Project
Illumina Body Map 2: averaged replicates
Navigation
Downloads
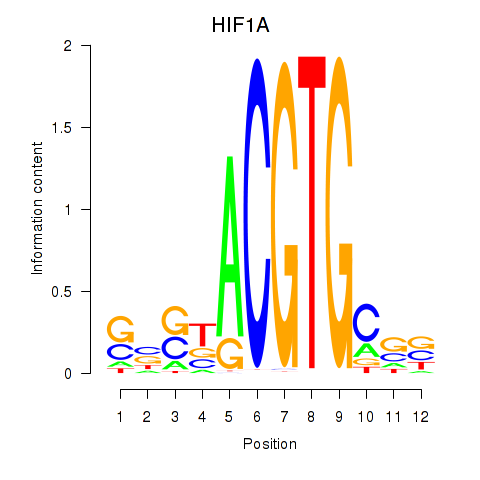
Results for HIF1A
Z-value: 0.86
Transcription factors associated with HIF1A
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HIF1A
|
ENSG00000100644.12 | hypoxia inducible factor 1 subunit alpha |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HIF1A | hg19_v2_chr14_+_62162258_62162269 | -0.40 | 2.3e-02 | Click! |
Activity profile of HIF1A motif
Sorted Z-values of HIF1A motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_220101944 | 1.86 |
ENST00000366926.3
ENST00000536992.1 |
SLC30A10
|
solute carrier family 30, member 10 |
| chr11_-_35440579 | 1.49 |
ENST00000606205.1
|
SLC1A2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
| chr11_-_35440796 | 1.45 |
ENST00000278379.3
|
SLC1A2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
| chr3_+_11034403 | 1.41 |
ENST00000287766.4
ENST00000425938.1 |
SLC6A1
|
solute carrier family 6 (neurotransmitter transporter), member 1 |
| chr7_+_100318423 | 1.32 |
ENST00000252723.2
|
EPO
|
erythropoietin |
| chr7_+_26331541 | 1.26 |
ENST00000416246.1
ENST00000338523.4 ENST00000412416.1 |
SNX10
|
sorting nexin 10 |
| chr12_+_6977323 | 1.17 |
ENST00000462761.1
|
TPI1
|
triosephosphate isomerase 1 |
| chr11_+_75479763 | 1.11 |
ENST00000228027.7
|
DGAT2
|
diacylglycerol O-acyltransferase 2 |
| chr7_+_26331678 | 1.10 |
ENST00000446848.2
|
SNX10
|
sorting nexin 10 |
| chr11_+_75479850 | 1.09 |
ENST00000376262.3
ENST00000604733.1 |
DGAT2
|
diacylglycerol O-acyltransferase 2 |
| chr12_+_6977258 | 1.00 |
ENST00000488464.2
ENST00000535434.1 ENST00000493987.1 |
TPI1
|
triosephosphate isomerase 1 |
| chr11_-_118901559 | 0.93 |
ENST00000330775.7
ENST00000545985.1 ENST00000357590.5 ENST00000538950.1 |
SLC37A4
|
solute carrier family 37 (glucose-6-phosphate transporter), member 4 |
| chr9_+_4490394 | 0.91 |
ENST00000262352.3
|
SLC1A1
|
solute carrier family 1 (neuronal/epithelial high affinity glutamate transporter, system Xag), member 1 |
| chr16_-_81129845 | 0.90 |
ENST00000569885.1
ENST00000566566.1 |
GCSH
|
glycine cleavage system protein H (aminomethyl carrier) |
| chr5_+_70883117 | 0.88 |
ENST00000340941.6
|
MCCC2
|
methylcrotonoyl-CoA carboxylase 2 (beta) |
| chr5_+_70883178 | 0.88 |
ENST00000323375.8
|
MCCC2
|
methylcrotonoyl-CoA carboxylase 2 (beta) |
| chr5_+_70883154 | 0.86 |
ENST00000509358.2
|
MCCC2
|
methylcrotonoyl-CoA carboxylase 2 (beta) |
| chr3_+_25831567 | 0.81 |
ENST00000280701.3
ENST00000420173.2 |
OXSM
|
3-oxoacyl-ACP synthase, mitochondrial |
| chr2_-_216946500 | 0.81 |
ENST00000265322.7
|
PECR
|
peroxisomal trans-2-enoyl-CoA reductase |
| chr7_+_65540853 | 0.75 |
ENST00000380839.4
ENST00000395332.3 ENST00000362000.5 ENST00000395331.3 |
ASL
|
argininosuccinate lyase |
| chr10_+_75910960 | 0.75 |
ENST00000539909.1
ENST00000286621.2 |
ADK
|
adenosine kinase |
| chr7_-_95951310 | 0.74 |
ENST00000542654.1
|
SLC25A13
|
solute carrier family 25 (aspartate/glutamate carrier), member 13 |
| chr7_+_65540780 | 0.74 |
ENST00000304874.9
|
ASL
|
argininosuccinate lyase |
| chrX_+_77359726 | 0.74 |
ENST00000442431.1
|
PGK1
|
phosphoglycerate kinase 1 |
| chrX_+_77359671 | 0.74 |
ENST00000373316.4
|
PGK1
|
phosphoglycerate kinase 1 |
| chr7_-_95951334 | 0.73 |
ENST00000265631.5
|
SLC25A13
|
solute carrier family 25 (aspartate/glutamate carrier), member 13 |
| chr5_+_174905532 | 0.72 |
ENST00000502393.1
ENST00000506963.1 |
SFXN1
|
sideroflexin 1 |
| chr14_+_92789498 | 0.71 |
ENST00000531433.1
|
SLC24A4
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 4 |
| chr7_-_95951432 | 0.71 |
ENST00000416240.2
|
SLC25A13
|
solute carrier family 25 (aspartate/glutamate carrier), member 13 |
| chr2_+_173420697 | 0.70 |
ENST00000282077.3
ENST00000392571.2 ENST00000410055.1 |
PDK1
|
pyruvate dehydrogenase kinase, isozyme 1 |
| chr1_+_65613852 | 0.69 |
ENST00000327299.7
|
AK4
|
adenylate kinase 4 |
| chr12_-_49318715 | 0.68 |
ENST00000444214.2
|
FKBP11
|
FK506 binding protein 11, 19 kDa |
| chr9_+_115983808 | 0.68 |
ENST00000374210.6
ENST00000374212.4 |
SLC31A1
|
solute carrier family 31 (copper transporter), member 1 |
| chr15_-_60884706 | 0.67 |
ENST00000449337.2
|
RORA
|
RAR-related orphan receptor A |
| chr16_-_81129951 | 0.65 |
ENST00000315467.3
|
GCSH
|
glycine cleavage system protein H (aminomethyl carrier) |
| chrX_+_105969893 | 0.64 |
ENST00000255499.2
|
RNF128
|
ring finger protein 128, E3 ubiquitin protein ligase |
| chr11_+_14665263 | 0.62 |
ENST00000282096.4
|
PDE3B
|
phosphodiesterase 3B, cGMP-inhibited |
| chr15_+_41056255 | 0.61 |
ENST00000561160.1
ENST00000559445.1 |
GCHFR
|
GTP cyclohydrolase I feedback regulator |
| chr4_-_39529049 | 0.61 |
ENST00000501493.2
ENST00000509391.1 ENST00000507089.1 |
UGDH
|
UDP-glucose 6-dehydrogenase |
| chr11_-_47664072 | 0.61 |
ENST00000542981.1
ENST00000530428.1 ENST00000302503.3 |
MTCH2
|
mitochondrial carrier 2 |
| chr15_+_41056218 | 0.60 |
ENST00000260447.4
|
GCHFR
|
GTP cyclohydrolase I feedback regulator |
| chr11_+_14665373 | 0.60 |
ENST00000455098.2
|
PDE3B
|
phosphodiesterase 3B, cGMP-inhibited |
| chr14_+_64970662 | 0.60 |
ENST00000556965.1
ENST00000554015.1 |
ZBTB1
|
zinc finger and BTB domain containing 1 |
| chr8_+_95908041 | 0.59 |
ENST00000396113.1
ENST00000519136.1 |
NDUFAF6
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 6 |
| chr11_-_18343669 | 0.59 |
ENST00000396253.3
ENST00000349215.3 ENST00000438420.2 |
HPS5
|
Hermansky-Pudlak syndrome 5 |
| chr12_+_56862301 | 0.59 |
ENST00000338146.5
|
SPRYD4
|
SPRY domain containing 4 |
| chr7_+_96747030 | 0.59 |
ENST00000360382.4
|
ACN9
|
ACN9 homolog (S. cerevisiae) |
| chr6_+_108487245 | 0.58 |
ENST00000368986.4
|
NR2E1
|
nuclear receptor subfamily 2, group E, member 1 |
| chr14_+_64970427 | 0.57 |
ENST00000553583.1
|
ZBTB1
|
zinc finger and BTB domain containing 1 |
| chr1_+_230203010 | 0.56 |
ENST00000541865.1
|
GALNT2
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 2 (GalNAc-T2) |
| chr1_-_231376836 | 0.55 |
ENST00000451322.1
|
C1orf131
|
chromosome 1 open reading frame 131 |
| chr20_+_44657845 | 0.55 |
ENST00000243964.3
|
SLC12A5
|
solute carrier family 12 (potassium/chloride transporter), member 5 |
| chr8_+_97274119 | 0.54 |
ENST00000455950.2
|
PTDSS1
|
phosphatidylserine synthase 1 |
| chr5_+_49962495 | 0.53 |
ENST00000515175.1
|
PARP8
|
poly (ADP-ribose) polymerase family, member 8 |
| chr1_-_8939265 | 0.53 |
ENST00000489867.1
|
ENO1
|
enolase 1, (alpha) |
| chr2_-_119916459 | 0.52 |
ENST00000272520.3
|
C1QL2
|
complement component 1, q subcomponent-like 2 |
| chr5_+_159343688 | 0.51 |
ENST00000306675.3
|
ADRA1B
|
adrenoceptor alpha 1B |
| chr13_-_52027134 | 0.50 |
ENST00000311234.4
ENST00000425000.1 ENST00000463928.1 ENST00000442263.3 ENST00000398119.2 |
INTS6
|
integrator complex subunit 6 |
| chr6_-_33679452 | 0.48 |
ENST00000374231.4
ENST00000607484.1 ENST00000374214.3 |
UQCC2
|
ubiquinol-cytochrome c reductase complex assembly factor 2 |
| chr9_-_139922726 | 0.48 |
ENST00000265662.5
ENST00000371605.3 |
ABCA2
|
ATP-binding cassette, sub-family A (ABC1), member 2 |
| chr1_-_231376867 | 0.48 |
ENST00000366649.2
ENST00000318906.2 ENST00000366651.3 |
C1orf131
|
chromosome 1 open reading frame 131 |
| chr1_-_6269448 | 0.48 |
ENST00000465335.1
|
RPL22
|
ribosomal protein L22 |
| chr9_-_123476612 | 0.47 |
ENST00000426959.1
|
MEGF9
|
multiple EGF-like-domains 9 |
| chr20_+_44657807 | 0.47 |
ENST00000372315.1
|
SLC12A5
|
solute carrier family 12 (potassium/chloride transporter), member 5 |
| chr11_-_74204742 | 0.46 |
ENST00000310109.4
|
LIPT2
|
lipoyl(octanoyl) transferase 2 (putative) |
| chr4_-_39529180 | 0.46 |
ENST00000515021.1
ENST00000510490.1 ENST00000316423.6 |
UGDH
|
UDP-glucose 6-dehydrogenase |
| chr14_-_53162361 | 0.46 |
ENST00000395686.3
|
ERO1L
|
ERO1-like (S. cerevisiae) |
| chr8_+_95907993 | 0.45 |
ENST00000523378.1
|
NDUFAF6
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 6 |
| chr14_-_64970494 | 0.45 |
ENST00000608382.1
|
ZBTB25
|
zinc finger and BTB domain containing 25 |
| chr12_-_121734489 | 0.45 |
ENST00000412367.2
ENST00000402834.4 ENST00000404169.3 |
CAMKK2
|
calcium/calmodulin-dependent protein kinase kinase 2, beta |
| chr3_-_52719546 | 0.45 |
ENST00000439181.1
ENST00000449505.1 |
PBRM1
|
polybromo 1 |
| chr17_-_67323232 | 0.44 |
ENST00000592568.1
ENST00000392676.3 |
ABCA5
|
ATP-binding cassette, sub-family A (ABC1), member 5 |
| chr1_+_230202936 | 0.44 |
ENST00000366672.4
|
GALNT2
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 2 (GalNAc-T2) |
| chr22_+_19702069 | 0.44 |
ENST00000412544.1
|
SEPT5
|
septin 5 |
| chr20_+_56964253 | 0.43 |
ENST00000395802.3
|
VAPB
|
VAMP (vesicle-associated membrane protein)-associated protein B and C |
| chr2_-_220118631 | 0.43 |
ENST00000248437.4
|
TUBA4A
|
tubulin, alpha 4a |
| chr1_+_214454492 | 0.43 |
ENST00000366957.5
ENST00000415093.2 |
SMYD2
|
SET and MYND domain containing 2 |
| chr9_+_133569108 | 0.43 |
ENST00000372358.5
ENST00000546165.1 ENST00000372352.3 ENST00000372351.3 ENST00000372350.3 ENST00000495699.2 |
EXOSC2
|
exosome component 2 |
| chr9_-_139922631 | 0.42 |
ENST00000341511.6
|
ABCA2
|
ATP-binding cassette, sub-family A (ABC1), member 2 |
| chr9_-_136857403 | 0.42 |
ENST00000406606.3
ENST00000371850.3 |
VAV2
|
vav 2 guanine nucleotide exchange factor |
| chr1_+_74663994 | 0.42 |
ENST00000472069.1
|
FPGT
|
fucose-1-phosphate guanylyltransferase |
| chr12_-_64616019 | 0.42 |
ENST00000311915.8
ENST00000398055.3 ENST00000544871.1 |
C12orf66
|
chromosome 12 open reading frame 66 |
| chr1_+_100436065 | 0.42 |
ENST00000370153.1
|
SLC35A3
|
solute carrier family 35 (UDP-N-acetylglucosamine (UDP-GlcNAc) transporter), member A3 |
| chr17_-_67323305 | 0.42 |
ENST00000392677.2
ENST00000593153.1 |
ABCA5
|
ATP-binding cassette, sub-family A (ABC1), member 5 |
| chr9_-_123476719 | 0.41 |
ENST00000373930.3
|
MEGF9
|
multiple EGF-like-domains 9 |
| chr20_+_33464407 | 0.41 |
ENST00000253382.5
|
ACSS2
|
acyl-CoA synthetase short-chain family member 2 |
| chr11_-_18343725 | 0.41 |
ENST00000531848.1
|
HPS5
|
Hermansky-Pudlak syndrome 5 |
| chr20_+_33464238 | 0.40 |
ENST00000360596.2
|
ACSS2
|
acyl-CoA synthetase short-chain family member 2 |
| chr3_-_58419537 | 0.40 |
ENST00000474765.1
ENST00000485460.1 ENST00000302746.6 ENST00000383714.4 |
PDHB
|
pyruvate dehydrogenase (lipoamide) beta |
| chr1_+_74663896 | 0.40 |
ENST00000370898.3
ENST00000467578.2 ENST00000370894.5 ENST00000482102.2 ENST00000609362.1 ENST00000534056.1 ENST00000557284.2 ENST00000370899.3 ENST00000370895.1 ENST00000534632.1 ENST00000370893.1 ENST00000370891.2 |
FPGT
FPGT-TNNI3K
TNNI3K
|
fucose-1-phosphate guanylyltransferase FPGT-TNNI3K readthrough TNNI3 interacting kinase |
| chrX_+_24483338 | 0.40 |
ENST00000379162.4
ENST00000441463.2 |
PDK3
|
pyruvate dehydrogenase kinase, isozyme 3 |
| chr11_+_12132117 | 0.40 |
ENST00000256194.4
|
MICAL2
|
microtubule associated monooxygenase, calponin and LIM domain containing 2 |
| chr17_+_34890807 | 0.40 |
ENST00000429467.2
ENST00000592983.1 |
PIGW
|
phosphatidylinositol glycan anchor biosynthesis, class W |
| chr19_-_5720123 | 0.40 |
ENST00000587365.1
ENST00000585374.1 ENST00000593119.1 |
LONP1
|
lon peptidase 1, mitochondrial |
| chr16_+_447226 | 0.40 |
ENST00000433358.1
|
NME4
|
NME/NM23 nucleoside diphosphate kinase 4 |
| chr11_+_18720316 | 0.39 |
ENST00000280734.2
|
TMEM86A
|
transmembrane protein 86A |
| chr11_-_57283159 | 0.39 |
ENST00000533263.1
ENST00000278426.3 |
SLC43A1
|
solute carrier family 43 (amino acid system L transporter), member 1 |
| chr15_+_52311398 | 0.39 |
ENST00000261845.5
|
MAPK6
|
mitogen-activated protein kinase 6 |
| chr11_-_73471655 | 0.39 |
ENST00000400470.2
|
RAB6A
|
RAB6A, member RAS oncogene family |
| chr6_+_64282447 | 0.39 |
ENST00000370650.2
ENST00000578299.1 |
PTP4A1
|
protein tyrosine phosphatase type IVA, member 1 |
| chr1_+_228353495 | 0.38 |
ENST00000366711.3
|
IBA57
|
IBA57, iron-sulfur cluster assembly homolog (S. cerevisiae) |
| chr17_+_27052892 | 0.38 |
ENST00000579671.1
ENST00000579060.1 |
NEK8
|
NIMA-related kinase 8 |
| chr12_+_46777450 | 0.38 |
ENST00000551503.1
|
RP11-96H19.1
|
RP11-96H19.1 |
| chr17_-_17942473 | 0.37 |
ENST00000585101.1
ENST00000474627.3 ENST00000444058.1 |
ATPAF2
|
ATP synthase mitochondrial F1 complex assembly factor 2 |
| chr16_+_447209 | 0.37 |
ENST00000382940.4
ENST00000219479.2 |
NME4
|
NME/NM23 nucleoside diphosphate kinase 4 |
| chr8_-_97273807 | 0.37 |
ENST00000517720.1
ENST00000287025.3 ENST00000523821.1 |
MTERFD1
|
MTERF domain containing 1 |
| chr2_-_220083076 | 0.37 |
ENST00000295750.4
|
ABCB6
|
ATP-binding cassette, sub-family B (MDR/TAP), member 6 |
| chr12_+_6643676 | 0.37 |
ENST00000396856.1
ENST00000396861.1 |
GAPDH
|
glyceraldehyde-3-phosphate dehydrogenase |
| chr2_-_73460334 | 0.37 |
ENST00000258083.2
|
PRADC1
|
protease-associated domain containing 1 |
| chr9_-_115983568 | 0.36 |
ENST00000446284.1
ENST00000414250.1 |
FKBP15
|
FK506 binding protein 15, 133kDa |
| chr10_+_101419187 | 0.36 |
ENST00000370489.4
|
ENTPD7
|
ectonucleoside triphosphate diphosphohydrolase 7 |
| chr2_+_170683979 | 0.36 |
ENST00000418381.1
|
UBR3
|
ubiquitin protein ligase E3 component n-recognin 3 (putative) |
| chr3_+_39851170 | 0.36 |
ENST00000425621.1
ENST00000396217.3 |
MYRIP
|
myosin VIIA and Rab interacting protein |
| chr6_+_64281906 | 0.36 |
ENST00000370651.3
|
PTP4A1
|
protein tyrosine phosphatase type IVA, member 1 |
| chr14_-_54908043 | 0.35 |
ENST00000556113.1
ENST00000553660.1 ENST00000395573.4 ENST00000557690.1 ENST00000216416.4 |
CNIH1
|
cornichon family AMPA receptor auxiliary protein 1 |
| chr8_+_98656336 | 0.35 |
ENST00000336273.3
|
MTDH
|
metadherin |
| chrX_-_84363974 | 0.35 |
ENST00000395409.3
ENST00000332921.5 ENST00000509231.1 |
SATL1
|
spermidine/spermine N1-acetyl transferase-like 1 |
| chr17_-_2615031 | 0.35 |
ENST00000576885.1
ENST00000574426.2 |
CLUH
|
clustered mitochondria (cluA/CLU1) homolog |
| chr11_+_126225789 | 0.35 |
ENST00000530591.1
ENST00000534083.1 |
ST3GAL4
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 4 |
| chr3_-_183543301 | 0.35 |
ENST00000318631.3
ENST00000431348.1 |
MAP6D1
|
MAP6 domain containing 1 |
| chr1_-_113498943 | 0.35 |
ENST00000369626.3
|
SLC16A1
|
solute carrier family 16 (monocarboxylate transporter), member 1 |
| chr20_+_33464368 | 0.34 |
ENST00000484354.1
ENST00000493805.2 ENST00000473172.1 |
ACSS2
|
acyl-CoA synthetase short-chain family member 2 |
| chr1_-_166136187 | 0.34 |
ENST00000338353.3
|
FAM78B
|
family with sequence similarity 78, member B |
| chr8_-_102218292 | 0.34 |
ENST00000518336.1
ENST00000520454.1 |
ZNF706
|
zinc finger protein 706 |
| chr3_-_127541679 | 0.34 |
ENST00000265052.5
|
MGLL
|
monoglyceride lipase |
| chr3_-_167813672 | 0.34 |
ENST00000470487.1
|
GOLIM4
|
golgi integral membrane protein 4 |
| chr11_-_14665163 | 0.33 |
ENST00000418988.2
|
PSMA1
|
proteasome (prosome, macropain) subunit, alpha type, 1 |
| chr1_+_171750776 | 0.33 |
ENST00000458517.1
ENST00000362019.3 ENST00000367737.5 ENST00000361735.3 |
METTL13
|
methyltransferase like 13 |
| chr1_-_113498616 | 0.33 |
ENST00000433570.4
ENST00000538576.1 ENST00000458229.1 |
SLC16A1
|
solute carrier family 16 (monocarboxylate transporter), member 1 |
| chr19_-_5719860 | 0.33 |
ENST00000590729.1
|
LONP1
|
lon peptidase 1, mitochondrial |
| chr8_-_9760839 | 0.33 |
ENST00000519461.1
ENST00000517675.1 |
LINC00599
|
long intergenic non-protein coding RNA 599 |
| chr8_+_98656693 | 0.33 |
ENST00000519934.1
|
MTDH
|
metadherin |
| chr17_-_36831156 | 0.33 |
ENST00000325814.5
|
C17orf96
|
chromosome 17 open reading frame 96 |
| chr18_+_43913919 | 0.33 |
ENST00000587853.1
|
RNF165
|
ring finger protein 165 |
| chr11_+_126225529 | 0.33 |
ENST00000227495.6
ENST00000444328.2 ENST00000356132.4 |
ST3GAL4
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 4 |
| chr20_+_36661910 | 0.32 |
ENST00000373433.4
|
RPRD1B
|
regulation of nuclear pre-mRNA domain containing 1B |
| chr4_+_83351715 | 0.32 |
ENST00000273920.3
|
ENOPH1
|
enolase-phosphatase 1 |
| chr2_+_68384976 | 0.32 |
ENST00000263657.2
|
PNO1
|
partner of NOB1 homolog (S. cerevisiae) |
| chr4_-_25162188 | 0.32 |
ENST00000302922.3
|
SEPSECS
|
Sep (O-phosphoserine) tRNA:Sec (selenocysteine) tRNA synthase |
| chr3_+_19988566 | 0.32 |
ENST00000273047.4
|
RAB5A
|
RAB5A, member RAS oncogene family |
| chr7_-_127032114 | 0.32 |
ENST00000436992.1
|
ZNF800
|
zinc finger protein 800 |
| chr1_-_8938736 | 0.31 |
ENST00000234590.4
|
ENO1
|
enolase 1, (alpha) |
| chr1_+_39571026 | 0.31 |
ENST00000524432.1
|
MACF1
|
microtubule-actin crosslinking factor 1 |
| chr3_+_19988885 | 0.31 |
ENST00000422242.1
|
RAB5A
|
RAB5A, member RAS oncogene family |
| chr2_+_170683942 | 0.31 |
ENST00000272793.5
|
UBR3
|
ubiquitin protein ligase E3 component n-recognin 3 (putative) |
| chr15_-_43622736 | 0.31 |
ENST00000544735.1
ENST00000567039.1 ENST00000305641.5 |
LCMT2
|
leucine carboxyl methyltransferase 2 |
| chr13_+_103451548 | 0.31 |
ENST00000419638.1
|
BIVM
|
basic, immunoglobulin-like variable motif containing |
| chr6_+_89791507 | 0.31 |
ENST00000354922.3
|
PNRC1
|
proline-rich nuclear receptor coactivator 1 |
| chr16_+_53164956 | 0.31 |
ENST00000563410.1
|
CHD9
|
chromodomain helicase DNA binding protein 9 |
| chr16_-_30022293 | 0.30 |
ENST00000565273.1
ENST00000567332.2 ENST00000350119.4 |
DOC2A
|
double C2-like domains, alpha |
| chr19_+_41903709 | 0.30 |
ENST00000542943.1
ENST00000457836.2 |
BCKDHA
|
branched chain keto acid dehydrogenase E1, alpha polypeptide |
| chr8_+_125551338 | 0.30 |
ENST00000276689.3
ENST00000518008.1 ENST00000522532.1 ENST00000517367.1 |
NDUFB9
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 9, 22kDa |
| chr19_-_5720248 | 0.30 |
ENST00000360614.3
|
LONP1
|
lon peptidase 1, mitochondrial |
| chr1_-_167522982 | 0.29 |
ENST00000370509.4
|
CREG1
|
cellular repressor of E1A-stimulated genes 1 |
| chr7_-_92465868 | 0.29 |
ENST00000424848.2
|
CDK6
|
cyclin-dependent kinase 6 |
| chrX_-_47509994 | 0.29 |
ENST00000343894.4
|
ELK1
|
ELK1, member of ETS oncogene family |
| chr17_-_2614927 | 0.29 |
ENST00000435359.1
|
CLUH
|
clustered mitochondria (cluA/CLU1) homolog |
| chr20_-_3065362 | 0.29 |
ENST00000380293.3
|
AVP
|
arginine vasopressin |
| chr15_+_43622843 | 0.29 |
ENST00000428046.3
ENST00000422466.2 ENST00000389651.4 |
ADAL
|
adenosine deaminase-like |
| chr14_-_53162215 | 0.28 |
ENST00000554251.1
|
ERO1L
|
ERO1-like (S. cerevisiae) |
| chrX_-_69509738 | 0.28 |
ENST00000374454.1
ENST00000239666.4 |
PDZD11
|
PDZ domain containing 11 |
| chr3_+_19988736 | 0.28 |
ENST00000443878.1
|
RAB5A
|
RAB5A, member RAS oncogene family |
| chr11_-_65548265 | 0.28 |
ENST00000532090.2
|
AP5B1
|
adaptor-related protein complex 5, beta 1 subunit |
| chr16_+_29466426 | 0.28 |
ENST00000567248.1
|
SLX1B
|
SLX1 structure-specific endonuclease subunit homolog B (S. cerevisiae) |
| chr16_-_23568651 | 0.28 |
ENST00000563232.1
ENST00000563459.1 ENST00000449606.1 |
EARS2
|
glutamyl-tRNA synthetase 2, mitochondrial |
| chr17_+_40719073 | 0.28 |
ENST00000435881.2
ENST00000246912.4 ENST00000346833.4 ENST00000591024.1 |
MLX
|
MLX, MAX dimerization protein |
| chr1_-_166135952 | 0.27 |
ENST00000354422.3
|
FAM78B
|
family with sequence similarity 78, member B |
| chr14_+_77787227 | 0.27 |
ENST00000216465.5
ENST00000361389.4 ENST00000554279.1 ENST00000557639.1 ENST00000349555.3 ENST00000556627.1 ENST00000557053.1 |
GSTZ1
|
glutathione S-transferase zeta 1 |
| chr8_+_55047763 | 0.27 |
ENST00000260102.4
ENST00000519831.1 |
MRPL15
|
mitochondrial ribosomal protein L15 |
| chr1_-_1709845 | 0.27 |
ENST00000341426.5
ENST00000344463.4 |
NADK
|
NAD kinase |
| chr1_-_1710229 | 0.26 |
ENST00000341991.3
|
NADK
|
NAD kinase |
| chr7_+_43152191 | 0.26 |
ENST00000395891.2
|
HECW1
|
HECT, C2 and WW domain containing E3 ubiquitin protein ligase 1 |
| chr10_-_101945771 | 0.26 |
ENST00000370408.2
ENST00000407654.3 |
ERLIN1
|
ER lipid raft associated 1 |
| chr19_-_663147 | 0.26 |
ENST00000606702.1
|
RNF126
|
ring finger protein 126 |
| chr3_+_52719936 | 0.26 |
ENST00000418458.1
ENST00000394799.2 |
GNL3
|
guanine nucleotide binding protein-like 3 (nucleolar) |
| chr2_+_216946589 | 0.26 |
ENST00000433112.1
ENST00000454545.1 ENST00000437356.2 ENST00000295658.4 ENST00000455479.1 ENST00000406027.2 |
TMEM169
|
transmembrane protein 169 |
| chr19_-_663171 | 0.26 |
ENST00000606896.1
ENST00000589762.2 |
RNF126
|
ring finger protein 126 |
| chr17_-_5342380 | 0.25 |
ENST00000225698.4
|
C1QBP
|
complement component 1, q subcomponent binding protein |
| chr1_-_166944652 | 0.25 |
ENST00000528703.1
ENST00000525740.1 ENST00000529387.1 ENST00000469934.2 ENST00000529071.1 ENST00000526687.1 |
ILDR2
|
immunoglobulin-like domain containing receptor 2 |
| chr11_-_62414070 | 0.25 |
ENST00000540933.1
ENST00000346178.4 ENST00000356638.3 ENST00000534779.1 ENST00000525994.1 |
GANAB
|
glucosidase, alpha; neutral AB |
| chr4_+_25162253 | 0.25 |
ENST00000512921.1
|
PI4K2B
|
phosphatidylinositol 4-kinase type 2 beta |
| chr2_-_220083692 | 0.24 |
ENST00000265316.3
|
ABCB6
|
ATP-binding cassette, sub-family B (MDR/TAP), member 6 |
| chr20_+_56964169 | 0.24 |
ENST00000475243.1
|
VAPB
|
VAMP (vesicle-associated membrane protein)-associated protein B and C |
| chr17_-_34890759 | 0.23 |
ENST00000431794.3
|
MYO19
|
myosin XIX |
| chr2_-_28113217 | 0.23 |
ENST00000444339.2
|
RBKS
|
ribokinase |
| chr13_+_27998681 | 0.23 |
ENST00000381140.4
|
GTF3A
|
general transcription factor IIIA |
| chr16_-_4466565 | 0.23 |
ENST00000572467.1
ENST00000423908.2 ENST00000572044.1 ENST00000571052.1 |
CORO7-PAM16
CORO7
|
CORO7-PAM16 readthrough coronin 7 |
| chr20_+_42086525 | 0.23 |
ENST00000244020.3
|
SRSF6
|
serine/arginine-rich splicing factor 6 |
| chr16_+_24621546 | 0.23 |
ENST00000566108.1
|
CTD-2540M10.1
|
CTD-2540M10.1 |
| chr11_+_73498973 | 0.23 |
ENST00000537007.1
|
MRPL48
|
mitochondrial ribosomal protein L48 |
| chr16_-_4897266 | 0.22 |
ENST00000591451.1
ENST00000436648.5 ENST00000381983.3 ENST00000588297.1 ENST00000321919.9 |
GLYR1
|
glyoxylate reductase 1 homolog (Arabidopsis) |
| chr3_-_71834207 | 0.22 |
ENST00000295619.3
|
PROK2
|
prokineticin 2 |
| chr18_+_43913973 | 0.22 |
ENST00000590330.1
|
RNF165
|
ring finger protein 165 |
| chr14_-_88459182 | 0.22 |
ENST00000544807.2
|
GALC
|
galactosylceramidase |
| chr6_-_153304148 | 0.22 |
ENST00000229758.3
|
FBXO5
|
F-box protein 5 |
| chr8_+_67341239 | 0.22 |
ENST00000320270.2
|
RRS1
|
RRS1 ribosome biogenesis regulator homolog (S. cerevisiae) |
| chr11_-_6624801 | 0.22 |
ENST00000534343.1
ENST00000254605.6 |
RRP8
|
ribosomal RNA processing 8, methyltransferase, homolog (yeast) |
| chr3_+_49507559 | 0.22 |
ENST00000421560.1
ENST00000308775.2 ENST00000545947.1 ENST00000541308.1 ENST00000539901.1 ENST00000538711.1 ENST00000418588.1 |
DAG1
|
dystroglycan 1 (dystrophin-associated glycoprotein 1) |
| chr1_-_43833628 | 0.22 |
ENST00000413844.2
ENST00000372458.3 |
ELOVL1
|
ELOVL fatty acid elongase 1 |
| chr3_-_156272924 | 0.22 |
ENST00000467789.1
ENST00000265044.2 |
SSR3
|
signal sequence receptor, gamma (translocon-associated protein gamma) |
Network of associatons between targets according to the STRING database.
First level regulatory network of HIF1A
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.2 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.5 | 2.2 | GO:0046166 | glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.5 | 1.5 | GO:0042450 | arginine biosynthetic process via ornithine(GO:0042450) |
| 0.4 | 1.3 | GO:0038162 | erythropoietin-mediated signaling pathway(GO:0038162) |
| 0.4 | 2.2 | GO:0071400 | cellular response to oleic acid(GO:0071400) |
| 0.4 | 1.2 | GO:0043095 | regulation of GTP cyclohydrolase I activity(GO:0043095) negative regulation of GTP cyclohydrolase I activity(GO:0043105) |
| 0.4 | 3.9 | GO:0070779 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.4 | 1.4 | GO:0014054 | positive regulation of gamma-aminobutyric acid secretion(GO:0014054) |
| 0.3 | 1.0 | GO:0070407 | oxidation-dependent protein catabolic process(GO:0070407) |
| 0.3 | 1.0 | GO:0040040 | thermosensory behavior(GO:0040040) |
| 0.3 | 1.5 | GO:0006546 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.3 | 1.2 | GO:2000174 | regulation of pro-T cell differentiation(GO:2000174) positive regulation of pro-T cell differentiation(GO:2000176) |
| 0.3 | 2.6 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 0.2 | 0.7 | GO:0044209 | AMP salvage(GO:0044209) |
| 0.2 | 0.7 | GO:0071629 | cytoplasm-associated proteasomal ubiquitin-dependent protein catabolic process(GO:0071629) |
| 0.2 | 0.7 | GO:0051780 | mevalonate transport(GO:0015728) behavioral response to nutrient(GO:0051780) |
| 0.2 | 2.0 | GO:0051790 | short-chain fatty acid biosynthetic process(GO:0051790) |
| 0.2 | 2.1 | GO:0071421 | manganese ion transmembrane transport(GO:0071421) |
| 0.2 | 0.5 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.1 | 0.6 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 0.1 | 1.1 | GO:0097411 | hypoxia-inducible factor-1alpha signaling pathway(GO:0097411) |
| 0.1 | 0.7 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.1 | 0.7 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.1 | 0.9 | GO:0032377 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.1 | 0.5 | GO:0001994 | norepinephrine-epinephrine vasoconstriction involved in regulation of systemic arterial blood pressure(GO:0001994) |
| 0.1 | 1.0 | GO:0002378 | immunoglobulin biosynthetic process(GO:0002378) protein O-linked glycosylation via serine(GO:0018242) |
| 0.1 | 0.8 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 0.1 | 0.4 | GO:0016340 | calcium-dependent cell-matrix adhesion(GO:0016340) |
| 0.1 | 0.4 | GO:0032917 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.1 | 0.6 | GO:0015788 | UDP-N-acetylglucosamine transport(GO:0015788) UDP-N-acetylglucosamine transmembrane transport(GO:1990569) |
| 0.1 | 0.5 | GO:0071051 | polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.1 | 1.1 | GO:0006065 | UDP-glucuronate biosynthetic process(GO:0006065) |
| 0.1 | 1.2 | GO:0033629 | negative regulation of cell adhesion mediated by integrin(GO:0033629) |
| 0.1 | 0.9 | GO:2000286 | receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 0.1 | 0.3 | GO:0007057 | spindle assembly involved in female meiosis I(GO:0007057) |
| 0.1 | 0.4 | GO:0019417 | sulfur oxidation(GO:0019417) |
| 0.1 | 0.3 | GO:0002125 | maternal aggressive behavior(GO:0002125) |
| 0.1 | 0.3 | GO:0046103 | adenosine catabolic process(GO:0006154) inosine biosynthetic process(GO:0046103) |
| 0.1 | 0.3 | GO:0006424 | glutamyl-tRNA aminoacylation(GO:0006424) |
| 0.1 | 0.5 | GO:0061762 | CAMKK-AMPK signaling cascade(GO:0061762) |
| 0.1 | 0.4 | GO:0019303 | D-ribose catabolic process(GO:0019303) |
| 0.1 | 0.6 | GO:0097056 | selenocysteinyl-tRNA(Sec) biosynthetic process(GO:0097056) |
| 0.1 | 0.9 | GO:0070131 | positive regulation of mitochondrial translation(GO:0070131) |
| 0.1 | 0.4 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.1 | 0.4 | GO:0009107 | lipoate biosynthetic process(GO:0009107) |
| 0.1 | 0.3 | GO:0046452 | dihydrofolate metabolic process(GO:0046452) |
| 0.1 | 0.3 | GO:0006500 | N-terminal protein palmitoylation(GO:0006500) |
| 0.1 | 0.7 | GO:0046940 | nucleoside monophosphate phosphorylation(GO:0046940) |
| 0.1 | 0.5 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.1 | 0.3 | GO:0070124 | mitochondrial translational initiation(GO:0070124) |
| 0.1 | 0.4 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.1 | 0.4 | GO:0043461 | proton-transporting ATP synthase complex assembly(GO:0043461) proton-transporting ATP synthase complex biogenesis(GO:0070272) |
| 0.1 | 0.8 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.1 | 1.5 | GO:0031639 | plasminogen activation(GO:0031639) |
| 0.1 | 2.2 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.1 | 0.2 | GO:0034402 | recruitment of 3'-end processing factors to RNA polymerase II holoenzyme complex(GO:0034402) |
| 0.1 | 0.2 | GO:1902595 | regulation of DNA replication origin binding(GO:1902595) |
| 0.1 | 0.3 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.1 | 0.7 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.1 | 0.2 | GO:0009443 | pyridoxal 5'-phosphate salvage(GO:0009443) |
| 0.1 | 0.5 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.1 | 0.4 | GO:0003172 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.1 | 0.7 | GO:0006987 | activation of signaling protein activity involved in unfolded protein response(GO:0006987) |
| 0.1 | 0.4 | GO:0090206 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.1 | 0.8 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.0 | 0.3 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.0 | 0.3 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.0 | 0.6 | GO:1904352 | positive regulation of protein catabolic process in the vacuole(GO:1904352) |
| 0.0 | 0.7 | GO:0021702 | cerebellar Purkinje cell layer formation(GO:0021694) cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.0 | 0.8 | GO:0015886 | heme transport(GO:0015886) |
| 0.0 | 0.6 | GO:0016080 | synaptic vesicle targeting(GO:0016080) |
| 0.0 | 0.3 | GO:0019896 | axonal transport of mitochondrion(GO:0019896) |
| 0.0 | 0.4 | GO:0071569 | protein ufmylation(GO:0071569) |
| 0.0 | 0.2 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.0 | 0.4 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.0 | 0.6 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.2 | GO:1904764 | negative regulation of fibril organization(GO:1902904) chaperone-mediated autophagy translocation complex disassembly(GO:1904764) |
| 0.0 | 0.2 | GO:0036135 | Schwann cell migration(GO:0036135) regulation of Schwann cell migration(GO:1900147) |
| 0.0 | 0.1 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.0 | 0.4 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
| 0.0 | 0.2 | GO:0046504 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.0 | 0.5 | GO:0071394 | cellular response to testosterone stimulus(GO:0071394) |
| 0.0 | 0.8 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.0 | 0.3 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.0 | 0.9 | GO:0010745 | negative regulation of macrophage derived foam cell differentiation(GO:0010745) |
| 0.0 | 0.5 | GO:1904628 | response to phorbol 13-acetate 12-myristate(GO:1904627) cellular response to phorbol 13-acetate 12-myristate(GO:1904628) |
| 0.0 | 0.2 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.0 | 0.4 | GO:0072599 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) protein targeting to ER(GO:0045047) establishment of protein localization to endoplasmic reticulum(GO:0072599) |
| 0.0 | 0.3 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.0 | 0.4 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 0.7 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.0 | 0.3 | GO:1902455 | negative regulation of stem cell population maintenance(GO:1902455) |
| 0.0 | 0.2 | GO:0090158 | endoplasmic reticulum membrane organization(GO:0090158) |
| 0.0 | 0.5 | GO:0000466 | maturation of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000466) |
| 0.0 | 0.2 | GO:0006452 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.0 | 0.1 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.0 | 0.2 | GO:0060501 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 0.0 | 0.4 | GO:0030050 | vesicle transport along actin filament(GO:0030050) |
| 0.0 | 0.1 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.0 | 0.4 | GO:0035330 | regulation of hippo signaling(GO:0035330) |
| 0.0 | 0.7 | GO:0097186 | amelogenesis(GO:0097186) |
| 0.0 | 0.1 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.0 | 0.1 | GO:0072619 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.0 | 0.2 | GO:0046015 | regulation of transcription by glucose(GO:0046015) |
| 0.0 | 0.2 | GO:0019368 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.4 | GO:0034656 | nucleobase-containing small molecule catabolic process(GO:0034656) |
| 0.0 | 0.5 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.0 | 0.1 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.0 | 0.2 | GO:0006086 | acetyl-CoA biosynthetic process from pyruvate(GO:0006086) regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.0 | 1.5 | GO:0006826 | iron ion transport(GO:0006826) |
| 0.0 | 0.1 | GO:0044829 | positive regulation by host of viral genome replication(GO:0044829) |
| 0.0 | 0.4 | GO:0031163 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 0.1 | GO:1901079 | positive regulation of relaxation of muscle(GO:1901079) |
| 0.0 | 0.3 | GO:0033235 | positive regulation of protein sumoylation(GO:0033235) |
| 0.0 | 1.3 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.7 | GO:0018146 | keratan sulfate biosynthetic process(GO:0018146) |
| 0.0 | 0.1 | GO:0043335 | protein unfolding(GO:0043335) |
| 0.0 | 0.1 | GO:0032261 | purine nucleotide salvage(GO:0032261) IMP salvage(GO:0032264) |
| 0.0 | 0.1 | GO:0009051 | pentose-phosphate shunt, oxidative branch(GO:0009051) |
| 0.0 | 0.2 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.0 | 0.1 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.0 | 0.0 | GO:0034729 | histone H3-K79 methylation(GO:0034729) |
| 0.0 | 0.3 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.0 | 0.7 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.3 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.0 | 0.5 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.5 | GO:0005960 | glycine cleavage complex(GO:0005960) |
| 0.4 | 2.6 | GO:1905202 | 3-methylcrotonyl-CoA carboxylase complex, mitochondrial(GO:0002169) methylcrotonoyl-CoA carboxylase complex(GO:1905202) |
| 0.3 | 1.0 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.2 | 2.4 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.2 | 0.7 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.1 | 0.4 | GO:0097543 | ciliary inversin compartment(GO:0097543) |
| 0.1 | 1.7 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.1 | 1.0 | GO:0098559 | cytoplasmic side of early endosome membrane(GO:0098559) |
| 0.1 | 0.7 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.1 | 0.4 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.1 | 0.5 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.1 | 0.4 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.1 | 0.7 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.1 | 2.9 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.2 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.0 | 1.2 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.1 | GO:0043224 | nuclear SCF ubiquitin ligase complex(GO:0043224) |
| 0.0 | 0.3 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.0 | 0.5 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 0.2 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.0 | 0.5 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.2 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 0.2 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 0.4 | GO:0016011 | dystroglycan complex(GO:0016011) |
| 0.0 | 1.9 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.2 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.0 | 0.3 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.2 | GO:0098575 | lumenal side of lysosomal membrane(GO:0098575) |
| 0.0 | 0.2 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.0 | 0.3 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.2 | GO:0005784 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.0 | 0.1 | GO:0071008 | U2-type post-mRNA release spliceosomal complex(GO:0071008) |
| 0.0 | 0.2 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.0 | 0.6 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 0.3 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 0.4 | GO:0032156 | septin cytoskeleton(GO:0032156) |
| 0.0 | 0.1 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.0 | 0.1 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 2.1 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.2 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.0 | 0.3 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.2 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.0 | 0.2 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 0.3 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.1 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.0 | 0.7 | GO:0090544 | BAF-type complex(GO:0090544) |
| 0.0 | 0.4 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 0.1 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 2.9 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 2.6 | GO:0055037 | recycling endosome(GO:0055037) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.2 | GO:0004807 | triose-phosphate isomerase activity(GO:0004807) |
| 0.5 | 1.5 | GO:0004618 | phosphoglycerate kinase activity(GO:0004618) |
| 0.4 | 1.3 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 0.4 | 2.2 | GO:0050252 | retinol O-fatty-acyltransferase activity(GO:0050252) |
| 0.4 | 2.6 | GO:0004485 | methylcrotonoyl-CoA carboxylase activity(GO:0004485) |
| 0.4 | 1.2 | GO:0004119 | cGMP-inhibited cyclic-nucleotide phosphodiesterase activity(GO:0004119) |
| 0.4 | 1.2 | GO:0044549 | GTP cyclohydrolase binding(GO:0044549) |
| 0.4 | 1.5 | GO:0004047 | aminomethyltransferase activity(GO:0004047) |
| 0.4 | 3.9 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.3 | 1.0 | GO:0070361 | mitochondrial light strand promoter anti-sense binding(GO:0070361) mitochondrial heavy strand promoter anti-sense binding(GO:0070362) mitochondrial heavy strand promoter sense binding(GO:0070364) |
| 0.3 | 1.1 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.3 | 2.2 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.3 | 0.8 | GO:0004315 | 3-oxoacyl-[acyl-carrier-protein] synthase activity(GO:0004315) |
| 0.3 | 0.8 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.3 | 0.8 | GO:0015439 | heme-transporting ATPase activity(GO:0015439) |
| 0.2 | 0.7 | GO:0015130 | mevalonate transmembrane transporter activity(GO:0015130) |
| 0.2 | 0.6 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.2 | 1.6 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.2 | 2.1 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.2 | 0.5 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.1 | 1.5 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.1 | 1.2 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.1 | 0.7 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.1 | 0.5 | GO:0004365 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.1 | 0.8 | GO:0070568 | guanylyltransferase activity(GO:0070568) |
| 0.1 | 0.4 | GO:0004145 | diamine N-acetyltransferase activity(GO:0004145) polyamine binding(GO:0019808) |
| 0.1 | 0.9 | GO:0015526 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.1 | 0.6 | GO:0005462 | UDP-N-acetylglucosamine transmembrane transporter activity(GO:0005462) |
| 0.1 | 0.7 | GO:0046899 | nucleoside triphosphate adenylate kinase activity(GO:0046899) |
| 0.1 | 0.3 | GO:0003863 | alpha-ketoacid dehydrogenase activity(GO:0003826) 3-methyl-2-oxobutanoate dehydrogenase (2-methylpropanoyl-transferring) activity(GO:0003863) |
| 0.1 | 0.4 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.1 | 0.3 | GO:0072510 | ferric iron transmembrane transporter activity(GO:0015091) trivalent inorganic cation transmembrane transporter activity(GO:0072510) |
| 0.1 | 0.3 | GO:0098770 | FBXO family protein binding(GO:0098770) |
| 0.1 | 0.7 | GO:0047288 | monosialoganglioside sialyltransferase activity(GO:0047288) |
| 0.1 | 0.3 | GO:0004818 | glutamate-tRNA ligase activity(GO:0004818) |
| 0.1 | 0.7 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.1 | 0.3 | GO:0030984 | kininogen binding(GO:0030984) |
| 0.1 | 1.0 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.1 | 0.7 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.1 | 2.5 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.1 | 0.8 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.1 | 0.3 | GO:0031893 | vasopressin receptor binding(GO:0031893) |
| 0.1 | 0.2 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.1 | 0.7 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.1 | 0.2 | GO:0008478 | pyridoxal kinase activity(GO:0008478) |
| 0.1 | 0.3 | GO:1990948 | ligase inhibitor activity(GO:0055104) ubiquitin ligase inhibitor activity(GO:1990948) |
| 0.0 | 0.1 | GO:0061711 | N(6)-L-threonylcarbamoyladenine synthase(GO:0061711) |
| 0.0 | 0.2 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.0 | 0.4 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.0 | 0.1 | GO:0004828 | serine-tRNA ligase activity(GO:0004828) |
| 0.0 | 0.2 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.0 | 0.4 | GO:0008312 | 7S RNA binding(GO:0008312) |
| 0.0 | 0.2 | GO:1901611 | phosphatidylglycerol binding(GO:1901611) |
| 0.0 | 0.1 | GO:0009383 | rRNA (cytosine-C5-)-methyltransferase activity(GO:0009383) |
| 0.0 | 0.4 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 0.5 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.0 | 1.2 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.2 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 0.2 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.0 | 1.3 | GO:0019200 | carbohydrate kinase activity(GO:0019200) |
| 0.0 | 0.4 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.0 | 0.2 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.0 | 0.1 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.0 | 0.1 | GO:0017057 | 6-phosphogluconolactonase activity(GO:0017057) |
| 0.0 | 1.0 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.3 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.2 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.0 | 0.2 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 0.0 | 0.7 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.3 | GO:0090599 | alpha-glucosidase activity(GO:0090599) |
| 0.0 | 0.2 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 0.3 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.1 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.0 | 0.2 | GO:0102338 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.2 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.0 | 0.1 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.0 | 0.4 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 1.1 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.2 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.0 | 0.3 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 0.1 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.0 | 0.3 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.0 | 0.1 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 0.0 | 0.1 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.6 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.5 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.5 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.0 | 0.4 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.4 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.4 | GO:0071949 | FAD binding(GO:0071949) |
| 0.0 | 0.2 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.0 | 0.3 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.2 | GO:0008565 | protein transporter activity(GO:0008565) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.5 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.4 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 1.2 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.9 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 0.7 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.6 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 0.2 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 2.0 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.9 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.1 | 1.7 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.1 | 7.0 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.1 | 1.2 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.1 | 1.6 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 4.3 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 1.2 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.5 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 1.2 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 1.1 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.7 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.9 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 2.3 | REACTOME TRIGLYCERIDE BIOSYNTHESIS | Genes involved in Triglyceride Biosynthesis |
| 0.0 | 1.5 | REACTOME REGULATION OF HYPOXIA INDUCIBLE FACTOR HIF BY OXYGEN | Genes involved in Regulation of Hypoxia-inducible Factor (HIF) by Oxygen |
| 0.0 | 1.2 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.7 | REACTOME TRANSFERRIN ENDOCYTOSIS AND RECYCLING | Genes involved in Transferrin endocytosis and recycling |
| 0.0 | 0.7 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 1.7 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.5 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.7 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.8 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.8 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 0.7 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.3 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.3 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.4 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.3 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.3 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 0.6 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.3 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 0.4 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.3 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |