Project
Illumina Body Map 2: averaged replicates
Navigation
Downloads
Results for HOXA2_HOXB1
Z-value: 2.19
Transcription factors associated with HOXA2_HOXB1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
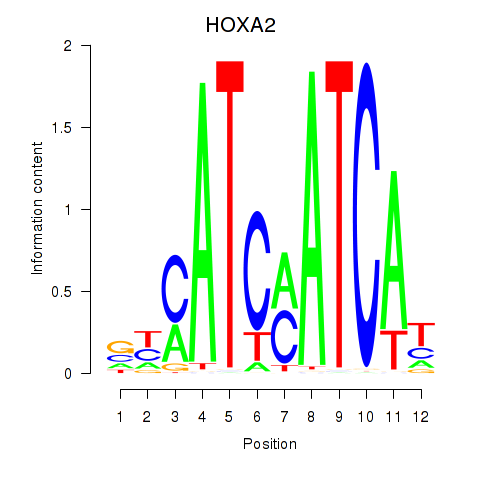
HOXA2
|
ENSG00000105996.5 | homeobox A2 |
|
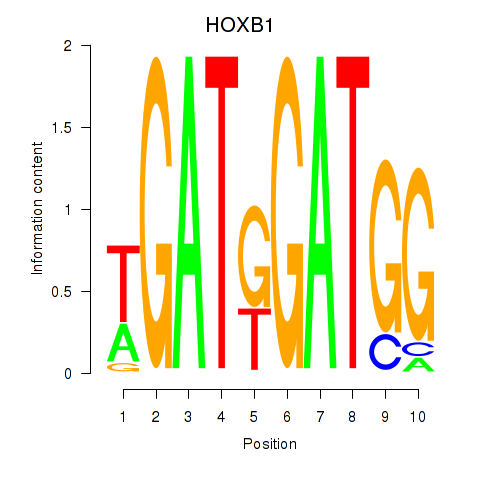
HOXB1
|
ENSG00000120094.6 | homeobox B1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXB1 | hg19_v2_chr17_-_46608272_46608385 | 0.25 | 1.6e-01 | Click! |
| HOXA2 | hg19_v2_chr7_-_27142290_27142430 | 0.07 | 7.1e-01 | Click! |
Activity profile of HOXA2_HOXB1 motif
Sorted Z-values of HOXA2_HOXB1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_-_93616340 | 14.32 |
ENST00000557420.1
ENST00000542321.2 |
RGMA
|
repulsive guidance molecule family member a |
| chr12_+_101988627 | 14.01 |
ENST00000547405.1
ENST00000452455.2 ENST00000441232.1 ENST00000360610.2 ENST00000392934.3 ENST00000547509.1 ENST00000361685.2 ENST00000549145.1 ENST00000553190.1 |
MYBPC1
|
myosin binding protein C, slow type |
| chr12_+_101988774 | 13.32 |
ENST00000545503.2
ENST00000536007.1 ENST00000541119.1 ENST00000361466.2 ENST00000551300.1 ENST00000550270.1 |
MYBPC1
|
myosin binding protein C, slow type |
| chr15_-_93616892 | 10.41 |
ENST00000556658.1
ENST00000538818.1 ENST00000425933.2 |
RGMA
|
repulsive guidance molecule family member a |
| chr16_-_49890016 | 8.37 |
ENST00000563137.2
|
ZNF423
|
zinc finger protein 423 |
| chr17_-_37353950 | 7.28 |
ENST00000394310.3
ENST00000394303.3 ENST00000344140.5 |
CACNB1
|
calcium channel, voltage-dependent, beta 1 subunit |
| chr6_+_151561085 | 6.42 |
ENST00000402676.2
|
AKAP12
|
A kinase (PRKA) anchor protein 12 |
| chr2_-_154335300 | 6.06 |
ENST00000325926.3
|
RPRM
|
reprimo, TP53 dependent G2 arrest mediator candidate |
| chr11_-_111782696 | 5.65 |
ENST00000227251.3
ENST00000526180.1 |
CRYAB
|
crystallin, alpha B |
| chr6_+_151561506 | 5.64 |
ENST00000253332.1
|
AKAP12
|
A kinase (PRKA) anchor protein 12 |
| chr11_-_111782484 | 5.39 |
ENST00000533971.1
|
CRYAB
|
crystallin, alpha B |
| chr12_-_15038779 | 4.99 |
ENST00000228938.5
ENST00000539261.1 |
MGP
|
matrix Gla protein |
| chr18_+_56887381 | 4.55 |
ENST00000256857.2
ENST00000529320.2 ENST00000420468.2 |
GRP
|
gastrin-releasing peptide |
| chr20_+_34802295 | 4.25 |
ENST00000432603.1
|
EPB41L1
|
erythrocyte membrane protein band 4.1-like 1 |
| chr17_+_37824411 | 4.25 |
ENST00000269582.2
|
PNMT
|
phenylethanolamine N-methyltransferase |
| chr16_-_79263322 | 4.14 |
ENST00000569677.1
|
RP11-679B19.2
|
RP11-679B19.2 |
| chr7_-_27170352 | 3.94 |
ENST00000428284.2
ENST00000360046.5 |
HOXA4
|
homeobox A4 |
| chr17_+_37824217 | 3.94 |
ENST00000394246.1
|
PNMT
|
phenylethanolamine N-methyltransferase |
| chr5_+_98109322 | 3.92 |
ENST00000513185.1
|
RGMB
|
repulsive guidance molecule family member b |
| chr11_-_2162162 | 3.90 |
ENST00000381389.1
|
IGF2
|
insulin-like growth factor 2 (somatomedin A) |
| chr3_+_189507523 | 3.66 |
ENST00000437221.1
ENST00000392463.2 ENST00000392461.3 ENST00000449992.1 ENST00000456148.1 |
TP63
|
tumor protein p63 |
| chr17_+_39975544 | 3.65 |
ENST00000544340.1
|
FKBP10
|
FK506 binding protein 10, 65 kDa |
| chr16_+_7560114 | 3.61 |
ENST00000570626.1
|
RBFOX1
|
RNA binding protein, fox-1 homolog (C. elegans) 1 |
| chr17_+_39975455 | 3.59 |
ENST00000455106.1
|
FKBP10
|
FK506 binding protein 10, 65 kDa |
| chr3_-_187388173 | 3.49 |
ENST00000287641.3
|
SST
|
somatostatin |
| chr17_+_37824700 | 3.49 |
ENST00000581428.1
|
PNMT
|
phenylethanolamine N-methyltransferase |
| chrX_-_13835398 | 3.33 |
ENST00000475307.1
|
GPM6B
|
glycoprotein M6B |
| chr14_-_54425475 | 3.32 |
ENST00000559642.1
|
BMP4
|
bone morphogenetic protein 4 |
| chrX_-_13835147 | 2.99 |
ENST00000493677.1
ENST00000355135.2 |
GPM6B
|
glycoprotein M6B |
| chr16_+_15596123 | 2.98 |
ENST00000452191.2
|
C16orf45
|
chromosome 16 open reading frame 45 |
| chr19_-_49944806 | 2.97 |
ENST00000221485.3
|
SLC17A7
|
solute carrier family 17 (vesicular glutamate transporter), member 7 |
| chrX_-_13835461 | 2.94 |
ENST00000316715.4
ENST00000356942.5 |
GPM6B
|
glycoprotein M6B |
| chr2_-_45236540 | 2.79 |
ENST00000303077.6
|
SIX2
|
SIX homeobox 2 |
| chr16_+_66429358 | 2.78 |
ENST00000539168.1
|
CDH5
|
cadherin 5, type 2 (vascular endothelium) |
| chr3_+_189507432 | 2.77 |
ENST00000354600.5
|
TP63
|
tumor protein p63 |
| chr10_+_24755416 | 2.71 |
ENST00000396446.1
ENST00000396445.1 ENST00000376451.2 |
KIAA1217
|
KIAA1217 |
| chr5_-_125930877 | 2.70 |
ENST00000510111.2
ENST00000509270.1 |
ALDH7A1
|
aldehyde dehydrogenase 7 family, member A1 |
| chr11_-_126870683 | 2.64 |
ENST00000525704.2
|
KIRREL3
|
kin of IRRE like 3 (Drosophila) |
| chr2_+_176995011 | 2.63 |
ENST00000548663.1
ENST00000450510.2 |
HOXD8
|
homeobox D8 |
| chr6_-_112575687 | 2.60 |
ENST00000521398.1
ENST00000424408.2 ENST00000243219.3 |
LAMA4
|
laminin, alpha 4 |
| chr14_-_21516590 | 2.50 |
ENST00000555026.1
|
NDRG2
|
NDRG family member 2 |
| chr6_-_112575912 | 2.36 |
ENST00000522006.1
ENST00000230538.7 ENST00000519932.1 |
LAMA4
|
laminin, alpha 4 |
| chr2_+_36923901 | 2.31 |
ENST00000457137.2
|
VIT
|
vitrin |
| chrX_-_10645773 | 2.29 |
ENST00000453318.2
|
MID1
|
midline 1 (Opitz/BBB syndrome) |
| chr7_-_86595190 | 2.27 |
ENST00000398276.2
ENST00000416314.1 ENST00000425689.1 |
KIAA1324L
|
KIAA1324-like |
| chr3_+_112929850 | 2.26 |
ENST00000464546.1
|
BOC
|
BOC cell adhesion associated, oncogene regulated |
| chr3_+_159481988 | 2.24 |
ENST00000472451.1
|
IQCJ-SCHIP1
|
IQCJ-SCHIP1 readthrough |
| chr6_+_155334780 | 2.21 |
ENST00000538270.1
ENST00000535231.1 |
TIAM2
|
T-cell lymphoma invasion and metastasis 2 |
| chr4_-_5021164 | 2.19 |
ENST00000506508.1
ENST00000509419.1 ENST00000307746.4 |
CYTL1
|
cytokine-like 1 |
| chr7_-_25790603 | 2.16 |
ENST00000456777.1
|
AC003090.1
|
AC003090.1 |
| chrX_+_99899180 | 2.16 |
ENST00000373004.3
|
SRPX2
|
sushi-repeat containing protein, X-linked 2 |
| chr10_-_100995540 | 2.15 |
ENST00000370546.1
ENST00000404542.1 |
HPSE2
|
heparanase 2 |
| chr6_-_112575758 | 2.13 |
ENST00000431543.2
ENST00000453937.2 ENST00000368638.4 ENST00000389463.4 |
LAMA4
|
laminin, alpha 4 |
| chr5_-_125930929 | 2.12 |
ENST00000553117.1
ENST00000447989.2 ENST00000409134.3 |
ALDH7A1
|
aldehyde dehydrogenase 7 family, member A1 |
| chr10_-_79397202 | 1.98 |
ENST00000372437.1
ENST00000372408.2 ENST00000372403.4 |
KCNMA1
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr16_+_25703274 | 1.98 |
ENST00000331351.5
|
HS3ST4
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 4 |
| chr12_-_28124903 | 1.89 |
ENST00000395872.1
ENST00000354417.3 ENST00000201015.4 |
PTHLH
|
parathyroid hormone-like hormone |
| chr2_-_23747214 | 1.88 |
ENST00000430988.1
|
AC011239.1
|
Uncharacterized protein |
| chr2_+_36923933 | 1.86 |
ENST00000497382.1
ENST00000404084.1 ENST00000379241.3 ENST00000401530.1 |
VIT
|
vitrin |
| chr20_+_34742650 | 1.86 |
ENST00000373945.1
ENST00000338074.2 |
EPB41L1
|
erythrocyte membrane protein band 4.1-like 1 |
| chr21_+_19617140 | 1.86 |
ENST00000299295.2
ENST00000338326.3 |
CHODL
|
chondrolectin |
| chr10_-_79397316 | 1.83 |
ENST00000372421.5
ENST00000457953.1 |
KCNMA1
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr3_+_148545586 | 1.80 |
ENST00000282957.4
ENST00000468341.1 |
CPB1
|
carboxypeptidase B1 (tissue) |
| chr10_-_105615164 | 1.80 |
ENST00000355946.2
ENST00000369774.4 |
SH3PXD2A
|
SH3 and PX domains 2A |
| chr2_+_36923830 | 1.76 |
ENST00000379242.3
ENST00000389975.3 |
VIT
|
vitrin |
| chr11_-_93583697 | 1.72 |
ENST00000409977.1
|
VSTM5
|
V-set and transmembrane domain containing 5 |
| chr10_+_95848824 | 1.72 |
ENST00000371385.3
ENST00000371375.1 |
PLCE1
|
phospholipase C, epsilon 1 |
| chr7_+_79765071 | 1.63 |
ENST00000457358.2
|
GNAI1
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 1 |
| chr13_-_36050819 | 1.61 |
ENST00000379919.4
|
MAB21L1
|
mab-21-like 1 (C. elegans) |
| chr10_+_124320156 | 1.61 |
ENST00000338354.3
ENST00000344338.3 ENST00000330163.4 ENST00000368909.3 ENST00000368955.3 ENST00000368956.2 |
DMBT1
|
deleted in malignant brain tumors 1 |
| chr3_-_46786245 | 1.55 |
ENST00000442359.2
|
PRSS45
|
protease, serine, 45 |
| chr10_-_100995603 | 1.53 |
ENST00000370552.3
ENST00000370549.1 |
HPSE2
|
heparanase 2 |
| chr20_+_52105495 | 1.50 |
ENST00000439873.2
|
AL354993.1
|
Cell growth-inhibiting protein 7; HCG1784586; Uncharacterized protein |
| chr7_-_150864635 | 1.49 |
ENST00000297537.4
|
GBX1
|
gastrulation brain homeobox 1 |
| chr10_-_79397547 | 1.48 |
ENST00000481070.1
|
KCNMA1
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr17_+_48609903 | 1.42 |
ENST00000268933.3
|
EPN3
|
epsin 3 |
| chr7_+_26591441 | 1.42 |
ENST00000420912.1
ENST00000457000.1 ENST00000430426.1 |
AC004947.2
|
AC004947.2 |
| chr12_-_49393092 | 1.40 |
ENST00000421952.2
|
DDN
|
dendrin |
| chr17_+_36584662 | 1.40 |
ENST00000431231.2
ENST00000437668.3 |
ARHGAP23
|
Rho GTPase activating protein 23 |
| chr9_+_124329336 | 1.38 |
ENST00000394340.3
ENST00000436835.1 ENST00000259371.2 |
DAB2IP
|
DAB2 interacting protein |
| chr7_+_20370746 | 1.36 |
ENST00000222573.4
|
ITGB8
|
integrin, beta 8 |
| chr10_-_79397479 | 1.27 |
ENST00000404771.3
|
KCNMA1
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr9_+_113431029 | 1.25 |
ENST00000189978.5
ENST00000374448.4 ENST00000374440.3 |
MUSK
|
muscle, skeletal, receptor tyrosine kinase |
| chr3_+_189507460 | 1.24 |
ENST00000434928.1
|
TP63
|
tumor protein p63 |
| chr7_+_86273218 | 1.23 |
ENST00000361669.2
|
GRM3
|
glutamate receptor, metabotropic 3 |
| chr10_-_98945264 | 1.23 |
ENST00000314867.5
|
SLIT1
|
slit homolog 1 (Drosophila) |
| chr7_-_14029515 | 1.22 |
ENST00000430479.1
ENST00000405218.2 ENST00000343495.5 |
ETV1
|
ets variant 1 |
| chr6_+_73331520 | 1.22 |
ENST00000342056.2
ENST00000355194.4 |
KCNQ5
|
potassium voltage-gated channel, KQT-like subfamily, member 5 |
| chr15_-_74501310 | 1.21 |
ENST00000423167.2
ENST00000432245.2 |
STRA6
|
stimulated by retinoic acid 6 |
| chr16_-_67427389 | 1.20 |
ENST00000562206.1
ENST00000290942.5 ENST00000393957.2 |
TPPP3
|
tubulin polymerization-promoting protein family member 3 |
| chr1_-_217250231 | 1.14 |
ENST00000493748.1
ENST00000463665.1 |
ESRRG
|
estrogen-related receptor gamma |
| chr1_+_178062855 | 1.12 |
ENST00000448150.3
|
RASAL2
|
RAS protein activator like 2 |
| chr8_-_38008783 | 1.11 |
ENST00000276449.4
|
STAR
|
steroidogenic acute regulatory protein |
| chr6_-_112575838 | 1.11 |
ENST00000455073.1
|
LAMA4
|
laminin, alpha 4 |
| chr11_-_126870655 | 1.10 |
ENST00000525144.2
|
KIRREL3
|
kin of IRRE like 3 (Drosophila) |
| chr3_+_150264555 | 1.08 |
ENST00000406576.3
ENST00000482093.1 ENST00000273435.5 |
EIF2A
|
eukaryotic translation initiation factor 2A, 65kDa |
| chr10_+_18689637 | 1.07 |
ENST00000377315.4
|
CACNB2
|
calcium channel, voltage-dependent, beta 2 subunit |
| chr17_-_57604200 | 1.07 |
ENST00000577478.1
|
RP11-567L7.6
|
RP11-567L7.6 |
| chr8_+_143530791 | 1.06 |
ENST00000517894.1
|
BAI1
|
brain-specific angiogenesis inhibitor 1 |
| chr15_+_48009541 | 1.05 |
ENST00000536845.2
ENST00000558816.1 |
SEMA6D
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D |
| chr2_+_46706725 | 1.04 |
ENST00000434431.1
|
TMEM247
|
transmembrane protein 247 |
| chr10_+_124320195 | 0.99 |
ENST00000359586.6
|
DMBT1
|
deleted in malignant brain tumors 1 |
| chr12_-_114843889 | 0.99 |
ENST00000405440.2
|
TBX5
|
T-box 5 |
| chr3_+_150264458 | 0.99 |
ENST00000487799.1
ENST00000460851.1 |
EIF2A
|
eukaryotic translation initiation factor 2A, 65kDa |
| chr12_+_7023491 | 0.98 |
ENST00000541477.1
ENST00000229277.1 |
ENO2
|
enolase 2 (gamma, neuronal) |
| chrX_+_125953746 | 0.94 |
ENST00000371125.3
|
CXorf64
|
chromosome X open reading frame 64 |
| chr1_-_67390474 | 0.92 |
ENST00000371023.3
ENST00000371022.3 ENST00000371026.3 ENST00000431318.1 |
WDR78
|
WD repeat domain 78 |
| chrX_-_10645724 | 0.91 |
ENST00000413894.1
|
MID1
|
midline 1 (Opitz/BBB syndrome) |
| chr7_-_120497178 | 0.90 |
ENST00000441017.1
ENST00000424710.1 ENST00000433758.1 |
TSPAN12
|
tetraspanin 12 |
| chr1_-_182642017 | 0.90 |
ENST00000367557.4
ENST00000258302.4 |
RGS8
|
regulator of G-protein signaling 8 |
| chr3_+_239652 | 0.86 |
ENST00000435603.1
|
CHL1
|
cell adhesion molecule L1-like |
| chr4_-_90757364 | 0.83 |
ENST00000508895.1
|
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor) |
| chr8_-_67874805 | 0.82 |
ENST00000563496.1
|
TCF24
|
transcription factor 24 |
| chr2_-_190044480 | 0.80 |
ENST00000374866.3
|
COL5A2
|
collagen, type V, alpha 2 |
| chr9_-_101471479 | 0.78 |
ENST00000259455.2
|
GABBR2
|
gamma-aminobutyric acid (GABA) B receptor, 2 |
| chr11_+_133902667 | 0.77 |
ENST00000533091.1
ENST00000527712.1 |
RP11-713P17.3
|
RP11-713P17.3 |
| chr9_+_113431059 | 0.77 |
ENST00000416899.2
|
MUSK
|
muscle, skeletal, receptor tyrosine kinase |
| chr1_-_205391178 | 0.75 |
ENST00000367153.4
ENST00000367151.2 ENST00000391936.2 ENST00000367149.3 |
LEMD1
|
LEM domain containing 1 |
| chr2_+_26624775 | 0.75 |
ENST00000288710.2
|
DRC1
|
dynein regulatory complex subunit 1 homolog (Chlamydomonas) |
| chr2_-_142888573 | 0.75 |
ENST00000434794.1
|
LRP1B
|
low density lipoprotein receptor-related protein 1B |
| chrX_+_82763265 | 0.75 |
ENST00000373200.2
|
POU3F4
|
POU class 3 homeobox 4 |
| chr18_+_18943554 | 0.74 |
ENST00000580732.2
|
GREB1L
|
growth regulation by estrogen in breast cancer-like |
| chr6_+_29426230 | 0.73 |
ENST00000442615.1
|
OR2H1
|
olfactory receptor, family 2, subfamily H, member 1 |
| chr6_+_32006042 | 0.72 |
ENST00000418967.2
|
CYP21A2
|
cytochrome P450, family 21, subfamily A, polypeptide 2 |
| chr2_+_242089833 | 0.71 |
ENST00000404405.3
ENST00000439916.1 ENST00000406106.3 ENST00000401987.1 |
PPP1R7
|
protein phosphatase 1, regulatory subunit 7 |
| chr4_-_113437328 | 0.65 |
ENST00000313341.3
|
NEUROG2
|
neurogenin 2 |
| chr4_-_90756769 | 0.64 |
ENST00000345009.4
ENST00000505199.1 ENST00000502987.1 |
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor) |
| chr14_-_81893734 | 0.62 |
ENST00000555447.1
|
STON2
|
stonin 2 |
| chr16_+_30080857 | 0.61 |
ENST00000565355.1
|
ALDOA
|
aldolase A, fructose-bisphosphate |
| chr1_-_65533390 | 0.61 |
ENST00000448344.1
|
RP4-535B20.1
|
RP4-535B20.1 |
| chr2_+_166326157 | 0.61 |
ENST00000421875.1
ENST00000314499.7 ENST00000409664.1 |
CSRNP3
|
cysteine-serine-rich nuclear protein 3 |
| chr15_-_95870348 | 0.59 |
ENST00000556899.1
|
CTD-2536I1.1
|
CTD-2536I1.1 |
| chr13_+_36050881 | 0.58 |
ENST00000537702.1
|
NBEA
|
neurobeachin |
| chr19_+_37997812 | 0.58 |
ENST00000542455.1
ENST00000587143.1 |
ZNF793
|
zinc finger protein 793 |
| chr15_-_74501360 | 0.57 |
ENST00000323940.5
|
STRA6
|
stimulated by retinoic acid 6 |
| chr12_-_86650045 | 0.56 |
ENST00000604798.1
|
MGAT4C
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme C (putative) |
| chr12_-_28125638 | 0.55 |
ENST00000545234.1
|
PTHLH
|
parathyroid hormone-like hormone |
| chr8_-_22550691 | 0.51 |
ENST00000519492.1
|
EGR3
|
early growth response 3 |
| chr9_-_73477826 | 0.49 |
ENST00000396285.1
ENST00000396292.4 ENST00000396280.5 ENST00000358082.3 ENST00000408909.2 ENST00000377097.3 |
TRPM3
|
transient receptor potential cation channel, subfamily M, member 3 |
| chr6_+_32006159 | 0.49 |
ENST00000478281.1
ENST00000471671.1 ENST00000435122.2 |
CYP21A2
|
cytochrome P450, family 21, subfamily A, polypeptide 2 |
| chr8_+_77316233 | 0.48 |
ENST00000603284.1
ENST00000603837.1 |
RP11-706J10.2
RP11-706J10.3
|
RP11-706J10.2 long intergenic non-protein coding RNA 1109 |
| chr19_+_35849723 | 0.47 |
ENST00000594310.1
|
FFAR3
|
free fatty acid receptor 3 |
| chr1_-_109584716 | 0.47 |
ENST00000531337.1
ENST00000529074.1 ENST00000369965.4 |
WDR47
|
WD repeat domain 47 |
| chr1_-_109584768 | 0.44 |
ENST00000357672.3
|
WDR47
|
WD repeat domain 47 |
| chr12_-_86650154 | 0.43 |
ENST00000552435.2
|
MGAT4C
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme C (putative) |
| chr2_-_200323414 | 0.40 |
ENST00000443023.1
|
SATB2
|
SATB homeobox 2 |
| chr14_-_24733444 | 0.39 |
ENST00000560478.1
ENST00000560443.1 |
TGM1
|
transglutaminase 1 |
| chr8_-_143961236 | 0.39 |
ENST00000377675.3
ENST00000517471.1 ENST00000292427.4 |
CYP11B1
|
cytochrome P450, family 11, subfamily B, polypeptide 1 |
| chr2_+_121493717 | 0.39 |
ENST00000418323.1
|
GLI2
|
GLI family zinc finger 2 |
| chr9_+_40028620 | 0.39 |
ENST00000426179.1
|
AL353791.1
|
AL353791.1 |
| chr12_-_88423164 | 0.38 |
ENST00000298699.2
ENST00000550553.1 |
C12orf50
|
chromosome 12 open reading frame 50 |
| chr7_-_144107320 | 0.37 |
ENST00000483238.1
ENST00000467773.1 |
NOBOX
|
NOBOX oogenesis homeobox |
| chr11_-_55905194 | 0.36 |
ENST00000301529.1
|
OR8J3
|
olfactory receptor, family 8, subfamily J, member 3 |
| chr8_+_77596014 | 0.36 |
ENST00000523885.1
|
ZFHX4
|
zinc finger homeobox 4 |
| chr1_+_68150744 | 0.35 |
ENST00000370986.4
ENST00000370985.3 |
GADD45A
|
growth arrest and DNA-damage-inducible, alpha |
| chrX_-_65253506 | 0.34 |
ENST00000427538.1
|
VSIG4
|
V-set and immunoglobulin domain containing 4 |
| chr7_-_100881041 | 0.32 |
ENST00000412417.1
ENST00000414035.1 |
CLDN15
|
claudin 15 |
| chr11_+_47430133 | 0.32 |
ENST00000531974.1
ENST00000531419.1 ENST00000531865.1 ENST00000362021.4 ENST00000354884.4 |
SLC39A13
|
solute carrier family 39 (zinc transporter), member 13 |
| chr1_-_109584608 | 0.30 |
ENST00000400794.3
ENST00000528747.1 ENST00000369962.3 ENST00000361054.3 |
WDR47
|
WD repeat domain 47 |
| chr20_+_30467600 | 0.30 |
ENST00000375934.4
ENST00000375922.4 |
TTLL9
|
tubulin tyrosine ligase-like family, member 9 |
| chr1_+_243419306 | 0.29 |
ENST00000355875.4
ENST00000391846.1 ENST00000366541.3 ENST00000343783.6 |
SDCCAG8
|
serologically defined colon cancer antigen 8 |
| chr12_+_10658489 | 0.29 |
ENST00000538173.1
|
EIF2S3L
|
Putative eukaryotic translation initiation factor 2 subunit 3-like protein |
| chrX_+_95939711 | 0.28 |
ENST00000373049.4
ENST00000324765.8 |
DIAPH2
|
diaphanous-related formin 2 |
| chr7_-_14029283 | 0.26 |
ENST00000433547.1
ENST00000405192.2 |
ETV1
|
ets variant 1 |
| chr18_-_53070913 | 0.26 |
ENST00000568186.1
ENST00000564228.1 |
TCF4
|
transcription factor 4 |
| chr9_-_73029540 | 0.26 |
ENST00000377126.2
|
KLF9
|
Kruppel-like factor 9 |
| chr7_-_78400364 | 0.26 |
ENST00000535697.1
|
MAGI2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr4_-_102267953 | 0.24 |
ENST00000523694.2
ENST00000507176.1 |
PPP3CA
|
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr9_-_127269661 | 0.24 |
ENST00000373588.4
|
NR5A1
|
nuclear receptor subfamily 5, group A, member 1 |
| chr3_-_98619999 | 0.23 |
ENST00000449482.1
|
DCBLD2
|
discoidin, CUB and LCCL domain containing 2 |
| chr17_+_43239231 | 0.21 |
ENST00000591576.1
ENST00000591070.1 ENST00000592695.1 |
HEXIM2
|
hexamethylene bis-acetamide inducible 2 |
| chr5_+_145826867 | 0.21 |
ENST00000296702.5
ENST00000394421.2 |
TCERG1
|
transcription elongation regulator 1 |
| chr12_+_7022909 | 0.19 |
ENST00000537688.1
|
ENO2
|
enolase 2 (gamma, neuronal) |
| chr15_-_95870310 | 0.18 |
ENST00000508732.2
|
CTD-2536I1.1
|
CTD-2536I1.1 |
| chr2_-_49381646 | 0.17 |
ENST00000346173.3
ENST00000406846.2 |
FSHR
|
follicle stimulating hormone receptor |
| chr5_-_145895753 | 0.17 |
ENST00000311104.2
|
GPR151
|
G protein-coupled receptor 151 |
| chr17_+_7758374 | 0.17 |
ENST00000301599.6
ENST00000574668.1 |
TMEM88
|
transmembrane protein 88 |
| chr8_-_143999259 | 0.17 |
ENST00000323110.2
|
CYP11B2
|
cytochrome P450, family 11, subfamily B, polypeptide 2 |
| chrX_+_95939638 | 0.17 |
ENST00000373061.3
ENST00000373054.4 ENST00000355827.4 |
DIAPH2
|
diaphanous-related formin 2 |
| chr12_+_57482877 | 0.16 |
ENST00000342556.6
ENST00000357680.4 |
NAB2
|
NGFI-A binding protein 2 (EGR1 binding protein 2) |
| chr2_-_44588694 | 0.16 |
ENST00000409957.1
|
PREPL
|
prolyl endopeptidase-like |
| chr12_-_86650077 | 0.16 |
ENST00000552808.2
ENST00000547225.1 |
MGAT4C
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme C (putative) |
| chr3_+_191046810 | 0.16 |
ENST00000392455.3
ENST00000392456.3 |
CCDC50
|
coiled-coil domain containing 50 |
| chr2_-_200322723 | 0.15 |
ENST00000417098.1
|
SATB2
|
SATB homeobox 2 |
| chr2_-_44588679 | 0.12 |
ENST00000409411.1
|
PREPL
|
prolyl endopeptidase-like |
| chr2_-_49381572 | 0.11 |
ENST00000454032.1
ENST00000304421.4 |
FSHR
|
follicle stimulating hormone receptor |
| chr17_+_43239191 | 0.10 |
ENST00000589230.1
|
HEXIM2
|
hexamethylene bis-acetamide inducible 2 |
| chrY_+_26997726 | 0.09 |
ENST00000382296.2
|
DAZ4
|
deleted in azoospermia 4 |
| chr19_+_44645731 | 0.07 |
ENST00000426739.2
|
ZNF234
|
zinc finger protein 234 |
| chr1_-_43833628 | 0.06 |
ENST00000413844.2
ENST00000372458.3 |
ELOVL1
|
ELOVL fatty acid elongase 1 |
| chr7_-_78400598 | 0.05 |
ENST00000536571.1
|
MAGI2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr9_+_125562370 | 0.05 |
ENST00000277309.2
|
OR1K1
|
olfactory receptor, family 1, subfamily K, member 1 |
| chr19_+_47421933 | 0.04 |
ENST00000404338.3
|
ARHGAP35
|
Rho GTPase activating protein 35 |
| chr16_-_54972581 | 0.03 |
ENST00000559802.1
ENST00000558156.1 |
CTD-3032H12.1
|
CTD-3032H12.1 |
| chr5_+_154181816 | 0.03 |
ENST00000518677.1
|
LARP1
|
La ribonucleoprotein domain family, member 1 |
| chr17_+_58755184 | 0.02 |
ENST00000589222.1
ENST00000407086.3 ENST00000390652.5 |
BCAS3
|
breast carcinoma amplified sequence 3 |
| chr1_+_36621174 | 0.01 |
ENST00000429533.2
|
MAP7D1
|
MAP7 domain containing 1 |
| chr9_-_26947220 | 0.01 |
ENST00000520884.1
|
PLAA
|
phospholipase A2-activating protein |
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXA2_HOXB1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.9 | 11.7 | GO:0042418 | epinephrine biosynthetic process(GO:0042418) |
| 1.9 | 7.7 | GO:0007499 | ectoderm and mesoderm interaction(GO:0007499) |
| 1.8 | 7.2 | GO:0000412 | histone peptidyl-prolyl isomerization(GO:0000412) |
| 1.2 | 3.7 | GO:0002121 | inter-male aggressive behavior(GO:0002121) |
| 1.2 | 27.3 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 1.2 | 4.8 | GO:0019285 | glycine betaine biosynthetic process from choline(GO:0019285) glycine betaine metabolic process(GO:0031455) glycine betaine biosynthetic process(GO:0031456) |
| 1.0 | 3.0 | GO:0042137 | sequestering of neurotransmitter(GO:0042137) |
| 1.0 | 24.7 | GO:0048681 | negative regulation of axon regeneration(GO:0048681) |
| 1.0 | 10.7 | GO:0051611 | negative regulation of neurotransmitter uptake(GO:0051581) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.9 | 4.6 | GO:1900738 | positive regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900738) |
| 0.8 | 12.1 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.7 | 3.0 | GO:1903976 | negative regulation of glial cell migration(GO:1903976) |
| 0.7 | 3.5 | GO:0008628 | hormone-mediated apoptotic signaling pathway(GO:0008628) |
| 0.7 | 3.3 | GO:2000005 | apoptotic process involved in endocardial cushion morphogenesis(GO:0003277) intermediate mesoderm morphogenesis(GO:0048390) intermediate mesoderm formation(GO:0048391) intermediate mesodermal cell differentiation(GO:0048392) regulation of cardiac muscle fiber development(GO:0055018) positive regulation of cardiac muscle fiber development(GO:0055020) bud dilation involved in lung branching(GO:0060503) BMP signaling pathway involved in ureter morphogenesis(GO:0061149) renal system segmentation(GO:0061150) BMP signaling pathway involved in renal system segmentation(GO:0061151) pulmonary artery endothelial tube morphogenesis(GO:0061155) regulation of transcription from RNA polymerase II promoter involved in mesonephros development(GO:0061216) BMP signaling pathway involved in nephric duct formation(GO:0071893) negative regulation of branch elongation involved in ureteric bud branching(GO:0072096) negative regulation of branch elongation involved in ureteric bud branching by BMP signaling pathway(GO:0072097) anterior/posterior pattern specification involved in ureteric bud development(GO:0072099) specification of ureteric bud anterior/posterior symmetry(GO:0072100) specification of ureteric bud anterior/posterior symmetry by BMP signaling pathway(GO:0072101) ureter epithelial cell differentiation(GO:0072192) negative regulation of mesenchymal cell proliferation involved in ureter development(GO:0072200) positive regulation of cell proliferation involved in outflow tract morphogenesis(GO:1901964) cardiac jelly development(GO:1905072) regulation of metanephric S-shaped body morphogenesis(GO:2000004) negative regulation of metanephric S-shaped body morphogenesis(GO:2000005) regulation of metanephric comma-shaped body morphogenesis(GO:2000006) negative regulation of metanephric comma-shaped body morphogenesis(GO:2000007) |
| 0.6 | 2.8 | GO:0072137 | condensed mesenchymal cell proliferation(GO:0072137) |
| 0.5 | 6.6 | GO:0034465 | response to carbon monoxide(GO:0034465) |
| 0.4 | 11.0 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.4 | 3.9 | GO:2000467 | positive regulation of glycogen (starch) synthase activity(GO:2000467) |
| 0.4 | 1.1 | GO:0018963 | phthalate metabolic process(GO:0018963) |
| 0.3 | 1.0 | GO:0003218 | cardiac left ventricle formation(GO:0003218) |
| 0.3 | 1.5 | GO:0019230 | proprioception(GO:0019230) |
| 0.3 | 1.8 | GO:0061143 | alveolar primary septum development(GO:0061143) |
| 0.3 | 1.7 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.3 | 1.4 | GO:0043553 | vascular endothelial growth factor receptor-2 signaling pathway(GO:0036324) negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.3 | 0.8 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.2 | 2.1 | GO:1990928 | response to amino acid starvation(GO:1990928) |
| 0.2 | 2.8 | GO:1901552 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.2 | 1.8 | GO:0072675 | osteoclast fusion(GO:0072675) |
| 0.2 | 2.0 | GO:0008582 | regulation of synaptic growth at neuromuscular junction(GO:0008582) |
| 0.2 | 0.4 | GO:0021965 | spinal cord ventral commissure morphogenesis(GO:0021965) |
| 0.2 | 3.2 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.2 | 2.6 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.1 | 0.8 | GO:0033563 | dorsal/ventral axon guidance(GO:0033563) |
| 0.1 | 2.5 | GO:0090360 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 0.1 | 1.1 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.1 | 4.5 | GO:0001502 | cartilage condensation(GO:0001502) |
| 0.1 | 0.5 | GO:0002879 | positive regulation of acute inflammatory response to non-antigenic stimulus(GO:0002879) |
| 0.1 | 2.6 | GO:0008595 | tripartite regional subdivision(GO:0007351) anterior/posterior axis specification, embryo(GO:0008595) |
| 0.1 | 7.3 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.1 | 8.4 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.1 | 2.2 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.1 | 1.6 | GO:1904776 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.1 | 2.4 | GO:0032331 | negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.1 | 4.9 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.1 | 1.8 | GO:0006705 | mineralocorticoid biosynthetic process(GO:0006705) mineralocorticoid metabolic process(GO:0008212) |
| 0.1 | 2.3 | GO:0045663 | positive regulation of myoblast differentiation(GO:0045663) |
| 0.1 | 0.3 | GO:0001545 | primary ovarian follicle growth(GO:0001545) |
| 0.1 | 1.2 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.1 | 0.5 | GO:0045586 | regulation of gamma-delta T cell differentiation(GO:0045586) |
| 0.1 | 1.5 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.1 | 0.2 | GO:1905205 | positive regulation of connective tissue replacement(GO:1905205) |
| 0.1 | 0.2 | GO:0007538 | primary sex determination(GO:0007538) |
| 0.1 | 0.4 | GO:0045229 | cell envelope organization(GO:0043163) external encapsulating structure organization(GO:0045229) |
| 0.1 | 0.3 | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 0.0 | 2.0 | GO:0030201 | heparan sulfate proteoglycan metabolic process(GO:0030201) |
| 0.0 | 0.5 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.0 | 3.7 | GO:0006027 | glycosaminoglycan catabolic process(GO:0006027) |
| 0.0 | 1.4 | GO:0001573 | ganglioside metabolic process(GO:0001573) |
| 0.0 | 2.2 | GO:0050650 | chondroitin sulfate proteoglycan biosynthetic process(GO:0050650) |
| 0.0 | 0.3 | GO:0051414 | response to cortisol(GO:0051414) |
| 0.0 | 0.5 | GO:2001054 | regulation of mesenchymal cell apoptotic process(GO:2001053) negative regulation of mesenchymal cell apoptotic process(GO:2001054) |
| 0.0 | 6.7 | GO:0045995 | regulation of embryonic development(GO:0045995) |
| 0.0 | 0.6 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 | 0.2 | GO:0016480 | negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 0.0 | 1.0 | GO:0014912 | negative regulation of smooth muscle cell migration(GO:0014912) |
| 0.0 | 5.9 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 | 0.8 | GO:0060285 | cilium-dependent cell motility(GO:0060285) |
| 0.0 | 2.0 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 1.2 | GO:0006735 | NADH regeneration(GO:0006735) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.0 | 0.9 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.9 | GO:0035640 | exploration behavior(GO:0035640) |
| 0.0 | 0.9 | GO:0003341 | cilium movement(GO:0003341) |
| 0.0 | 0.8 | GO:0007194 | negative regulation of adenylate cyclase activity(GO:0007194) |
| 0.0 | 3.0 | GO:0030509 | BMP signaling pathway(GO:0030509) |
| 0.0 | 0.3 | GO:0071850 | mitotic cell cycle arrest(GO:0071850) |
| 0.0 | 1.1 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.0 | 1.2 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.0 | 6.1 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.0 | 0.3 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 0.5 | GO:0016048 | detection of temperature stimulus(GO:0016048) |
| 0.0 | 1.1 | GO:0016525 | negative regulation of angiogenesis(GO:0016525) |
| 0.0 | 0.7 | GO:0035306 | positive regulation of dephosphorylation(GO:0035306) positive regulation of protein dephosphorylation(GO:0035307) |
| 0.0 | 0.3 | GO:1904030 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 27.3 | GO:0032982 | myosin filament(GO:0032982) |
| 0.7 | 11.0 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.5 | 1.4 | GO:1990032 | parallel fiber(GO:1990032) |
| 0.5 | 1.4 | GO:0034686 | integrin alphav-beta8 complex(GO:0034686) |
| 0.3 | 2.1 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.3 | 0.8 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.2 | 3.0 | GO:0060203 | clathrin-sculpted glutamate transport vesicle(GO:0060199) clathrin-sculpted glutamate transport vesicle membrane(GO:0060203) |
| 0.2 | 0.8 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.2 | 5.9 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.2 | 2.6 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.2 | 8.2 | GO:0005605 | basal lamina(GO:0005605) |
| 0.1 | 1.2 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.1 | 28.6 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 8.4 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.1 | 1.1 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.1 | 1.2 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.1 | 9.1 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.1 | 2.3 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.1 | 1.4 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 6.6 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 0.9 | GO:0005858 | axonemal dynein complex(GO:0005858) |
| 0.0 | 0.3 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 10.9 | GO:0045121 | membrane raft(GO:0045121) |
| 0.0 | 0.2 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 2.0 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 1.8 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 9.8 | GO:0005938 | cell cortex(GO:0005938) |
| 0.0 | 2.8 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 2.3 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 10.7 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 3.7 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 6.0 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 1.2 | GO:0030118 | clathrin coat(GO:0030118) |
| 0.0 | 1.4 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 4.2 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.3 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 1.2 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 1.1 | GO:0005930 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.9 | 11.7 | GO:0004603 | phenylethanolamine N-methyltransferase activity(GO:0004603) |
| 1.4 | 24.7 | GO:1990459 | transferrin receptor binding(GO:1990459) |
| 1.1 | 27.3 | GO:0031432 | titin binding(GO:0031432) |
| 0.9 | 3.7 | GO:0030305 | heparanase activity(GO:0030305) |
| 0.7 | 6.6 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.6 | 12.1 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.5 | 7.7 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.4 | 11.0 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.4 | 1.4 | GO:1990175 | EH domain binding(GO:1990175) |
| 0.3 | 3.0 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.3 | 8.4 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.3 | 1.5 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.2 | 1.4 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.2 | 3.3 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.2 | 2.0 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.2 | 7.2 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.2 | 1.2 | GO:0001641 | group II metabotropic glutamate receptor activity(GO:0001641) |
| 0.2 | 1.4 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.2 | 0.6 | GO:0047783 | steroid 11-beta-monooxygenase activity(GO:0004507) corticosterone 18-monooxygenase activity(GO:0047783) |
| 0.2 | 0.8 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.1 | 1.1 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.1 | 4.8 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.1 | 1.2 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.1 | 1.2 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.1 | 4.6 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.1 | 14.0 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 1.6 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.1 | 1.8 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.1 | 3.9 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.1 | 0.8 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.1 | 2.4 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.1 | 1.9 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 0.3 | GO:0004963 | follicle-stimulating hormone receptor activity(GO:0004963) |
| 0.1 | 2.6 | GO:0038187 | signaling pattern recognition receptor activity(GO:0008329) pattern recognition receptor activity(GO:0038187) |
| 0.1 | 0.3 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.1 | 0.6 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 3.0 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.0 | 1.0 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 1.8 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.9 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.9 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 1.7 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 1.8 | GO:0051183 | vitamin transporter activity(GO:0051183) |
| 0.0 | 3.5 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.7 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 1.1 | GO:0017127 | cholesterol transporter activity(GO:0017127) |
| 0.0 | 0.3 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 2.1 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.0 | 3.2 | GO:0051219 | phosphoprotein binding(GO:0051219) |
| 0.0 | 5.9 | GO:0005539 | glycosaminoglycan binding(GO:0005539) |
| 0.0 | 2.0 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.0 | 4.2 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.2 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 2.8 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 2.2 | GO:0019838 | growth factor binding(GO:0019838) |
| 0.0 | 0.0 | GO:0010698 | acetyltransferase activator activity(GO:0010698) |
| 0.0 | 0.3 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.0 | 0.3 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 0.7 | GO:0070888 | E-box binding(GO:0070888) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 32.0 | PID BMP PATHWAY | BMP receptor signaling |
| 0.1 | 8.2 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.1 | 5.1 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.1 | 4.0 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.1 | 8.0 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 5.0 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.1 | 5.9 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 4.7 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 1.4 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 1.5 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 10.7 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 2.1 | PID TCPTP PATHWAY | Signaling events mediated by TCPTP |
| 0.0 | 1.5 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 0.8 | NABA COLLAGENS | Genes encoding collagen proteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 11.7 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.4 | 27.3 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.3 | 29.5 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.1 | 4.6 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.1 | 3.9 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 9.2 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 3.7 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.1 | 3.0 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.1 | 3.2 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.1 | 2.0 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.1 | 6.1 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.1 | 1.8 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.1 | 2.0 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.1 | 1.6 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.1 | 2.8 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 2.5 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 1.1 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.0 | 1.0 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 1.8 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 1.4 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.3 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 1.0 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 1.2 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 2.4 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 1.5 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 4.8 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 3.5 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 2.2 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 1.4 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |