Project
Illumina Body Map 2: averaged replicates
Navigation
Downloads
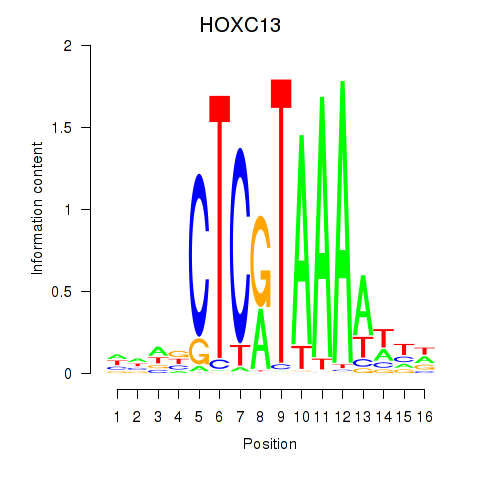
Results for HOXC13
Z-value: 1.41
Transcription factors associated with HOXC13
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXC13
|
ENSG00000123364.3 | homeobox C13 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXC13 | hg19_v2_chr12_+_54332535_54332636 | 0.30 | 1.0e-01 | Click! |
Activity profile of HOXC13 motif
Sorted Z-values of HOXC13 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_71532668 | 7.31 |
ENST00000510437.1
|
IGJ
|
immunoglobulin J polypeptide, linker protein for immunoglobulin alpha and mu polypeptides |
| chr4_-_71532601 | 5.35 |
ENST00000510614.1
|
IGJ
|
immunoglobulin J polypeptide, linker protein for immunoglobulin alpha and mu polypeptides |
| chr14_-_106668095 | 3.76 |
ENST00000390606.2
|
IGHV3-20
|
immunoglobulin heavy variable 3-20 |
| chr14_-_106725723 | 3.08 |
ENST00000390609.2
|
IGHV3-23
|
immunoglobulin heavy variable 3-23 |
| chr18_+_61554932 | 2.65 |
ENST00000299502.4
ENST00000457692.1 ENST00000413956.1 |
SERPINB2
|
serpin peptidase inhibitor, clade B (ovalbumin), member 2 |
| chr4_-_71532339 | 2.44 |
ENST00000254801.4
|
IGJ
|
immunoglobulin J polypeptide, linker protein for immunoglobulin alpha and mu polypeptides |
| chr22_+_23101182 | 2.41 |
ENST00000390312.2
|
IGLV2-14
|
immunoglobulin lambda variable 2-14 |
| chr22_+_23114284 | 2.29 |
ENST00000390313.2
|
IGLV3-12
|
immunoglobulin lambda variable 3-12 |
| chr2_+_89986318 | 2.27 |
ENST00000491977.1
|
IGKV2D-29
|
immunoglobulin kappa variable 2D-29 |
| chr7_+_80275953 | 2.26 |
ENST00000538969.1
ENST00000544133.1 ENST00000433696.2 |
CD36
|
CD36 molecule (thrombospondin receptor) |
| chr15_+_57891609 | 2.12 |
ENST00000569089.1
|
MYZAP
|
myocardial zonula adherens protein |
| chr7_+_80275752 | 2.10 |
ENST00000419819.2
|
CD36
|
CD36 molecule (thrombospondin receptor) |
| chr7_+_80275621 | 2.07 |
ENST00000426978.1
ENST00000432207.1 |
CD36
|
CD36 molecule (thrombospondin receptor) |
| chr7_+_80275663 | 2.06 |
ENST00000413265.1
|
CD36
|
CD36 molecule (thrombospondin receptor) |
| chr10_+_51549498 | 1.94 |
ENST00000358559.2
ENST00000298239.6 |
MSMB
|
microseminoprotein, beta- |
| chr11_+_118175132 | 1.94 |
ENST00000361763.4
|
CD3E
|
CD3e molecule, epsilon (CD3-TCR complex) |
| chr9_-_37034028 | 1.92 |
ENST00000520281.1
ENST00000446742.1 ENST00000522003.1 ENST00000523145.1 ENST00000414447.1 ENST00000377847.2 ENST00000377853.2 ENST00000377852.2 ENST00000523241.1 ENST00000520154.1 ENST00000358127.4 |
PAX5
|
paired box 5 |
| chr14_-_106963409 | 1.92 |
ENST00000390621.2
|
IGHV1-45
|
immunoglobulin heavy variable 1-45 |
| chr1_-_169680745 | 1.76 |
ENST00000236147.4
|
SELL
|
selectin L |
| chr5_+_35852797 | 1.68 |
ENST00000508941.1
|
IL7R
|
interleukin 7 receptor |
| chr1_+_109656719 | 1.67 |
ENST00000457623.2
ENST00000529753.1 |
KIAA1324
|
KIAA1324 |
| chr1_+_192544857 | 1.67 |
ENST00000367459.3
ENST00000469578.2 |
RGS1
|
regulator of G-protein signaling 1 |
| chr14_-_106816253 | 1.63 |
ENST00000390615.2
|
IGHV3-33
|
immunoglobulin heavy variable 3-33 |
| chr7_+_142031986 | 1.61 |
ENST00000547918.2
|
TRBV7-1
|
T cell receptor beta variable 7-1 (non-functional) |
| chr12_+_47473369 | 1.61 |
ENST00000546455.1
|
PCED1B
|
PC-esterase domain containing 1B |
| chr12_-_114841703 | 1.59 |
ENST00000526441.1
|
TBX5
|
T-box 5 |
| chr1_+_109656579 | 1.58 |
ENST00000526264.1
ENST00000369939.3 |
KIAA1324
|
KIAA1324 |
| chr3_+_130300585 | 1.57 |
ENST00000511332.1
|
COL6A6
|
collagen, type VI, alpha 6 |
| chr6_-_33041378 | 1.56 |
ENST00000428995.1
|
HLA-DPA1
|
major histocompatibility complex, class II, DP alpha 1 |
| chr6_-_32784687 | 1.52 |
ENST00000447394.1
ENST00000438763.2 |
HLA-DOB
|
major histocompatibility complex, class II, DO beta |
| chr21_+_43823983 | 1.47 |
ENST00000291535.6
ENST00000450356.1 ENST00000319294.6 ENST00000398367.1 |
UBASH3A
|
ubiquitin associated and SH3 domain containing A |
| chr17_-_74639730 | 1.43 |
ENST00000589992.1
|
ST6GALNAC1
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 1 |
| chr16_+_12059050 | 1.39 |
ENST00000396495.3
|
TNFRSF17
|
tumor necrosis factor receptor superfamily, member 17 |
| chr18_-_70532906 | 1.39 |
ENST00000299430.2
ENST00000397929.1 |
NETO1
|
neuropilin (NRP) and tolloid (TLL)-like 1 |
| chr20_+_36946029 | 1.38 |
ENST00000417318.1
|
BPI
|
bactericidal/permeability-increasing protein |
| chr1_+_158815588 | 1.37 |
ENST00000438394.1
|
MNDA
|
myeloid cell nuclear differentiation antigen |
| chr17_-_74639886 | 1.37 |
ENST00000156626.7
|
ST6GALNAC1
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 1 |
| chr6_+_127898312 | 1.33 |
ENST00000329722.7
|
C6orf58
|
chromosome 6 open reading frame 58 |
| chr3_-_172241250 | 1.30 |
ENST00000420541.2
ENST00000241261.2 |
TNFSF10
|
tumor necrosis factor (ligand) superfamily, member 10 |
| chr14_+_97925151 | 1.29 |
ENST00000554862.1
ENST00000554260.1 ENST00000499910.2 |
CTD-2506J14.1
|
CTD-2506J14.1 |
| chr6_-_49681235 | 1.25 |
ENST00000339139.4
|
CRISP2
|
cysteine-rich secretory protein 2 |
| chr8_-_95274536 | 1.21 |
ENST00000297596.2
ENST00000396194.2 |
GEM
|
GTP binding protein overexpressed in skeletal muscle |
| chr8_-_86290333 | 1.20 |
ENST00000521846.1
ENST00000523022.1 ENST00000524324.1 ENST00000519991.1 ENST00000520663.1 ENST00000517590.1 ENST00000522579.1 ENST00000522814.1 ENST00000522662.1 ENST00000523858.1 ENST00000519129.1 |
CA1
|
carbonic anhydrase I |
| chr3_-_112320749 | 1.16 |
ENST00000610103.1
|
RP11-572C15.6
|
RP11-572C15.6 |
| chr15_+_94774767 | 1.14 |
ENST00000543482.1
|
MCTP2
|
multiple C2 domains, transmembrane 2 |
| chr19_+_859654 | 1.13 |
ENST00000592860.1
|
CFD
|
complement factor D (adipsin) |
| chr15_+_77287715 | 1.09 |
ENST00000559161.1
|
PSTPIP1
|
proline-serine-threonine phosphatase interacting protein 1 |
| chr15_+_71228826 | 1.09 |
ENST00000558456.1
ENST00000560158.2 ENST00000558808.1 ENST00000559806.1 ENST00000559069.1 |
LRRC49
|
leucine rich repeat containing 49 |
| chr14_+_22409308 | 1.08 |
ENST00000390441.2
|
TRAV9-2
|
T cell receptor alpha variable 9-2 |
| chr12_-_54689532 | 1.06 |
ENST00000540264.2
ENST00000312156.4 |
NFE2
|
nuclear factor, erythroid 2 |
| chr19_-_18632861 | 1.05 |
ENST00000262809.4
|
ELL
|
elongation factor RNA polymerase II |
| chr6_+_126221034 | 0.98 |
ENST00000433571.1
|
NCOA7
|
nuclear receptor coactivator 7 |
| chr16_-_68033356 | 0.98 |
ENST00000393847.1
ENST00000573808.1 ENST00000572624.1 |
DPEP2
|
dipeptidase 2 |
| chr6_+_151358048 | 0.98 |
ENST00000450635.1
|
MTHFD1L
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1-like |
| chr2_+_103035102 | 0.97 |
ENST00000264260.2
|
IL18RAP
|
interleukin 18 receptor accessory protein |
| chr9_+_67977438 | 0.94 |
ENST00000456982.1
|
RP11-195B21.3
|
Protein LOC644249 |
| chr11_+_107643129 | 0.93 |
ENST00000447610.1
|
AP001024.2
|
Uncharacterized protein |
| chr10_+_30723533 | 0.92 |
ENST00000413724.1
|
MAP3K8
|
mitogen-activated protein kinase kinase kinase 8 |
| chr16_+_33605231 | 0.92 |
ENST00000570121.2
|
IGHV3OR16-12
|
immunoglobulin heavy variable 3/OR16-12 (non-functional) |
| chr1_+_174844645 | 0.89 |
ENST00000486220.1
|
RABGAP1L
|
RAB GTPase activating protein 1-like |
| chr1_+_109656532 | 0.88 |
ENST00000531664.1
ENST00000534476.1 |
KIAA1324
|
KIAA1324 |
| chr4_-_147866960 | 0.87 |
ENST00000513335.1
|
TTC29
|
tetratricopeptide repeat domain 29 |
| chr19_+_41698927 | 0.86 |
ENST00000310054.4
|
CYP2S1
|
cytochrome P450, family 2, subfamily S, polypeptide 1 |
| chr12_-_47473425 | 0.85 |
ENST00000550413.1
|
AMIGO2
|
adhesion molecule with Ig-like domain 2 |
| chr10_+_70847852 | 0.85 |
ENST00000242465.3
|
SRGN
|
serglycin |
| chr12_+_96883347 | 0.84 |
ENST00000524981.4
ENST00000298953.3 |
C12orf55
|
chromosome 12 open reading frame 55 |
| chr6_-_112080256 | 0.84 |
ENST00000462856.2
ENST00000229471.4 |
FYN
|
FYN oncogene related to SRC, FGR, YES |
| chr13_-_46964177 | 0.83 |
ENST00000389908.3
|
KIAA0226L
|
KIAA0226-like |
| chr8_-_27850180 | 0.81 |
ENST00000380385.2
ENST00000301906.4 ENST00000354914.3 |
SCARA5
|
scavenger receptor class A, member 5 (putative) |
| chr3_-_160116995 | 0.80 |
ENST00000465537.1
ENST00000486856.1 ENST00000468218.1 ENST00000478370.1 |
IFT80
|
intraflagellar transport 80 homolog (Chlamydomonas) |
| chr19_+_36545833 | 0.80 |
ENST00000401500.2
ENST00000270301.7 ENST00000427823.2 |
WDR62
|
WD repeat domain 62 |
| chr12_-_9885888 | 0.79 |
ENST00000327839.3
|
CLECL1
|
C-type lectin-like 1 |
| chr15_+_71184931 | 0.79 |
ENST00000560369.1
ENST00000260382.5 |
LRRC49
|
leucine rich repeat containing 49 |
| chr7_+_106505696 | 0.78 |
ENST00000440650.2
ENST00000496166.1 ENST00000473541.1 |
PIK3CG
|
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit gamma |
| chr9_+_112887772 | 0.77 |
ENST00000259318.7
|
AKAP2
|
A kinase (PRKA) anchor protein 2 |
| chr7_-_86595190 | 0.76 |
ENST00000398276.2
ENST00000416314.1 ENST00000425689.1 |
KIAA1324L
|
KIAA1324-like |
| chr19_+_7011509 | 0.74 |
ENST00000377296.3
|
AC025278.1
|
Uncharacterized protein |
| chr3_-_160117035 | 0.74 |
ENST00000489004.1
ENST00000496589.1 |
IFT80
|
intraflagellar transport 80 homolog (Chlamydomonas) |
| chr3_+_167582561 | 0.73 |
ENST00000463642.1
ENST00000464514.1 |
RP11-298O21.6
RP11-298O21.7
|
RP11-298O21.6 RP11-298O21.7 |
| chr9_-_130533615 | 0.73 |
ENST00000373277.4
|
SH2D3C
|
SH2 domain containing 3C |
| chr4_+_124571409 | 0.71 |
ENST00000514823.1
ENST00000511919.1 ENST00000508111.1 |
RP11-93L9.1
|
long intergenic non-protein coding RNA 1091 |
| chr15_+_74421961 | 0.71 |
ENST00000565159.1
ENST00000567206.1 |
ISLR2
|
immunoglobulin superfamily containing leucine-rich repeat 2 |
| chr11_+_62037622 | 0.71 |
ENST00000227918.2
ENST00000525380.1 |
SCGB2A2
|
secretoglobin, family 2A, member 2 |
| chr20_-_44420507 | 0.70 |
ENST00000243938.4
|
WFDC3
|
WAP four-disulfide core domain 3 |
| chr10_-_21186144 | 0.70 |
ENST00000377119.1
|
NEBL
|
nebulette |
| chr12_-_91546926 | 0.70 |
ENST00000550758.1
|
DCN
|
decorin |
| chr15_+_71185148 | 0.70 |
ENST00000443425.2
ENST00000560755.1 |
LRRC49
|
leucine rich repeat containing 49 |
| chr7_+_106505912 | 0.69 |
ENST00000359195.3
|
PIK3CG
|
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit gamma |
| chr7_-_29152509 | 0.69 |
ENST00000448959.1
|
CPVL
|
carboxypeptidase, vitellogenic-like |
| chr4_-_120243545 | 0.68 |
ENST00000274024.3
|
FABP2
|
fatty acid binding protein 2, intestinal |
| chr8_-_27850141 | 0.68 |
ENST00000524352.1
|
SCARA5
|
scavenger receptor class A, member 5 (putative) |
| chr7_-_4877677 | 0.67 |
ENST00000538469.1
|
RADIL
|
Ras association and DIL domains |
| chr3_-_49837254 | 0.66 |
ENST00000412678.2
ENST00000343366.4 ENST00000487256.1 |
CDHR4
|
cadherin-related family member 4 |
| chr5_-_39270725 | 0.65 |
ENST00000512138.1
ENST00000512982.1 ENST00000540520.1 |
FYB
|
FYN binding protein |
| chr12_-_9885702 | 0.65 |
ENST00000542530.1
|
CLECL1
|
C-type lectin-like 1 |
| chr1_+_174128639 | 0.65 |
ENST00000251507.4
|
RABGAP1L
|
RAB GTPase activating protein 1-like |
| chr12_-_53207842 | 0.64 |
ENST00000458244.2
|
KRT4
|
keratin 4 |
| chr1_+_158979680 | 0.63 |
ENST00000368131.4
ENST00000340979.6 |
IFI16
|
interferon, gamma-inducible protein 16 |
| chr12_+_75874580 | 0.62 |
ENST00000456650.3
|
GLIPR1
|
GLI pathogenesis-related 1 |
| chr12_-_123728548 | 0.62 |
ENST00000545406.1
|
MPHOSPH9
|
M-phase phosphoprotein 9 |
| chr1_-_155880672 | 0.62 |
ENST00000609492.1
ENST00000368322.3 |
RIT1
|
Ras-like without CAAX 1 |
| chr5_-_180076613 | 0.62 |
ENST00000261937.6
ENST00000393347.3 |
FLT4
|
fms-related tyrosine kinase 4 |
| chr1_+_158979686 | 0.62 |
ENST00000368132.3
ENST00000295809.7 |
IFI16
|
interferon, gamma-inducible protein 16 |
| chr14_-_92333873 | 0.61 |
ENST00000435962.2
|
TC2N
|
tandem C2 domains, nuclear |
| chr17_+_35732955 | 0.60 |
ENST00000300618.4
|
C17orf78
|
chromosome 17 open reading frame 78 |
| chr6_-_138060103 | 0.60 |
ENST00000411615.1
ENST00000419220.1 |
RP11-356I2.1
|
RP11-356I2.1 |
| chr11_+_101785727 | 0.59 |
ENST00000263468.8
|
KIAA1377
|
KIAA1377 |
| chr9_-_139137648 | 0.58 |
ENST00000358701.5
|
QSOX2
|
quiescin Q6 sulfhydryl oxidase 2 |
| chr2_+_33683109 | 0.57 |
ENST00000437184.1
|
RASGRP3
|
RAS guanyl releasing protein 3 (calcium and DAG-regulated) |
| chr7_-_92777606 | 0.57 |
ENST00000437805.1
ENST00000446959.1 ENST00000439952.1 ENST00000414791.1 ENST00000446033.1 ENST00000411955.1 ENST00000318238.4 |
SAMD9L
|
sterile alpha motif domain containing 9-like |
| chr22_+_38142235 | 0.57 |
ENST00000407319.2
ENST00000403663.2 ENST00000428075.1 |
TRIOBP
|
TRIO and F-actin binding protein |
| chr12_-_91539918 | 0.56 |
ENST00000548218.1
|
DCN
|
decorin |
| chr6_-_112081113 | 0.56 |
ENST00000517419.1
|
FYN
|
FYN oncogene related to SRC, FGR, YES |
| chr6_-_128222103 | 0.56 |
ENST00000434358.1
ENST00000543064.1 ENST00000368248.2 |
THEMIS
|
thymocyte selection associated |
| chr17_-_39140549 | 0.55 |
ENST00000377755.4
|
KRT40
|
keratin 40 |
| chr1_+_206579736 | 0.55 |
ENST00000439126.1
|
SRGAP2
|
SLIT-ROBO Rho GTPase activating protein 2 |
| chr7_+_6797288 | 0.55 |
ENST00000433859.2
ENST00000359718.3 |
RSPH10B2
|
radial spoke head 10 homolog B2 (Chlamydomonas) |
| chr3_-_108672742 | 0.54 |
ENST00000261047.3
|
GUCA1C
|
guanylate cyclase activator 1C |
| chr3_+_160117062 | 0.54 |
ENST00000497311.1
|
SMC4
|
structural maintenance of chromosomes 4 |
| chr8_+_77318769 | 0.53 |
ENST00000518732.1
|
RP11-706J10.1
|
long intergenic non-protein coding RNA 1111 |
| chr1_+_174128704 | 0.53 |
ENST00000457696.1
|
RABGAP1L
|
RAB GTPase activating protein 1-like |
| chr12_-_92539614 | 0.52 |
ENST00000256015.3
|
BTG1
|
B-cell translocation gene 1, anti-proliferative |
| chr1_+_171107241 | 0.52 |
ENST00000236166.3
|
FMO6P
|
flavin containing monooxygenase 6 pseudogene |
| chr19_+_21106081 | 0.52 |
ENST00000300540.3
ENST00000595854.1 ENST00000601284.1 ENST00000328178.8 ENST00000599885.1 ENST00000596476.1 ENST00000345030.6 |
ZNF85
|
zinc finger protein 85 |
| chr4_+_8321882 | 0.52 |
ENST00000509453.1
ENST00000503186.1 |
RP11-774O3.2
RP11-774O3.1
|
RP11-774O3.2 RP11-774O3.1 |
| chr5_-_180076580 | 0.51 |
ENST00000502649.1
|
FLT4
|
fms-related tyrosine kinase 4 |
| chr3_-_108672609 | 0.51 |
ENST00000393963.3
ENST00000471108.1 |
GUCA1C
|
guanylate cyclase activator 1C |
| chr10_-_6019984 | 0.49 |
ENST00000525219.2
|
IL15RA
|
interleukin 15 receptor, alpha |
| chr7_-_38389573 | 0.49 |
ENST00000390344.2
|
TRGV5
|
T cell receptor gamma variable 5 |
| chr19_-_48753064 | 0.49 |
ENST00000520153.1
ENST00000357778.5 ENST00000520015.1 |
CARD8
|
caspase recruitment domain family, member 8 |
| chr11_-_102576537 | 0.49 |
ENST00000260229.4
|
MMP27
|
matrix metallopeptidase 27 |
| chr3_+_50211240 | 0.49 |
ENST00000420831.1
|
SEMA3F
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3F |
| chr1_+_43766668 | 0.48 |
ENST00000441333.2
ENST00000538015.1 |
TIE1
|
tyrosine kinase with immunoglobulin-like and EGF-like domains 1 |
| chr12_-_57630873 | 0.48 |
ENST00000556732.1
|
NDUFA4L2
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 4-like 2 |
| chr16_-_9770700 | 0.48 |
ENST00000561538.1
|
RP11-297M9.1
|
Uncharacterized protein |
| chrY_+_14958970 | 0.47 |
ENST00000453031.1
|
USP9Y
|
ubiquitin specific peptidase 9, Y-linked |
| chr15_+_26640219 | 0.46 |
ENST00000451579.1
|
AC009878.2
|
AC009878.2 |
| chr18_-_24722995 | 0.46 |
ENST00000581714.1
|
CHST9
|
carbohydrate (N-acetylgalactosamine 4-0) sulfotransferase 9 |
| chr5_-_169725231 | 0.46 |
ENST00000046794.5
|
LCP2
|
lymphocyte cytosolic protein 2 (SH2 domain containing leukocyte protein of 76kDa) |
| chr5_+_56910048 | 0.45 |
ENST00000509844.1
|
CTD-2023N9.3
|
CTD-2023N9.3 |
| chr14_-_88200641 | 0.45 |
ENST00000556168.1
|
RP11-1152H15.1
|
RP11-1152H15.1 |
| chr22_+_32439019 | 0.44 |
ENST00000266088.4
|
SLC5A1
|
solute carrier family 5 (sodium/glucose cotransporter), member 1 |
| chr9_-_95244781 | 0.44 |
ENST00000375544.3
ENST00000375543.1 ENST00000395538.3 ENST00000450139.2 |
ASPN
|
asporin |
| chr4_+_147145709 | 0.44 |
ENST00000504313.1
|
RP11-6L6.2
|
Uncharacterized protein |
| chr1_+_43766642 | 0.44 |
ENST00000372476.3
|
TIE1
|
tyrosine kinase with immunoglobulin-like and EGF-like domains 1 |
| chr6_+_33239787 | 0.43 |
ENST00000439602.2
ENST00000474973.1 |
RPS18
|
ribosomal protein S18 |
| chr21_+_25801088 | 0.43 |
ENST00000415182.1
|
AP000476.1
|
AP000476.1 |
| chr17_+_35732916 | 0.43 |
ENST00000586700.1
|
C17orf78
|
chromosome 17 open reading frame 78 |
| chr12_-_89413456 | 0.42 |
ENST00000500381.2
|
RP11-13A1.1
|
RP11-13A1.1 |
| chr6_+_21666633 | 0.42 |
ENST00000606851.1
|
CASC15
|
cancer susceptibility candidate 15 (non-protein coding) |
| chr11_+_61891445 | 0.42 |
ENST00000394818.3
ENST00000533896.1 ENST00000278849.4 |
INCENP
|
inner centromere protein antigens 135/155kDa |
| chr5_+_140180635 | 0.42 |
ENST00000522353.2
ENST00000532566.2 |
PCDHA3
|
protocadherin alpha 3 |
| chr20_+_37590942 | 0.42 |
ENST00000373325.2
ENST00000252011.3 ENST00000373323.4 |
DHX35
|
DEAH (Asp-Glu-Ala-His) box polypeptide 35 |
| chr2_+_48796120 | 0.41 |
ENST00000394754.1
|
STON1-GTF2A1L
|
STON1-GTF2A1L readthrough |
| chr12_+_93115281 | 0.41 |
ENST00000549856.1
|
PLEKHG7
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 7 |
| chr1_+_166958346 | 0.41 |
ENST00000367872.4
|
MAEL
|
maelstrom spermatogenic transposon silencer |
| chr3_+_111630451 | 0.41 |
ENST00000495180.1
|
PHLDB2
|
pleckstrin homology-like domain, family B, member 2 |
| chr1_-_109656439 | 0.40 |
ENST00000369949.4
|
C1orf194
|
chromosome 1 open reading frame 194 |
| chr4_+_88720698 | 0.40 |
ENST00000226284.5
|
IBSP
|
integrin-binding sialoprotein |
| chr7_-_41742697 | 0.40 |
ENST00000242208.4
|
INHBA
|
inhibin, beta A |
| chr11_-_114430570 | 0.39 |
ENST00000251921.2
|
NXPE1
|
neurexophilin and PC-esterase domain family, member 1 |
| chr2_+_33359646 | 0.39 |
ENST00000390003.4
ENST00000418533.2 |
LTBP1
|
latent transforming growth factor beta binding protein 1 |
| chr11_+_3011093 | 0.38 |
ENST00000332881.2
|
AC131971.1
|
HCG1782999; PRO0943; Uncharacterized protein |
| chr3_+_57875738 | 0.38 |
ENST00000417128.1
ENST00000438794.1 |
SLMAP
|
sarcolemma associated protein |
| chr2_+_162016804 | 0.38 |
ENST00000392749.2
ENST00000440506.1 |
TANK
|
TRAF family member-associated NFKB activator |
| chr2_+_33359687 | 0.38 |
ENST00000402934.1
ENST00000404525.1 ENST00000407925.1 |
LTBP1
|
latent transforming growth factor beta binding protein 1 |
| chr6_-_36515177 | 0.38 |
ENST00000229812.7
|
STK38
|
serine/threonine kinase 38 |
| chr19_-_54618650 | 0.38 |
ENST00000391757.1
|
TFPT
|
TCF3 (E2A) fusion partner (in childhood Leukemia) |
| chr19_-_48759119 | 0.37 |
ENST00000522889.1
ENST00000520753.1 ENST00000519940.1 ENST00000519332.1 ENST00000521437.1 ENST00000520007.1 ENST00000521613.1 |
CARD8
|
caspase recruitment domain family, member 8 |
| chr18_-_73967160 | 0.37 |
ENST00000579714.1
|
RP11-94B19.7
|
RP11-94B19.7 |
| chr6_-_130031358 | 0.37 |
ENST00000368149.2
|
ARHGAP18
|
Rho GTPase activating protein 18 |
| chr1_-_168464875 | 0.37 |
ENST00000422253.1
|
RP5-968D22.3
|
RP5-968D22.3 |
| chr7_-_16872932 | 0.36 |
ENST00000419572.2
ENST00000412973.1 |
AGR2
|
anterior gradient 2 |
| chr18_-_5544241 | 0.36 |
ENST00000341928.2
ENST00000540638.2 |
EPB41L3
|
erythrocyte membrane protein band 4.1-like 3 |
| chr17_-_8286484 | 0.36 |
ENST00000582556.1
ENST00000584164.1 ENST00000293842.5 ENST00000584343.1 ENST00000578812.1 ENST00000583011.1 |
RPL26
|
ribosomal protein L26 |
| chr11_+_4510109 | 0.36 |
ENST00000307632.3
|
OR52K1
|
olfactory receptor, family 52, subfamily K, member 1 |
| chr4_-_164395014 | 0.36 |
ENST00000280605.3
|
TKTL2
|
transketolase-like 2 |
| chr3_-_123339343 | 0.35 |
ENST00000578202.1
|
MYLK
|
myosin light chain kinase |
| chr3_+_101498269 | 0.34 |
ENST00000491511.2
|
NXPE3
|
neurexophilin and PC-esterase domain family, member 3 |
| chr2_+_162016916 | 0.34 |
ENST00000405852.1
|
TANK
|
TRAF family member-associated NFKB activator |
| chr9_-_5304432 | 0.34 |
ENST00000416837.1
ENST00000308420.3 |
RLN2
|
relaxin 2 |
| chr18_-_52989525 | 0.33 |
ENST00000457482.3
|
TCF4
|
transcription factor 4 |
| chr3_-_99569821 | 0.32 |
ENST00000487087.1
|
FILIP1L
|
filamin A interacting protein 1-like |
| chrX_+_13707235 | 0.32 |
ENST00000464506.1
|
RAB9A
|
RAB9A, member RAS oncogene family |
| chr17_+_68047418 | 0.32 |
ENST00000586373.1
ENST00000588782.1 |
LINC01028
|
long intergenic non-protein coding RNA 1028 |
| chr16_+_86612112 | 0.32 |
ENST00000320241.3
|
FOXL1
|
forkhead box L1 |
| chr2_-_225811747 | 0.31 |
ENST00000409592.3
|
DOCK10
|
dedicator of cytokinesis 10 |
| chr7_-_25219897 | 0.31 |
ENST00000283905.3
ENST00000409280.1 ENST00000415598.1 |
C7orf31
|
chromosome 7 open reading frame 31 |
| chr11_-_58378759 | 0.30 |
ENST00000601906.1
|
AP001350.1
|
Uncharacterized protein |
| chr11_+_63606373 | 0.30 |
ENST00000402010.2
ENST00000315032.8 ENST00000377809.4 ENST00000413835.2 ENST00000377810.3 |
MARK2
|
MAP/microtubule affinity-regulating kinase 2 |
| chr15_+_77713299 | 0.30 |
ENST00000559099.1
|
HMG20A
|
high mobility group 20A |
| chrX_-_38186775 | 0.30 |
ENST00000339363.3
ENST00000309513.3 ENST00000338898.3 ENST00000342811.3 ENST00000378505.2 |
RPGR
|
retinitis pigmentosa GTPase regulator |
| chr22_+_39916558 | 0.30 |
ENST00000337304.2
ENST00000396680.1 |
ATF4
|
activating transcription factor 4 |
| chr7_+_63709496 | 0.29 |
ENST00000255746.4
|
ZNF679
|
zinc finger protein 679 |
| chr19_+_21264980 | 0.29 |
ENST00000596053.1
ENST00000597086.1 ENST00000596143.1 ENST00000596367.1 ENST00000601416.1 |
ZNF714
|
zinc finger protein 714 |
| chr5_-_176433565 | 0.29 |
ENST00000428382.2
|
UIMC1
|
ubiquitin interaction motif containing 1 |
| chr2_+_162016827 | 0.29 |
ENST00000429217.1
ENST00000406287.1 ENST00000402568.1 |
TANK
|
TRAF family member-associated NFKB activator |
| chr19_+_36545781 | 0.29 |
ENST00000388999.3
|
WDR62
|
WD repeat domain 62 |
| chr9_+_40028620 | 0.28 |
ENST00000426179.1
|
AL353791.1
|
AL353791.1 |
| chr12_+_93096759 | 0.28 |
ENST00000544406.2
|
C12orf74
|
chromosome 12 open reading frame 74 |
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXC13
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 8.5 | GO:2000334 | blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.5 | 1.6 | GO:0003218 | cardiac left ventricle formation(GO:0003218) |
| 0.5 | 1.5 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) |
| 0.4 | 1.9 | GO:0002669 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.3 | 1.2 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.3 | 0.8 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 0.3 | 1.4 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.3 | 1.0 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.2 | 1.5 | GO:0010897 | negative regulation of triglyceride catabolic process(GO:0010897) |
| 0.2 | 1.7 | GO:0061737 | leukotriene signaling pathway(GO:0061737) |
| 0.2 | 4.1 | GO:2000786 | positive regulation of autophagosome assembly(GO:2000786) |
| 0.2 | 1.0 | GO:0009257 | 10-formyltetrahydrofolate biosynthetic process(GO:0009257) |
| 0.1 | 0.5 | GO:0003363 | lamellipodium assembly involved in ameboidal cell migration(GO:0003363) extension of a leading process involved in cell motility in cerebral cortex radial glia guided migration(GO:0021816) |
| 0.1 | 9.7 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.1 | 0.4 | GO:0060279 | positive regulation of ovulation(GO:0060279) |
| 0.1 | 2.8 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.1 | 0.9 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.1 | 1.7 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.1 | 1.4 | GO:1902951 | negative regulation of dendritic spine maintenance(GO:1902951) |
| 0.1 | 0.3 | GO:0061394 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) |
| 0.1 | 0.6 | GO:0030047 | actin modification(GO:0030047) |
| 0.1 | 0.4 | GO:0070171 | negative regulation of tooth mineralization(GO:0070171) |
| 0.1 | 1.0 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.1 | 1.3 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.1 | 1.8 | GO:0051573 | negative regulation of histone H3-K9 methylation(GO:0051573) |
| 0.1 | 0.6 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.1 | 0.6 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.1 | 1.4 | GO:0043031 | negative regulation of macrophage activation(GO:0043031) |
| 0.1 | 1.1 | GO:0031284 | positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.1 | 0.2 | GO:0046603 | negative regulation of mitotic centrosome separation(GO:0046603) |
| 0.1 | 0.2 | GO:2000097 | regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.1 | 0.9 | GO:0050713 | negative regulation of interleukin-1 beta secretion(GO:0050713) |
| 0.1 | 1.5 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.1 | 0.4 | GO:1903899 | lung goblet cell differentiation(GO:0060480) positive regulation of PERK-mediated unfolded protein response(GO:1903899) |
| 0.1 | 0.2 | GO:0061534 | gamma-aminobutyric acid secretion, neurotransmission(GO:0061534) |
| 0.1 | 0.4 | GO:1902231 | regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902162) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.1 | 1.1 | GO:0001946 | lymphangiogenesis(GO:0001946) regulation of blood vessel remodeling(GO:0060312) positive regulation of protein kinase C signaling(GO:0090037) |
| 0.1 | 1.4 | GO:0030889 | negative regulation of B cell proliferation(GO:0030889) |
| 0.0 | 0.4 | GO:0001951 | intestinal D-glucose absorption(GO:0001951) |
| 0.0 | 1.3 | GO:0097296 | activation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097296) |
| 0.0 | 0.2 | GO:0072429 | response to intra-S DNA damage checkpoint signaling(GO:0072429) |
| 0.0 | 0.3 | GO:0061146 | Peyer's patch morphogenesis(GO:0061146) |
| 0.0 | 1.1 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 0.5 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.0 | 0.2 | GO:0015788 | UDP-N-acetylglucosamine transport(GO:0015788) UDP-N-acetylglucosamine transmembrane transport(GO:1990569) |
| 0.0 | 0.4 | GO:0006235 | dTTP biosynthetic process(GO:0006235) pyrimidine deoxyribonucleoside triphosphate biosynthetic process(GO:0009212) |
| 0.0 | 1.0 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 0.6 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 0.3 | GO:0070537 | histone H2A K63-linked deubiquitination(GO:0070537) |
| 0.0 | 1.5 | GO:0034755 | iron ion transmembrane transport(GO:0034755) |
| 0.0 | 0.1 | GO:0045726 | positive regulation of integrin biosynthetic process(GO:0045726) |
| 0.0 | 0.5 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.0 | 0.5 | GO:0035290 | trunk segmentation(GO:0035290) trunk neural crest cell migration(GO:0036484) ventral trunk neural crest cell migration(GO:0036486) |
| 0.0 | 0.2 | GO:0060741 | prostate gland stromal morphogenesis(GO:0060741) |
| 0.0 | 0.3 | GO:0052405 | modulation of molecular function in other organism(GO:0044359) negative regulation of molecular function in other organism(GO:0044362) negative regulation of molecular function in other organism involved in symbiotic interaction(GO:0052204) modulation of molecular function in other organism involved in symbiotic interaction(GO:0052205) negative regulation by host of symbiont molecular function(GO:0052405) modification by host of symbiont molecular function(GO:0052428) |
| 0.0 | 0.2 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.0 | 0.6 | GO:0043383 | negative T cell selection(GO:0043383) |
| 0.0 | 0.2 | GO:0021847 | ventricular zone neuroblast division(GO:0021847) |
| 0.0 | 1.1 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.0 | 3.1 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.0 | 1.6 | GO:0033198 | response to ATP(GO:0033198) |
| 0.0 | 0.3 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.0 | 0.4 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.0 | 2.9 | GO:0035722 | interleukin-12-mediated signaling pathway(GO:0035722) cellular response to interleukin-12(GO:0071349) |
| 0.0 | 5.2 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.0 | 0.3 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.0 | 1.2 | GO:0032731 | positive regulation of interleukin-1 beta production(GO:0032731) |
| 0.0 | 0.2 | GO:2000660 | negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.0 | 0.3 | GO:0002220 | innate immune response activating cell surface receptor signaling pathway(GO:0002220) stimulatory C-type lectin receptor signaling pathway(GO:0002223) |
| 0.0 | 0.3 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.0 | 0.1 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.0 | 0.3 | GO:2000269 | regulation of fibroblast apoptotic process(GO:2000269) |
| 0.0 | 1.8 | GO:0042073 | intraciliary transport(GO:0042073) |
| 0.0 | 1.1 | GO:0007099 | centriole replication(GO:0007099) |
| 0.0 | 2.5 | GO:0031295 | T cell costimulation(GO:0031295) |
| 0.0 | 0.3 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.0 | 0.7 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.0 | 0.2 | GO:0018406 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.0 | 2.1 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.0 | 0.5 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.0 | 0.3 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.0 | 0.2 | GO:0015781 | pyrimidine nucleotide-sugar transport(GO:0015781) |
| 0.0 | 0.1 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.0 | 2.3 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.0 | 0.4 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.0 | 0.4 | GO:0043101 | purine-containing compound salvage(GO:0043101) |
| 0.0 | 0.3 | GO:0046602 | regulation of mitotic centrosome separation(GO:0046602) |
| 0.0 | 0.7 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.1 | GO:0032466 | negative regulation of cytokinesis(GO:0032466) |
| 0.0 | 0.2 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.0 | 0.1 | GO:0001692 | histamine metabolic process(GO:0001692) |
| 0.0 | 0.5 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.2 | GO:0046541 | saliva secretion(GO:0046541) |
| 0.0 | 0.1 | GO:0060087 | relaxation of vascular smooth muscle(GO:0060087) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.5 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.2 | 8.5 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.2 | 9.7 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.1 | 1.9 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.1 | 3.1 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.1 | 0.4 | GO:0043512 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.1 | 0.4 | GO:0030849 | autosome(GO:0030849) |
| 0.1 | 1.4 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.1 | 0.5 | GO:0036398 | TCR signalosome(GO:0036398) |
| 0.1 | 1.3 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.1 | 0.3 | GO:0031251 | PAN complex(GO:0031251) |
| 0.1 | 0.4 | GO:1902560 | GMP reductase complex(GO:1902560) |
| 0.1 | 0.5 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.1 | 0.9 | GO:0072559 | NLRP3 inflammasome complex(GO:0072559) |
| 0.0 | 0.3 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.0 | 1.5 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.3 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.0 | 0.5 | GO:0097486 | multivesicular body lumen(GO:0097486) |
| 0.0 | 0.4 | GO:0000801 | central element(GO:0000801) |
| 0.0 | 1.1 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 0.5 | GO:0000796 | condensin complex(GO:0000796) |
| 0.0 | 0.2 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.0 | 1.0 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.0 | 0.5 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.0 | 1.0 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.0 | 1.1 | GO:0071011 | precatalytic spliceosome(GO:0071011) |
| 0.0 | 1.1 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
| 0.0 | 0.4 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 0.5 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 0.3 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 0.0 | 0.8 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 5.2 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 0.6 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.6 | GO:0033202 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.0 | 1.1 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 0.1 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.0 | 0.2 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 3.9 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.0 | 0.3 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 2.3 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.0 | 0.2 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 0.0 | GO:0097232 | lamellar body membrane(GO:0097232) alveolar lamellar body membrane(GO:0097233) |
| 0.0 | 3.2 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.0 | 0.4 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 1.6 | GO:0005581 | collagen trimer(GO:0005581) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 8.5 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.4 | 1.7 | GO:0004917 | interleukin-7 receptor activity(GO:0004917) |
| 0.4 | 1.1 | GO:0036328 | VEGF-C-activated receptor activity(GO:0036328) |
| 0.3 | 2.8 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.2 | 1.8 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.2 | 3.3 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.2 | 1.0 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.2 | 3.1 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.2 | 1.0 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.2 | 1.1 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.2 | 1.2 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.1 | 9.7 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.1 | 0.9 | GO:0032089 | NACHT domain binding(GO:0032089) |
| 0.1 | 1.5 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.1 | 0.9 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.1 | 0.9 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.1 | 0.6 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.1 | 0.3 | GO:0004947 | bradykinin receptor activity(GO:0004947) |
| 0.1 | 0.3 | GO:0015633 | zinc transporting ATPase activity(GO:0015633) |
| 0.1 | 0.4 | GO:0004802 | transketolase activity(GO:0004802) |
| 0.1 | 0.4 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.1 | 0.5 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.1 | 0.4 | GO:0003920 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 0.1 | 0.5 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.1 | 0.2 | GO:0046848 | hydroxyapatite binding(GO:0046848) |
| 0.0 | 0.4 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.0 | 1.7 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.2 | GO:0005462 | UDP-N-acetylglucosamine transmembrane transporter activity(GO:0005462) |
| 0.0 | 0.2 | GO:0051800 | phosphatidylinositol-3,4-bisphosphate 3-phosphatase activity(GO:0051800) |
| 0.0 | 8.4 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 0.4 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.2 | GO:0004167 | dopachrome isomerase activity(GO:0004167) |
| 0.0 | 0.6 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 1.4 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.0 | 1.0 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.7 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.6 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 0.8 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 0.1 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.0 | 1.3 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.1 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.0 | 0.2 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.2 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.0 | 0.5 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.2 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.0 | 1.5 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 2.5 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 1.1 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 1.4 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.0 | 0.6 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.2 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.7 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 1.5 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.4 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 1.8 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.5 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.0 | 0.4 | GO:0042923 | neuropeptide binding(GO:0042923) |
| 0.0 | 0.3 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.3 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.3 | GO:0004690 | cyclic nucleotide-dependent protein kinase activity(GO:0004690) cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.4 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 1.0 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.3 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.3 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 2.9 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 1.8 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 2.9 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 1.4 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 2.6 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 1.1 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 1.6 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 3.1 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 1.3 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 1.6 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.7 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 0.3 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.7 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.2 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.5 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.1 | 1.2 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.1 | 1.4 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.1 | 8.5 | REACTOME ANTIGEN PROCESSING CROSS PRESENTATION | Genes involved in Antigen processing-Cross presentation |
| 0.1 | 1.7 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 1.1 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 1.0 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 1.1 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.0 | 0.9 | REACTOME CD28 DEPENDENT PI3K AKT SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.0 | 1.7 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 1.3 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 1.9 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.0 | 1.4 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.6 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.0 | 0.3 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.7 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 1.0 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.0 | 0.9 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 0.3 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.4 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 1.3 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |