 ENST00000503627.1 ENST00000535709.1 ENST00000324022.10 ENST00000360771.4 ENST00000283126.7 |
GYPA
GYPB
|
glycophorin A (MNS blood group) glycophorin B (MNS blood group) |
| chr1_+_179561011 | 0.62 |
ENST00000294848.8
ENST00000444136.1 |
TDRD5
|
tudor domain containing 5 |
| chr21_+_42694732 | 0.61 |
ENST00000398646.3
|
FAM3B
|
family with sequence similarity 3, member B |
| chr2_+_86116396 | 0.61 |
ENST00000455121.3
|
AC105053.4
|
AC105053.4 |
| chrX_-_48326683 | 0.60 |
ENST00000440085.1
|
SLC38A5
|
solute carrier family 38, member 5 |
| chr13_+_115047097 | 0.59 |
ENST00000351487.5
|
UPF3A
|
UPF3 regulator of nonsense transcripts homolog A (yeast) |
| chr14_+_65878650 | 0.59 |
ENST00000555559.1
|
FUT8
|
fucosyltransferase 8 (alpha (1,6) fucosyltransferase) |
| chr16_+_333152 | 0.58 |
ENST00000219406.6
ENST00000404312.1 ENST00000456379.1 |
PDIA2
|
protein disulfide isomerase family A, member 2 |
| chr20_+_36012051 | 0.57 |
ENST00000373567.2
|
SRC
|
v-src avian sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog |
| chr1_-_201476274 | 0.54 |
ENST00000340006.2
|
CSRP1
|
cysteine and glycine-rich protein 1 |
| chr2_+_25264933 | 0.53 |
ENST00000401432.3
ENST00000403714.3 |
EFR3B
|
EFR3 homolog B (S. cerevisiae) |
| chr9_+_100174344 | 0.52 |
ENST00000422139.2
|
TDRD7
|
tudor domain containing 7 |
| chr22_-_50523760 | 0.52 |
ENST00000395876.2
|
MLC1
|
megalencephalic leukoencephalopathy with subcortical cysts 1 |
| chr4_+_6202448 | 0.51 |
ENST00000508601.1
|
RP11-586D19.1
|
RP11-586D19.1 |
| chr4_+_9446156 | 0.51 |
ENST00000334879.1
|
DEFB131
|
defensin, beta 131 |
| chr9_+_103204553 | 0.51 |
ENST00000502978.1
ENST00000334943.6 |
MSANTD3-TMEFF1
TMEFF1
|
MSANTD3-TMEFF1 readthrough transmembrane protein with EGF-like and two follistatin-like domains 1 |
| chr3_+_38537763 | 0.51 |
ENST00000287675.5
ENST00000358249.2 ENST00000422077.2 |
EXOG
|
endo/exonuclease (5'-3'), endonuclease G-like |
| chr7_+_99202003 | 0.50 |
ENST00000609449.1
|
GS1-259H13.2
|
GS1-259H13.2 |
| chr2_-_86564776 | 0.50 |
ENST00000165698.5
ENST00000541910.1 ENST00000535845.1 |
REEP1
|
receptor accessory protein 1 |
| chr19_-_53632925 | 0.49 |
ENST00000595174.1
|
ZNF415
|
zinc finger protein 415 |
| chr22_+_42095497 | 0.48 |
ENST00000401548.3
ENST00000540833.1 ENST00000400107.1 ENST00000300398.4 |
MEI1
|
meiosis inhibitor 1 |
| chr4_-_76439483 | 0.48 |
ENST00000380840.2
ENST00000513257.1 ENST00000507014.1 |
RCHY1
|
ring finger and CHY zinc finger domain containing 1, E3 ubiquitin protein ligase |
| chr7_-_130598059 | 0.47 |
ENST00000432045.2
|
MIR29B1
|
microRNA 29a |
| chr1_-_1293904 | 0.46 |
ENST00000309212.6
ENST00000342753.4 ENST00000445648.2 |
MXRA8
|
matrix-remodelling associated 8 |
| chr15_+_68871569 | 0.46 |
ENST00000566799.1
|
CORO2B
|
coronin, actin binding protein, 2B |
| chrX_-_16888276 | 0.46 |
ENST00000493145.1
|
RBBP7
|
retinoblastoma binding protein 7 |
| chr19_-_55954230 | 0.46 |
ENST00000376325.4
|
SHISA7
|
shisa family member 7 |
| chr14_+_77647966 | 0.45 |
ENST00000554766.1
|
TMEM63C
|
transmembrane protein 63C |
| chr14_+_65878565 | 0.45 |
ENST00000556518.1
ENST00000557164.1 |
FUT8
|
fucosyltransferase 8 (alpha (1,6) fucosyltransferase) |
| chr10_-_71169031 | 0.45 |
ENST00000373307.1
|
TACR2
|
tachykinin receptor 2 |
| chr17_-_79791118 | 0.45 |
ENST00000576431.1
ENST00000575061.1 ENST00000455127.2 ENST00000572645.1 ENST00000538396.1 ENST00000573478.1 |
FAM195B
|
family with sequence similarity 195, member B |
| chr2_+_175199674 | 0.44 |
ENST00000394967.2
|
SP9
|
Sp9 transcription factor |
| chr1_+_44457261 | 0.44 |
ENST00000372318.3
|
CCDC24
|
coiled-coil domain containing 24 |
| chr7_-_6570817 | 0.44 |
ENST00000435185.1
|
GRID2IP
|
glutamate receptor, ionotropic, delta 2 (Grid2) interacting protein |
| chr22_-_50523843 | 0.43 |
ENST00000535444.1
ENST00000431262.2 |
MLC1
|
megalencephalic leukoencephalopathy with subcortical cysts 1 |
| chr4_-_76439596 | 0.43 |
ENST00000451788.1
ENST00000512706.1 |
RCHY1
|
ring finger and CHY zinc finger domain containing 1, E3 ubiquitin protein ligase |
| chr19_+_58095501 | 0.43 |
ENST00000536878.2
ENST00000597850.1 ENST00000597219.1 ENST00000598689.1 ENST00000599456.1 ENST00000307468.4 |
ZIK1
|
zinc finger protein interacting with K protein 1 |
| chr1_+_45140360 | 0.42 |
ENST00000418644.1
ENST00000458657.2 ENST00000441519.1 ENST00000535358.1 ENST00000445071.1 |
C1orf228
|
chromosome 1 open reading frame 228 |
| chr5_+_115298165 | 0.41 |
ENST00000357872.4
|
AQPEP
|
Aminopeptidase Q |
| chr3_-_189839467 | 0.41 |
ENST00000426003.1
|
LEPREL1
|
leprecan-like 1 |
| chr22_+_23235872 | 0.40 |
ENST00000390320.2
|
IGLJ1
|
immunoglobulin lambda joining 1 |
| chr3_+_39851170 | 0.40 |
ENST00000425621.1
ENST00000396217.3 |
MYRIP
|
myosin VIIA and Rab interacting protein |
| chr1_+_150254936 | 0.39 |
ENST00000447007.1
ENST00000369095.1 ENST00000369094.1 |
C1orf51
|
chromosome 1 open reading frame 51 |
| chr12_-_6756559 | 0.38 |
ENST00000536350.1
ENST00000414226.2 ENST00000546114.1 |
ACRBP
|
acrosin binding protein |
| chrX_+_153524024 | 0.37 |
ENST00000369915.3
ENST00000217905.7 |
TKTL1
|
transketolase-like 1 |
| chr19_-_55953704 | 0.37 |
ENST00000416792.1
|
SHISA7
|
shisa family member 7 |
| chr4_+_76439665 | 0.37 |
ENST00000508105.1
ENST00000311638.3 ENST00000380837.3 ENST00000507556.1 ENST00000504190.1 ENST00000507885.1 ENST00000502620.1 ENST00000514480.1 |
THAP6
|
THAP domain containing 6 |
| chr2_-_220108309 | 0.36 |
ENST00000409640.1
|
GLB1L
|
galactosidase, beta 1-like |
| chr20_+_306221 | 0.35 |
ENST00000342665.2
|
SOX12
|
SRY (sex determining region Y)-box 12 |
| chr2_-_105372203 | 0.34 |
ENST00000424321.1
|
AC068057.2
|
long intergenic non-protein coding RNA 1114 |
| chr12_-_6756609 | 0.34 |
ENST00000229243.2
|
ACRBP
|
acrosin binding protein |
| chrX_-_153523462 | 0.33 |
ENST00000361930.3
ENST00000369926.1 |
TEX28
|
testis expressed 28 |
| chr17_-_41465674 | 0.33 |
ENST00000592135.1
ENST00000587874.1 ENST00000588654.1 ENST00000592094.1 |
LINC00910
|
long intergenic non-protein coding RNA 910 |
| chr14_-_67826538 | 0.32 |
ENST00000553687.1
|
ATP6V1D
|
ATPase, H+ transporting, lysosomal 34kDa, V1 subunit D |
| chr19_+_37837218 | 0.32 |
ENST00000591134.1
|
HKR1
|
HKR1, GLI-Kruppel zinc finger family member |
| chrX_-_16888448 | 0.32 |
ENST00000468092.1
ENST00000404022.1 ENST00000380087.2 |
RBBP7
|
retinoblastoma binding protein 7 |
| chr11_-_6440624 | 0.32 |
ENST00000311051.3
|
APBB1
|
amyloid beta (A4) precursor protein-binding, family B, member 1 (Fe65) |
| chr5_+_112227311 | 0.32 |
ENST00000391338.1
|
ZRSR1
|
zinc finger (CCCH type), RNA-binding motif and serine/arginine rich 1 |
| chr19_-_19739321 | 0.32 |
ENST00000588461.1
|
LPAR2
|
lysophosphatidic acid receptor 2 |
| chr11_+_134146635 | 0.30 |
ENST00000431683.2
|
GLB1L3
|
galactosidase, beta 1-like 3 |
| chr22_-_42310570 | 0.30 |
ENST00000457093.1
|
SHISA8
|
shisa family member 8 |
| chr6_+_46620676 | 0.30 |
ENST00000371347.5
ENST00000411689.2 |
SLC25A27
|
solute carrier family 25, member 27 |
| chr8_-_65711310 | 0.29 |
ENST00000310193.3
|
CYP7B1
|
cytochrome P450, family 7, subfamily B, polypeptide 1 |
| chr2_+_202655184 | 0.29 |
ENST00000410091.3
|
CDK15
|
cyclin-dependent kinase 15 |
| chr5_+_179125907 | 0.29 |
ENST00000247461.4
ENST00000452673.2 ENST00000502498.1 ENST00000507307.1 ENST00000513246.1 ENST00000502673.1 ENST00000506654.1 ENST00000512607.2 ENST00000510810.1 |
CANX
|
calnexin |
| chr11_+_64059464 | 0.28 |
ENST00000394525.2
|
KCNK4
|
potassium channel, subfamily K, member 4 |
| chr8_+_38089198 | 0.28 |
ENST00000528358.1
ENST00000529642.1 ENST00000532222.1 ENST00000520272.2 |
DDHD2
|
DDHD domain containing 2 |
| chr19_+_45281118 | 0.28 |
ENST00000270279.3
ENST00000341505.4 |
CBLC
|
Cbl proto-oncogene C, E3 ubiquitin protein ligase |
| chr14_-_81687197 | 0.28 |
ENST00000553612.1
|
GTF2A1
|
general transcription factor IIA, 1, 19/37kDa |
| chr1_-_147632428 | 0.28 |
ENST00000599640.1
|
BX842679.1
|
Uncharacterized protein FLJ46360 |
| chr7_+_148959262 | 0.27 |
ENST00000434415.1
|
ZNF783
|
zinc finger family member 783 |
| chr2_-_74648702 | 0.27 |
ENST00000518863.1
|
C2orf81
|
chromosome 2 open reading frame 81 |
| chr4_-_135248604 | 0.26 |
ENST00000515491.1
ENST00000504728.1 ENST00000506638.1 |
RP11-400D2.2
|
RP11-400D2.2 |
| chr4_+_113152881 | 0.26 |
ENST00000274000.5
|
AP1AR
|
adaptor-related protein complex 1 associated regulatory protein |
| chr2_+_201981119 | 0.26 |
ENST00000395148.2
|
CFLAR
|
CASP8 and FADD-like apoptosis regulator |
| chr19_-_36822551 | 0.24 |
ENST00000591372.1
|
LINC00665
|
long intergenic non-protein coding RNA 665 |
| chr4_+_113152978 | 0.24 |
ENST00000309703.6
|
AP1AR
|
adaptor-related protein complex 1 associated regulatory protein |
| chr11_+_6866883 | 0.24 |
ENST00000299454.4
ENST00000379831.2 |
OR10A5
|
olfactory receptor, family 10, subfamily A, member 5 |
| chr17_-_17109579 | 0.23 |
ENST00000321560.3
|
PLD6
|
phospholipase D family, member 6 |
| chr7_-_53879623 | 0.22 |
ENST00000380970.2
|
GS1-179L18.1
|
GS1-179L18.1 |
| chr18_+_11752040 | 0.22 |
ENST00000423027.3
|
GNAL
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide, olfactory type |
| chr20_+_306177 | 0.21 |
ENST00000544632.1
|
SOX12
|
SRY (sex determining region Y)-box 12 |
| chr2_-_169769787 | 0.20 |
ENST00000451987.1
|
SPC25
|
SPC25, NDC80 kinetochore complex component |
| chr11_+_9595180 | 0.19 |
ENST00000450114.2
|
WEE1
|
WEE1 G2 checkpoint kinase |
| chr11_-_777467 | 0.19 |
ENST00000397472.2
ENST00000524550.1 ENST00000319863.8 ENST00000526325.1 ENST00000442059.2 |
PDDC1
|
Parkinson disease 7 domain containing 1 |
| chrX_+_133594168 | 0.19 |
ENST00000298556.7
|
HPRT1
|
hypoxanthine phosphoribosyltransferase 1 |
| chr6_+_46620705 | 0.19 |
ENST00000452689.2
|
SLC25A27
|
solute carrier family 25, member 27 |
| chr6_-_114292449 | 0.18 |
ENST00000519065.1
|
HDAC2
|
histone deacetylase 2 |
| chr5_+_95066823 | 0.18 |
ENST00000506817.1
ENST00000379982.3 |
RHOBTB3
|
Rho-related BTB domain containing 3 |
| chr8_-_142012169 | 0.17 |
ENST00000517453.1
|
PTK2
|
protein tyrosine kinase 2 |
| chr11_-_119066545 | 0.17 |
ENST00000415318.1
|
CCDC153
|
coiled-coil domain containing 153 |
| chr5_+_115298145 | 0.17 |
ENST00000395528.2
|
AQPEP
|
Aminopeptidase Q |
| chr11_-_795170 | 0.16 |
ENST00000481290.1
|
SLC25A22
|
solute carrier family 25 (mitochondrial carrier: glutamate), member 22 |
| chr1_+_44445549 | 0.16 |
ENST00000356836.6
|
B4GALT2
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 2 |
| chr14_-_67826486 | 0.15 |
ENST00000555431.1
ENST00000554236.1 ENST00000555474.1 |
ATP6V1D
|
ATPase, H+ transporting, lysosomal 34kDa, V1 subunit D |
| chrX_-_153523796 | 0.14 |
ENST00000428625.1
|
TEX28
|
testis expressed 28 |
| chr2_+_113403434 | 0.14 |
ENST00000272542.3
|
SLC20A1
|
solute carrier family 20 (phosphate transporter), member 1 |
| chr4_-_52904425 | 0.14 |
ENST00000535450.1
|
SGCB
|
sarcoglycan, beta (43kDa dystrophin-associated glycoprotein) |
| chr13_+_80055284 | 0.14 |
ENST00000218652.7
|
NDFIP2
|
Nedd4 family interacting protein 2 |
| chr21_-_33985127 | 0.14 |
ENST00000290155.3
|
C21orf59
|
chromosome 21 open reading frame 59 |
| chr6_-_74161977 | 0.14 |
ENST00000370318.1
ENST00000370315.3 |
MB21D1
|
Mab-21 domain containing 1 |
| chr22_+_41777927 | 0.13 |
ENST00000266304.4
|
TEF
|
thyrotrophic embryonic factor |
| chr14_+_90422239 | 0.12 |
ENST00000393452.3
ENST00000554180.1 ENST00000393454.2 ENST00000553617.1 ENST00000335725.4 ENST00000357382.3 ENST00000556867.1 ENST00000553527.1 |
TDP1
|
tyrosyl-DNA phosphodiesterase 1 |
| chr15_+_91498089 | 0.12 |
ENST00000394258.2
ENST00000555155.1 |
RCCD1
|
RCC1 domain containing 1 |
| chr10_-_131309525 | 0.12 |
ENST00000601031.1
|
AL355531.2
|
Uncharacterized protein |
| chr17_+_46048471 | 0.12 |
ENST00000578018.1
ENST00000579175.1 |
CDK5RAP3
|
CDK5 regulatory subunit associated protein 3 |
| chr14_+_23025534 | 0.12 |
ENST00000557595.1
|
AE000662.92
|
Uncharacterized protein |
| chr6_-_114292284 | 0.11 |
ENST00000520895.1
ENST00000521163.1 ENST00000524334.1 ENST00000368632.2 ENST00000398283.2 |
HDAC2
|
histone deacetylase 2 |
| chr17_-_43025005 | 0.11 |
ENST00000587309.1
ENST00000593135.1 ENST00000339151.4 |
KIF18B
|
kinesin family member 18B |
| chr22_+_30752606 | 0.11 |
ENST00000399824.2
ENST00000405659.1 ENST00000338306.3 |
CCDC157
|
coiled-coil domain containing 157 |
| chr11_+_537494 | 0.11 |
ENST00000270115.7
|
LRRC56
|
leucine rich repeat containing 56 |
| chr11_-_124267264 | 0.11 |
ENST00000354597.3
|
OR8B3
|
olfactory receptor, family 8, subfamily B, member 3 |
| chr1_-_31661000 | 0.11 |
ENST00000263693.1
ENST00000398657.2 ENST00000526106.1 |
NKAIN1
|
Na+/K+ transporting ATPase interacting 1 |
| chr4_+_76439095 | 0.10 |
ENST00000506261.1
|
THAP6
|
THAP domain containing 6 |
| chr9_-_139658965 | 0.10 |
ENST00000316144.5
|
LCN15
|
lipocalin 15 |
| chr11_+_74951948 | 0.10 |
ENST00000562197.2
|
TPBGL
|
trophoblast glycoprotein-like |
| chr17_+_46048497 | 0.10 |
ENST00000583352.1
|
CDK5RAP3
|
CDK5 regulatory subunit associated protein 3 |
| chr22_-_25170683 | 0.10 |
ENST00000332271.5
|
PIWIL3
|
piwi-like RNA-mediated gene silencing 3 |
| chr19_+_8740061 | 0.10 |
ENST00000593792.1
|
CTD-2586B10.1
|
CTD-2586B10.1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of HES7_HES5
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 10.7 | GO:1905098 | negative regulation of guanyl-nucleotide exchange factor activity(GO:1905098) |
| 0.9 | 3.7 | GO:0036071 | N-glycan fucosylation(GO:0036071) |
| 0.6 | 1.9 | GO:0016539 | intein-mediated protein splicing(GO:0016539) protein splicing(GO:0030908) |
| 0.5 | 5.3 | GO:0099566 | regulation of postsynaptic cytosolic calcium ion concentration(GO:0099566) |
| 0.5 | 3.2 | GO:0086021 | SA node cell to atrial cardiac muscle cell communication by electrical coupling(GO:0086021) |
| 0.5 | 1.5 | GO:1903251 | multi-ciliated epithelial cell differentiation(GO:1903251) |
| 0.4 | 8.6 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.3 | 2.6 | GO:0043932 | ossification involved in bone remodeling(GO:0043932) |
| 0.3 | 0.9 | GO:0040040 | thermosensory behavior(GO:0040040) |
| 0.3 | 0.9 | GO:0021718 | superior olivary nucleus development(GO:0021718) superior olivary nucleus maturation(GO:0021722) |
| 0.3 | 1.9 | GO:0070370 | heat acclimation(GO:0010286) cellular heat acclimation(GO:0070370) |
| 0.3 | 4.1 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.3 | 0.8 | GO:0042668 | auditory receptor cell fate determination(GO:0042668) |
| 0.2 | 0.5 | GO:0048203 | vesicle targeting, trans-Golgi to endosome(GO:0048203) |
| 0.1 | 3.6 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.1 | 1.5 | GO:0061088 | regulation of sequestering of zinc ion(GO:0061088) |
| 0.1 | 0.3 | GO:0050760 | negative regulation of thymidylate synthase biosynthetic process(GO:0050760) |
| 0.1 | 0.8 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.1 | 0.5 | GO:0051562 | negative regulation of mitochondrial calcium ion concentration(GO:0051562) |
| 0.1 | 0.5 | GO:0035106 | negative regulation of luteinizing hormone secretion(GO:0033685) operant conditioning(GO:0035106) |
| 0.1 | 1.6 | GO:0015816 | glycine transport(GO:0015816) |
| 0.1 | 1.8 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.1 | 1.3 | GO:0030050 | vesicle transport along actin filament(GO:0030050) |
| 0.1 | 2.1 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.1 | 1.4 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.1 | 0.3 | GO:0071469 | detection of mechanical stimulus involved in sensory perception of touch(GO:0050976) cellular response to alkaline pH(GO:0071469) |
| 0.1 | 0.2 | GO:1901069 | guanosine-containing compound catabolic process(GO:1901069) |
| 0.1 | 2.6 | GO:0007202 | activation of phospholipase C activity(GO:0007202) |
| 0.0 | 0.3 | GO:0006344 | maintenance of chromatin silencing(GO:0006344) |
| 0.0 | 0.8 | GO:0072520 | seminiferous tubule development(GO:0072520) |
| 0.0 | 1.1 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 0.6 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.0 | 0.6 | GO:0007028 | cytoplasm organization(GO:0007028) |
| 0.0 | 1.0 | GO:0047484 | regulation of response to osmotic stress(GO:0047484) |
| 0.0 | 0.4 | GO:0006772 | thiamine metabolic process(GO:0006772) |
| 0.0 | 3.2 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 1.3 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.0 | 0.7 | GO:0002031 | G-protein coupled receptor internalization(GO:0002031) |
| 0.0 | 1.1 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 1.3 | GO:0048873 | homeostasis of number of cells within a tissue(GO:0048873) |
| 0.0 | 0.1 | GO:1902595 | regulation of DNA replication origin binding(GO:1902595) |
| 0.0 | 0.9 | GO:0070987 | error-free translesion synthesis(GO:0070987) |
| 0.0 | 0.3 | GO:0035754 | B cell chemotaxis(GO:0035754) |
| 0.0 | 0.4 | GO:1901838 | positive regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901838) |
| 0.0 | 0.5 | GO:0045141 | meiotic telomere clustering(GO:0045141) |
| 0.0 | 0.5 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.0 | 0.1 | GO:0007518 | myoblast fate determination(GO:0007518) |
| 0.0 | 0.5 | GO:0060856 | establishment of blood-brain barrier(GO:0060856) |
| 0.0 | 0.4 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.7 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.0 | 0.1 | GO:0097084 | vascular smooth muscle cell development(GO:0097084) |
| 0.0 | 0.2 | GO:0071569 | protein ufmylation(GO:0071569) |
| 0.0 | 0.5 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.0 | 1.7 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.2 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.0 | 0.7 | GO:0048488 | synaptic vesicle endocytosis(GO:0048488) |
| 0.0 | 0.3 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 0.6 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 0.2 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.0 | 0.0 | GO:0042360 | vitamin E metabolic process(GO:0042360) |
| 0.0 | 0.1 | GO:1901675 | negative regulation of histone H3-K27 acetylation(GO:1901675) |
| 0.0 | 0.3 | GO:0090141 | positive regulation of mitochondrial fission(GO:0090141) |
| 0.0 | 0.4 | GO:0060292 | long term synaptic depression(GO:0060292) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 5.3 | GO:0044308 | axonal spine(GO:0044308) |
| 0.5 | 1.8 | GO:0030849 | autosome(GO:0030849) |
| 0.2 | 0.9 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.1 | 0.8 | GO:0034991 | nuclear meiotic cohesin complex(GO:0034991) |
| 0.1 | 3.2 | GO:0005922 | connexon complex(GO:0005922) |
| 0.1 | 8.7 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.1 | 1.1 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.1 | 2.2 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.3 | GO:0089701 | U2AF(GO:0089701) |
| 0.0 | 1.3 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 0.7 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 8.5 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.3 | GO:1990812 | growth cone filopodium(GO:1990812) |
| 0.0 | 0.2 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.0 | 3.8 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 1.1 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 1.1 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.3 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 0.3 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.0 | 0.7 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
| 0.0 | 0.7 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.0 | 0.6 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 2.6 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.6 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.0 | 3.5 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 4.1 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 12.6 | GO:0005874 | microtubule(GO:0005874) |
| 0.0 | 0.1 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 1.7 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 1.5 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 0.1 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.0 | 1.4 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 1.3 | GO:0042734 | presynaptic membrane(GO:0042734) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 3.7 | GO:0046921 | glycoprotein 6-alpha-L-fucosyltransferase activity(GO:0008424) alpha-(1->6)-fucosyltransferase activity(GO:0046921) |
| 0.5 | 5.3 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.5 | 3.2 | GO:0086020 | gap junction channel activity involved in SA node cell-atrial cardiac muscle cell electrical coupling(GO:0086020) |
| 0.5 | 1.5 | GO:0015633 | zinc transporting ATPase activity(GO:0015633) |
| 0.3 | 0.9 | GO:0016534 | cyclin-dependent protein kinase 5 activator activity(GO:0016534) |
| 0.3 | 4.1 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.3 | 8.0 | GO:0031005 | filamin binding(GO:0031005) |
| 0.2 | 2.6 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.2 | 0.7 | GO:0050254 | rhodopsin kinase activity(GO:0050254) |
| 0.2 | 1.5 | GO:0004844 | uracil DNA N-glycosylase activity(GO:0004844) deaminated base DNA N-glycosylase activity(GO:0097506) |
| 0.2 | 3.6 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.1 | 4.6 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.1 | 0.7 | GO:0052812 | phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) |
| 0.1 | 0.5 | GO:0035650 | AP-1 adaptor complex binding(GO:0035650) |
| 0.1 | 1.9 | GO:0005113 | patched binding(GO:0005113) |
| 0.1 | 1.6 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.1 | 0.3 | GO:0098782 | mechanically-gated potassium channel activity(GO:0098782) |
| 0.1 | 0.4 | GO:0004802 | transketolase activity(GO:0004802) |
| 0.1 | 0.5 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.1 | 0.6 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.1 | 0.9 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.1 | 0.5 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.1 | 0.6 | GO:0071253 | connexin binding(GO:0071253) |
| 0.1 | 2.6 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.1 | 0.8 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.1 | 1.8 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.1 | 1.3 | GO:0047617 | very long-chain fatty acid-CoA ligase activity(GO:0031957) acyl-CoA hydrolase activity(GO:0047617) |
| 0.1 | 0.7 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.0 | 0.3 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.0 | 0.2 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.0 | 0.1 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.0 | 0.6 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.0 | 0.1 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.0 | 0.1 | GO:0070259 | tyrosyl-DNA phosphodiesterase activity(GO:0070259) |
| 0.0 | 0.9 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.0 | 1.0 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 2.7 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 2.3 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.0 | 0.1 | GO:0005280 | hydrogen:amino acid symporter activity(GO:0005280) |
| 0.0 | 1.3 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 0.3 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.0 | 0.3 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 0.1 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.0 | 9.4 | GO:0015631 | tubulin binding(GO:0015631) |
| 0.0 | 0.5 | GO:0008409 | 5'-3' exonuclease activity(GO:0008409) |
| 0.0 | 0.1 | GO:0022865 | transmembrane electron transfer carrier(GO:0022865) |
| 0.0 | 0.6 | GO:0046875 | ephrin receptor binding(GO:0046875) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 10.9 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.1 | 2.6 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.1 | 6.1 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.0 | 1.9 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 2.6 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.0 | 2.3 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 0.7 | PID ERBB1 RECEPTOR PROXIMAL PATHWAY | EGF receptor (ErbB1) signaling pathway |
| 0.0 | 1.0 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 0.2 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.2 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.6 | PID AURORA B PATHWAY | Aurora B signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 5.8 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 5.3 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.1 | 3.2 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.1 | 3.9 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.1 | 1.5 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 1.7 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.9 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 1.5 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.6 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.0 | 0.7 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 1.9 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 1.1 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 1.7 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.3 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 1.0 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.3 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 1.9 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.5 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.0 | 0.3 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.0 | 3.1 | REACTOME G ALPHA Q SIGNALLING EVENTS | Genes involved in G alpha (q) signalling events |
| 0.0 | 0.8 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |