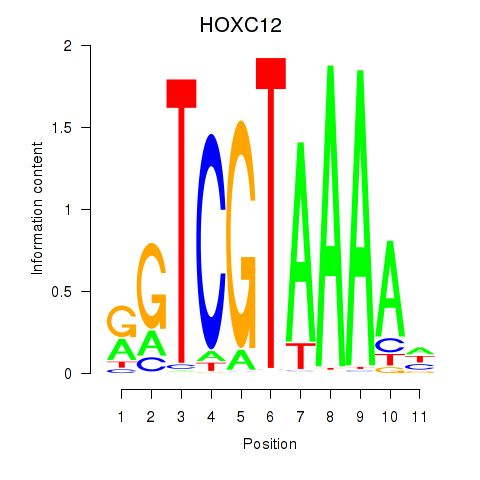
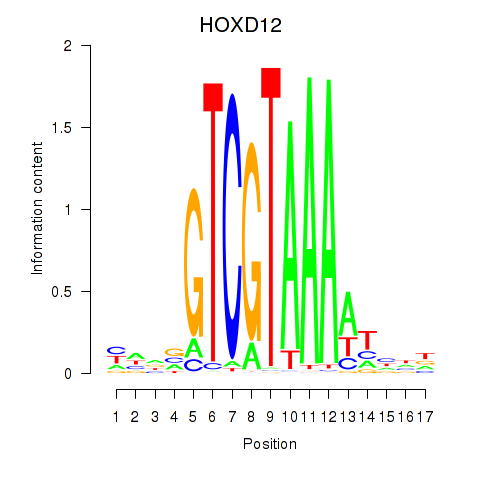
Project
Illumina Body Map 2: averaged replicates
Navigation
Downloads
Results for HOXC12_HOXD12
Z-value: 1.35
Transcription factors associated with HOXC12_HOXD12
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXC12
|
ENSG00000123407.3 | homeobox C12 |
|
HOXD12
|
ENSG00000170178.5 | homeobox D12 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXC12 | hg19_v2_chr12_+_54348618_54348693 | 0.27 | 1.3e-01 | Click! |
| HOXD12 | hg19_v2_chr2_+_176964458_176964540 | -0.08 | 6.6e-01 | Click! |
Activity profile of HOXC12_HOXD12 motif
Sorted Z-values of HOXC12_HOXD12 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chrY_+_2709527 | 7.05 |
ENST00000250784.8
|
RPS4Y1
|
ribosomal protein S4, Y-linked 1 |
| chr12_+_54384370 | 3.09 |
ENST00000504315.1
|
HOXC6
|
homeobox C6 |
| chrY_+_2709906 | 2.80 |
ENST00000430575.1
|
RPS4Y1
|
ribosomal protein S4, Y-linked 1 |
| chr19_+_7011509 | 2.31 |
ENST00000377296.3
|
AC025278.1
|
Uncharacterized protein |
| chr20_+_36946029 | 2.26 |
ENST00000417318.1
|
BPI
|
bactericidal/permeability-increasing protein |
| chr2_-_152589670 | 2.10 |
ENST00000604864.1
ENST00000603639.1 |
NEB
|
nebulin |
| chr14_+_39944025 | 1.96 |
ENST00000554328.1
ENST00000556620.1 ENST00000557197.1 |
RP11-111A21.1
|
RP11-111A21.1 |
| chr7_-_27219849 | 1.71 |
ENST00000396344.4
|
HOXA10
|
homeobox A10 |
| chrX_-_142605301 | 1.58 |
ENST00000370503.2
|
SPANXN3
|
SPANX family, member N3 |
| chr20_+_36932521 | 1.36 |
ENST00000262865.4
|
BPI
|
bactericidal/permeability-increasing protein |
| chr8_-_33370607 | 1.35 |
ENST00000360742.5
ENST00000523305.1 |
TTI2
|
TELO2 interacting protein 2 |
| chr1_+_156308245 | 1.29 |
ENST00000368253.2
ENST00000470342.1 ENST00000368254.1 |
TSACC
|
TSSK6 activating co-chaperone |
| chr2_+_132160448 | 1.25 |
ENST00000437751.1
|
AC073869.19
|
long intergenic non-protein coding RNA 1120 |
| chr4_-_141419484 | 1.22 |
ENST00000515402.1
ENST00000515354.1 |
RP11-542P2.1
|
Glycosyltransferase 54 domain-containing protein |
| chr16_-_54304608 | 1.20 |
ENST00000561336.1
|
RP11-324D17.1
|
HCG2045435; Uncharacterized protein |
| chr12_-_13248562 | 1.18 |
ENST00000457134.2
ENST00000537302.1 |
GSG1
|
germ cell associated 1 |
| chr6_-_49681235 | 1.15 |
ENST00000339139.4
|
CRISP2
|
cysteine-rich secretory protein 2 |
| chr17_-_28257080 | 1.14 |
ENST00000579954.1
ENST00000540801.1 ENST00000269033.3 ENST00000590153.1 ENST00000582084.1 |
SSH2
|
slingshot protein phosphatase 2 |
| chr1_-_26394114 | 1.12 |
ENST00000374272.3
|
TRIM63
|
tripartite motif containing 63, E3 ubiquitin protein ligase |
| chrX_-_77582980 | 1.12 |
ENST00000373304.3
|
CYSLTR1
|
cysteinyl leukotriene receptor 1 |
| chr12_+_54402790 | 1.10 |
ENST00000040584.4
|
HOXC8
|
homeobox C8 |
| chr3_-_51909600 | 1.08 |
ENST00000446461.1
|
IQCF5
|
IQ motif containing F5 |
| chr2_+_190541153 | 1.07 |
ENST00000313581.4
ENST00000438402.2 ENST00000431575.2 ENST00000281412.6 |
ANKAR
|
ankyrin and armadillo repeat containing |
| chr4_-_164395014 | 1.05 |
ENST00000280605.3
|
TKTL2
|
transketolase-like 2 |
| chr1_-_220220000 | 1.04 |
ENST00000366923.3
|
EPRS
|
glutamyl-prolyl-tRNA synthetase |
| chr7_+_76101379 | 1.03 |
ENST00000429179.1
|
DTX2
|
deltex homolog 2 (Drosophila) |
| chr9_-_7961080 | 1.02 |
ENST00000435444.1
|
RP11-29B9.2
|
RP11-29B9.2 |
| chr12_+_54410664 | 1.02 |
ENST00000303406.4
|
HOXC4
|
homeobox C4 |
| chr13_+_53030107 | 1.01 |
ENST00000490903.1
ENST00000480747.1 |
CKAP2
|
cytoskeleton associated protein 2 |
| chr11_-_5531159 | 1.00 |
ENST00000445998.1
|
UBQLN3
|
ubiquilin 3 |
| chr6_+_106988986 | 0.99 |
ENST00000457437.1
ENST00000535438.1 |
AIM1
|
absent in melanoma 1 |
| chr20_-_18810797 | 0.97 |
ENST00000278779.4
|
C20orf78
|
chromosome 20 open reading frame 78 |
| chr1_+_156308403 | 0.97 |
ENST00000481479.1
ENST00000368252.1 ENST00000466306.1 ENST00000368251.1 |
TSACC
|
TSSK6 activating co-chaperone |
| chr2_+_149974684 | 0.97 |
ENST00000450639.1
|
LYPD6B
|
LY6/PLAUR domain containing 6B |
| chr2_+_176995011 | 0.95 |
ENST00000548663.1
ENST00000450510.2 |
HOXD8
|
homeobox D8 |
| chr12_-_13248732 | 0.94 |
ENST00000396302.3
|
GSG1
|
germ cell associated 1 |
| chr13_+_113030625 | 0.94 |
ENST00000283550.3
|
SPACA7
|
sperm acrosome associated 7 |
| chr1_-_220219775 | 0.91 |
ENST00000609181.1
|
EPRS
|
glutamyl-prolyl-tRNA synthetase |
| chr13_+_53029564 | 0.90 |
ENST00000468284.1
ENST00000378034.3 ENST00000258607.5 ENST00000378037.5 |
CKAP2
|
cytoskeleton associated protein 2 |
| chr12_-_13248705 | 0.87 |
ENST00000396310.2
|
GSG1
|
germ cell associated 1 |
| chr12_+_70219052 | 0.87 |
ENST00000552032.2
ENST00000547771.2 |
MYRFL
|
myelin regulatory factor-like |
| chrX_-_133792480 | 0.86 |
ENST00000359237.4
|
PLAC1
|
placenta-specific 1 |
| chr3_+_32737027 | 0.86 |
ENST00000454516.2
|
CNOT10
|
CCR4-NOT transcription complex, subunit 10 |
| chr22_-_20307532 | 0.86 |
ENST00000405465.3
ENST00000248879.3 |
DGCR6L
|
DiGeorge syndrome critical region gene 6-like |
| chr5_-_176433565 | 0.86 |
ENST00000428382.2
|
UIMC1
|
ubiquitin interaction motif containing 1 |
| chr10_+_124670121 | 0.86 |
ENST00000368894.1
|
FAM24A
|
family with sequence similarity 24, member A |
| chr7_-_27219632 | 0.85 |
ENST00000470747.4
|
RP1-170O19.20
|
Uncharacterized protein |
| chr6_-_31681839 | 0.85 |
ENST00000409239.1
ENST00000461287.1 |
LY6G6E
XXbac-BPG32J3.20
|
lymphocyte antigen 6 complex, locus G6E (pseudogene) Uncharacterized protein |
| chr14_+_22180536 | 0.84 |
ENST00000390424.2
|
TRAV2
|
T cell receptor alpha variable 2 |
| chr10_-_1071796 | 0.82 |
ENST00000277517.1
|
IDI2
|
isopentenyl-diphosphate delta isomerase 2 |
| chr12_+_123259063 | 0.81 |
ENST00000392441.4
ENST00000539171.1 |
CCDC62
|
coiled-coil domain containing 62 |
| chr7_-_27179814 | 0.81 |
ENST00000522788.1
ENST00000521779.1 |
HOXA3
|
homeobox A3 |
| chrY_+_26997726 | 0.80 |
ENST00000382296.2
|
DAZ4
|
deleted in azoospermia 4 |
| chr1_-_91182794 | 0.80 |
ENST00000370445.4
|
BARHL2
|
BarH-like homeobox 2 |
| chr7_-_27169801 | 0.80 |
ENST00000511914.1
|
HOXA4
|
homeobox A4 |
| chr4_-_141419376 | 0.77 |
ENST00000511113.1
ENST00000503109.2 |
RP11-542P2.1
|
Glycosyltransferase 54 domain-containing protein |
| chr4_+_146601356 | 0.75 |
ENST00000438731.1
ENST00000511965.1 |
C4orf51
|
chromosome 4 open reading frame 51 |
| chr7_-_144107320 | 0.74 |
ENST00000483238.1
ENST00000467773.1 |
NOBOX
|
NOBOX oogenesis homeobox |
| chr22_+_29168652 | 0.74 |
ENST00000249064.4
ENST00000444523.1 ENST00000448492.2 ENST00000421503.2 |
CCDC117
|
coiled-coil domain containing 117 |
| chr4_-_46911248 | 0.72 |
ENST00000355591.3
ENST00000505102.1 |
COX7B2
|
cytochrome c oxidase subunit VIIb2 |
| chr8_-_52721975 | 0.72 |
ENST00000356297.4
ENST00000543296.1 |
PXDNL
|
peroxidasin homolog (Drosophila)-like |
| chr3_-_108672742 | 0.71 |
ENST00000261047.3
|
GUCA1C
|
guanylate cyclase activator 1C |
| chr14_+_22947861 | 0.70 |
ENST00000390482.1
|
TRAJ57
|
T cell receptor alpha joining 57 |
| chr14_-_64846033 | 0.70 |
ENST00000556556.1
|
CTD-2555O16.1
|
CTD-2555O16.1 |
| chr17_-_42767092 | 0.68 |
ENST00000588687.1
|
CCDC43
|
coiled-coil domain containing 43 |
| chr9_+_134065506 | 0.67 |
ENST00000483497.2
|
NUP214
|
nucleoporin 214kDa |
| chr7_+_6797288 | 0.67 |
ENST00000433859.2
ENST00000359718.3 |
RSPH10B2
|
radial spoke head 10 homolog B2 (Chlamydomonas) |
| chr5_-_176433693 | 0.67 |
ENST00000507513.1
ENST00000511320.1 |
UIMC1
|
ubiquitin interaction motif containing 1 |
| chr1_+_178694300 | 0.67 |
ENST00000367635.3
|
RALGPS2
|
Ral GEF with PH domain and SH3 binding motif 2 |
| chrX_-_55291163 | 0.66 |
ENST00000519203.1
ENST00000374951.1 |
PAGE3
|
P antigen family, member 3 (prostate associated) |
| chr7_+_138145145 | 0.65 |
ENST00000415680.2
|
TRIM24
|
tripartite motif containing 24 |
| chr14_-_60952739 | 0.64 |
ENST00000555476.1
ENST00000321731.3 |
C14orf39
|
chromosome 14 open reading frame 39 |
| chr5_+_35852797 | 0.64 |
ENST00000508941.1
|
IL7R
|
interleukin 7 receptor |
| chr8_+_33613265 | 0.64 |
ENST00000517292.1
|
RP11-317N12.1
|
RP11-317N12.1 |
| chr17_-_46667594 | 0.63 |
ENST00000476342.1
ENST00000460160.1 ENST00000472863.1 |
HOXB3
|
homeobox B3 |
| chr19_-_40790729 | 0.63 |
ENST00000423127.1
ENST00000583859.1 ENST00000456441.1 ENST00000452077.1 |
AKT2
|
v-akt murine thymoma viral oncogene homolog 2 |
| chr6_-_51952418 | 0.61 |
ENST00000371117.3
|
PKHD1
|
polycystic kidney and hepatic disease 1 (autosomal recessive) |
| chr18_+_9885961 | 0.61 |
ENST00000306084.6
|
TXNDC2
|
thioredoxin domain containing 2 (spermatozoa) |
| chr5_-_180670370 | 0.61 |
ENST00000502844.1
|
GNB2L1
|
guanine nucleotide binding protein (G protein), beta polypeptide 2-like 1 |
| chr8_-_38008783 | 0.61 |
ENST00000276449.4
|
STAR
|
steroidogenic acute regulatory protein |
| chr7_+_138915102 | 0.60 |
ENST00000486663.1
|
UBN2
|
ubinuclein 2 |
| chr14_+_22386325 | 0.60 |
ENST00000390439.2
|
TRAV13-2
|
T cell receptor alpha variable 13-2 |
| chr2_-_211179883 | 0.59 |
ENST00000352451.3
|
MYL1
|
myosin, light chain 1, alkali; skeletal, fast |
| chr18_+_9885760 | 0.59 |
ENST00000536353.2
ENST00000584255.1 |
TXNDC2
|
thioredoxin domain containing 2 (spermatozoa) |
| chr10_-_81270678 | 0.58 |
ENST00000538322.1
|
AL133481.1
|
Uncharacterized protein |
| chr8_-_144699668 | 0.58 |
ENST00000425753.2
|
TSTA3
|
tissue specific transplantation antigen P35B |
| chr8_-_144699628 | 0.57 |
ENST00000529048.1
ENST00000529064.1 |
TSTA3
|
tissue specific transplantation antigen P35B |
| chrX_-_15619076 | 0.57 |
ENST00000252519.3
|
ACE2
|
angiotensin I converting enzyme 2 |
| chr18_+_9885934 | 0.56 |
ENST00000357775.5
|
TXNDC2
|
thioredoxin domain containing 2 (spermatozoa) |
| chr11_-_32457075 | 0.56 |
ENST00000448076.3
|
WT1
|
Wilms tumor 1 |
| chr7_-_121944491 | 0.56 |
ENST00000331178.4
ENST00000427185.2 ENST00000442488.2 |
FEZF1
|
FEZ family zinc finger 1 |
| chr2_-_136875712 | 0.56 |
ENST00000241393.3
|
CXCR4
|
chemokine (C-X-C motif) receptor 4 |
| chr12_-_53074182 | 0.56 |
ENST00000252244.3
|
KRT1
|
keratin 1 |
| chr1_+_158900568 | 0.56 |
ENST00000458222.1
|
PYHIN1
|
pyrin and HIN domain family, member 1 |
| chr2_-_88285309 | 0.55 |
ENST00000420840.2
|
RGPD2
|
RANBP2-like and GRIP domain containing 2 |
| chr11_-_16419067 | 0.55 |
ENST00000533411.1
|
SOX6
|
SRY (sex determining region Y)-box 6 |
| chr14_+_101123580 | 0.54 |
ENST00000556697.1
ENST00000360899.2 ENST00000553623.1 |
LINC00523
|
long intergenic non-protein coding RNA 523 |
| chr2_-_225362533 | 0.54 |
ENST00000451538.1
|
CUL3
|
cullin 3 |
| chr6_-_36515177 | 0.54 |
ENST00000229812.7
|
STK38
|
serine/threonine kinase 38 |
| chr2_+_101869262 | 0.54 |
ENST00000289382.3
|
CNOT11
|
CCR4-NOT transcription complex, subunit 11 |
| chr5_+_169010638 | 0.53 |
ENST00000265295.4
ENST00000506574.1 ENST00000515224.1 ENST00000508247.1 ENST00000513941.1 |
SPDL1
|
spindle apparatus coiled-coil protein 1 |
| chr17_+_45286706 | 0.53 |
ENST00000393450.1
ENST00000572303.1 |
MYL4
|
myosin, light chain 4, alkali; atrial, embryonic |
| chr17_+_6679528 | 0.52 |
ENST00000321535.4
|
FBXO39
|
F-box protein 39 |
| chr17_+_19091325 | 0.52 |
ENST00000584923.1
|
SNORD3A
|
small nucleolar RNA, C/D box 3A |
| chr6_-_51952367 | 0.51 |
ENST00000340994.4
|
PKHD1
|
polycystic kidney and hepatic disease 1 (autosomal recessive) |
| chrX_+_23018058 | 0.51 |
ENST00000327968.5
|
DDX53
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 53 |
| chr17_-_46667628 | 0.51 |
ENST00000498678.1
|
HOXB3
|
homeobox B3 |
| chr16_+_31085714 | 0.50 |
ENST00000300850.5
ENST00000564189.1 ENST00000428260.1 |
ZNF646
|
zinc finger protein 646 |
| chr5_-_39270725 | 0.50 |
ENST00000512138.1
ENST00000512982.1 ENST00000540520.1 |
FYB
|
FYN binding protein |
| chr3_-_18466026 | 0.49 |
ENST00000417717.2
|
SATB1
|
SATB homeobox 1 |
| chr3_-_138739768 | 0.49 |
ENST00000329447.5
|
PRR23B
|
proline rich 23B |
| chr15_+_101459420 | 0.49 |
ENST00000388948.3
ENST00000284395.5 ENST00000534045.1 ENST00000532029.2 |
LRRK1
|
leucine-rich repeat kinase 1 |
| chr16_-_20364122 | 0.49 |
ENST00000396138.4
ENST00000577168.1 |
UMOD
|
uromodulin |
| chr14_+_22968672 | 0.48 |
ENST00000390497.1
|
TRAJ40
|
T cell receptor alpha joining 40 |
| chr17_+_29664830 | 0.47 |
ENST00000444181.2
ENST00000417592.2 |
NF1
|
neurofibromin 1 |
| chr1_+_117297007 | 0.47 |
ENST00000369478.3
ENST00000369477.1 |
CD2
|
CD2 molecule |
| chr5_-_176433582 | 0.46 |
ENST00000506128.1
|
UIMC1
|
ubiquitin interaction motif containing 1 |
| chr12_-_95945246 | 0.45 |
ENST00000258499.3
|
USP44
|
ubiquitin specific peptidase 44 |
| chr22_+_38035459 | 0.45 |
ENST00000357436.4
|
SH3BP1
|
SH3-domain binding protein 1 |
| chr14_+_90422239 | 0.45 |
ENST00000393452.3
ENST00000554180.1 ENST00000393454.2 ENST00000553617.1 ENST00000335725.4 ENST00000357382.3 ENST00000556867.1 ENST00000553527.1 |
TDP1
|
tyrosyl-DNA phosphodiesterase 1 |
| chr16_-_66907139 | 0.45 |
ENST00000561579.2
|
NAE1
|
NEDD8 activating enzyme E1 subunit 1 |
| chr18_-_46895066 | 0.45 |
ENST00000583225.1
ENST00000584983.1 ENST00000583280.1 ENST00000581738.1 |
DYM
|
dymeclin |
| chr7_-_142169013 | 0.44 |
ENST00000454561.2
|
TRBV5-4
|
T cell receptor beta variable 5-4 |
| chr7_+_76090993 | 0.44 |
ENST00000425780.1
ENST00000456590.1 ENST00000451769.1 ENST00000324432.5 ENST00000307569.8 ENST00000457529.1 ENST00000446600.1 ENST00000413936.2 ENST00000423646.1 ENST00000438930.1 ENST00000430490.2 |
DTX2
|
deltex homolog 2 (Drosophila) |
| chr1_+_151735431 | 0.44 |
ENST00000321531.5
ENST00000315067.8 |
OAZ3
|
ornithine decarboxylase antizyme 3 |
| chr12_+_110940005 | 0.44 |
ENST00000409246.1
ENST00000392672.4 ENST00000409300.1 ENST00000409425.1 |
RAD9B
|
RAD9 homolog B (S. pombe) |
| chr15_+_66797627 | 0.43 |
ENST00000565627.1
ENST00000564179.1 |
ZWILCH
|
zwilch kinetochore protein |
| chr13_+_113030658 | 0.43 |
ENST00000414180.1
ENST00000443541.1 |
SPACA7
|
sperm acrosome associated 7 |
| chr2_+_135596106 | 0.43 |
ENST00000356140.5
|
ACMSD
|
aminocarboxymuconate semialdehyde decarboxylase |
| chr7_-_6006768 | 0.43 |
ENST00000441023.2
|
RSPH10B
|
radial spoke head 10 homolog B (Chlamydomonas) |
| chr18_+_61223393 | 0.42 |
ENST00000269491.1
ENST00000382768.1 |
SERPINB12
|
serpin peptidase inhibitor, clade B (ovalbumin), member 12 |
| chr15_-_34331243 | 0.42 |
ENST00000306730.3
|
AVEN
|
apoptosis, caspase activation inhibitor |
| chrX_-_100548045 | 0.42 |
ENST00000372907.3
ENST00000372905.2 |
TAF7L
|
TAF7-like RNA polymerase II, TATA box binding protein (TBP)-associated factor, 50kDa |
| chr1_+_47603109 | 0.42 |
ENST00000371890.3
ENST00000294337.3 ENST00000371891.3 |
CYP4A22
|
cytochrome P450, family 4, subfamily A, polypeptide 22 |
| chr15_+_66797455 | 0.42 |
ENST00000446801.2
|
ZWILCH
|
zwilch kinetochore protein |
| chr2_-_136873735 | 0.42 |
ENST00000409817.1
|
CXCR4
|
chemokine (C-X-C motif) receptor 4 |
| chr7_-_142124565 | 0.41 |
ENST00000390376.2
|
TRBV6-8
|
T cell receptor beta variable 6-8 |
| chr18_+_12703002 | 0.41 |
ENST00000590217.1
|
PSMG2
|
proteasome (prosome, macropain) assembly chaperone 2 |
| chr7_+_63709496 | 0.40 |
ENST00000255746.4
|
ZNF679
|
zinc finger protein 679 |
| chr13_-_24895566 | 0.40 |
ENST00000422229.2
|
AL359736.1
|
protein PCOTH isoform 1 |
| chr19_+_13885252 | 0.40 |
ENST00000221576.4
|
C19orf53
|
chromosome 19 open reading frame 53 |
| chr22_-_36925124 | 0.40 |
ENST00000457241.1
|
EIF3D
|
eukaryotic translation initiation factor 3, subunit D |
| chr8_+_1993173 | 0.40 |
ENST00000523438.1
|
MYOM2
|
myomesin 2 |
| chr1_-_247697141 | 0.40 |
ENST00000366487.3
|
OR2C3
|
olfactory receptor, family 2, subfamily C, member 3 |
| chr8_+_1993152 | 0.39 |
ENST00000262113.4
|
MYOM2
|
myomesin 2 |
| chr1_+_202431859 | 0.38 |
ENST00000391959.3
ENST00000367270.4 |
PPP1R12B
|
protein phosphatase 1, regulatory subunit 12B |
| chr14_+_31028348 | 0.38 |
ENST00000550944.1
ENST00000438909.2 ENST00000553504.1 |
G2E3
|
G2/M-phase specific E3 ubiquitin protein ligase |
| chr12_+_14956506 | 0.37 |
ENST00000330828.2
|
C12orf60
|
chromosome 12 open reading frame 60 |
| chr7_-_27183263 | 0.37 |
ENST00000222726.3
|
HOXA5
|
homeobox A5 |
| chr12_-_10875831 | 0.37 |
ENST00000279550.7
ENST00000228251.4 |
YBX3
|
Y box binding protein 3 |
| chr16_+_67143828 | 0.37 |
ENST00000563853.2
ENST00000569914.1 ENST00000569600.1 |
C16orf70
|
chromosome 16 open reading frame 70 |
| chr18_-_69449517 | 0.37 |
ENST00000584810.1
|
RP11-723G8.2
|
Uncharacterized protein |
| chr4_+_122722466 | 0.37 |
ENST00000243498.5
ENST00000379663.3 ENST00000509800.1 |
EXOSC9
|
exosome component 9 |
| chrX_+_107037451 | 0.36 |
ENST00000372379.2
|
NCBP2L
|
nuclear cap binding protein subunit 2-like |
| chr12_-_59175485 | 0.36 |
ENST00000550678.1
ENST00000552201.1 |
RP11-767I20.1
|
RP11-767I20.1 |
| chr1_+_100818009 | 0.36 |
ENST00000370125.2
ENST00000361544.6 ENST00000370124.3 |
CDC14A
|
cell division cycle 14A |
| chr5_+_169011033 | 0.36 |
ENST00000513795.1
|
SPDL1
|
spindle apparatus coiled-coil protein 1 |
| chr8_-_66701319 | 0.36 |
ENST00000379419.4
|
PDE7A
|
phosphodiesterase 7A |
| chr17_-_60883993 | 0.36 |
ENST00000583803.1
ENST00000456609.2 |
MARCH10
|
membrane-associated ring finger (C3HC4) 10, E3 ubiquitin protein ligase |
| chr4_-_46911223 | 0.35 |
ENST00000396533.1
|
COX7B2
|
cytochrome c oxidase subunit VIIb2 |
| chr15_+_72410629 | 0.35 |
ENST00000340912.4
ENST00000544171.1 |
SENP8
|
SUMO/sentrin specific peptidase family member 8 |
| chr22_-_36925186 | 0.35 |
ENST00000541106.1
ENST00000455547.1 ENST00000432675.1 |
EIF3D
|
eukaryotic translation initiation factor 3, subunit D |
| chr12_+_110940111 | 0.35 |
ENST00000409778.3
|
RAD9B
|
RAD9 homolog B (S. pombe) |
| chr11_-_19223523 | 0.35 |
ENST00000265968.3
|
CSRP3
|
cysteine and glycine-rich protein 3 (cardiac LIM protein) |
| chr17_-_34890617 | 0.35 |
ENST00000586886.1
ENST00000585719.1 ENST00000585818.1 |
MYO19
|
myosin XIX |
| chr5_+_39520499 | 0.34 |
ENST00000604954.1
|
CTD-2078B5.2
|
CTD-2078B5.2 |
| chr17_-_42976736 | 0.34 |
ENST00000591382.1
ENST00000593072.1 ENST00000592576.1 ENST00000402521.3 |
EFTUD2
|
elongation factor Tu GTP binding domain containing 2 |
| chr1_+_100817262 | 0.34 |
ENST00000455467.1
|
CDC14A
|
cell division cycle 14A |
| chr1_+_166958497 | 0.34 |
ENST00000367870.2
|
MAEL
|
maelstrom spermatogenic transposon silencer |
| chr6_+_4087664 | 0.34 |
ENST00000430835.2
|
C6orf201
|
chromosome 6 open reading frame 201 |
| chr19_+_21106028 | 0.34 |
ENST00000597314.1
ENST00000601924.1 |
ZNF85
|
zinc finger protein 85 |
| chr15_+_26360970 | 0.34 |
ENST00000556159.1
ENST00000557523.1 |
LINC00929
|
long intergenic non-protein coding RNA 929 |
| chr7_+_138145076 | 0.33 |
ENST00000343526.4
|
TRIM24
|
tripartite motif containing 24 |
| chr11_+_110001723 | 0.33 |
ENST00000528673.1
|
ZC3H12C
|
zinc finger CCCH-type containing 12C |
| chr1_-_7913089 | 0.33 |
ENST00000361696.5
|
UTS2
|
urotensin 2 |
| chr4_-_83765613 | 0.33 |
ENST00000503937.1
|
SEC31A
|
SEC31 homolog A (S. cerevisiae) |
| chr3_-_108672609 | 0.33 |
ENST00000393963.3
ENST00000471108.1 |
GUCA1C
|
guanylate cyclase activator 1C |
| chr20_-_1569278 | 0.33 |
ENST00000262929.5
ENST00000567028.1 |
SIRPB1
RP4-576H24.4
|
signal-regulatory protein beta 1 Uncharacterized protein |
| chr16_+_67906919 | 0.33 |
ENST00000358933.5
|
EDC4
|
enhancer of mRNA decapping 4 |
| chr16_+_67143880 | 0.32 |
ENST00000219139.3
ENST00000566026.1 |
C16orf70
|
chromosome 16 open reading frame 70 |
| chr13_+_50570019 | 0.32 |
ENST00000442421.1
|
TRIM13
|
tripartite motif containing 13 |
| chr8_-_101718991 | 0.32 |
ENST00000517990.1
|
PABPC1
|
poly(A) binding protein, cytoplasmic 1 |
| chr1_-_173176452 | 0.32 |
ENST00000281834.3
|
TNFSF4
|
tumor necrosis factor (ligand) superfamily, member 4 |
| chr16_-_88878305 | 0.32 |
ENST00000569616.1
ENST00000563655.1 ENST00000567713.1 ENST00000426324.2 ENST00000378364.3 |
APRT
|
adenine phosphoribosyltransferase |
| chr1_-_169680745 | 0.32 |
ENST00000236147.4
|
SELL
|
selectin L |
| chr21_+_17214724 | 0.32 |
ENST00000449491.1
|
USP25
|
ubiquitin specific peptidase 25 |
| chr21_-_45960078 | 0.32 |
ENST00000400375.1
|
KRTAP10-1
|
keratin associated protein 10-1 |
| chr2_+_170335924 | 0.32 |
ENST00000554017.1
ENST00000392663.2 ENST00000513963.1 |
BBS5
RP11-724O16.1
|
Bardet-Biedl syndrome 5 Bardet-Biedl syndrome 5 protein; Uncharacterized protein |
| chr14_+_22987424 | 0.32 |
ENST00000390511.1
|
TRAJ26
|
T cell receptor alpha joining 26 |
| chr3_+_26735991 | 0.32 |
ENST00000456208.2
|
LRRC3B
|
leucine rich repeat containing 3B |
| chr7_-_105221898 | 0.31 |
ENST00000486180.1
ENST00000485614.1 ENST00000480514.1 |
EFCAB10
|
EF-hand calcium binding domain 10 |
| chr12_-_10007448 | 0.31 |
ENST00000538152.1
|
CLEC2B
|
C-type lectin domain family 2, member B |
| chr3_-_179322436 | 0.31 |
ENST00000392659.2
ENST00000476781.1 |
MRPL47
|
mitochondrial ribosomal protein L47 |
| chr7_+_80275953 | 0.31 |
ENST00000538969.1
ENST00000544133.1 ENST00000433696.2 |
CD36
|
CD36 molecule (thrombospondin receptor) |
| chr8_-_87755878 | 0.31 |
ENST00000320005.5
|
CNGB3
|
cyclic nucleotide gated channel beta 3 |
| chr4_-_153601136 | 0.31 |
ENST00000504064.1
ENST00000304385.3 |
TMEM154
|
transmembrane protein 154 |
| chr19_-_55453077 | 0.31 |
ENST00000328092.5
ENST00000590030.1 |
NLRP7
|
NLR family, pyrin domain containing 7 |
| chr7_-_142143973 | 0.31 |
ENST00000390373.2
|
TRBV6-7
|
T cell receptor beta variable 6-7 (non-functional) |
| chr15_+_26360932 | 0.31 |
ENST00000556213.1
|
LINC00929
|
long intergenic non-protein coding RNA 929 |
| chr19_-_49496557 | 0.31 |
ENST00000323798.3
ENST00000541188.1 ENST00000544287.1 ENST00000540532.1 ENST00000263276.6 |
GYS1
|
glycogen synthase 1 (muscle) |
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXC12_HOXD12
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.0 | GO:0006433 | glutamyl-tRNA aminoacylation(GO:0006424) prolyl-tRNA aminoacylation(GO:0006433) |
| 0.4 | 2.1 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.3 | 1.2 | GO:0042351 | 'de novo' GDP-L-fucose biosynthetic process(GO:0042351) |
| 0.3 | 1.0 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.2 | 2.0 | GO:0070537 | histone H2A K63-linked deubiquitination(GO:0070537) |
| 0.2 | 1.1 | GO:0014878 | response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) |
| 0.2 | 2.3 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.2 | 0.8 | GO:0050992 | dimethylallyl diphosphate biosynthetic process(GO:0050992) dimethylallyl diphosphate metabolic process(GO:0050993) |
| 0.2 | 0.9 | GO:0090214 | spongiotrophoblast layer developmental growth(GO:0090214) |
| 0.2 | 0.5 | GO:0061534 | gamma-aminobutyric acid secretion, neurotransmission(GO:0061534) |
| 0.2 | 3.4 | GO:0043031 | negative regulation of macrophage activation(GO:0043031) |
| 0.2 | 0.6 | GO:0018963 | insecticide metabolic process(GO:0017143) phthalate metabolic process(GO:0018963) |
| 0.1 | 0.6 | GO:0072302 | negative regulation of metanephric glomerulus development(GO:0072299) negative regulation of metanephric glomerular mesangial cell proliferation(GO:0072302) |
| 0.1 | 0.4 | GO:0060435 | bronchiole development(GO:0060435) |
| 0.1 | 1.0 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.1 | 0.6 | GO:0003051 | angiotensin-mediated drinking behavior(GO:0003051) |
| 0.1 | 0.8 | GO:0071461 | cellular response to redox state(GO:0071461) |
| 0.1 | 0.3 | GO:0046005 | positive regulation of circadian sleep/wake cycle, REM sleep(GO:0046005) |
| 0.1 | 1.0 | GO:0044800 | fusion of virus membrane with host plasma membrane(GO:0019064) membrane fusion involved in viral entry into host cell(GO:0039663) multi-organism membrane fusion(GO:0044800) |
| 0.1 | 0.5 | GO:0090202 | transcriptional activation by promoter-enhancer looping(GO:0071733) gene looping(GO:0090202) dsDNA loop formation(GO:0090579) |
| 0.1 | 0.3 | GO:2000523 | regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 0.1 | 0.5 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 0.1 | 0.3 | GO:0044209 | AMP salvage(GO:0044209) |
| 0.1 | 1.8 | GO:0006662 | glycerol ether metabolic process(GO:0006662) |
| 0.1 | 0.5 | GO:0072233 | thick ascending limb development(GO:0072023) metanephric thick ascending limb development(GO:0072233) |
| 0.1 | 0.6 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.1 | 0.4 | GO:0034473 | U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) |
| 0.1 | 0.5 | GO:0021779 | oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.1 | 0.3 | GO:0060279 | positive regulation of ovulation(GO:0060279) |
| 0.1 | 0.6 | GO:0071486 | cellular response to high light intensity(GO:0071486) retinal rod cell apoptotic process(GO:0097473) |
| 0.1 | 0.4 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.1 | 0.7 | GO:0032484 | Ral protein signal transduction(GO:0032484) regulation of Ral protein signal transduction(GO:0032485) |
| 0.1 | 1.0 | GO:2000334 | blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.1 | 0.4 | GO:0003095 | pressure natriuresis(GO:0003095) |
| 0.1 | 0.5 | GO:0015887 | biotin transport(GO:0015878) pantothenate transmembrane transport(GO:0015887) |
| 0.1 | 1.4 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.1 | 0.8 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.1 | 0.7 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.1 | 0.5 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.1 | 0.2 | GO:1903926 | signal transduction involved in intra-S DNA damage checkpoint(GO:0072428) response to bisphenol A(GO:1903925) cellular response to bisphenol A(GO:1903926) |
| 0.1 | 0.3 | GO:0010157 | response to chlorate(GO:0010157) |
| 0.1 | 0.9 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.1 | 0.3 | GO:0009236 | cobalamin biosynthetic process(GO:0009236) |
| 0.1 | 0.1 | GO:0021550 | medulla oblongata development(GO:0021550) |
| 0.1 | 10.2 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.1 | 0.4 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.1 | 0.5 | GO:0030885 | regulation of myeloid dendritic cell activation(GO:0030885) |
| 0.1 | 0.3 | GO:0030573 | bile acid catabolic process(GO:0030573) |
| 0.1 | 0.2 | GO:0045041 | protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.1 | 0.2 | GO:0032687 | negative regulation of interferon-alpha production(GO:0032687) |
| 0.1 | 0.8 | GO:0031284 | positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.1 | 1.0 | GO:0007351 | tripartite regional subdivision(GO:0007351) anterior/posterior axis specification, embryo(GO:0008595) |
| 0.0 | 0.8 | GO:0050713 | negative regulation of interleukin-1 beta secretion(GO:0050713) |
| 0.0 | 0.4 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 1.2 | GO:0000076 | DNA replication checkpoint(GO:0000076) |
| 0.0 | 0.4 | GO:2000660 | negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.0 | 1.7 | GO:0060065 | uterus development(GO:0060065) |
| 0.0 | 0.3 | GO:0035995 | detection of muscle stretch(GO:0035995) |
| 0.0 | 0.7 | GO:0051256 | mitotic spindle midzone assembly(GO:0051256) |
| 0.0 | 0.8 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.0 | 1.2 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.0 | 1.9 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.0 | 0.2 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.0 | 0.3 | GO:0051177 | meiotic sister chromatid cohesion(GO:0051177) |
| 0.0 | 0.6 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 0.3 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.0 | 0.4 | GO:2000623 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 | 0.2 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.0 | 0.2 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.0 | 0.3 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.0 | 0.6 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.2 | GO:0035992 | tendon cell differentiation(GO:0035990) tendon formation(GO:0035992) |
| 0.0 | 0.3 | GO:0045602 | negative regulation of endothelial cell differentiation(GO:0045602) |
| 0.0 | 0.2 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.0 | 0.5 | GO:0000733 | DNA strand renaturation(GO:0000733) |
| 0.0 | 0.1 | GO:0033037 | polysaccharide localization(GO:0033037) |
| 0.0 | 0.1 | GO:0061156 | pulmonary artery morphogenesis(GO:0061156) regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.0 | 0.1 | GO:0051771 | negative regulation of nitric-oxide synthase biosynthetic process(GO:0051771) |
| 0.0 | 0.5 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.0 | 0.2 | GO:0045448 | regulation of mitotic cell cycle, embryonic(GO:0009794) mitotic cell cycle, embryonic(GO:0045448) |
| 0.0 | 0.2 | GO:1904796 | regulation of core promoter binding(GO:1904796) |
| 0.0 | 0.6 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.0 | 0.3 | GO:0098914 | membrane repolarization during atrial cardiac muscle cell action potential(GO:0098914) |
| 0.0 | 0.1 | GO:0072752 | cellular response to rapamycin(GO:0072752) |
| 0.0 | 0.3 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.0 | 0.1 | GO:0086097 | phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) |
| 0.0 | 0.2 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.0 | 0.4 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.5 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 0.2 | GO:0042220 | response to cocaine(GO:0042220) |
| 0.0 | 0.1 | GO:0090071 | negative regulation of ribosome biogenesis(GO:0090071) |
| 0.0 | 0.4 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.0 | 0.2 | GO:0021562 | vestibulocochlear nerve development(GO:0021562) |
| 0.0 | 0.2 | GO:1900264 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.0 | 0.4 | GO:0033689 | negative regulation of osteoblast proliferation(GO:0033689) |
| 0.0 | 0.2 | GO:0006561 | proline biosynthetic process(GO:0006561) |
| 0.0 | 0.1 | GO:0042427 | serotonin biosynthetic process(GO:0042427) |
| 0.0 | 0.1 | GO:0044029 | DNA hypomethylation(GO:0044028) hypomethylation of CpG island(GO:0044029) |
| 0.0 | 0.5 | GO:0090266 | regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.0 | 1.1 | GO:0010824 | regulation of centrosome duplication(GO:0010824) |
| 0.0 | 1.1 | GO:0002437 | inflammatory response to antigenic stimulus(GO:0002437) |
| 0.0 | 0.2 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.0 | 1.4 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.0 | 1.1 | GO:0010591 | regulation of lamellipodium assembly(GO:0010591) |
| 0.0 | 0.3 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 0.3 | GO:1902187 | negative regulation of viral release from host cell(GO:1902187) |
| 0.0 | 0.1 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) |
| 0.0 | 0.7 | GO:0042744 | hydrogen peroxide catabolic process(GO:0042744) |
| 0.0 | 0.3 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.2 | GO:0002759 | regulation of antimicrobial humoral response(GO:0002759) |
| 0.0 | 0.8 | GO:0001709 | cell fate determination(GO:0001709) |
| 0.0 | 4.5 | GO:0009952 | anterior/posterior pattern specification(GO:0009952) |
| 0.0 | 0.2 | GO:0017183 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.0 | 0.1 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.0 | 0.2 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 0.1 | GO:2000035 | regulation of stem cell division(GO:2000035) |
| 0.0 | 0.5 | GO:0000042 | protein targeting to Golgi(GO:0000042) |
| 0.0 | 0.2 | GO:0014067 | negative regulation of phosphatidylinositol 3-kinase signaling(GO:0014067) |
| 0.0 | 0.1 | GO:0090625 | mRNA cleavage involved in gene silencing by siRNA(GO:0090625) |
| 0.0 | 0.1 | GO:0060011 | Sertoli cell proliferation(GO:0060011) |
| 0.0 | 0.1 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 0.0 | 0.5 | GO:0036035 | osteoclast development(GO:0036035) |
| 0.0 | 1.1 | GO:0030049 | muscle filament sliding(GO:0030049) actin-myosin filament sliding(GO:0033275) |
| 0.0 | 0.7 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.2 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.0 | 0.5 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.0 | 0.7 | GO:0006409 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.0 | 0.1 | GO:0097461 | ferric iron import into cell(GO:0097461) ferric iron import across plasma membrane(GO:0098706) |
| 0.0 | 0.2 | GO:0097296 | activation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097296) |
| 0.0 | 0.1 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.0 | 0.2 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.0 | 0.1 | GO:0001880 | Mullerian duct regression(GO:0001880) |
| 0.0 | 0.3 | GO:0046339 | diacylglycerol metabolic process(GO:0046339) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.0 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.2 | 0.9 | GO:1990423 | RZZ complex(GO:1990423) |
| 0.2 | 0.8 | GO:0030849 | autosome(GO:0030849) |
| 0.2 | 1.0 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.2 | 2.0 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 0.1 | 0.7 | GO:1990876 | cytoplasmic side of nuclear pore(GO:1990876) |
| 0.1 | 9.8 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 1.8 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.1 | 1.0 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.1 | 0.3 | GO:0043511 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.1 | 0.7 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.1 | 0.4 | GO:0005846 | nuclear cap binding complex(GO:0005846) |
| 0.1 | 0.5 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.1 | 0.3 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.0 | 0.2 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.0 | 1.1 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 0.0 | 0.9 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.8 | GO:0072559 | NLRP3 inflammasome complex(GO:0072559) |
| 0.0 | 1.4 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 0.7 | GO:0060091 | kinocilium(GO:0060091) |
| 0.0 | 0.3 | GO:0000799 | nuclear condensin complex(GO:0000799) |
| 0.0 | 0.5 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.0 | 0.1 | GO:0097057 | TRAF2-GSTP1 complex(GO:0097057) |
| 0.0 | 0.6 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 0.1 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.0 | 0.3 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.0 | 0.2 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.0 | 0.1 | GO:1902912 | pyruvate kinase complex(GO:1902912) |
| 0.0 | 4.8 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.0 | 0.2 | GO:0097362 | MCM8-MCM9 complex(GO:0097362) |
| 0.0 | 0.2 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 0.3 | GO:0014802 | terminal cisterna(GO:0014802) |
| 0.0 | 0.4 | GO:0000243 | commitment complex(GO:0000243) |
| 0.0 | 1.0 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
| 0.0 | 1.4 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.0 | 0.3 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.0 | 0.4 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 0.1 | GO:0002944 | cyclin K-CDK12 complex(GO:0002944) |
| 0.0 | 0.5 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 0.1 | GO:0045272 | plasma membrane respiratory chain complex I(GO:0045272) |
| 0.0 | 1.0 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.3 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 1.1 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.3 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 0.1 | GO:0001674 | female germ cell nucleus(GO:0001674) |
| 0.0 | 0.1 | GO:0044354 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.0 | 0.4 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.5 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.1 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.0 | 0.2 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 0.0 | GO:0033597 | mitotic checkpoint complex(GO:0033597) bub1-bub3 complex(GO:1990298) |
| 0.0 | 0.1 | GO:0030678 | mitochondrial ribonuclease P complex(GO:0030678) |
| 0.0 | 0.2 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.0 | 0.2 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.0 | 0.1 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.1 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.0 | 0.2 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 1.1 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.3 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.9 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.3 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.8 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.6 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.0 | GO:0004827 | glutamate-tRNA ligase activity(GO:0004818) proline-tRNA ligase activity(GO:0004827) |
| 0.4 | 1.2 | GO:0042356 | GDP-4-dehydro-D-rhamnose reductase activity(GO:0042356) GDP-L-fucose synthase activity(GO:0050577) |
| 0.3 | 1.1 | GO:0004802 | transketolase activity(GO:0004802) |
| 0.3 | 1.0 | GO:0098808 | mRNA cap binding(GO:0098808) |
| 0.2 | 0.8 | GO:0004452 | isopentenyl-diphosphate delta-isomerase activity(GO:0004452) |
| 0.2 | 1.1 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.2 | 0.6 | GO:0004917 | interleukin-7 receptor activity(GO:0004917) |
| 0.2 | 0.5 | GO:0019781 | NEDD8 activating enzyme activity(GO:0019781) |
| 0.1 | 1.0 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.1 | 0.4 | GO:0030627 | pre-mRNA 5'-splice site binding(GO:0030627) |
| 0.1 | 0.8 | GO:0032089 | NACHT domain binding(GO:0032089) |
| 0.1 | 1.8 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.1 | 9.9 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.1 | 0.6 | GO:0044729 | hemi-methylated DNA-binding(GO:0044729) |
| 0.1 | 0.5 | GO:0031208 | POZ domain binding(GO:0031208) |
| 0.1 | 0.5 | GO:0032038 | myosin II heavy chain binding(GO:0032038) |
| 0.1 | 1.6 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.1 | 0.5 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.1 | 0.5 | GO:0070259 | tyrosyl-DNA phosphodiesterase activity(GO:0070259) |
| 0.1 | 1.0 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.1 | 0.9 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.1 | 0.4 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.1 | 1.0 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.1 | 0.4 | GO:0018685 | alkane 1-monooxygenase activity(GO:0018685) |
| 0.1 | 0.3 | GO:0002060 | purine nucleobase binding(GO:0002060) |
| 0.1 | 0.3 | GO:1904455 | ubiquitin-specific protease activity involved in negative regulation of ERAD pathway(GO:1904455) |
| 0.1 | 3.9 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.1 | 1.4 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.1 | 0.6 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.1 | 0.5 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.1 | 0.2 | GO:0016418 | S-acetyltransferase activity(GO:0016418) |
| 0.1 | 0.8 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.1 | 0.4 | GO:0016713 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced iron-sulfur protein as one donor, and incorporation of one atom of oxygen(GO:0016713) |
| 0.1 | 0.5 | GO:0008523 | sodium-dependent multivitamin transmembrane transporter activity(GO:0008523) |
| 0.1 | 0.2 | GO:0003896 | DNA primase activity(GO:0003896) |
| 0.1 | 1.0 | GO:0099602 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.1 | 0.3 | GO:0047787 | delta4-3-oxosteroid 5beta-reductase activity(GO:0047787) |
| 0.1 | 1.8 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.1 | 0.3 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.1 | 0.2 | GO:0019150 | D-ribulokinase activity(GO:0019150) |
| 0.0 | 2.0 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.3 | GO:0005119 | smoothened binding(GO:0005119) |
| 0.0 | 1.3 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.6 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.0 | 0.2 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.0 | 0.2 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.0 | 0.2 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.0 | 0.5 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.0 | 0.3 | GO:0086089 | voltage-gated potassium channel activity involved in atrial cardiac muscle cell action potential repolarization(GO:0086089) |
| 0.0 | 0.7 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.3 | GO:0005223 | intracellular cGMP activated cation channel activity(GO:0005223) |
| 0.0 | 0.3 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.0 | 0.6 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.9 | GO:0031432 | titin binding(GO:0031432) |
| 0.0 | 0.3 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.0 | 3.8 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.1 | GO:0031711 | angiotensin type I receptor activity(GO:0001596) bradykinin receptor binding(GO:0031711) |
| 0.0 | 0.1 | GO:0004510 | tryptophan 5-monooxygenase activity(GO:0004510) |
| 0.0 | 1.1 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.2 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 0.2 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.0 | 0.5 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 2.1 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.5 | GO:0019864 | IgG binding(GO:0019864) |
| 0.0 | 0.3 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.0 | 0.1 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 0.0 | 0.3 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.0 | 0.1 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.0 | 0.1 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.0 | 0.5 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 0.1 | GO:0070026 | nitric oxide binding(GO:0070026) |
| 0.0 | 0.1 | GO:0050659 | N-acetylgalactosamine 4-sulfate 6-O-sulfotransferase activity(GO:0050659) |
| 0.0 | 0.1 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.0 | 0.4 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.2 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.0 | 0.2 | GO:0019826 | oxygen sensor activity(GO:0019826) |
| 0.0 | 0.2 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.0 | 1.2 | GO:0070063 | RNA polymerase binding(GO:0070063) |
| 0.0 | 0.1 | GO:0052851 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.0 | 0.3 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 0.1 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.0 | 2.3 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 0.1 | GO:0005497 | androgen binding(GO:0005497) |
| 0.0 | 0.4 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.0 | 1.1 | GO:0008408 | 3'-5' exonuclease activity(GO:0008408) |
| 0.0 | 0.1 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.9 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.0 | 0.2 | GO:0043142 | single-stranded DNA-dependent ATPase activity(GO:0043142) |
| 0.0 | 0.3 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.4 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.1 | GO:0034597 | phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.0 | 0.6 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.1 | GO:0005221 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.0 | 0.1 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.0 | 1.2 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.4 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 1.5 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 2.2 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 0.7 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.9 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 1.7 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 0.6 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 0.8 | PID S1P S1P3 PATHWAY | S1P3 pathway |
| 0.0 | 0.7 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 0.5 | PID MYC PATHWAY | C-MYC pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.0 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.1 | 10.9 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.1 | 1.1 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.0 | 2.0 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 3.2 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.4 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.0 | 0.6 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 1.6 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 0.5 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.4 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.3 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.5 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 1.2 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.8 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 1.0 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 1.1 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.4 | REACTOME POL SWITCHING | Genes involved in Polymerase switching |
| 0.0 | 0.5 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.0 | 0.7 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 0.4 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.2 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 0.7 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 0.7 | REACTOME REGULATION OF GLUCOKINASE BY GLUCOKINASE REGULATORY PROTEIN | Genes involved in Regulation of Glucokinase by Glucokinase Regulatory Protein |
| 0.0 | 0.6 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.0 | 0.3 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.2 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.4 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.5 | REACTOME DOUBLE STRAND BREAK REPAIR | Genes involved in Double-Strand Break Repair |
| 0.0 | 0.2 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.0 | 0.2 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |