Project
Illumina Body Map 2: averaged replicates
Navigation
Downloads
Results for HOXB6_PRRX2
Z-value: 4.61
Transcription factors associated with HOXB6_PRRX2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
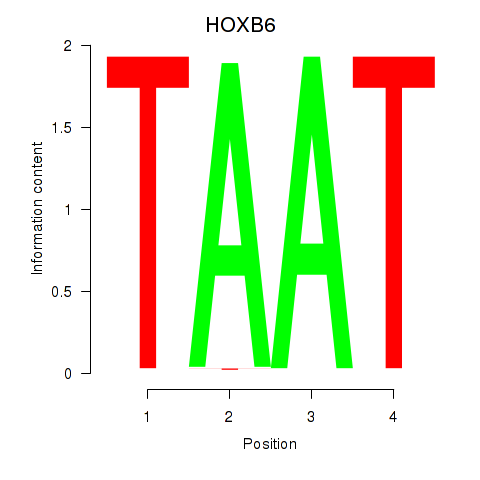
HOXB6
|
ENSG00000108511.8 | homeobox B6 |
|
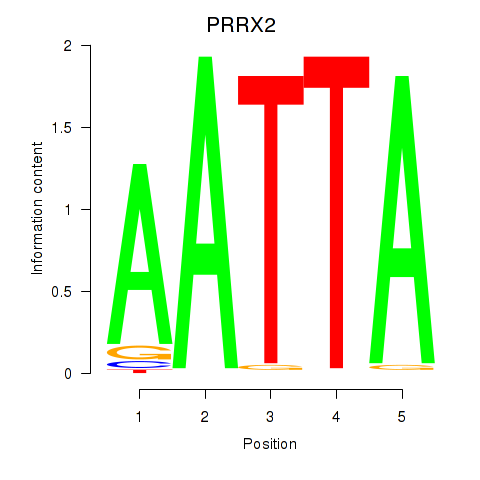
PRRX2
|
ENSG00000167157.9 | paired related homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXB6 | hg19_v2_chr17_-_46682321_46682362 | -0.20 | 2.8e-01 | Click! |
| PRRX2 | hg19_v2_chr9_+_132427883_132427951 | 0.02 | 9.1e-01 | Click! |
Activity profile of HOXB6_PRRX2 motif
Sorted Z-values of HOXB6_PRRX2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_+_7382745 | 31.40 |
ENST00000436368.2
ENST00000311745.5 ENST00000355637.4 ENST00000340209.4 |
RBFOX1
|
RNA binding protein, fox-1 homolog (C. elegans) 1 |
| chr12_-_16761007 | 24.82 |
ENST00000354662.1
ENST00000441439.2 |
LMO3
|
LIM domain only 3 (rhombotin-like 2) |
| chr2_-_50201327 | 24.41 |
ENST00000412315.1
|
NRXN1
|
neurexin 1 |
| chr7_+_107110488 | 17.72 |
ENST00000304402.4
|
GPR22
|
G protein-coupled receptor 22 |
| chr12_-_16761117 | 17.53 |
ENST00000538051.1
|
LMO3
|
LIM domain only 3 (rhombotin-like 2) |
| chr3_-_181160240 | 16.54 |
ENST00000460993.1
|
RP11-275H4.1
|
RP11-275H4.1 |
| chr2_+_162272605 | 14.60 |
ENST00000389554.3
|
TBR1
|
T-box, brain, 1 |
| chr18_+_32290218 | 13.99 |
ENST00000348997.5
ENST00000588949.1 ENST00000597599.1 |
DTNA
|
dystrobrevin, alpha |
| chr5_-_88120083 | 13.73 |
ENST00000509373.1
|
MEF2C
|
myocyte enhancer factor 2C |
| chr1_-_217250231 | 13.39 |
ENST00000493748.1
ENST00000463665.1 |
ESRRG
|
estrogen-related receptor gamma |
| chr13_-_84456527 | 13.33 |
ENST00000377084.2
|
SLITRK1
|
SLIT and NTRK-like family, member 1 |
| chr5_-_88120151 | 12.71 |
ENST00000506716.1
|
MEF2C
|
myocyte enhancer factor 2C |
| chr8_+_85618155 | 12.47 |
ENST00000523850.1
ENST00000521376.1 |
RALYL
|
RALY RNA binding protein-like |
| chr11_-_16419067 | 12.19 |
ENST00000533411.1
|
SOX6
|
SRY (sex determining region Y)-box 6 |
| chr2_-_224467093 | 12.00 |
ENST00000305409.2
|
SCG2
|
secretogranin II |
| chr1_-_216896780 | 11.46 |
ENST00000459955.1
ENST00000366937.1 ENST00000408911.3 ENST00000391890.3 |
ESRRG
|
estrogen-related receptor gamma |
| chr2_-_183291741 | 10.86 |
ENST00000351439.5
ENST00000409365.1 |
PDE1A
|
phosphodiesterase 1A, calmodulin-dependent |
| chr11_-_8285405 | 10.81 |
ENST00000335790.3
ENST00000534484.1 |
LMO1
|
LIM domain only 1 (rhombotin 1) |
| chr7_-_31380502 | 10.76 |
ENST00000297142.3
|
NEUROD6
|
neuronal differentiation 6 |
| chr3_-_73483055 | 10.69 |
ENST00000479530.1
|
PDZRN3
|
PDZ domain containing ring finger 3 |
| chr3_+_35721106 | 10.38 |
ENST00000474696.1
ENST00000412048.1 ENST00000396482.2 ENST00000432682.1 |
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa |
| chr12_-_16759440 | 10.38 |
ENST00000537304.1
|
LMO3
|
LIM domain only 3 (rhombotin-like 2) |
| chr18_-_53089538 | 10.35 |
ENST00000566777.1
|
TCF4
|
transcription factor 4 |
| chr1_+_50569575 | 10.10 |
ENST00000371827.1
|
ELAVL4
|
ELAV like neuron-specific RNA binding protein 4 |
| chr12_-_16759711 | 10.07 |
ENST00000447609.1
|
LMO3
|
LIM domain only 3 (rhombotin-like 2) |
| chr4_-_176733377 | 9.84 |
ENST00000505375.1
|
GPM6A
|
glycoprotein M6A |
| chr3_+_35721182 | 9.81 |
ENST00000413378.1
ENST00000417925.1 |
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa |
| chr1_+_65730385 | 9.57 |
ENST00000263441.7
ENST00000395325.3 |
DNAJC6
|
DnaJ (Hsp40) homolog, subfamily C, member 6 |
| chr3_+_158787041 | 9.49 |
ENST00000471575.1
ENST00000476809.1 ENST00000485419.1 |
IQCJ-SCHIP1
|
IQCJ-SCHIP1 readthrough |
| chr12_-_16762802 | 9.44 |
ENST00000534946.1
|
LMO3
|
LIM domain only 3 (rhombotin-like 2) |
| chr3_+_159557637 | 9.22 |
ENST00000445224.2
|
SCHIP1
|
schwannomin interacting protein 1 |
| chr3_+_173116225 | 9.22 |
ENST00000457714.1
|
NLGN1
|
neuroligin 1 |
| chr2_-_77749474 | 9.22 |
ENST00000409093.1
ENST00000409088.3 |
LRRTM4
|
leucine rich repeat transmembrane neuronal 4 |
| chr11_+_131240373 | 9.21 |
ENST00000374791.3
ENST00000436745.1 |
NTM
|
neurotrimin |
| chr3_+_111717511 | 8.94 |
ENST00000478951.1
ENST00000393917.2 |
TAGLN3
|
transgelin 3 |
| chr5_-_160973649 | 8.89 |
ENST00000393959.1
ENST00000517547.1 |
GABRB2
|
gamma-aminobutyric acid (GABA) A receptor, beta 2 |
| chr12_-_16762971 | 8.87 |
ENST00000540590.1
|
LMO3
|
LIM domain only 3 (rhombotin-like 2) |
| chr2_-_77749446 | 8.83 |
ENST00000409911.1
|
LRRTM4
|
leucine rich repeat transmembrane neuronal 4 |
| chr11_-_83393429 | 8.72 |
ENST00000426717.2
|
DLG2
|
discs, large homolog 2 (Drosophila) |
| chr12_-_86650045 | 8.71 |
ENST00000604798.1
|
MGAT4C
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme C (putative) |
| chr3_+_111717600 | 8.65 |
ENST00000273368.4
|
TAGLN3
|
transgelin 3 |
| chr11_-_126810521 | 8.57 |
ENST00000530572.1
|
RP11-688I9.4
|
RP11-688I9.4 |
| chr11_-_8290263 | 8.53 |
ENST00000428101.2
|
LMO1
|
LIM domain only 1 (rhombotin 1) |
| chr13_-_36050819 | 8.48 |
ENST00000379919.4
|
MAB21L1
|
mab-21-like 1 (C. elegans) |
| chr4_+_41258786 | 8.47 |
ENST00000503431.1
ENST00000284440.4 ENST00000508768.1 ENST00000512788.1 |
UCHL1
|
ubiquitin carboxyl-terminal esterase L1 (ubiquitin thiolesterase) |
| chr1_+_50574585 | 8.46 |
ENST00000371824.1
ENST00000371823.4 |
ELAVL4
|
ELAV like neuron-specific RNA binding protein 4 |
| chr8_+_77316233 | 8.42 |
ENST00000603284.1
ENST00000603837.1 |
RP11-706J10.2
RP11-706J10.3
|
RP11-706J10.2 long intergenic non-protein coding RNA 1109 |
| chr13_+_36050881 | 8.27 |
ENST00000537702.1
|
NBEA
|
neurobeachin |
| chr5_+_173930676 | 8.24 |
ENST00000504512.1
|
RP11-267A15.1
|
RP11-267A15.1 |
| chr4_-_176733897 | 8.22 |
ENST00000393658.2
|
GPM6A
|
glycoprotein M6A |
| chr12_-_87232644 | 8.20 |
ENST00000549405.2
|
MGAT4C
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme C (putative) |
| chr2_+_210444298 | 8.20 |
ENST00000445941.1
|
MAP2
|
microtubule-associated protein 2 |
| chr5_+_174151536 | 8.19 |
ENST00000239243.6
ENST00000507785.1 |
MSX2
|
msh homeobox 2 |
| chrX_-_33357558 | 8.17 |
ENST00000288447.4
|
DMD
|
dystrophin |
| chrX_+_28605516 | 8.16 |
ENST00000378993.1
|
IL1RAPL1
|
interleukin 1 receptor accessory protein-like 1 |
| chr5_-_24645078 | 7.85 |
ENST00000264463.4
|
CDH10
|
cadherin 10, type 2 (T2-cadherin) |
| chr3_+_35721130 | 7.84 |
ENST00000432450.1
|
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa |
| chr18_-_40857493 | 7.62 |
ENST00000255224.3
|
SYT4
|
synaptotagmin IV |
| chr17_-_10421853 | 7.60 |
ENST00000226207.5
|
MYH1
|
myosin, heavy chain 1, skeletal muscle, adult |
| chr18_-_3874271 | 7.59 |
ENST00000400149.3
ENST00000400155.1 ENST00000400150.3 |
DLGAP1
|
discs, large (Drosophila) homolog-associated protein 1 |
| chr10_+_24755416 | 7.41 |
ENST00000396446.1
ENST00000396445.1 ENST00000376451.2 |
KIAA1217
|
KIAA1217 |
| chr18_-_53089723 | 7.37 |
ENST00000561992.1
ENST00000562512.2 |
TCF4
|
transcription factor 4 |
| chr10_-_69455873 | 7.32 |
ENST00000433211.2
|
CTNNA3
|
catenin (cadherin-associated protein), alpha 3 |
| chr9_+_87286997 | 7.32 |
ENST00000395866.2
|
NTRK2
|
neurotrophic tyrosine kinase, receptor, type 2 |
| chr9_+_130026756 | 7.31 |
ENST00000314904.5
ENST00000373387.4 |
GARNL3
|
GTPase activating Rap/RanGAP domain-like 3 |
| chr2_-_224467002 | 7.30 |
ENST00000421386.1
ENST00000433889.1 |
SCG2
|
secretogranin II |
| chr11_+_131240593 | 7.30 |
ENST00000539799.1
|
NTM
|
neurotrimin |
| chr5_+_161494521 | 7.29 |
ENST00000356592.3
|
GABRG2
|
gamma-aminobutyric acid (GABA) A receptor, gamma 2 |
| chr1_+_170632250 | 7.27 |
ENST00000367760.3
|
PRRX1
|
paired related homeobox 1 |
| chr18_+_32173276 | 7.23 |
ENST00000591816.1
ENST00000588125.1 ENST00000598334.1 ENST00000588684.1 ENST00000554864.3 ENST00000399121.5 ENST00000595022.1 ENST00000269190.7 ENST00000399097.3 |
DTNA
|
dystrobrevin, alpha |
| chr1_-_102312517 | 7.22 |
ENST00000338858.5
|
OLFM3
|
olfactomedin 3 |
| chr5_-_124084493 | 7.19 |
ENST00000509799.1
|
ZNF608
|
zinc finger protein 608 |
| chr2_-_213403565 | 7.16 |
ENST00000342788.4
ENST00000436443.1 |
ERBB4
|
v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 4 |
| chr13_-_36429763 | 7.15 |
ENST00000379893.1
|
DCLK1
|
doublecortin-like kinase 1 |
| chr5_+_161494770 | 7.14 |
ENST00000414552.2
ENST00000361925.4 |
GABRG2
|
gamma-aminobutyric acid (GABA) A receptor, gamma 2 |
| chr2_+_182850551 | 6.96 |
ENST00000452904.1
ENST00000409137.3 ENST00000280295.3 |
PPP1R1C
|
protein phosphatase 1, regulatory (inhibitor) subunit 1C |
| chr1_-_216896674 | 6.91 |
ENST00000475275.1
ENST00000469486.1 ENST00000481543.1 |
ESRRG
|
estrogen-related receptor gamma |
| chr16_+_7560114 | 6.88 |
ENST00000570626.1
|
RBFOX1
|
RNA binding protein, fox-1 homolog (C. elegans) 1 |
| chr7_-_84122033 | 6.85 |
ENST00000424555.1
|
SEMA3A
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chr7_+_123241908 | 6.85 |
ENST00000434204.1
ENST00000437535.1 ENST00000451215.1 |
ASB15
|
ankyrin repeat and SOCS box containing 15 |
| chr2_+_166428839 | 6.84 |
ENST00000342316.4
|
CSRNP3
|
cysteine-serine-rich nuclear protein 3 |
| chr2_+_210444142 | 6.81 |
ENST00000360351.4
ENST00000361559.4 |
MAP2
|
microtubule-associated protein 2 |
| chr18_+_59000815 | 6.80 |
ENST00000262717.4
|
CDH20
|
cadherin 20, type 2 |
| chr12_+_79371565 | 6.74 |
ENST00000551304.1
|
SYT1
|
synaptotagmin I |
| chr8_+_80523321 | 6.71 |
ENST00000518111.1
|
STMN2
|
stathmin-like 2 |
| chr3_+_111718036 | 6.62 |
ENST00000455401.2
|
TAGLN3
|
transgelin 3 |
| chr2_-_175711133 | 6.59 |
ENST00000409597.1
ENST00000413882.1 |
CHN1
|
chimerin 1 |
| chr2_+_210444748 | 6.59 |
ENST00000392194.1
|
MAP2
|
microtubule-associated protein 2 |
| chr12_-_99548645 | 6.54 |
ENST00000549025.2
|
ANKS1B
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr18_-_24237339 | 6.52 |
ENST00000580191.1
|
KCTD1
|
potassium channel tetramerization domain containing 1 |
| chr12_+_41831485 | 6.49 |
ENST00000539469.2
ENST00000298919.7 |
PDZRN4
|
PDZ domain containing ring finger 4 |
| chr7_+_114055052 | 6.45 |
ENST00000462331.1
ENST00000408937.3 ENST00000403559.4 ENST00000350908.4 ENST00000393498.2 ENST00000393495.3 ENST00000378237.3 ENST00000393489.3 |
FOXP2
|
forkhead box P2 |
| chr12_-_86650077 | 6.41 |
ENST00000552808.2
ENST00000547225.1 |
MGAT4C
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme C (putative) |
| chr13_-_103053946 | 6.35 |
ENST00000376131.4
|
FGF14
|
fibroblast growth factor 14 |
| chr12_-_16758873 | 6.29 |
ENST00000535535.1
|
LMO3
|
LIM domain only 3 (rhombotin-like 2) |
| chr9_+_12693336 | 6.27 |
ENST00000381137.2
ENST00000388918.5 |
TYRP1
|
tyrosinase-related protein 1 |
| chr8_+_79503458 | 6.25 |
ENST00000518467.1
|
PKIA
|
protein kinase (cAMP-dependent, catalytic) inhibitor alpha |
| chr19_+_30719410 | 6.19 |
ENST00000585628.1
ENST00000591488.1 |
ZNF536
|
zinc finger protein 536 |
| chr9_-_13165457 | 6.13 |
ENST00000542239.1
ENST00000538841.1 ENST00000433359.2 |
MPDZ
|
multiple PDZ domain protein |
| chr12_-_16760021 | 6.10 |
ENST00000540445.1
|
LMO3
|
LIM domain only 3 (rhombotin-like 2) |
| chr2_+_47454054 | 6.05 |
ENST00000426892.1
|
AC106869.2
|
AC106869.2 |
| chr11_-_83984231 | 5.88 |
ENST00000330014.6
ENST00000537455.1 ENST00000376106.3 ENST00000418306.2 ENST00000531015.1 |
DLG2
|
discs, large homolog 2 (Drosophila) |
| chr12_-_16758304 | 5.76 |
ENST00000320122.6
|
LMO3
|
LIM domain only 3 (rhombotin-like 2) |
| chr1_-_203055129 | 5.76 |
ENST00000241651.4
|
MYOG
|
myogenin (myogenic factor 4) |
| chr3_+_63428982 | 5.75 |
ENST00000479198.1
ENST00000460711.1 ENST00000465156.1 |
SYNPR
|
synaptoporin |
| chr11_-_40315640 | 5.69 |
ENST00000278198.2
|
LRRC4C
|
leucine rich repeat containing 4C |
| chr3_+_115342349 | 5.64 |
ENST00000393780.3
|
GAP43
|
growth associated protein 43 |
| chr10_+_24528108 | 5.62 |
ENST00000438429.1
|
KIAA1217
|
KIAA1217 |
| chr5_+_36606700 | 5.56 |
ENST00000416645.2
|
SLC1A3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3 |
| chr18_+_32073253 | 5.52 |
ENST00000283365.9
ENST00000596745.1 ENST00000315456.6 |
DTNA
|
dystrobrevin, alpha |
| chr18_-_3845321 | 5.49 |
ENST00000539435.1
ENST00000400147.2 |
DLGAP1
|
discs, large (Drosophila) homolog-associated protein 1 |
| chr21_+_17442799 | 5.48 |
ENST00000602580.1
ENST00000458468.1 ENST00000602935.1 |
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr1_-_165414414 | 5.46 |
ENST00000359842.5
|
RXRG
|
retinoid X receptor, gamma |
| chr10_+_119302508 | 5.44 |
ENST00000442245.4
|
EMX2
|
empty spiracles homeobox 2 |
| chr12_-_16758059 | 5.42 |
ENST00000261169.6
|
LMO3
|
LIM domain only 3 (rhombotin-like 2) |
| chr12_-_16758835 | 5.41 |
ENST00000541295.1
|
LMO3
|
LIM domain only 3 (rhombotin-like 2) |
| chr3_+_111718173 | 5.40 |
ENST00000494932.1
|
TAGLN3
|
transgelin 3 |
| chr11_-_83393457 | 5.39 |
ENST00000404783.3
|
DLG2
|
discs, large homolog 2 (Drosophila) |
| chr11_-_84634447 | 5.37 |
ENST00000532653.1
|
DLG2
|
discs, large homolog 2 (Drosophila) |
| chr2_-_77749387 | 5.36 |
ENST00000409884.1
|
LRRTM4
|
leucine rich repeat transmembrane neuronal 4 |
| chr6_-_123958141 | 5.33 |
ENST00000334268.4
|
TRDN
|
triadin |
| chr7_-_14026123 | 5.32 |
ENST00000420159.2
ENST00000399357.3 ENST00000403527.1 |
ETV1
|
ets variant 1 |
| chr2_-_77749336 | 5.24 |
ENST00000409282.1
|
LRRTM4
|
leucine rich repeat transmembrane neuronal 4 |
| chr11_-_84634217 | 5.20 |
ENST00000524982.1
|
DLG2
|
discs, large homolog 2 (Drosophila) |
| chr8_-_42358742 | 5.18 |
ENST00000517366.1
|
SLC20A2
|
solute carrier family 20 (phosphate transporter), member 2 |
| chr4_+_88754113 | 5.16 |
ENST00000560249.1
ENST00000540395.1 ENST00000511670.1 ENST00000361056.3 |
MEPE
|
matrix extracellular phosphoglycoprotein |
| chr4_+_114066764 | 5.15 |
ENST00000511380.1
|
ANK2
|
ankyrin 2, neuronal |
| chr1_+_107682629 | 5.12 |
ENST00000370074.4
ENST00000370073.2 ENST00000370071.2 ENST00000542803.1 ENST00000370061.3 ENST00000370072.3 ENST00000370070.2 |
NTNG1
|
netrin G1 |
| chr6_+_54173227 | 5.12 |
ENST00000259782.4
ENST00000370864.3 |
TINAG
|
tubulointerstitial nephritis antigen |
| chrX_+_135279179 | 5.08 |
ENST00000370676.3
|
FHL1
|
four and a half LIM domains 1 |
| chr3_+_35722844 | 5.03 |
ENST00000436702.1
ENST00000438071.1 |
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa |
| chr2_-_80531399 | 5.01 |
ENST00000409148.1
ENST00000415098.1 ENST00000452811.1 |
LRRTM1
|
leucine rich repeat transmembrane neuronal 1 |
| chr5_-_1882858 | 5.01 |
ENST00000511126.1
ENST00000231357.2 |
IRX4
|
iroquois homeobox 4 |
| chr2_+_210443993 | 4.97 |
ENST00000392193.1
|
MAP2
|
microtubule-associated protein 2 |
| chr5_-_41794313 | 4.86 |
ENST00000512084.1
|
OXCT1
|
3-oxoacid CoA transferase 1 |
| chr4_-_87770416 | 4.86 |
ENST00000273905.6
|
SLC10A6
|
solute carrier family 10 (sodium/bile acid cotransporter), member 6 |
| chr12_-_117799446 | 4.85 |
ENST00000317775.6
ENST00000344089.3 |
NOS1
|
nitric oxide synthase 1 (neuronal) |
| chr2_-_152589670 | 4.82 |
ENST00000604864.1
ENST00000603639.1 |
NEB
|
nebulin |
| chr16_-_29910853 | 4.81 |
ENST00000308713.5
|
SEZ6L2
|
seizure related 6 homolog (mouse)-like 2 |
| chr3_+_69985734 | 4.79 |
ENST00000314557.6
ENST00000394351.3 |
MITF
|
microphthalmia-associated transcription factor |
| chr2_+_36923901 | 4.78 |
ENST00000457137.2
|
VIT
|
vitrin |
| chr18_-_3874247 | 4.78 |
ENST00000581699.1
|
DLGAP1
|
discs, large (Drosophila) homolog-associated protein 1 |
| chrX_+_135252050 | 4.74 |
ENST00000449474.1
ENST00000345434.3 |
FHL1
|
four and a half LIM domains 1 |
| chr8_+_104831472 | 4.72 |
ENST00000262231.10
ENST00000507740.1 |
RIMS2
|
regulating synaptic membrane exocytosis 2 |
| chr10_-_21186144 | 4.67 |
ENST00000377119.1
|
NEBL
|
nebulette |
| chrX_+_135251783 | 4.64 |
ENST00000394153.2
|
FHL1
|
four and a half LIM domains 1 |
| chr13_-_67802549 | 4.58 |
ENST00000328454.5
ENST00000377865.2 |
PCDH9
|
protocadherin 9 |
| chr5_-_96478466 | 4.55 |
ENST00000274382.4
|
LIX1
|
Lix1 homolog (chicken) |
| chr12_+_81101277 | 4.54 |
ENST00000228641.3
|
MYF6
|
myogenic factor 6 (herculin) |
| chr7_-_107880508 | 4.54 |
ENST00000425651.2
|
NRCAM
|
neuronal cell adhesion molecule |
| chr1_-_102312600 | 4.51 |
ENST00000359814.3
|
OLFM3
|
olfactomedin 3 |
| chr8_+_70476088 | 4.51 |
ENST00000525999.1
|
SULF1
|
sulfatase 1 |
| chr3_+_35722487 | 4.49 |
ENST00000441454.1
|
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa |
| chr6_-_46293378 | 4.48 |
ENST00000330430.6
|
RCAN2
|
regulator of calcineurin 2 |
| chr8_-_40755333 | 4.47 |
ENST00000297737.6
ENST00000315769.7 |
ZMAT4
|
zinc finger, matrin-type 4 |
| chr5_-_38557561 | 4.47 |
ENST00000511561.1
|
LIFR
|
leukemia inhibitory factor receptor alpha |
| chr7_-_81635106 | 4.43 |
ENST00000443883.1
|
CACNA2D1
|
calcium channel, voltage-dependent, alpha 2/delta subunit 1 |
| chr18_-_3845293 | 4.41 |
ENST00000400145.2
|
DLGAP1
|
discs, large (Drosophila) homolog-associated protein 1 |
| chr9_+_77230499 | 4.41 |
ENST00000396204.2
|
RORB
|
RAR-related orphan receptor B |
| chr12_-_86650154 | 4.40 |
ENST00000552435.2
|
MGAT4C
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme C (putative) |
| chr6_-_76072719 | 4.38 |
ENST00000370020.1
|
FILIP1
|
filamin A interacting protein 1 |
| chr4_+_41614909 | 4.37 |
ENST00000509454.1
ENST00000396595.3 ENST00000381753.4 |
LIMCH1
|
LIM and calponin homology domains 1 |
| chr4_+_88754069 | 4.37 |
ENST00000395102.4
ENST00000497649.2 |
MEPE
|
matrix extracellular phosphoglycoprotein |
| chr2_-_180427304 | 4.37 |
ENST00000336917.5
|
ZNF385B
|
zinc finger protein 385B |
| chr6_-_56716686 | 4.35 |
ENST00000520645.1
|
DST
|
dystonin |
| chr16_-_14109841 | 4.34 |
ENST00000576797.1
ENST00000575424.1 |
CTD-2135D7.5
|
CTD-2135D7.5 |
| chr1_-_190446759 | 4.34 |
ENST00000367462.3
|
BRINP3
|
bone morphogenetic protein/retinoic acid inducible neural-specific 3 |
| chr6_+_69942915 | 4.32 |
ENST00000604969.1
ENST00000603207.1 |
BAI3
|
brain-specific angiogenesis inhibitor 3 |
| chr10_+_45406627 | 4.32 |
ENST00000389583.4
|
TMEM72
|
transmembrane protein 72 |
| chr3_-_196911002 | 4.32 |
ENST00000452595.1
|
DLG1
|
discs, large homolog 1 (Drosophila) |
| chr10_+_69869237 | 4.30 |
ENST00000373675.3
|
MYPN
|
myopalladin |
| chr22_-_36236623 | 4.30 |
ENST00000405409.2
|
RBFOX2
|
RNA binding protein, fox-1 homolog (C. elegans) 2 |
| chr2_-_142888573 | 4.29 |
ENST00000434794.1
|
LRP1B
|
low density lipoprotein receptor-related protein 1B |
| chr2_-_158182410 | 4.28 |
ENST00000419116.2
ENST00000410096.1 |
ERMN
|
ermin, ERM-like protein |
| chr3_+_69985792 | 4.28 |
ENST00000531774.1
|
MITF
|
microphthalmia-associated transcription factor |
| chrM_+_9207 | 4.26 |
ENST00000362079.2
|
MT-CO3
|
mitochondrially encoded cytochrome c oxidase III |
| chr6_+_155537771 | 4.26 |
ENST00000275246.7
|
TIAM2
|
T-cell lymphoma invasion and metastasis 2 |
| chr7_-_14026063 | 4.25 |
ENST00000443608.1
ENST00000438956.1 |
ETV1
|
ets variant 1 |
| chr15_+_48051920 | 4.24 |
ENST00000559196.1
|
SEMA6D
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D |
| chr2_-_80531720 | 4.24 |
ENST00000416268.1
|
LRRTM1
|
leucine rich repeat transmembrane neuronal 1 |
| chr3_+_155838337 | 4.23 |
ENST00000490337.1
ENST00000389636.5 |
KCNAB1
|
potassium voltage-gated channel, shaker-related subfamily, beta member 1 |
| chr5_+_137203465 | 4.22 |
ENST00000239926.4
|
MYOT
|
myotilin |
| chr15_+_34260921 | 4.18 |
ENST00000560035.1
|
CHRM5
|
cholinergic receptor, muscarinic 5 |
| chr18_-_21891460 | 4.18 |
ENST00000357041.4
|
OSBPL1A
|
oxysterol binding protein-like 1A |
| chr4_-_186877806 | 4.18 |
ENST00000355634.5
|
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr4_-_87028478 | 4.18 |
ENST00000515400.1
ENST00000395157.3 |
MAPK10
|
mitogen-activated protein kinase 10 |
| chr1_-_91317072 | 4.17 |
ENST00000435649.2
ENST00000443802.1 |
RP4-665J23.1
|
RP4-665J23.1 |
| chr16_+_72459838 | 4.17 |
ENST00000564508.1
|
AC004158.3
|
AC004158.3 |
| chr8_-_93107443 | 4.16 |
ENST00000360348.2
ENST00000520428.1 ENST00000518992.1 ENST00000520556.1 ENST00000518317.1 ENST00000521319.1 ENST00000521375.1 ENST00000518449.1 |
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chrX_-_13835147 | 4.14 |
ENST00000493677.1
ENST00000355135.2 |
GPM6B
|
glycoprotein M6B |
| chr6_-_76203345 | 4.14 |
ENST00000393004.2
|
FILIP1
|
filamin A interacting protein 1 |
| chr15_+_48498480 | 4.12 |
ENST00000380993.3
ENST00000396577.3 |
SLC12A1
|
solute carrier family 12 (sodium/potassium/chloride transporter), member 1 |
| chr14_+_62462541 | 4.11 |
ENST00000430451.2
|
SYT16
|
synaptotagmin XVI |
| chr6_-_76203454 | 4.11 |
ENST00000237172.7
|
FILIP1
|
filamin A interacting protein 1 |
| chr17_-_27333311 | 4.10 |
ENST00000317338.12
ENST00000585644.1 |
SEZ6
|
seizure related 6 homolog (mouse) |
| chr15_+_51669444 | 4.10 |
ENST00000396399.2
|
GLDN
|
gliomedin |
| chr2_+_170366203 | 4.09 |
ENST00000284669.1
|
KLHL41
|
kelch-like family member 41 |
| chr9_+_103204553 | 4.09 |
ENST00000502978.1
ENST00000334943.6 |
MSANTD3-TMEFF1
TMEFF1
|
MSANTD3-TMEFF1 readthrough transmembrane protein with EGF-like and two follistatin-like domains 1 |
| chrX_+_135251835 | 4.06 |
ENST00000456445.1
|
FHL1
|
four and a half LIM domains 1 |
| chr12_-_28124903 | 4.06 |
ENST00000395872.1
ENST00000354417.3 ENST00000201015.4 |
PTHLH
|
parathyroid hormone-like hormone |
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXB6_PRRX2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.1 | 112.9 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 4.8 | 33.8 | GO:0003138 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 4.8 | 14.4 | GO:0048174 | negative regulation of short-term neuronal synaptic plasticity(GO:0048174) |
| 4.4 | 17.7 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 4.3 | 17.2 | GO:1903045 | neural crest cell migration involved in sympathetic nervous system development(GO:1903045) |
| 3.8 | 15.0 | GO:1901846 | positive regulation of cell communication by electrical coupling involved in cardiac conduction(GO:1901846) |
| 3.6 | 17.8 | GO:0099543 | retrograde trans-synaptic signaling by soluble gas(GO:0098923) trans-synaptic signaling by soluble gas(GO:0099543) |
| 3.3 | 19.7 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 3.2 | 16.2 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 3.1 | 18.8 | GO:0021779 | oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 2.9 | 17.7 | GO:0021764 | amygdala development(GO:0021764) |
| 2.9 | 8.8 | GO:0043438 | acetoacetic acid metabolic process(GO:0043438) |
| 2.9 | 25.8 | GO:0097116 | gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 2.7 | 8.2 | GO:0007412 | axon target recognition(GO:0007412) |
| 2.7 | 8.2 | GO:0051795 | positive regulation of catagen(GO:0051795) activation of meiosis(GO:0090427) |
| 2.5 | 15.2 | GO:0099551 | synaptic signaling via neuropeptide(GO:0099538) trans-synaptic signaling by neuropeptide(GO:0099540) trans-synaptic signaling by neuropeptide, modulating synaptic transmission(GO:0099551) |
| 2.4 | 7.2 | GO:0061026 | cardiac muscle tissue regeneration(GO:0061026) |
| 2.3 | 7.0 | GO:0060304 | regulation of phosphatidylinositol dephosphorylation(GO:0060304) |
| 1.9 | 24.7 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 1.8 | 41.5 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 1.8 | 12.3 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 1.7 | 5.2 | GO:0061033 | secretion by lung epithelial cell involved in lung growth(GO:0061033) |
| 1.7 | 6.7 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
| 1.6 | 14.6 | GO:1901843 | positive regulation of high voltage-gated calcium channel activity(GO:1901843) |
| 1.4 | 5.8 | GO:0098582 | innate vocalization behavior(GO:0098582) |
| 1.3 | 14.8 | GO:0048664 | neuron fate determination(GO:0048664) |
| 1.3 | 7.9 | GO:0010650 | positive regulation of cell communication by electrical coupling(GO:0010650) |
| 1.3 | 6.5 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 1.3 | 16.8 | GO:0002091 | negative regulation of receptor internalization(GO:0002091) |
| 1.3 | 17.9 | GO:0048245 | eosinophil chemotaxis(GO:0048245) |
| 1.3 | 17.9 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 1.3 | 3.8 | GO:0031548 | regulation of brain-derived neurotrophic factor receptor signaling pathway(GO:0031548) |
| 1.2 | 54.2 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 1.2 | 11.9 | GO:0051611 | negative regulation of neurotransmitter uptake(GO:0051581) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 1.2 | 5.8 | GO:0021763 | subthalamic nucleus development(GO:0021763) prolactin secreting cell differentiation(GO:0060127) superior vena cava morphogenesis(GO:0060578) |
| 1.1 | 9.0 | GO:0035995 | detection of muscle stretch(GO:0035995) |
| 1.1 | 7.5 | GO:0048165 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 1.1 | 13.7 | GO:0061669 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 1.0 | 8.3 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 1.0 | 3.1 | GO:1903422 | negative regulation of synaptic vesicle recycling(GO:1903422) |
| 1.0 | 11.1 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 1.0 | 20.1 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 1.0 | 7.9 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 1.0 | 4.8 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.9 | 7.6 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.9 | 3.6 | GO:0061348 | chemoattraction of serotonergic neuron axon(GO:0036517) planar cell polarity pathway involved in outflow tract morphogenesis(GO:0061347) planar cell polarity pathway involved in ventricular septum morphogenesis(GO:0061348) planar cell polarity pathway involved in cardiac right atrium morphogenesis(GO:0061349) planar cell polarity pathway involved in cardiac muscle tissue morphogenesis(GO:0061350) planar cell polarity pathway involved in pericardium morphogenesis(GO:0061354) chemoattraction of axon(GO:0061642) negative regulation of cell proliferation in midbrain(GO:1904934) planar cell polarity pathway involved in midbrain dopaminergic neuron differentiation(GO:1904955) |
| 0.9 | 7.2 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.9 | 7.1 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.9 | 2.7 | GO:1901877 | regulation of calcium ion binding(GO:1901876) negative regulation of calcium ion binding(GO:1901877) |
| 0.9 | 8.8 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.9 | 5.2 | GO:0060316 | positive regulation of ryanodine-sensitive calcium-release channel activity(GO:0060316) |
| 0.9 | 9.6 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.8 | 3.4 | GO:0016062 | adaptation of rhodopsin mediated signaling(GO:0016062) light adaption(GO:0036367) |
| 0.8 | 2.5 | GO:0002121 | inter-male aggressive behavior(GO:0002121) |
| 0.8 | 3.4 | GO:1902725 | negative regulation of satellite cell differentiation(GO:1902725) |
| 0.8 | 2.5 | GO:0060829 | regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060827) negative regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060829) |
| 0.8 | 8.2 | GO:0097105 | presynaptic membrane assembly(GO:0097105) |
| 0.8 | 4.1 | GO:0008628 | hormone-mediated apoptotic signaling pathway(GO:0008628) |
| 0.8 | 2.4 | GO:0060024 | rhythmic synaptic transmission(GO:0060024) |
| 0.8 | 9.5 | GO:0021800 | cerebral cortex tangential migration(GO:0021800) |
| 0.8 | 7.7 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.8 | 11.6 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.8 | 8.4 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 0.8 | 3.0 | GO:0097156 | fasciculation of motor neuron axon(GO:0097156) |
| 0.8 | 3.8 | GO:2000639 | regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.8 | 9.0 | GO:0006537 | glutamate biosynthetic process(GO:0006537) |
| 0.7 | 14.7 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.7 | 2.2 | GO:0097325 | melanocyte proliferation(GO:0097325) |
| 0.7 | 1.4 | GO:0035992 | tendon cell differentiation(GO:0035990) tendon formation(GO:0035992) |
| 0.7 | 4.9 | GO:0003199 | endocardial cushion to mesenchymal transition involved in heart valve formation(GO:0003199) |
| 0.7 | 3.4 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.7 | 7.4 | GO:0060686 | negative regulation of prostatic bud formation(GO:0060686) |
| 0.7 | 10.6 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.6 | 1.9 | GO:1902897 | regulation of postsynaptic density protein 95 clustering(GO:1902897) |
| 0.6 | 2.6 | GO:1990926 | short-term synaptic potentiation(GO:1990926) |
| 0.6 | 50.0 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.6 | 3.8 | GO:0018032 | protein amidation(GO:0018032) |
| 0.6 | 18.5 | GO:0021542 | dentate gyrus development(GO:0021542) |
| 0.6 | 1.2 | GO:1901979 | regulation of inward rectifier potassium channel activity(GO:1901979) |
| 0.6 | 4.2 | GO:0007406 | negative regulation of neuroblast proliferation(GO:0007406) |
| 0.6 | 9.0 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.6 | 1.8 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.6 | 2.3 | GO:2000866 | positive regulation of estrogen secretion(GO:2000863) positive regulation of estradiol secretion(GO:2000866) |
| 0.6 | 2.3 | GO:0060133 | somatotropin secreting cell development(GO:0060133) |
| 0.6 | 4.6 | GO:0021730 | trigeminal sensory nucleus development(GO:0021730) principal sensory nucleus of trigeminal nerve development(GO:0021740) negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.6 | 4.5 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.6 | 5.0 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.6 | 4.4 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.5 | 2.7 | GO:1900039 | positive regulation of cellular response to hypoxia(GO:1900039) |
| 0.5 | 0.5 | GO:1990709 | presynaptic active zone organization(GO:1990709) |
| 0.5 | 41.2 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.5 | 30.1 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.5 | 1.1 | GO:0051389 | inactivation of MAPKK activity(GO:0051389) |
| 0.5 | 6.2 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.5 | 24.6 | GO:0008038 | neuron recognition(GO:0008038) |
| 0.5 | 5.1 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.5 | 6.6 | GO:0086073 | bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 0.5 | 4.5 | GO:0060373 | regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 0.5 | 27.5 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.5 | 1.5 | GO:0031106 | septin ring assembly(GO:0000921) septin ring organization(GO:0031106) |
| 0.5 | 11.4 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.5 | 8.2 | GO:0008627 | intrinsic apoptotic signaling pathway in response to osmotic stress(GO:0008627) |
| 0.5 | 1.9 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.5 | 2.4 | GO:0008616 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.5 | 2.9 | GO:0032571 | response to vitamin K(GO:0032571) regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904207) positive regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904209) bone regeneration(GO:1990523) |
| 0.5 | 7.1 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 0.5 | 2.4 | GO:0003051 | angiotensin-mediated drinking behavior(GO:0003051) |
| 0.5 | 0.5 | GO:0032148 | activation of protein kinase B activity(GO:0032148) |
| 0.4 | 6.7 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.4 | 2.2 | GO:0010513 | positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) |
| 0.4 | 12.7 | GO:0060074 | synapse maturation(GO:0060074) |
| 0.4 | 1.7 | GO:0086069 | bundle of His cell to Purkinje myocyte communication(GO:0086069) |
| 0.4 | 2.9 | GO:0071442 | positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.4 | 1.2 | GO:0018315 | molybdenum incorporation into molybdenum-molybdopterin complex(GO:0018315) metal incorporation into metallo-molybdopterin complex(GO:0042040) glycine receptor clustering(GO:0072579) |
| 0.4 | 33.4 | GO:0046426 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.4 | 7.6 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 0.4 | 4.3 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 0.4 | 1.2 | GO:0042245 | RNA repair(GO:0042245) |
| 0.4 | 29.3 | GO:1901016 | regulation of potassium ion transmembrane transporter activity(GO:1901016) |
| 0.4 | 3.5 | GO:0035494 | SNARE complex disassembly(GO:0035494) |
| 0.4 | 9.7 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.4 | 3.8 | GO:0003025 | regulation of systemic arterial blood pressure by baroreceptor feedback(GO:0003025) |
| 0.4 | 8.8 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.4 | 2.6 | GO:0090324 | negative regulation of oxidative phosphorylation(GO:0090324) |
| 0.4 | 4.8 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.4 | 5.2 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.4 | 1.1 | GO:0071963 | establishment or maintenance of cell polarity regulating cell shape(GO:0071963) |
| 0.4 | 2.2 | GO:0098886 | modification of dendritic spine(GO:0098886) |
| 0.4 | 1.4 | GO:0033122 | negative regulation of cyclic nucleotide catabolic process(GO:0030806) negative regulation of cAMP catabolic process(GO:0030821) negative regulation of purine nucleotide catabolic process(GO:0033122) |
| 0.4 | 3.5 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.4 | 2.1 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.3 | 8.0 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.3 | 5.5 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.3 | 3.1 | GO:2000543 | positive regulation of gastrulation(GO:2000543) |
| 0.3 | 5.1 | GO:0010454 | negative regulation of cell fate commitment(GO:0010454) |
| 0.3 | 1.7 | GO:0006127 | glycerophosphate shuttle(GO:0006127) |
| 0.3 | 5.4 | GO:2001014 | regulation of skeletal muscle cell differentiation(GO:2001014) |
| 0.3 | 2.4 | GO:0046373 | arabinose metabolic process(GO:0019566) L-arabinose metabolic process(GO:0046373) |
| 0.3 | 2.4 | GO:1900825 | regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900825) |
| 0.3 | 4.0 | GO:0098828 | positive regulation of inhibitory postsynaptic potential(GO:0097151) modulation of inhibitory postsynaptic potential(GO:0098828) |
| 0.3 | 1.0 | GO:0021812 | neuronal-glial interaction involved in cerebral cortex radial glia guided migration(GO:0021812) |
| 0.3 | 19.0 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.3 | 3.3 | GO:0060449 | bud elongation involved in lung branching(GO:0060449) |
| 0.3 | 1.6 | GO:0035948 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) |
| 0.3 | 3.3 | GO:0032596 | protein transport into membrane raft(GO:0032596) |
| 0.3 | 6.2 | GO:0044557 | relaxation of smooth muscle(GO:0044557) |
| 0.3 | 1.6 | GO:1900454 | positive regulation of long term synaptic depression(GO:1900454) |
| 0.3 | 8.7 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.3 | 3.2 | GO:0021984 | adenohypophysis development(GO:0021984) |
| 0.3 | 1.9 | GO:0036343 | psychomotor behavior(GO:0036343) motor behavior(GO:0061744) |
| 0.3 | 1.6 | GO:0070495 | regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 0.3 | 3.4 | GO:0019896 | axonal transport of mitochondrion(GO:0019896) |
| 0.3 | 2.4 | GO:0021681 | cerebellar granular layer development(GO:0021681) |
| 0.3 | 0.6 | GO:0033563 | dorsal/ventral axon guidance(GO:0033563) |
| 0.3 | 8.4 | GO:0070168 | negative regulation of biomineral tissue development(GO:0070168) |
| 0.3 | 1.2 | GO:0060529 | ectoderm and mesoderm interaction(GO:0007499) squamous basal epithelial stem cell differentiation involved in prostate gland acinus development(GO:0060529) |
| 0.3 | 5.7 | GO:0030949 | positive regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030949) |
| 0.3 | 4.2 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.3 | 63.1 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.3 | 6.3 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.3 | 15.5 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.3 | 1.5 | GO:0072137 | condensed mesenchymal cell proliferation(GO:0072137) |
| 0.3 | 4.1 | GO:0007617 | mating behavior(GO:0007617) multi-organism reproductive behavior(GO:0044705) |
| 0.3 | 1.2 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.3 | 1.8 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.3 | 1.5 | GO:1903860 | negative regulation of dendrite extension(GO:1903860) |
| 0.3 | 2.3 | GO:0072205 | metanephric part of ureteric bud development(GO:0035502) metanephric collecting duct development(GO:0072205) |
| 0.3 | 1.7 | GO:0061146 | Peyer's patch morphogenesis(GO:0061146) |
| 0.3 | 1.4 | GO:2000821 | regulation of grooming behavior(GO:2000821) |
| 0.3 | 1.1 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.3 | 2.2 | GO:0070459 | prolactin secretion(GO:0070459) |
| 0.3 | 0.8 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.3 | 2.7 | GO:0060012 | synaptic transmission, glycinergic(GO:0060012) |
| 0.3 | 6.2 | GO:0051654 | establishment of mitochondrion localization(GO:0051654) |
| 0.3 | 0.3 | GO:0097102 | endothelial tip cell fate specification(GO:0097102) |
| 0.3 | 1.6 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.3 | 1.8 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.3 | 1.8 | GO:0032468 | Golgi calcium ion homeostasis(GO:0032468) |
| 0.3 | 2.0 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.3 | 2.3 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.3 | 1.3 | GO:0048263 | determination of dorsal identity(GO:0048263) |
| 0.2 | 12.9 | GO:1901379 | regulation of potassium ion transmembrane transport(GO:1901379) |
| 0.2 | 4.4 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.2 | 1.0 | GO:2000807 | positive regulation of presynaptic membrane organization(GO:1901631) regulation of synaptic vesicle clustering(GO:2000807) positive regulation of synaptic vesicle clustering(GO:2000809) |
| 0.2 | 2.5 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.2 | 1.7 | GO:1900452 | regulation of long term synaptic depression(GO:1900452) |
| 0.2 | 0.7 | GO:2000393 | negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 0.2 | 2.8 | GO:0038044 | transforming growth factor-beta secretion(GO:0038044) |
| 0.2 | 3.7 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.2 | 5.1 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.2 | 11.3 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.2 | 1.2 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.2 | 8.2 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.2 | 0.9 | GO:1903824 | regulation of telomere maintenance via recombination(GO:0032207) negative regulation of telomere maintenance via recombination(GO:0032208) negative regulation of single strand break repair(GO:1903517) negative regulation of beta-galactosidase activity(GO:1903770) telomere single strand break repair(GO:1903823) negative regulation of telomere single strand break repair(GO:1903824) |
| 0.2 | 1.8 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.2 | 3.1 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.2 | 3.5 | GO:0035640 | exploration behavior(GO:0035640) |
| 0.2 | 1.9 | GO:0031284 | positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.2 | 2.8 | GO:0038166 | angiotensin-activated signaling pathway(GO:0038166) |
| 0.2 | 3.1 | GO:0015705 | iodide transport(GO:0015705) |
| 0.2 | 3.8 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.2 | 6.5 | GO:0070884 | regulation of calcineurin-NFAT signaling cascade(GO:0070884) |
| 0.2 | 0.8 | GO:1902358 | sulfate transmembrane transport(GO:1902358) |
| 0.2 | 1.9 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.2 | 6.4 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.2 | 35.5 | GO:0048813 | dendrite morphogenesis(GO:0048813) |
| 0.2 | 2.2 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.2 | 1.8 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.2 | 1.0 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.2 | 1.4 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.2 | 2.0 | GO:0035507 | regulation of myosin-light-chain-phosphatase activity(GO:0035507) |
| 0.2 | 0.6 | GO:0070375 | ERK5 cascade(GO:0070375) |
| 0.2 | 5.2 | GO:0010880 | regulation of release of sequestered calcium ion into cytosol by sarcoplasmic reticulum(GO:0010880) |
| 0.2 | 3.8 | GO:2000394 | positive regulation of lamellipodium morphogenesis(GO:2000394) |
| 0.2 | 1.1 | GO:0042427 | serotonin biosynthetic process(GO:0042427) |
| 0.2 | 2.4 | GO:1903546 | protein localization to photoreceptor outer segment(GO:1903546) |
| 0.2 | 1.5 | GO:0010669 | epithelial structure maintenance(GO:0010669) |
| 0.2 | 1.5 | GO:0009253 | peptidoglycan metabolic process(GO:0000270) peptidoglycan catabolic process(GO:0009253) |
| 0.2 | 0.2 | GO:0044851 | anagen(GO:0042640) hair cycle phase(GO:0044851) |
| 0.2 | 1.8 | GO:1902365 | regulation of spindle elongation(GO:0032887) regulation of mitotic spindle elongation(GO:0032888) anastral spindle assembly(GO:0055048) protein localization to spindle pole body(GO:0071988) regulation of protein localization to spindle pole body(GO:1902363) positive regulation of protein localization to spindle pole body(GO:1902365) positive regulation of mitotic spindle elongation(GO:1902846) |
| 0.2 | 0.7 | GO:0009956 | radial pattern formation(GO:0009956) |
| 0.2 | 20.7 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.2 | 0.9 | GO:0060355 | positive regulation of cell adhesion molecule production(GO:0060355) |
| 0.2 | 1.9 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.2 | 3.2 | GO:0051531 | NFAT protein import into nucleus(GO:0051531) |
| 0.2 | 2.8 | GO:0097369 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.2 | 1.3 | GO:0010025 | wax biosynthetic process(GO:0010025) wax metabolic process(GO:0010166) |
| 0.2 | 5.4 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.2 | 0.6 | GO:0090027 | negative regulation of monocyte chemotaxis(GO:0090027) |
| 0.2 | 2.6 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.2 | 2.2 | GO:0035873 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) |
| 0.2 | 5.9 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.2 | 7.8 | GO:0006688 | glycosphingolipid biosynthetic process(GO:0006688) |
| 0.2 | 2.1 | GO:0007635 | chemosensory behavior(GO:0007635) |
| 0.2 | 2.2 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.2 | 3.0 | GO:0032331 | negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.2 | 2.0 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.2 | 2.8 | GO:0048016 | inositol phosphate-mediated signaling(GO:0048016) |
| 0.2 | 0.8 | GO:1903593 | regulation of histamine secretion by mast cell(GO:1903593) |
| 0.2 | 0.6 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.2 | 0.8 | GO:2000507 | positive regulation of energy homeostasis(GO:2000507) |
| 0.2 | 1.4 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.2 | 0.5 | GO:0002940 | tRNA N2-guanine methylation(GO:0002940) |
| 0.1 | 2.5 | GO:1990035 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.1 | 4.7 | GO:0010506 | regulation of autophagy(GO:0010506) |
| 0.1 | 0.6 | GO:2000230 | negative regulation of pancreatic stellate cell proliferation(GO:2000230) |
| 0.1 | 0.7 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.1 | 2.6 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.1 | 0.9 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.1 | 0.4 | GO:0045918 | negative regulation of cytolysis(GO:0045918) |
| 0.1 | 0.7 | GO:0000117 | regulation of transcription involved in G2/M transition of mitotic cell cycle(GO:0000117) |
| 0.1 | 0.9 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 0.1 | 2.4 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.1 | 1.6 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.1 | 10.0 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.1 | 1.1 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) |
| 0.1 | 0.3 | GO:1903774 | positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 0.1 | 1.3 | GO:0010727 | negative regulation of hydrogen peroxide metabolic process(GO:0010727) |
| 0.1 | 0.5 | GO:0034238 | macrophage fusion(GO:0034238) regulation of macrophage fusion(GO:0034239) positive regulation of macrophage fusion(GO:0034241) |
| 0.1 | 3.0 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.1 | 2.4 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.1 | 0.8 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.1 | 7.9 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.1 | 1.1 | GO:1904220 | regulation of serine C-palmitoyltransferase activity(GO:1904220) |
| 0.1 | 0.9 | GO:0098915 | membrane repolarization during ventricular cardiac muscle cell action potential(GO:0098915) |
| 0.1 | 0.9 | GO:1904381 | Golgi apparatus mannose trimming(GO:1904381) |
| 0.1 | 0.5 | GO:0006422 | aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.1 | 1.5 | GO:0090103 | cochlea morphogenesis(GO:0090103) |
| 0.1 | 5.1 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.1 | 0.8 | GO:1904590 | negative regulation of protein import into nucleus(GO:0042308) negative regulation of protein import(GO:1904590) |
| 0.1 | 1.0 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.1 | 1.1 | GO:0021914 | negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021914) |
| 0.1 | 7.2 | GO:2000144 | positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.1 | 0.8 | GO:0021891 | olfactory bulb interneuron development(GO:0021891) |
| 0.1 | 1.2 | GO:0071670 | smooth muscle cell chemotaxis(GO:0071670) |
| 0.1 | 1.6 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.1 | 0.6 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.1 | 0.8 | GO:0052199 | negative regulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052199) |
| 0.1 | 2.5 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.1 | 24.3 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.1 | 0.2 | GO:0090149 | mitochondrial membrane fission(GO:0090149) |
| 0.1 | 0.7 | GO:0032528 | microvillus organization(GO:0032528) |
| 0.1 | 1.2 | GO:0010832 | negative regulation of myotube differentiation(GO:0010832) |
| 0.1 | 0.4 | GO:1990418 | response to insulin-like growth factor stimulus(GO:1990418) |
| 0.1 | 4.2 | GO:0014047 | glutamate secretion(GO:0014047) |
| 0.1 | 0.4 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.1 | 1.4 | GO:0016264 | gap junction assembly(GO:0016264) |
| 0.1 | 0.4 | GO:0046462 | monoacylglycerol metabolic process(GO:0046462) monoacylglycerol catabolic process(GO:0052651) |
| 0.1 | 4.8 | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.1 | 2.6 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.1 | 1.0 | GO:0009972 | cytidine catabolic process(GO:0006216) cytidine deamination(GO:0009972) cytidine metabolic process(GO:0046087) |
| 0.1 | 2.1 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.1 | 0.6 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.1 | 0.9 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.1 | 1.6 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.1 | 1.1 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.1 | 2.1 | GO:0006309 | apoptotic DNA fragmentation(GO:0006309) |
| 0.1 | 0.1 | GO:0034724 | DNA replication-independent nucleosome organization(GO:0034724) |
| 0.1 | 3.4 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.1 | 0.4 | GO:0021847 | ventricular zone neuroblast division(GO:0021847) |
| 0.1 | 0.7 | GO:0060011 | Sertoli cell proliferation(GO:0060011) |
| 0.1 | 4.0 | GO:0014912 | negative regulation of smooth muscle cell migration(GO:0014912) |
| 0.1 | 3.9 | GO:0003301 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.1 | 0.7 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.1 | 2.0 | GO:0007628 | adult walking behavior(GO:0007628) |
| 0.1 | 0.7 | GO:0032916 | transforming growth factor beta3 production(GO:0032907) regulation of transforming growth factor beta3 production(GO:0032910) positive regulation of transforming growth factor beta3 production(GO:0032916) negative regulation of lung blood pressure(GO:0061767) |
| 0.1 | 1.8 | GO:0051775 | response to redox state(GO:0051775) |
| 0.1 | 3.0 | GO:1902895 | positive regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902895) |
| 0.1 | 5.4 | GO:0050771 | negative regulation of axonogenesis(GO:0050771) |
| 0.1 | 0.7 | GO:2001214 | positive regulation of vasculogenesis(GO:2001214) |
| 0.1 | 1.2 | GO:0008306 | associative learning(GO:0008306) |
| 0.1 | 1.5 | GO:0071850 | mitotic cell cycle arrest(GO:0071850) |
| 0.1 | 0.7 | GO:0009214 | cAMP catabolic process(GO:0006198) cyclic nucleotide catabolic process(GO:0009214) |
| 0.1 | 6.5 | GO:0050806 | positive regulation of synaptic transmission(GO:0050806) |
| 0.1 | 0.9 | GO:0042090 | interleukin-12 biosynthetic process(GO:0042090) regulation of interleukin-12 biosynthetic process(GO:0045075) |
| 0.1 | 0.5 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.1 | 0.3 | GO:0051957 | positive regulation of amino acid transport(GO:0051957) |
| 0.1 | 0.1 | GO:0097476 | spinal cord motor neuron migration(GO:0097476) |
| 0.1 | 0.2 | GO:0023016 | signal transduction by trans-phosphorylation(GO:0023016) |
| 0.1 | 0.7 | GO:0009629 | response to gravity(GO:0009629) |
| 0.1 | 0.3 | GO:0060723 | spongiotrophoblast cell proliferation(GO:0060720) regulation of spongiotrophoblast cell proliferation(GO:0060721) cell proliferation involved in embryonic placenta development(GO:0060722) regulation of cell proliferation involved in embryonic placenta development(GO:0060723) |
| 0.1 | 0.7 | GO:0032233 | positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.1 | 3.7 | GO:0045599 | negative regulation of fat cell differentiation(GO:0045599) |
| 0.1 | 0.7 | GO:0035860 | glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.1 | 0.6 | GO:0072217 | negative regulation of mesenchymal to epithelial transition involved in metanephros morphogenesis(GO:0003340) negative regulation of metanephros development(GO:0072217) |
| 0.1 | 1.4 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.1 | 4.7 | GO:0019933 | cAMP-mediated signaling(GO:0019933) |
| 0.1 | 0.4 | GO:0045653 | negative regulation of megakaryocyte differentiation(GO:0045653) |
| 0.1 | 0.2 | GO:0061394 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) |
| 0.1 | 1.2 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.1 | 3.4 | GO:0046676 | negative regulation of insulin secretion(GO:0046676) |
| 0.1 | 0.2 | GO:0000957 | mitochondrial RNA catabolic process(GO:0000957) regulation of mitochondrial RNA catabolic process(GO:0000960) |
| 0.1 | 0.2 | GO:0032747 | positive regulation of interleukin-23 production(GO:0032747) |
| 0.1 | 0.5 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.1 | 3.8 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.1 | 0.4 | GO:2000169 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) regulation of peptidyl-cysteine S-nitrosylation(GO:2000169) |
| 0.1 | 1.2 | GO:0015747 | urate transport(GO:0015747) |
| 0.1 | 8.1 | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.1 | 22.0 | GO:0065004 | protein-DNA complex assembly(GO:0065004) |
| 0.1 | 22.1 | GO:0006936 | muscle contraction(GO:0006936) |
| 0.1 | 0.3 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.1 | 1.7 | GO:0030325 | adrenal gland development(GO:0030325) |
| 0.1 | 2.0 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.1 | 0.4 | GO:0035865 | cellular response to potassium ion(GO:0035865) |
| 0.1 | 2.3 | GO:0000281 | mitotic cytokinesis(GO:0000281) |
| 0.1 | 4.5 | GO:0015698 | inorganic anion transport(GO:0015698) |
| 0.1 | 1.8 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.1 | 0.9 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.1 | 2.0 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.1 | 12.4 | GO:0031032 | actomyosin structure organization(GO:0031032) |
| 0.1 | 0.8 | GO:0045777 | positive regulation of blood pressure(GO:0045777) |
| 0.1 | 1.6 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.1 | 0.4 | GO:0009099 | branched-chain amino acid biosynthetic process(GO:0009082) leucine biosynthetic process(GO:0009098) valine biosynthetic process(GO:0009099) |
| 0.1 | 0.7 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.1 | 0.4 | GO:0008626 | granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 0.1 | 0.4 | GO:2000074 | regulation of type B pancreatic cell development(GO:2000074) |
| 0.1 | 0.4 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.1 | 1.6 | GO:0022400 | regulation of rhodopsin mediated signaling pathway(GO:0022400) |
| 0.1 | 0.8 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.1 | 0.2 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.1 | 9.2 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.1 | 0.6 | GO:0000380 | alternative mRNA splicing, via spliceosome(GO:0000380) |
| 0.1 | 0.3 | GO:1902109 | negative regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902109) |
| 0.1 | 1.0 | GO:0044342 | type B pancreatic cell proliferation(GO:0044342) |
| 0.1 | 0.2 | GO:1904428 | negative regulation of tubulin deacetylation(GO:1904428) |
| 0.1 | 1.1 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.0 | 5.5 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.0 | 21.3 | GO:0043413 | protein glycosylation(GO:0006486) macromolecule glycosylation(GO:0043413) |
| 0.0 | 4.0 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 0.4 | GO:1902307 | positive regulation of sodium ion transmembrane transport(GO:1902307) positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.0 | 0.4 | GO:0034214 | protein hexamerization(GO:0034214) |
| 0.0 | 6.7 | GO:0046718 | viral entry into host cell(GO:0046718) |
| 0.0 | 4.7 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 0.8 | GO:0001514 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.0 | 1.6 | GO:0043330 | response to exogenous dsRNA(GO:0043330) |
| 0.0 | 2.1 | GO:0051693 | actin filament capping(GO:0051693) |
| 0.0 | 0.3 | GO:0006196 | AMP catabolic process(GO:0006196) |
| 0.0 | 0.5 | GO:0050885 | neuromuscular process controlling balance(GO:0050885) |
| 0.0 | 3.0 | GO:0007040 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.0 | 0.5 | GO:0060065 | uterus development(GO:0060065) |
| 0.0 | 0.5 | GO:0097205 | glomerular filtration(GO:0003094) renal filtration(GO:0097205) |
| 0.0 | 0.8 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 1.2 | GO:0042744 | hydrogen peroxide catabolic process(GO:0042744) |
| 0.0 | 0.4 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.0 | 2.8 | GO:0030838 | positive regulation of actin filament polymerization(GO:0030838) |
| 0.0 | 1.7 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.5 | GO:0060628 | regulation of ER to Golgi vesicle-mediated transport(GO:0060628) |
| 0.0 | 2.0 | GO:0042461 | photoreceptor cell development(GO:0042461) |
| 0.0 | 0.7 | GO:0043578 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.5 | GO:0043552 | positive regulation of phosphatidylinositol 3-kinase activity(GO:0043552) |
| 0.0 | 0.7 | GO:0048665 | neuron fate specification(GO:0048665) |
| 0.0 | 0.4 | GO:0010990 | regulation of SMAD protein complex assembly(GO:0010990) negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.0 | 0.4 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 0.0 | 1.0 | GO:1901186 | positive regulation of epidermal growth factor receptor signaling pathway(GO:0045742) positive regulation of ERBB signaling pathway(GO:1901186) |
| 0.0 | 0.1 | GO:0097061 | dendritic spine organization(GO:0097061) |
| 0.0 | 0.2 | GO:0097210 | response to gonadotropin-releasing hormone(GO:0097210) cellular response to gonadotropin-releasing hormone(GO:0097211) |
| 0.0 | 5.9 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 0.5 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 0.3 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.0 | 2.5 | GO:0008630 | intrinsic apoptotic signaling pathway in response to DNA damage(GO:0008630) |
| 0.0 | 2.0 | GO:0000910 | cytokinesis(GO:0000910) |
| 0.0 | 0.4 | GO:0007039 | protein catabolic process in the vacuole(GO:0007039) |
| 0.0 | 0.6 | GO:0050872 | white fat cell differentiation(GO:0050872) |
| 0.0 | 1.2 | GO:0009408 | response to heat(GO:0009408) |
| 0.0 | 0.6 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 0.2 | GO:0009649 | entrainment of circadian clock(GO:0009649) |
| 0.0 | 0.2 | GO:0033189 | response to vitamin A(GO:0033189) |
| 0.0 | 0.7 | GO:1903779 | regulation of cardiac conduction(GO:1903779) |
| 0.0 | 1.7 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.7 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.0 | 0.4 | GO:0015824 | proline transport(GO:0015824) |
| 0.0 | 0.6 | GO:0050974 | detection of mechanical stimulus involved in sensory perception(GO:0050974) |
| 0.0 | 0.2 | GO:0046716 | muscle cell cellular homeostasis(GO:0046716) |
| 0.0 | 1.5 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 0.1 | GO:2000628 | regulation of miRNA metabolic process(GO:2000628) |
| 0.0 | 0.4 | GO:1905145 | acetylcholine receptor signaling pathway(GO:0095500) postsynaptic signal transduction(GO:0098926) signal transduction involved in cellular response to ammonium ion(GO:1903831) response to acetylcholine(GO:1905144) cellular response to acetylcholine(GO:1905145) |
| 0.0 | 0.2 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 | 0.1 | GO:0006480 | N-terminal protein amino acid methylation(GO:0006480) |
| 0.0 | 0.4 | GO:0001542 | ovulation from ovarian follicle(GO:0001542) |
| 0.0 | 0.3 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.0 | 0.4 | GO:0046135 | pyrimidine nucleoside catabolic process(GO:0046135) |
| 0.0 | 0.9 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 0.4 | GO:0035090 | maintenance of apical/basal cell polarity(GO:0035090) maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.0 | 2.8 | GO:0019886 | antigen processing and presentation of exogenous peptide antigen via MHC class II(GO:0019886) |
| 0.0 | 0.6 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.0 | 0.6 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.7 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 2.2 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.0 | 0.2 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.0 | 0.2 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.0 | 1.1 | GO:0008277 | regulation of G-protein coupled receptor protein signaling pathway(GO:0008277) |
| 0.0 | 0.8 | GO:0050434 | positive regulation of viral transcription(GO:0050434) |
| 0.0 | 0.2 | GO:0016048 | detection of temperature stimulus(GO:0016048) |
| 0.0 | 0.4 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.2 | GO:0035878 | nail development(GO:0035878) |
| 0.0 | 0.2 | GO:0021884 | forebrain neuron development(GO:0021884) |
| 0.0 | 0.6 | GO:0061512 | protein localization to cilium(GO:0061512) |
| 0.0 | 0.3 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.0 | 0.4 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
| 0.0 | 0.3 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 | 0.3 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.8 | GO:0045652 | regulation of megakaryocyte differentiation(GO:0045652) |
| 0.0 | 1.1 | GO:0018210 | peptidyl-threonine phosphorylation(GO:0018107) peptidyl-threonine modification(GO:0018210) |
| 0.0 | 0.1 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.0 | 0.5 | GO:0061621 | NADH regeneration(GO:0006735) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.0 | 1.1 | GO:1903955 | positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.0 | 0.1 | GO:0060004 | reflex(GO:0060004) |
| 0.0 | 5.6 | GO:0000209 | protein polyubiquitination(GO:0000209) |
| 0.0 | 0.1 | GO:2001222 | regulation of neuron migration(GO:2001222) |
| 0.0 | 0.4 | GO:0051353 | positive regulation of oxidoreductase activity(GO:0051353) |
| 0.0 | 0.5 | GO:0032210 | regulation of telomere maintenance via telomerase(GO:0032210) |
| 0.0 | 0.0 | GO:0002331 | pre-B cell allelic exclusion(GO:0002331) |
| 0.0 | 0.3 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.0 | 0.8 | GO:0006446 | regulation of translational initiation(GO:0006446) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.9 | 14.4 | GO:0032127 | dense core granule membrane(GO:0032127) |
| 2.9 | 31.4 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 2.2 | 19.7 | GO:0060200 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 1.7 | 20.5 | GO:0016013 | syntrophin complex(GO:0016013) |
| 1.7 | 3.4 | GO:0019897 | extrinsic component of plasma membrane(GO:0019897) |
| 1.5 | 9.0 | GO:0031673 | H zone(GO:0031673) |
| 1.4 | 22.4 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 1.3 | 6.5 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 1.2 | 3.7 | GO:0034685 | integrin alphav-beta6 complex(GO:0034685) |
| 1.2 | 47.6 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 1.2 | 7.2 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 1.2 | 21.9 | GO:0033270 | paranode region of axon(GO:0033270) |
| 1.0 | 7.3 | GO:0005916 | fascia adherens(GO:0005916) |
| 1.0 | 4.9 | GO:1990425 | ryanodine receptor complex(GO:1990425) |
| 1.0 | 19.3 | GO:0031045 | dense core granule(GO:0031045) |
| 0.9 | 41.9 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.9 | 11.5 | GO:0043219 | lateral loop(GO:0043219) |
| 0.9 | 3.4 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 0.8 | 6.3 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.8 | 7.8 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.7 | 2.2 | GO:0097444 | spine apparatus(GO:0097444) |
| 0.7 | 5.0 | GO:0098843 | postsynaptic endocytic zone(GO:0098843) |
| 0.7 | 10.7 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.6 | 16.8 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.6 | 6.8 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.6 | 2.4 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.6 | 41.9 | GO:0031430 | M band(GO:0031430) |
| 0.6 | 12.3 | GO:0032982 | myosin filament(GO:0032982) |
| 0.6 | 8.8 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.6 | 1.7 | GO:1990032 | parallel fiber(GO:1990032) |
| 0.5 | 2.2 | GO:0072534 | perineuronal net(GO:0072534) |
| 0.5 | 13.2 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.5 | 14.7 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.5 | 3.8 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.5 | 9.4 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.5 | 3.1 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.5 | 3.6 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.5 | 30.3 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.5 | 9.2 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.5 | 3.9 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.5 | 25.5 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.5 | 7.5 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.5 | 4.6 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.5 | 170.4 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.4 | 13.8 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.4 | 15.5 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.4 | 8.7 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 0.4 | 2.0 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.4 | 4.3 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.4 | 1.1 | GO:0005595 | collagen type XII trimer(GO:0005595) |
| 0.4 | 1.5 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.4 | 8.4 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.4 | 3.2 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.3 | 1.0 | GO:0043257 | laminin-8 complex(GO:0043257) |
| 0.3 | 2.6 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) |
| 0.3 | 2.7 | GO:0016011 | dystroglycan complex(GO:0016011) sarcoglycan complex(GO:0016012) |
| 0.3 | 2.2 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.3 | 49.2 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.2 | 11.0 | GO:0030669 | clathrin-coated endocytic vesicle membrane(GO:0030669) |
| 0.2 | 5.4 | GO:0045263 | proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.2 | 1.7 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.2 | 3.1 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.2 | 3.5 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.2 | 2.8 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.2 | 2.5 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.2 | 10.1 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.2 | 11.3 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.2 | 8.4 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.2 | 32.8 | GO:0031674 | I band(GO:0031674) |
| 0.2 | 0.6 | GO:0002947 | tumor necrosis factor receptor superfamily complex(GO:0002947) |
| 0.2 | 69.8 | GO:0098794 | postsynapse(GO:0098794) |
| 0.2 | 65.7 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.2 | 0.8 | GO:0043034 | costamere(GO:0043034) |
| 0.2 | 11.5 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.2 | 11.9 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.2 | 6.7 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.2 | 2.5 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.2 | 1.9 | GO:0045275 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.2 | 1.8 | GO:0055028 | cortical microtubule(GO:0055028) |
| 0.2 | 1.9 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.2 | 1.7 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.2 | 1.7 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.2 | 8.0 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.2 | 1.1 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.2 | 30.4 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.2 | 22.5 | GO:0030426 | growth cone(GO:0030426) |
| 0.1 | 1.1 | GO:0030478 | actin cap(GO:0030478) |
| 0.1 | 0.8 | GO:0042025 | host cell nucleus(GO:0042025) |
| 0.1 | 1.1 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.1 | 0.5 | GO:0000333 | telomerase catalytic core complex(GO:0000333) |
| 0.1 | 9.1 | GO:0043204 | perikaryon(GO:0043204) |
| 0.1 | 0.8 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.1 | 0.8 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.1 | 4.0 | GO:0031984 | organelle subcompartment(GO:0031984) |
| 0.1 | 8.6 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.1 | 11.8 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.1 | 0.8 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.1 | 0.8 | GO:0097422 | tubular endosome(GO:0097422) |
| 0.1 | 29.9 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.1 | 4.1 | GO:0032420 | stereocilium(GO:0032420) |
| 0.1 | 3.1 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.1 | 2.1 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.1 | 1.4 | GO:0031010 | ISWI-type complex(GO:0031010) |
| 0.1 | 21.9 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.1 | 22.0 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.1 | 0.8 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.1 | 23.3 | GO:0030424 | axon(GO:0030424) |
| 0.1 | 1.1 | GO:0031211 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.1 | 0.9 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.1 | 0.2 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 0.1 | 1.2 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.1 | 1.4 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.1 | 0.2 | GO:0097451 | astrocyte end-foot(GO:0097450) glial limiting end-foot(GO:0097451) |
| 0.1 | 1.0 | GO:0030658 | transport vesicle membrane(GO:0030658) |
| 0.1 | 2.3 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.1 | 0.9 | GO:0097433 | dense body(GO:0097433) |
| 0.1 | 1.4 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.1 | 3.1 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.1 | 7.7 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 0.7 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.1 | 5.5 | GO:0005604 | basement membrane(GO:0005604) |
| 0.1 | 1.4 | GO:0005922 | connexon complex(GO:0005922) |
| 0.1 | 1.2 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.1 | 0.6 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.1 | 2.1 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.1 | 1.3 | GO:0005861 | troponin complex(GO:0005861) |
| 0.1 | 5.4 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.1 | 2.9 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 1.4 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.0 | 0.1 | GO:0035101 | FACT complex(GO:0035101) |
| 0.0 | 7.5 | GO:0043005 | neuron projection(GO:0043005) |
| 0.0 | 0.7 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 0.9 | GO:0098827 | endoplasmic reticulum subcompartment(GO:0098827) |
| 0.0 | 0.5 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.0 | 0.2 | GO:0001674 | female germ cell nucleus(GO:0001674) |
| 0.0 | 27.5 | GO:0015629 | actin cytoskeleton(GO:0015629) |
| 0.0 | 3.1 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 0.5 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.3 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 0.4 | GO:0030118 | clathrin coat(GO:0030118) |
| 0.0 | 3.3 | GO:0045202 | synapse(GO:0045202) |
| 0.0 | 1.3 | GO:0031231 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.0 | 0.2 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.0 | 0.9 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.0 | 3.8 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 0.3 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 9.9 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 1.6 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.8 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 1.0 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 0.8 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
| 0.0 | 10.4 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.0 | 10.1 | GO:0030055 | cell-substrate junction(GO:0030055) |
| 0.0 | 1.9 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 0.6 | GO:0010369 | chromocenter(GO:0010369) |
| 0.0 | 26.1 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 0.5 | GO:0000800 | lateral element(GO:0000800) |
| 0.0 | 0.9 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.4 | GO:0034709 | methylosome(GO:0034709) |
| 0.0 | 0.5 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.0 | 0.3 | GO:0090543 | Flemming body(GO:0090543) |
| 0.0 | 0.5 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 0.4 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.8 | GO:0044297 | cell body(GO:0044297) |
| 0.0 | 0.3 | GO:0005845 | mRNA cap binding complex(GO:0005845) |
| 0.0 | 0.5 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 1.7 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 10.4 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 1.3 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 1.9 | GO:0070821 | tertiary granule membrane(GO:0070821) |
| 0.0 | 1.0 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.1 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 1.6 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.0 | 0.8 | GO:0035580 | specific granule lumen(GO:0035580) |
| 0.0 | 0.3 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.7 | 37.9 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 3.1 | 34.0 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 2.9 | 26.4 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 2.9 | 20.1 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 2.6 | 7.7 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 2.5 | 15.2 | GO:0060175 | brain-derived neurotrophic factor-activated receptor activity(GO:0060175) |
| 2.1 | 8.5 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 2.1 | 8.3 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 1.9 | 7.6 | GO:0008260 | 3-oxoacid CoA-transferase activity(GO:0008260) |
| 1.8 | 26.4 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 1.7 | 13.5 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 1.6 | 9.5 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 1.5 | 25.8 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 1.5 | 4.4 | GO:0086057 | voltage-gated calcium channel activity involved in bundle of His cell action potential(GO:0086057) |
| 1.5 | 32.1 | GO:0003680 | AT DNA binding(GO:0003680) |
| 1.4 | 47.1 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 1.4 | 13.7 | GO:0099529 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 1.4 | 5.5 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 1.3 | 25.8 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 1.3 | 3.8 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 1.2 | 9.7 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) |
| 1.1 | 4.5 | GO:0004923 | leukemia inhibitory factor receptor activity(GO:0004923) |
| 1.0 | 7.2 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 1.0 | 23.4 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 1.0 | 49.7 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.9 | 6.5 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.9 | 3.6 | GO:1902379 | receptor tyrosine kinase-like orphan receptor binding(GO:0005115) chemoattractant activity involved in axon guidance(GO:1902379) |
| 0.8 | 2.5 | GO:0015275 | stretch-activated, cation-selective, calcium channel activity(GO:0015275) |
| 0.8 | 4.1 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.8 | 4.9 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.8 | 5.6 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.8 | 6.2 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.8 | 21.4 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.8 | 3.8 | GO:0047391 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) |
| 0.8 | 3.8 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 0.8 | 3.8 | GO:0086056 | voltage-gated calcium channel activity involved in cardiac muscle cell action potential(GO:0086007) voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.7 | 7.5 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.7 | 3.0 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 0.7 | 12.0 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.7 | 5.3 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.7 | 2.0 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.6 | 3.8 | GO:0004598 | peptidylglycine monooxygenase activity(GO:0004504) peptidylamidoglycolate lyase activity(GO:0004598) |
| 0.6 | 7.5 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.6 | 8.8 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.6 | 5.2 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.6 | 19.7 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.6 | 19.9 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.6 | 8.8 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.5 | 3.2 | GO:0051430 | corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.5 | 13.8 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.5 | 7.5 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.5 | 29.2 | GO:0070888 | E-box binding(GO:0070888) |
| 0.5 | 2.0 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.5 | 2.4 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.5 | 1.4 | GO:0005137 | interleukin-5 receptor binding(GO:0005137) |
| 0.5 | 1.9 | GO:0016639 | oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 0.5 | 2.3 | GO:0055100 | adiponectin binding(GO:0055100) |
| 0.5 | 4.6 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.5 | 1.8 | GO:0017129 | triglyceride binding(GO:0017129) |
| 0.5 | 0.9 | GO:0017098 | sulfonylurea receptor binding(GO:0017098) |
| 0.4 | 1.3 | GO:0005427 | proton-dependent oligopeptide secondary active transmembrane transporter activity(GO:0005427) secondary active oligopeptide transmembrane transporter activity(GO:0015322) |
| 0.4 | 2.2 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.4 | 8.8 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.4 | 1.7 | GO:0052591 | sn-glycerol-3-phosphate:ubiquinone oxidoreductase activity(GO:0052590) sn-glycerol-3-phosphate:ubiquinone-8 oxidoreductase activity(GO:0052591) |
| 0.4 | 14.4 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.4 | 1.3 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.4 | 20.2 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.4 | 0.8 | GO:0046790 | virion binding(GO:0046790) |
| 0.4 | 0.8 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) |
| 0.4 | 1.2 | GO:0061599 | molybdopterin adenylyltransferase activity(GO:0061598) molybdopterin molybdotransferase activity(GO:0061599) |
| 0.4 | 2.4 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.4 | 2.8 | GO:0001595 | angiotensin receptor activity(GO:0001595) angiotensin type II receptor activity(GO:0004945) |
| 0.4 | 1.2 | GO:0019808 | polyamine binding(GO:0019808) |
| 0.4 | 3.1 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.4 | 9.0 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.4 | 1.5 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.4 | 2.6 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.4 | 2.5 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.4 | 2.9 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 0.4 | 24.3 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.4 | 66.9 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.4 | 12.2 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.4 | 7.2 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.4 | 1.1 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.4 | 2.1 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.3 | 4.2 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.3 | 4.5 | GO:0043176 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.3 | 2.8 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.3 | 1.7 | GO:0050115 | myosin-light-chain-phosphatase activity(GO:0050115) |
| 0.3 | 6.8 | GO:0019966 | interleukin-1 binding(GO:0019966) |
| 0.3 | 2.4 | GO:0046556 | alpha-L-arabinofuranosidase activity(GO:0046556) |
| 0.3 | 3.4 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.3 | 1.3 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 0.3 | 2.7 | GO:0015111 | iodide transmembrane transporter activity(GO:0015111) |
| 0.3 | 2.6 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.3 | 4.2 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.3 | 7.7 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.3 | 6.3 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.3 | 7.0 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.3 | 0.9 | GO:0047275 | glucosaminylgalactosylglucosylceramide beta-galactosyltransferase activity(GO:0047275) |
| 0.3 | 11.3 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.3 | 3.4 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.3 | 2.1 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.3 | 41.1 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.3 | 3.0 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.3 | 1.2 | GO:0035514 | DNA demethylase activity(GO:0035514) |
| 0.3 | 9.1 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.3 | 0.6 | GO:0030305 | heparanase activity(GO:0030305) |
| 0.3 | 14.7 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.3 | 3.2 | GO:0005167 | neurotrophin TRK receptor binding(GO:0005167) |
| 0.3 | 5.1 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.3 | 15.4 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.3 | 1.1 | GO:0004510 | tryptophan 5-monooxygenase activity(GO:0004510) |
| 0.3 | 0.8 | GO:0050429 | calcium-dependent phospholipase C activity(GO:0050429) |
| 0.3 | 1.7 | GO:0071253 | connexin binding(GO:0071253) |
| 0.3 | 1.9 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.3 | 2.5 | GO:0034603 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 0.3 | 59.8 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.3 | 1.1 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.3 | 1.6 | GO:0005223 | intracellular cGMP activated cation channel activity(GO:0005223) |
| 0.3 | 2.9 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.3 | 2.8 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.3 | 1.3 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.3 | 2.3 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.3 | 9.0 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.2 | 8.7 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.2 | 1.0 | GO:0031697 | beta-1 adrenergic receptor binding(GO:0031697) |
| 0.2 | 1.7 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.2 | 0.7 | GO:0004531 | deoxyribonuclease II activity(GO:0004531) |
| 0.2 | 6.9 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.2 | 5.2 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.2 | 2.1 | GO:0016594 | glycine binding(GO:0016594) |
| 0.2 | 0.9 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.2 | 1.9 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.2 | 4.7 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.2 | 0.9 | GO:0016608 | growth hormone-releasing hormone activity(GO:0016608) |
| 0.2 | 1.3 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.2 | 0.4 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.2 | 2.8 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.2 | 59.9 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.2 | 2.4 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.2 | 1.6 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.2 | 2.7 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.2 | 1.9 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.2 | 5.5 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.2 | 5.1 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.2 | 1.7 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.2 | 1.1 | GO:0098960 | postsynaptic neurotransmitter receptor activity(GO:0098960) |
| 0.2 | 4.7 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.2 | 8.8 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.2 | 6.4 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.2 | 1.9 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.2 | 47.3 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.2 | 2.2 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.2 | 1.3 | GO:0033857 | diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.2 | 5.6 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.2 | 0.5 | GO:0004618 | phosphoglycerate kinase activity(GO:0004618) |
| 0.2 | 12.2 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.2 | 1.1 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.2 | 1.7 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.2 | 19.2 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.2 | 10.0 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.2 | 0.6 | GO:0034040 | lipid-transporting ATPase activity(GO:0034040) |
| 0.1 | 0.6 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.1 | 1.6 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.1 | 0.9 | GO:0005534 | galactose binding(GO:0005534) |
| 0.1 | 0.6 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 0.1 | 7.2 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.1 | 0.9 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.1 | 2.6 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.1 | 3.5 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.1 | 1.5 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.1 | 13.9 | GO:0002039 | p53 binding(GO:0002039) |
| 0.1 | 47.7 | GO:0044822 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.1 | 1.1 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.1 | 1.0 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.1 | 1.0 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.1 | 3.6 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.1 | 0.4 | GO:0034437 | glycoprotein transporter activity(GO:0034437) |
| 0.1 | 0.8 | GO:0015086 | cadmium ion transmembrane transporter activity(GO:0015086) cobalt ion transmembrane transporter activity(GO:0015087) lead ion transmembrane transporter activity(GO:0015094) ferrous iron uptake transmembrane transporter activity(GO:0015639) |
| 0.1 | 0.9 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.1 | 0.4 | GO:0046848 | hydroxyapatite binding(GO:0046848) |
| 0.1 | 1.7 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.1 | 1.9 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.1 | 1.6 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.1 | 1.0 | GO:0030553 | cGMP binding(GO:0030553) |
| 0.1 | 3.1 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.1 | 3.5 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.1 | 2.9 | GO:0042393 | histone binding(GO:0042393) |
| 0.1 | 20.4 | GO:0005057 | receptor signaling protein activity(GO:0005057) |
| 0.1 | 4.6 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.1 | 0.5 | GO:0008665 | 2'-phosphotransferase activity(GO:0008665) |
| 0.1 | 7.4 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.1 | 1.4 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.1 | 0.9 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.1 | 0.8 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.1 | 9.5 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.1 | 3.0 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.1 | 0.5 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.1 | 1.0 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.1 | 10.0 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.1 | 1.1 | GO:0042805 | actinin binding(GO:0042805) |
| 0.1 | 0.8 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.1 | 0.9 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.1 | 0.5 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.1 | 1.0 | GO:0050694 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.1 | 1.5 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.1 | 3.3 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.1 | 2.5 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.1 | 0.7 | GO:0016167 | glial cell-derived neurotrophic factor receptor activity(GO:0016167) |
| 0.1 | 0.6 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.1 | 0.4 | GO:0004167 | dopachrome isomerase activity(GO:0004167) |
| 0.1 | 0.2 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.1 | 0.4 | GO:0048273 | mitogen-activated protein kinase p38 binding(GO:0048273) |
| 0.1 | 2.2 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.1 | 5.0 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.1 | 0.9 | GO:0003720 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.1 | 3.3 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.1 | 0.5 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.1 | 0.7 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.1 | 0.8 | GO:0019534 | toxin transporter activity(GO:0019534) |
| 0.1 | 0.7 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.1 | 0.2 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.1 | 0.2 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.1 | 2.4 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.1 | 1.4 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.1 | 0.8 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.1 | 0.8 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.1 | 2.0 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 0.5 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.1 | 1.1 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.1 | 0.9 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.1 | 0.4 | GO:0052655 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.1 | 1.0 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.1 | 1.8 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.1 | 1.0 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.1 | 52.8 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.1 | 0.4 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.1 | 0.8 | GO:0005542 | folic acid binding(GO:0005542) |
| 0.1 | 0.7 | GO:0016303 | 1-phosphatidylinositol-3-kinase activity(GO:0016303) phosphatidylinositol kinase activity(GO:0052742) |
| 0.1 | 5.1 | GO:0015078 | hydrogen ion transmembrane transporter activity(GO:0015078) |
| 0.1 | 1.1 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.1 | 0.2 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.1 | 2.7 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.1 | 0.6 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.1 | 0.2 | GO:0004968 | gonadotropin-releasing hormone receptor activity(GO:0004968) |
| 0.0 | 0.3 | GO:0016402 | pristanoyl-CoA oxidase activity(GO:0016402) |
| 0.0 | 2.1 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 0.9 | GO:0015464 | acetylcholine receptor activity(GO:0015464) |
| 0.0 | 0.0 | GO:0061752 | telomeric repeat-containing RNA binding(GO:0061752) |
| 0.0 | 7.7 | GO:0016779 | nucleotidyltransferase activity(GO:0016779) |
| 0.0 | 0.6 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 1.9 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 2.3 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.0 | 0.3 | GO:0001190 | transcriptional activator activity, RNA polymerase II transcription factor binding(GO:0001190) transcriptional repressor activity, RNA polymerase II activating transcription factor binding(GO:0098811) |
| 0.0 | 0.8 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.4 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 15.9 | GO:0015631 | tubulin binding(GO:0015631) |
| 0.0 | 0.2 | GO:0023024 | MHC class I protein complex binding(GO:0023024) |
| 0.0 | 0.7 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.0 | 1.9 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.9 | GO:0004690 | cyclic nucleotide-dependent protein kinase activity(GO:0004690) |
| 0.0 | 0.7 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.4 | GO:0022820 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.0 | 11.4 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 0.8 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 3.5 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.3 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.4 | GO:0015038 | glutathione disulfide oxidoreductase activity(GO:0015038) |
| 0.0 | 0.3 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.2 | GO:0047522 | 13-prostaglandin reductase activity(GO:0036132) 15-oxoprostaglandin 13-oxidase activity(GO:0047522) |
| 0.0 | 0.4 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 10.2 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 0.8 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 1.5 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 3.0 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.3 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.5 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.1 | GO:0071885 | N-terminal protein N-methyltransferase activity(GO:0071885) |
| 0.0 | 1.0 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.4 | GO:0015172 | L-glutamate transmembrane transporter activity(GO:0005313) acidic amino acid transmembrane transporter activity(GO:0015172) |
| 0.0 | 1.7 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.0 | 7.9 | GO:0042277 | peptide binding(GO:0042277) |
| 0.0 | 1.5 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.0 | 0.3 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.5 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 2.2 | GO:0008081 | phosphoric diester hydrolase activity(GO:0008081) |
| 0.0 | 10.5 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.0 | 1.1 | GO:0002020 | protease binding(GO:0002020) |
| 0.0 | 0.5 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 2.8 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 0.8 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.6 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 1.2 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 2.0 | GO:0046983 | protein dimerization activity(GO:0046983) |
| 0.0 | 1.4 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.4 | GO:0022839 | ion gated channel activity(GO:0022839) |
| 0.0 | 0.4 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.0 | 4.9 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 0.4 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 1.1 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.6 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.1 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.6 | GO:0031072 | heat shock protein binding(GO:0031072) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 49.4 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.4 | 27.0 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.3 | 13.5 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.3 | 25.1 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.3 | 4.8 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.3 | 10.7 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.2 | 10.1 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.2 | 8.8 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.2 | 2.8 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.2 | 1.6 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.2 | 8.3 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.2 | 7.2 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.2 | 3.9 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.2 | 7.8 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.2 | 3.7 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.2 | 18.3 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.2 | 7.0 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.2 | 3.3 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.1 | 12.3 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.1 | 5.3 | PID FGF PATHWAY | FGF signaling pathway |
| 0.1 | 5.4 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.1 | 12.9 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.1 | 7.2 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.1 | 15.4 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.1 | 7.7 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 17.7 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.1 | 11.2 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.1 | 1.4 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.1 | 8.4 | PID BMP PATHWAY | BMP receptor signaling |
| 0.1 | 3.4 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.1 | 0.9 | ST GAQ PATHWAY | G alpha q Pathway |
| 0.1 | 2.5 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.1 | 5.1 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.1 | 4.5 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.1 | 2.4 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
| 0.1 | 2.8 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.1 | 5.1 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.1 | 21.2 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 1.1 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.1 | 2.6 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.1 | 5.3 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.1 | 4.9 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.1 | 2.9 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.1 | 2.4 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.1 | 1.8 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.1 | 1.5 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.1 | 0.9 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.1 | 1.5 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.1 | 15.4 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 2.5 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 0.7 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 2.8 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.6 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 1.4 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 2.5 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 2.3 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 0.6 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.6 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 1.4 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.0 | 2.5 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 2.7 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.0 | 1.2 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.5 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 1.0 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.0 | 2.9 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.3 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.0 | 1.7 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.5 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.5 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.6 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 0.2 | PID SHP2 PATHWAY | SHP2 signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 50.1 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 1.2 | 28.4 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.9 | 29.2 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.8 | 68.0 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.7 | 18.3 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.6 | 16.2 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.6 | 28.7 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.5 | 25.8 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.4 | 10.7 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.4 | 8.0 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.3 | 16.8 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.3 | 14.4 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.3 | 22.8 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.3 | 34.4 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.3 | 4.9 | REACTOME NITRIC OXIDE STIMULATES GUANYLATE CYCLASE | Genes involved in Nitric oxide stimulates guanylate cyclase |
| 0.3 | 4.5 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.3 | 7.8 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.3 | 6.9 | REACTOME ACTIVATED POINT MUTANTS OF FGFR2 | Genes involved in Activated point mutants of FGFR2 |
| 0.3 | 8.6 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.3 | 7.8 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.2 | 2.8 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.2 | 4.1 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.2 | 16.9 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.2 | 4.7 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.2 | 1.6 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.2 | 15.0 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.2 | 9.6 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.2 | 6.7 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.2 | 4.1 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.2 | 2.4 | REACTOME OPSINS | Genes involved in Opsins |
| 0.2 | 4.2 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.2 | 3.6 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.2 | 6.2 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.2 | 6.9 | REACTOME GRB2 EVENTS IN ERBB2 SIGNALING | Genes involved in GRB2 events in ERBB2 signaling |
| 0.2 | 31.1 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.1 | 2.7 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.1 | 2.5 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.1 | 6.4 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.1 | 4.6 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.1 | 3.5 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.1 | 4.7 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.1 | 0.9 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.1 | 2.6 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.1 | 3.5 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 3.1 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.1 | 1.7 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.1 | 4.3 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.1 | 0.8 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.1 | 3.8 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.1 | 3.8 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.1 | 0.8 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.1 | 1.4 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.1 | 1.8 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.1 | 4.1 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.1 | 1.7 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.1 | 1.0 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.1 | 1.1 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.1 | 1.2 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.1 | 3.6 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 1.7 | REACTOME ENERGY DEPENDENT REGULATION OF MTOR BY LKB1 AMPK | Genes involved in Energy dependent regulation of mTOR by LKB1-AMPK |
| 0.1 | 1.4 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.1 | 16.9 | REACTOME FATTY ACID TRIACYLGLYCEROL AND KETONE BODY METABOLISM | Genes involved in Fatty acid, triacylglycerol, and ketone body metabolism |
| 0.0 | 1.4 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 14.9 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 1.2 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 2.7 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 2.3 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 2.1 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.0 | 0.9 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 1.3 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.6 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 1.2 | REACTOME MUSCLE CONTRACTION | Genes involved in Muscle contraction |
| 0.0 | 0.5 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.6 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.7 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.5 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.0 | 2.8 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 0.4 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.5 | REACTOME REGULATORY RNA PATHWAYS | Genes involved in Regulatory RNA pathways |
| 0.0 | 1.8 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.0 | 0.6 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.6 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 1.0 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.5 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.0 | 0.6 | REACTOME EXTRACELLULAR MATRIX ORGANIZATION | Genes involved in Extracellular matrix organization |
| 0.0 | 0.4 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.5 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.3 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.4 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 1.2 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |