Project
Illumina Body Map 2: averaged replicates
Navigation
Downloads
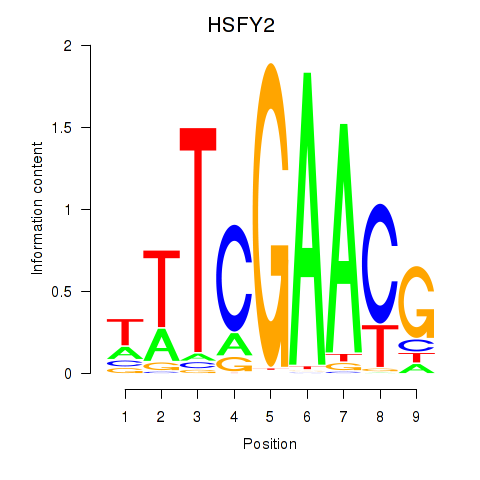
Results for HSFY2
Z-value: 1.07
Transcription factors associated with HSFY2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HSFY2
|
ENSG00000169953.11 | heat shock transcription factor Y-linked 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HSFY2 | hg19_v2_chrY_-_20935572_20935621 | 0.12 | 5.0e-01 | Click! |
Activity profile of HSFY2 motif
Sorted Z-values of HSFY2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_95025661 | 1.48 |
ENST00000542556.1
ENST00000265627.5 ENST00000427422.1 ENST00000451904.1 |
PON1
PON3
|
paraoxonase 1 paraoxonase 3 |
| chr4_+_81256871 | 1.47 |
ENST00000358105.3
ENST00000508675.1 |
C4orf22
|
chromosome 4 open reading frame 22 |
| chr13_+_76378407 | 1.44 |
ENST00000447038.1
|
LMO7
|
LIM domain 7 |
| chr10_-_101825151 | 1.37 |
ENST00000441382.1
|
CPN1
|
carboxypeptidase N, polypeptide 1 |
| chr2_-_152589670 | 1.36 |
ENST00000604864.1
ENST00000603639.1 |
NEB
|
nebulin |
| chr3_+_186383741 | 1.35 |
ENST00000232003.4
|
HRG
|
histidine-rich glycoprotein |
| chr13_+_76378357 | 1.33 |
ENST00000489941.2
ENST00000525373.1 |
LMO7
|
LIM domain 7 |
| chr1_-_145076186 | 1.33 |
ENST00000369348.3
|
PDE4DIP
|
phosphodiesterase 4D interacting protein |
| chr1_-_57431679 | 1.30 |
ENST00000371237.4
ENST00000535057.1 ENST00000543257.1 |
C8B
|
complement component 8, beta polypeptide |
| chr13_+_76378305 | 1.25 |
ENST00000526371.1
ENST00000526528.1 |
LMO7
|
LIM domain 7 |
| chr12_+_101962128 | 1.24 |
ENST00000550514.1
|
MYBPC1
|
myosin binding protein C, slow type |
| chr12_+_85430110 | 1.24 |
ENST00000393212.3
ENST00000393217.2 |
LRRIQ1
|
leucine-rich repeats and IQ motif containing 1 |
| chr5_-_135290651 | 1.18 |
ENST00000522943.1
ENST00000514447.2 |
LECT2
|
leukocyte cell-derived chemotaxin 2 |
| chrX_+_46940254 | 1.10 |
ENST00000336169.3
|
RGN
|
regucalcin |
| chr1_-_57045228 | 1.09 |
ENST00000371250.3
|
PPAP2B
|
phosphatidic acid phosphatase type 2B |
| chr19_+_50706866 | 1.07 |
ENST00000440075.2
ENST00000376970.2 ENST00000425460.1 ENST00000599920.1 ENST00000601313.1 |
MYH14
|
myosin, heavy chain 14, non-muscle |
| chr2_+_1418154 | 1.07 |
ENST00000423320.1
ENST00000382198.1 |
TPO
|
thyroid peroxidase |
| chr1_-_204121013 | 1.04 |
ENST00000367201.3
|
ETNK2
|
ethanolamine kinase 2 |
| chr18_-_48351743 | 1.04 |
ENST00000588444.1
ENST00000256425.2 ENST00000428869.2 |
MRO
|
maestro |
| chr6_+_53976235 | 0.96 |
ENST00000502396.1
ENST00000358276.5 |
MLIP
|
muscular LMNA-interacting protein |
| chr8_-_120259055 | 0.94 |
ENST00000522828.1
ENST00000523307.1 ENST00000524129.1 ENST00000521048.1 ENST00000522187.1 |
RP11-4K16.2
|
RP11-4K16.2 |
| chr4_-_100575781 | 0.90 |
ENST00000511828.1
|
RP11-766F14.2
|
Protein LOC285556 |
| chr12_+_100897130 | 0.90 |
ENST00000551379.1
ENST00000188403.7 ENST00000551184.1 |
NR1H4
|
nuclear receptor subfamily 1, group H, member 4 |
| chr2_+_234959323 | 0.89 |
ENST00000373368.1
ENST00000168148.3 |
SPP2
|
secreted phosphoprotein 2, 24kDa |
| chr6_+_53976285 | 0.88 |
ENST00000514433.1
|
MLIP
|
muscular LMNA-interacting protein |
| chr6_+_53976211 | 0.87 |
ENST00000503951.1
|
MLIP
|
muscular LMNA-interacting protein |
| chr5_-_122372354 | 0.84 |
ENST00000306442.4
|
PPIC
|
peptidylprolyl isomerase C (cyclophilin C) |
| chr6_-_52705641 | 0.84 |
ENST00000370989.2
|
GSTA5
|
glutathione S-transferase alpha 5 |
| chr1_-_145075847 | 0.84 |
ENST00000530740.1
ENST00000369359.4 |
PDE4DIP
|
phosphodiesterase 4D interacting protein |
| chr10_+_5005598 | 0.84 |
ENST00000442997.1
|
AKR1C1
|
aldo-keto reductase family 1, member C1 |
| chr1_+_145726886 | 0.81 |
ENST00000443667.1
|
PDZK1
|
PDZ domain containing 1 |
| chr18_+_616711 | 0.80 |
ENST00000579494.1
|
CLUL1
|
clusterin-like 1 (retinal) |
| chr15_-_45670924 | 0.80 |
ENST00000396659.3
|
GATM
|
glycine amidinotransferase (L-arginine:glycine amidinotransferase) |
| chr15_+_84115868 | 0.78 |
ENST00000427482.2
|
SH3GL3
|
SH3-domain GRB2-like 3 |
| chr13_-_46679185 | 0.77 |
ENST00000439329.3
|
CPB2
|
carboxypeptidase B2 (plasma) |
| chr6_+_163148161 | 0.77 |
ENST00000337019.3
ENST00000366889.2 |
PACRG
|
PARK2 co-regulated |
| chr9_+_131684027 | 0.77 |
ENST00000426694.1
|
PHYHD1
|
phytanoyl-CoA dioxygenase domain containing 1 |
| chr9_+_131684562 | 0.75 |
ENST00000421063.2
|
PHYHD1
|
phytanoyl-CoA dioxygenase domain containing 1 |
| chr6_+_46661575 | 0.75 |
ENST00000450697.1
|
TDRD6
|
tudor domain containing 6 |
| chr4_+_152330390 | 0.75 |
ENST00000503146.1
ENST00000435205.1 |
FAM160A1
|
family with sequence similarity 160, member A1 |
| chr19_-_55677920 | 0.75 |
ENST00000524407.2
ENST00000526003.1 ENST00000534170.1 |
DNAAF3
|
dynein, axonemal, assembly factor 3 |
| chr21_+_41239243 | 0.75 |
ENST00000328619.5
|
PCP4
|
Purkinje cell protein 4 |
| chr16_+_31494323 | 0.74 |
ENST00000569576.1
ENST00000330498.3 |
SLC5A2
|
solute carrier family 5 (sodium/glucose cotransporter), member 2 |
| chr11_+_112047087 | 0.74 |
ENST00000526088.1
ENST00000532593.1 ENST00000531169.1 |
BCO2
|
beta-carotene oxygenase 2 |
| chr10_-_91403625 | 0.73 |
ENST00000322191.6
ENST00000342512.3 ENST00000371774.2 |
PANK1
|
pantothenate kinase 1 |
| chr1_+_209859510 | 0.73 |
ENST00000367028.2
ENST00000261465.1 |
HSD11B1
|
hydroxysteroid (11-beta) dehydrogenase 1 |
| chr22_-_36013368 | 0.72 |
ENST00000442617.1
ENST00000397326.2 ENST00000397328.1 ENST00000451685.1 |
MB
|
myoglobin |
| chr15_-_45694380 | 0.72 |
ENST00000561148.1
|
GATM
|
glycine amidinotransferase (L-arginine:glycine amidinotransferase) |
| chr8_+_134029937 | 0.72 |
ENST00000518108.1
|
TG
|
thyroglobulin |
| chr17_+_7517264 | 0.72 |
ENST00000593717.1
ENST00000572182.1 ENST00000574539.1 ENST00000576728.1 ENST00000575314.1 ENST00000570547.1 ENST00000572262.1 ENST00000576478.1 |
AC007421.1
SHBG
|
AC007421.1 sex hormone-binding globulin |
| chr2_-_28113965 | 0.71 |
ENST00000302188.3
|
RBKS
|
ribokinase |
| chr6_+_8652370 | 0.70 |
ENST00000503668.1
|
HULC
|
hepatocellular carcinoma up-regulated long non-coding RNA |
| chr4_+_52917451 | 0.69 |
ENST00000295213.4
ENST00000419395.2 |
SPATA18
|
spermatogenesis associated 18 |
| chr11_+_86667117 | 0.68 |
ENST00000531827.1
|
RP11-736K20.6
|
RP11-736K20.6 |
| chr8_+_22224811 | 0.67 |
ENST00000381237.1
|
SLC39A14
|
solute carrier family 39 (zinc transporter), member 14 |
| chr6_+_163148973 | 0.67 |
ENST00000366888.2
|
PACRG
|
PARK2 co-regulated |
| chr14_-_25519095 | 0.67 |
ENST00000419632.2
ENST00000358326.2 ENST00000396700.1 ENST00000548724.1 |
STXBP6
|
syntaxin binding protein 6 (amisyn) |
| chr11_+_111126707 | 0.66 |
ENST00000280325.4
|
C11orf53
|
chromosome 11 open reading frame 53 |
| chr5_-_35991486 | 0.66 |
ENST00000503189.1
|
UGT3A1
|
UDP glycosyltransferase 3 family, polypeptide A1 |
| chr15_-_58571445 | 0.66 |
ENST00000558231.1
|
ALDH1A2
|
aldehyde dehydrogenase 1 family, member A2 |
| chr7_-_121784285 | 0.66 |
ENST00000417368.2
|
AASS
|
aminoadipate-semialdehyde synthase |
| chr10_+_35416223 | 0.65 |
ENST00000489321.1
ENST00000427847.2 ENST00000345491.3 ENST00000395895.2 ENST00000374728.3 ENST00000487132.1 |
CREM
|
cAMP responsive element modulator |
| chr6_+_125524785 | 0.65 |
ENST00000392482.2
|
TPD52L1
|
tumor protein D52-like 1 |
| chrX_-_15511438 | 0.65 |
ENST00000380420.5
|
PIR
|
pirin (iron-binding nuclear protein) |
| chr4_+_187148556 | 0.65 |
ENST00000264690.6
ENST00000446598.2 ENST00000414291.1 ENST00000513864.1 |
KLKB1
|
kallikrein B, plasma (Fletcher factor) 1 |
| chr1_-_213020991 | 0.65 |
ENST00000332912.3
|
C1orf227
|
chromosome 1 open reading frame 227 |
| chrX_+_43515467 | 0.65 |
ENST00000338702.3
ENST00000542639.1 |
MAOA
|
monoamine oxidase A |
| chr2_+_234959376 | 0.64 |
ENST00000425558.1
|
SPP2
|
secreted phosphoprotein 2, 24kDa |
| chr8_+_20811140 | 0.63 |
ENST00000523035.1
|
RP11-369E15.4
|
RP11-369E15.4 |
| chr3_-_124653579 | 0.61 |
ENST00000478191.1
ENST00000311075.3 |
MUC13
|
mucin 13, cell surface associated |
| chr1_-_118727781 | 0.61 |
ENST00000336338.5
|
SPAG17
|
sperm associated antigen 17 |
| chr1_+_111888890 | 0.61 |
ENST00000369738.4
|
PIFO
|
primary cilia formation |
| chr9_+_133971909 | 0.60 |
ENST00000247291.3
ENST00000372302.1 ENST00000372300.1 ENST00000372298.1 |
AIF1L
|
allograft inflammatory factor 1-like |
| chr3_-_165555200 | 0.60 |
ENST00000479451.1
ENST00000540653.1 ENST00000488954.1 ENST00000264381.3 |
BCHE
|
butyrylcholinesterase |
| chr18_-_61329118 | 0.59 |
ENST00000332821.8
ENST00000283752.5 |
SERPINB3
|
serpin peptidase inhibitor, clade B (ovalbumin), member 3 |
| chr1_-_169337176 | 0.59 |
ENST00000472647.1
ENST00000367811.3 |
NME7
|
NME/NM23 family member 7 |
| chr14_+_94385235 | 0.59 |
ENST00000557719.1
ENST00000267594.5 |
FAM181A
|
family with sequence similarity 181, member A |
| chr8_-_133637624 | 0.59 |
ENST00000522789.1
|
LRRC6
|
leucine rich repeat containing 6 |
| chr6_-_46424599 | 0.58 |
ENST00000405162.1
|
RCAN2
|
regulator of calcineurin 2 |
| chr10_+_7745232 | 0.58 |
ENST00000358415.4
|
ITIH2
|
inter-alpha-trypsin inhibitor heavy chain 2 |
| chr19_-_38878632 | 0.58 |
ENST00000586599.1
ENST00000334928.6 ENST00000587676.1 |
GGN
|
gametogenetin |
| chr7_-_32338917 | 0.57 |
ENST00000396193.1
|
PDE1C
|
phosphodiesterase 1C, calmodulin-dependent 70kDa |
| chr8_-_36636676 | 0.57 |
ENST00000524132.1
ENST00000519451.1 |
RP11-962G15.1
|
RP11-962G15.1 |
| chr10_-_124459284 | 0.57 |
ENST00000432000.1
ENST00000329446.4 |
C10orf120
|
chromosome 10 open reading frame 120 |
| chr12_+_75728419 | 0.57 |
ENST00000378695.4
ENST00000312442.2 |
GLIPR1L1
|
GLI pathogenesis-related 1 like 1 |
| chr1_-_85358850 | 0.57 |
ENST00000370611.3
|
LPAR3
|
lysophosphatidic acid receptor 3 |
| chr1_+_111889212 | 0.56 |
ENST00000369737.4
|
PIFO
|
primary cilia formation |
| chr20_-_3219828 | 0.55 |
ENST00000539553.2
|
SLC4A11
|
solute carrier family 4, sodium borate transporter, member 11 |
| chr12_+_96337061 | 0.55 |
ENST00000266736.2
|
AMDHD1
|
amidohydrolase domain containing 1 |
| chr16_-_20367584 | 0.54 |
ENST00000570689.1
|
UMOD
|
uromodulin |
| chr4_+_184826418 | 0.54 |
ENST00000308497.4
ENST00000438269.1 |
STOX2
|
storkhead box 2 |
| chr2_-_11810284 | 0.54 |
ENST00000306928.5
|
NTSR2
|
neurotensin receptor 2 |
| chr10_+_7745303 | 0.54 |
ENST00000429820.1
ENST00000379587.4 |
ITIH2
|
inter-alpha-trypsin inhibitor heavy chain 2 |
| chr7_-_50132801 | 0.53 |
ENST00000419417.1
|
ZPBP
|
zona pellucida binding protein |
| chr3_-_149293990 | 0.53 |
ENST00000472417.1
|
WWTR1
|
WW domain containing transcription regulator 1 |
| chr15_+_71389281 | 0.53 |
ENST00000355327.3
|
THSD4
|
thrombospondin, type I, domain containing 4 |
| chr4_+_152330409 | 0.52 |
ENST00000513086.1
|
FAM160A1
|
family with sequence similarity 160, member A1 |
| chr1_+_2938044 | 0.51 |
ENST00000378404.2
|
ACTRT2
|
actin-related protein T2 |
| chrX_-_73072534 | 0.51 |
ENST00000429829.1
|
XIST
|
X inactive specific transcript (non-protein coding) |
| chr14_+_85996471 | 0.51 |
ENST00000330753.4
|
FLRT2
|
fibronectin leucine rich transmembrane protein 2 |
| chr1_+_61542922 | 0.51 |
ENST00000407417.3
|
NFIA
|
nuclear factor I/A |
| chr9_+_71939488 | 0.51 |
ENST00000455972.1
|
FAM189A2
|
family with sequence similarity 189, member A2 |
| chr16_-_20416053 | 0.51 |
ENST00000302451.4
|
PDILT
|
protein disulfide isomerase-like, testis expressed |
| chr21_+_44590245 | 0.51 |
ENST00000398132.1
|
CRYAA
|
crystallin, alpha A |
| chr17_+_11757333 | 0.50 |
ENST00000579703.1
|
DNAH9
|
dynein, axonemal, heavy chain 9 |
| chr10_-_129691195 | 0.50 |
ENST00000368671.3
|
CLRN3
|
clarin 3 |
| chr3_+_52821841 | 0.50 |
ENST00000405128.3
|
ITIH1
|
inter-alpha-trypsin inhibitor heavy chain 1 |
| chr6_-_28321971 | 0.50 |
ENST00000396838.2
ENST00000426434.1 ENST00000434036.1 ENST00000439628.1 |
ZSCAN31
|
zinc finger and SCAN domain containing 31 |
| chr6_+_50061315 | 0.50 |
ENST00000415106.1
|
RP11-397G17.1
|
RP11-397G17.1 |
| chr5_-_135290705 | 0.50 |
ENST00000274507.1
|
LECT2
|
leukocyte cell-derived chemotaxin 2 |
| chr7_-_50132860 | 0.50 |
ENST00000046087.2
|
ZPBP
|
zona pellucida binding protein |
| chr11_+_101983176 | 0.49 |
ENST00000524575.1
|
YAP1
|
Yes-associated protein 1 |
| chr1_-_84326618 | 0.49 |
ENST00000417565.1
|
RP11-475O6.1
|
RP11-475O6.1 |
| chr18_+_616672 | 0.49 |
ENST00000338387.7
|
CLUL1
|
clusterin-like 1 (retinal) |
| chr1_-_117210290 | 0.49 |
ENST00000369483.1
ENST00000369486.3 |
IGSF3
|
immunoglobulin superfamily, member 3 |
| chrX_-_55020511 | 0.48 |
ENST00000375006.3
ENST00000374992.2 |
PFKFB1
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 |
| chr9_+_133971863 | 0.48 |
ENST00000372309.3
|
AIF1L
|
allograft inflammatory factor 1-like |
| chrX_-_33146477 | 0.48 |
ENST00000378677.2
|
DMD
|
dystrophin |
| chr3_+_100428188 | 0.48 |
ENST00000418917.2
ENST00000490574.1 |
TFG
|
TRK-fused gene |
| chr4_-_65275100 | 0.47 |
ENST00000509536.1
|
TECRL
|
trans-2,3-enoyl-CoA reductase-like |
| chr22_+_30792846 | 0.47 |
ENST00000312932.9
ENST00000428195.1 |
SEC14L2
|
SEC14-like 2 (S. cerevisiae) |
| chr17_-_50237343 | 0.47 |
ENST00000575181.1
ENST00000570565.1 |
CA10
|
carbonic anhydrase X |
| chr8_-_17579726 | 0.47 |
ENST00000381861.3
|
MTUS1
|
microtubule associated tumor suppressor 1 |
| chr9_-_34329198 | 0.47 |
ENST00000379166.2
ENST00000345050.2 |
KIF24
|
kinesin family member 24 |
| chr4_-_89080003 | 0.47 |
ENST00000237612.3
|
ABCG2
|
ATP-binding cassette, sub-family G (WHITE), member 2 |
| chr3_+_100328433 | 0.47 |
ENST00000273352.3
|
GPR128
|
G protein-coupled receptor 128 |
| chr5_+_42756903 | 0.46 |
ENST00000361970.5
ENST00000388827.4 |
CCDC152
|
coiled-coil domain containing 152 |
| chr3_+_2933893 | 0.46 |
ENST00000397459.2
|
CNTN4
|
contactin 4 |
| chr6_-_39902185 | 0.46 |
ENST00000373195.3
ENST00000308559.7 ENST00000373188.2 |
MOCS1
|
molybdenum cofactor synthesis 1 |
| chr12_-_71533055 | 0.45 |
ENST00000552128.1
|
TSPAN8
|
tetraspanin 8 |
| chr4_-_100356291 | 0.45 |
ENST00000476959.1
ENST00000482593.1 |
ADH7
|
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
| chr12_+_81331594 | 0.45 |
ENST00000549175.1
|
ACSS3
|
acyl-CoA synthetase short-chain family member 3 |
| chr11_-_7695437 | 0.44 |
ENST00000533558.1
ENST00000527542.1 ENST00000531096.1 |
CYB5R2
|
cytochrome b5 reductase 2 |
| chr19_+_3721719 | 0.44 |
ENST00000589378.1
ENST00000382008.3 |
TJP3
|
tight junction protein 3 |
| chr14_+_39944025 | 0.43 |
ENST00000554328.1
ENST00000556620.1 ENST00000557197.1 |
RP11-111A21.1
|
RP11-111A21.1 |
| chr11_-_108464465 | 0.43 |
ENST00000525344.1
|
EXPH5
|
exophilin 5 |
| chr6_-_39399087 | 0.43 |
ENST00000229913.5
ENST00000541946.1 ENST00000394362.1 |
KIF6
|
kinesin family member 6 |
| chr19_-_39466396 | 0.43 |
ENST00000292852.4
|
FBXO17
|
F-box protein 17 |
| chr8_+_22601 | 0.43 |
ENST00000522481.3
ENST00000518652.1 |
AC144568.2
|
Uncharacterized protein |
| chr8_+_124864227 | 0.42 |
ENST00000522917.1
|
FER1L6
|
fer-1-like 6 (C. elegans) |
| chr8_+_1993152 | 0.42 |
ENST00000262113.4
|
MYOM2
|
myomesin 2 |
| chr12_-_4758159 | 0.42 |
ENST00000545990.2
|
AKAP3
|
A kinase (PRKA) anchor protein 3 |
| chr19_+_45349630 | 0.42 |
ENST00000252483.5
|
PVRL2
|
poliovirus receptor-related 2 (herpesvirus entry mediator B) |
| chr16_+_48278178 | 0.42 |
ENST00000285737.4
ENST00000535754.1 |
LONP2
|
lon peptidase 2, peroxisomal |
| chr10_+_96522361 | 0.42 |
ENST00000371321.3
|
CYP2C19
|
cytochrome P450, family 2, subfamily C, polypeptide 19 |
| chr10_-_5046042 | 0.42 |
ENST00000421196.3
ENST00000455190.1 |
AKR1C2
|
aldo-keto reductase family 1, member C2 |
| chr20_+_39969519 | 0.41 |
ENST00000373257.3
|
LPIN3
|
lipin 3 |
| chr12_-_91572278 | 0.41 |
ENST00000425043.1
ENST00000420120.2 ENST00000441303.2 ENST00000456569.2 |
DCN
|
decorin |
| chr14_-_25519317 | 0.41 |
ENST00000323944.5
|
STXBP6
|
syntaxin binding protein 6 (amisyn) |
| chr2_+_223536428 | 0.41 |
ENST00000446656.3
|
MOGAT1
|
monoacylglycerol O-acyltransferase 1 |
| chr14_+_101293687 | 0.40 |
ENST00000455286.1
|
MEG3
|
maternally expressed 3 (non-protein coding) |
| chr11_-_66725837 | 0.40 |
ENST00000393958.2
ENST00000393960.1 ENST00000524491.1 ENST00000355677.3 |
PC
|
pyruvate carboxylase |
| chr19_-_19144243 | 0.40 |
ENST00000594445.1
ENST00000452918.2 ENST00000600377.1 ENST00000337018.6 |
SUGP2
|
SURP and G patch domain containing 2 |
| chr15_+_45694567 | 0.40 |
ENST00000559860.1
|
SPATA5L1
|
spermatogenesis associated 5-like 1 |
| chr3_+_100428316 | 0.40 |
ENST00000479672.1
ENST00000476228.1 ENST00000463568.1 |
TFG
|
TRK-fused gene |
| chr2_+_201170596 | 0.40 |
ENST00000439084.1
ENST00000409718.1 |
SPATS2L
|
spermatogenesis associated, serine-rich 2-like |
| chr1_+_207226574 | 0.40 |
ENST00000367080.3
ENST00000367079.2 |
PFKFB2
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2 |
| chr7_-_124405681 | 0.40 |
ENST00000303921.2
|
GPR37
|
G protein-coupled receptor 37 (endothelin receptor type B-like) |
| chr10_+_104614008 | 0.39 |
ENST00000369883.3
|
C10orf32
|
chromosome 10 open reading frame 32 |
| chr8_-_101661887 | 0.39 |
ENST00000311812.2
|
SNX31
|
sorting nexin 31 |
| chr9_+_470288 | 0.39 |
ENST00000382303.1
|
KANK1
|
KN motif and ankyrin repeat domains 1 |
| chr17_-_50237374 | 0.38 |
ENST00000442502.2
|
CA10
|
carbonic anhydrase X |
| chr11_+_34654011 | 0.38 |
ENST00000531794.1
|
EHF
|
ets homologous factor |
| chr15_+_67547113 | 0.38 |
ENST00000512104.1
ENST00000358767.3 ENST00000546225.1 |
IQCH
|
IQ motif containing H |
| chr7_-_82792215 | 0.38 |
ENST00000333891.9
ENST00000423517.2 |
PCLO
|
piccolo presynaptic cytomatrix protein |
| chr12_+_8309630 | 0.38 |
ENST00000396570.3
|
ZNF705A
|
zinc finger protein 705A |
| chr7_-_102789503 | 0.38 |
ENST00000465647.1
ENST00000418294.1 |
NAPEPLD
|
N-acyl phosphatidylethanolamine phospholipase D |
| chr3_+_174158732 | 0.38 |
ENST00000434257.1
|
NAALADL2
|
N-acetylated alpha-linked acidic dipeptidase-like 2 |
| chr7_-_102789629 | 0.37 |
ENST00000417955.1
ENST00000341533.4 ENST00000425379.1 |
NAPEPLD
|
N-acyl phosphatidylethanolamine phospholipase D |
| chr7_-_93520191 | 0.37 |
ENST00000545378.1
|
TFPI2
|
tissue factor pathway inhibitor 2 |
| chr19_-_893200 | 0.37 |
ENST00000269814.4
ENST00000395808.3 ENST00000312090.6 ENST00000325464.1 |
MED16
|
mediator complex subunit 16 |
| chr11_-_63376013 | 0.37 |
ENST00000540943.1
|
PLA2G16
|
phospholipase A2, group XVI |
| chr1_+_196788887 | 0.36 |
ENST00000320493.5
ENST00000367424.4 ENST00000367421.3 |
CFHR1
CFHR2
|
complement factor H-related 1 complement factor H-related 2 |
| chr14_+_104394770 | 0.36 |
ENST00000409874.4
ENST00000339063.5 |
TDRD9
|
tudor domain containing 9 |
| chr20_+_10199468 | 0.36 |
ENST00000254976.2
ENST00000304886.2 |
SNAP25
|
synaptosomal-associated protein, 25kDa |
| chr2_+_108863651 | 0.36 |
ENST00000329106.2
ENST00000376700.1 |
SULT1C3
|
sulfotransferase family, cytosolic, 1C, member 3 |
| chr7_-_50633078 | 0.36 |
ENST00000444124.2
|
DDC
|
dopa decarboxylase (aromatic L-amino acid decarboxylase) |
| chr6_-_39902160 | 0.36 |
ENST00000340692.5
|
MOCS1
|
molybdenum cofactor synthesis 1 |
| chr10_-_69455873 | 0.36 |
ENST00000433211.2
|
CTNNA3
|
catenin (cadherin-associated protein), alpha 3 |
| chr2_+_201170770 | 0.36 |
ENST00000409988.3
ENST00000409385.1 |
SPATS2L
|
spermatogenesis associated, serine-rich 2-like |
| chr6_-_72129806 | 0.35 |
ENST00000413945.1
ENST00000602878.1 ENST00000436803.1 ENST00000421704.1 ENST00000441570.1 |
LINC00472
|
long intergenic non-protein coding RNA 472 |
| chr2_+_233497931 | 0.35 |
ENST00000264059.3
|
EFHD1
|
EF-hand domain family, member D1 |
| chr6_-_155635583 | 0.35 |
ENST00000367166.4
|
TFB1M
|
transcription factor B1, mitochondrial |
| chr11_-_72145669 | 0.35 |
ENST00000543042.1
ENST00000294053.3 |
CLPB
|
ClpB caseinolytic peptidase B homolog (E. coli) |
| chr14_-_21490958 | 0.35 |
ENST00000554104.1
|
NDRG2
|
NDRG family member 2 |
| chr4_-_186317034 | 0.35 |
ENST00000505916.1
|
LRP2BP
|
LRP2 binding protein |
| chr14_+_85996507 | 0.34 |
ENST00000554746.1
|
FLRT2
|
fibronectin leucine rich transmembrane protein 2 |
| chr10_-_70092671 | 0.34 |
ENST00000358769.2
ENST00000432941.1 ENST00000495025.2 |
PBLD
|
phenazine biosynthesis-like protein domain containing |
| chr11_-_72145426 | 0.34 |
ENST00000535990.1
ENST00000437826.2 ENST00000340729.5 |
CLPB
|
ClpB caseinolytic peptidase B homolog (E. coli) |
| chr6_-_47009996 | 0.34 |
ENST00000371243.2
|
GPR110
|
G protein-coupled receptor 110 |
| chr4_-_114438763 | 0.34 |
ENST00000509594.1
|
CAMK2D
|
calcium/calmodulin-dependent protein kinase II delta |
| chr1_-_169455169 | 0.33 |
ENST00000367804.4
ENST00000236137.5 |
SLC19A2
|
solute carrier family 19 (thiamine transporter), member 2 |
| chr18_-_59274139 | 0.33 |
ENST00000586949.1
|
RP11-879F14.2
|
RP11-879F14.2 |
| chr15_+_84116106 | 0.33 |
ENST00000535412.1
ENST00000324537.5 |
SH3GL3
|
SH3-domain GRB2-like 3 |
| chr20_+_33292068 | 0.33 |
ENST00000374810.3
ENST00000374809.2 ENST00000451665.1 |
TP53INP2
|
tumor protein p53 inducible nuclear protein 2 |
| chr2_-_97509729 | 0.33 |
ENST00000418232.1
|
ANKRD23
|
ankyrin repeat domain 23 |
| chr19_+_50433476 | 0.33 |
ENST00000596658.1
|
ATF5
|
activating transcription factor 5 |
| chr3_-_72897545 | 0.33 |
ENST00000325599.8
|
SHQ1
|
SHQ1, H/ACA ribonucleoprotein assembly factor |
| chr11_+_125034640 | 0.32 |
ENST00000542175.1
|
PKNOX2
|
PBX/knotted 1 homeobox 2 |
| chr17_-_30470154 | 0.32 |
ENST00000398832.2
|
AC090616.2
|
Uncharacterized protein |
Network of associatons between targets according to the STRING database.
First level regulatory network of HSFY2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | GO:1903625 | negative regulation of DNA catabolic process(GO:1903625) |
| 0.4 | 1.5 | GO:0006601 | creatine biosynthetic process(GO:0006601) |
| 0.4 | 1.5 | GO:0034444 | regulation of plasma lipoprotein particle oxidation(GO:0034444) negative regulation of plasma lipoprotein particle oxidation(GO:0034445) phenylpropanoid catabolic process(GO:0046271) |
| 0.3 | 1.4 | GO:2001027 | negative regulation of endothelial cell chemotaxis(GO:2001027) |
| 0.3 | 1.4 | GO:0030070 | insulin processing(GO:0030070) |
| 0.3 | 1.4 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.2 | 0.7 | GO:0042214 | terpene metabolic process(GO:0042214) |
| 0.2 | 0.9 | GO:0034255 | nitrogen catabolite regulation of transcription from RNA polymerase II promoter(GO:0001079) nitrogen catabolite activation of transcription from RNA polymerase II promoter(GO:0001080) regulation of urea metabolic process(GO:0034255) intracellular bile acid receptor signaling pathway(GO:0038185) interleukin-17 secretion(GO:0072615) nitrogen catabolite regulation of transcription(GO:0090293) nitrogen catabolite activation of transcription(GO:0090294) regulation of nitrogen cycle metabolic process(GO:1903314) positive regulation of glutamate metabolic process(GO:2000213) regulation of ammonia assimilation cycle(GO:2001248) positive regulation of ammonia assimilation cycle(GO:2001250) |
| 0.2 | 0.6 | GO:0002541 | activation of plasma proteins involved in acute inflammatory response(GO:0002541) |
| 0.2 | 0.6 | GO:0014016 | neuroblast differentiation(GO:0014016) |
| 0.2 | 0.8 | GO:0003330 | regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) |
| 0.2 | 0.4 | GO:0052314 | phytoalexin metabolic process(GO:0052314) |
| 0.2 | 0.6 | GO:1990926 | short-term synaptic potentiation(GO:1990926) |
| 0.2 | 1.3 | GO:0071395 | response to jasmonic acid(GO:0009753) cellular response to jasmonic acid stimulus(GO:0071395) |
| 0.1 | 0.6 | GO:0035425 | autocrine signaling(GO:0035425) |
| 0.1 | 0.7 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.1 | 1.0 | GO:1903301 | positive regulation of glucokinase activity(GO:0033133) positive regulation of hexokinase activity(GO:1903301) |
| 0.1 | 0.6 | GO:1901837 | negative regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901837) |
| 0.1 | 0.6 | GO:0010430 | fatty acid omega-oxidation(GO:0010430) |
| 0.1 | 0.7 | GO:0046726 | positive regulation by virus of viral protein levels in host cell(GO:0046726) |
| 0.1 | 0.9 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.1 | 0.7 | GO:0019303 | D-ribose catabolic process(GO:0019303) |
| 0.1 | 0.4 | GO:2000393 | negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 0.1 | 0.7 | GO:0035900 | response to isolation stress(GO:0035900) |
| 0.1 | 0.8 | GO:0019556 | histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 0.1 | 0.7 | GO:0033512 | L-lysine catabolic process to acetyl-CoA via saccharopine(GO:0033512) |
| 0.1 | 0.5 | GO:0072233 | thick ascending limb development(GO:0072023) metanephric thick ascending limb development(GO:0072233) |
| 0.1 | 0.3 | GO:2000532 | renal albumin absorption(GO:0097018) regulation of renal albumin absorption(GO:2000532) |
| 0.1 | 0.4 | GO:0019074 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.1 | 0.2 | GO:0051792 | medium-chain fatty acid biosynthetic process(GO:0051792) |
| 0.1 | 0.7 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.1 | 0.3 | GO:0060370 | susceptibility to T cell mediated cytotoxicity(GO:0060370) |
| 0.1 | 0.7 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.1 | 1.2 | GO:0032324 | molybdopterin cofactor biosynthetic process(GO:0032324) molybdopterin cofactor metabolic process(GO:0043545) prosthetic group metabolic process(GO:0051189) |
| 0.1 | 0.3 | GO:0090071 | negative regulation of ribosome biogenesis(GO:0090071) |
| 0.1 | 1.7 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.1 | 0.2 | GO:2000854 | positive regulation of corticosterone secretion(GO:2000854) |
| 0.1 | 0.3 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.1 | 0.4 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.1 | 0.3 | GO:0071934 | thiamine transmembrane transport(GO:0071934) |
| 0.1 | 0.1 | GO:0005988 | lactose metabolic process(GO:0005988) lactose biosynthetic process(GO:0005989) |
| 0.1 | 0.7 | GO:0035799 | ureter maturation(GO:0035799) |
| 0.1 | 0.3 | GO:0090210 | vitamin E metabolic process(GO:0042360) regulation of establishment of blood-brain barrier(GO:0090210) negative regulation of establishment of blood-brain barrier(GO:0090212) |
| 0.1 | 0.3 | GO:0034476 | U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) |
| 0.1 | 0.4 | GO:0036343 | psychomotor behavior(GO:0036343) |
| 0.1 | 0.4 | GO:0016098 | monoterpenoid metabolic process(GO:0016098) |
| 0.1 | 1.0 | GO:0072307 | metanephric nephron tubule epithelial cell differentiation(GO:0072257) regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072307) |
| 0.1 | 0.3 | GO:0090370 | negative regulation of cholesterol efflux(GO:0090370) |
| 0.1 | 0.3 | GO:0003175 | tricuspid valve development(GO:0003175) |
| 0.1 | 1.1 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.1 | 1.3 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.1 | 0.2 | GO:0019417 | sulfur oxidation(GO:0019417) |
| 0.1 | 0.3 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.0 | 0.7 | GO:0015705 | iodide transport(GO:0015705) |
| 0.0 | 0.2 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.0 | 0.2 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.0 | 0.7 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 0.0 | 1.5 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 0.2 | GO:0033686 | positive regulation of luteinizing hormone secretion(GO:0033686) |
| 0.0 | 0.3 | GO:0019918 | peptidyl-arginine methylation, to symmetrical-dimethyl arginine(GO:0019918) |
| 0.0 | 0.5 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 0.6 | GO:0042420 | dopamine catabolic process(GO:0042420) |
| 0.0 | 1.1 | GO:0006590 | thyroid hormone generation(GO:0006590) |
| 0.0 | 0.6 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.0 | 0.3 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.0 | 0.4 | GO:0045915 | positive regulation of catecholamine metabolic process(GO:0045915) positive regulation of dopamine metabolic process(GO:0045964) |
| 0.0 | 0.5 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.2 | GO:0070901 | mitochondrial tRNA methylation(GO:0070901) |
| 0.0 | 0.2 | GO:0061034 | olfactory bulb mitral cell layer development(GO:0061034) |
| 0.0 | 0.3 | GO:0010615 | positive regulation of cardiac muscle adaptation(GO:0010615) positive regulation of cardiac muscle hypertrophy in response to stress(GO:1903244) |
| 0.0 | 0.5 | GO:0032808 | lacrimal gland development(GO:0032808) |
| 0.0 | 0.5 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.0 | 0.4 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.0 | 1.3 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.0 | 0.3 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.7 | GO:0015684 | ferrous iron transport(GO:0015684) ferrous iron transmembrane transport(GO:1903874) |
| 0.0 | 0.6 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.0 | 1.0 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.0 | 0.3 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.0 | 0.4 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.3 | GO:0021860 | pyramidal neuron development(GO:0021860) |
| 0.0 | 0.1 | GO:0042727 | flavin-containing compound biosynthetic process(GO:0042727) |
| 0.0 | 0.2 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.0 | 0.1 | GO:0032764 | negative regulation of mast cell cytokine production(GO:0032764) negative regulation of isotype switching to IgE isotypes(GO:0048294) |
| 0.0 | 0.8 | GO:0006704 | glucocorticoid biosynthetic process(GO:0006704) |
| 0.0 | 0.2 | GO:0030638 | polyketide metabolic process(GO:0030638) daunorubicin metabolic process(GO:0044597) doxorubicin metabolic process(GO:0044598) |
| 0.0 | 0.2 | GO:0000730 | DNA recombinase assembly(GO:0000730) double-strand break repair via synthesis-dependent strand annealing(GO:0045003) |
| 0.0 | 0.2 | GO:2001295 | malonyl-CoA biosynthetic process(GO:2001295) |
| 0.0 | 0.5 | GO:0042268 | regulation of cytolysis(GO:0042268) |
| 0.0 | 0.2 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.0 | 0.1 | GO:0043686 | co-translational protein modification(GO:0043686) |
| 0.0 | 0.1 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) pulmonary vein morphogenesis(GO:0060577) |
| 0.0 | 0.2 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.0 | 0.4 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.0 | 0.5 | GO:0072189 | ureter development(GO:0072189) |
| 0.0 | 0.5 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.4 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.0 | 0.8 | GO:0015879 | carnitine transport(GO:0015879) |
| 0.0 | 1.1 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.0 | 0.4 | GO:0086073 | bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 0.0 | 0.2 | GO:1902572 | negative regulation of serine-type endopeptidase activity(GO:1900004) negative regulation of serine-type peptidase activity(GO:1902572) |
| 0.0 | 0.2 | GO:0015887 | biotin transport(GO:0015878) pantothenate transmembrane transport(GO:0015887) |
| 0.0 | 0.6 | GO:0019511 | peptidyl-proline hydroxylation(GO:0019511) |
| 0.0 | 0.2 | GO:2000113 | negative regulation of cellular macromolecule biosynthetic process(GO:2000113) |
| 0.0 | 0.1 | GO:0072425 | signal transduction involved in G2 DNA damage checkpoint(GO:0072425) signal transduction involved in mitotic G2 DNA damage checkpoint(GO:0072434) |
| 0.0 | 0.7 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.0 | 1.3 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.0 | 1.5 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.0 | 0.4 | GO:1901725 | regulation of histone deacetylase activity(GO:1901725) |
| 0.0 | 0.2 | GO:0071169 | establishment of protein localization to chromatin(GO:0071169) |
| 0.0 | 0.2 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 0.2 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.2 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.0 | 0.3 | GO:1904322 | response to forskolin(GO:1904321) cellular response to forskolin(GO:1904322) |
| 0.0 | 0.2 | GO:1903976 | negative regulation of glial cell migration(GO:1903976) |
| 0.0 | 0.4 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.3 | GO:0055059 | asymmetric neuroblast division(GO:0055059) |
| 0.0 | 0.2 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.0 | 0.3 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.0 | 1.6 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.3 | GO:0090361 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 0.0 | 0.1 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 0.0 | 0.4 | GO:0031998 | regulation of fatty acid beta-oxidation(GO:0031998) |
| 0.0 | 1.4 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.0 | 0.0 | GO:0035928 | rRNA import into mitochondrion(GO:0035928) |
| 0.0 | 0.2 | GO:0070172 | positive regulation of tooth mineralization(GO:0070172) |
| 0.0 | 0.3 | GO:0060297 | regulation of sarcomere organization(GO:0060297) |
| 0.0 | 0.1 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.0 | 0.2 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.4 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 0.3 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 0.1 | GO:0032990 | cell part morphogenesis(GO:0032990) cell projection morphogenesis(GO:0048858) |
| 0.0 | 0.6 | GO:0071498 | cellular response to fluid shear stress(GO:0071498) |
| 0.0 | 0.3 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.0 | 0.6 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 0.1 | GO:0016480 | negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 0.0 | 0.7 | GO:0070884 | regulation of calcineurin-NFAT signaling cascade(GO:0070884) |
| 0.0 | 0.3 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.2 | GO:0001302 | replicative cell aging(GO:0001302) |
| 0.0 | 0.2 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 0.0 | 0.1 | GO:2000298 | regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000298) |
| 0.0 | 0.1 | GO:0051586 | positive regulation of neurotransmitter uptake(GO:0051582) positive regulation of dopamine uptake involved in synaptic transmission(GO:0051586) positive regulation of catecholamine uptake involved in synaptic transmission(GO:0051944) |
| 0.0 | 0.3 | GO:0021930 | olfactory bulb interneuron development(GO:0021891) cell proliferation in external granule layer(GO:0021924) cerebellar granule cell precursor proliferation(GO:0021930) |
| 0.0 | 0.1 | GO:0001705 | ectoderm formation(GO:0001705) ectodermal cell fate commitment(GO:0001712) |
| 0.0 | 0.1 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.0 | 0.2 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.3 | GO:0031167 | rRNA methylation(GO:0031167) |
| 0.0 | 0.1 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 0.2 | GO:0036500 | ATF6-mediated unfolded protein response(GO:0036500) |
| 0.0 | 0.2 | GO:0060124 | positive regulation of growth hormone secretion(GO:0060124) |
| 0.0 | 0.1 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.0 | 0.1 | GO:0070124 | mitochondrial translational initiation(GO:0070124) |
| 0.0 | 0.5 | GO:0001893 | maternal placenta development(GO:0001893) |
| 0.0 | 0.2 | GO:0007059 | chromosome segregation(GO:0007059) |
| 0.0 | 0.1 | GO:0060474 | positive regulation of sperm motility involved in capacitation(GO:0060474) |
| 0.0 | 0.4 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.0 | 0.3 | GO:0031639 | plasminogen activation(GO:0031639) |
| 0.0 | 0.1 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 | 0.2 | GO:0060484 | lung-associated mesenchyme development(GO:0060484) |
| 0.0 | 0.2 | GO:0002053 | positive regulation of mesenchymal cell proliferation(GO:0002053) |
| 0.0 | 0.1 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.0 | 0.0 | GO:0071030 | nuclear mRNA surveillance of spliceosomal pre-mRNA splicing(GO:0071030) nuclear retention of unspliced pre-mRNA at the site of transcription(GO:0071048) |
| 0.0 | 0.0 | GO:1904116 | response to vasopressin(GO:1904116) cellular response to vasopressin(GO:1904117) |
| 0.0 | 0.3 | GO:0035810 | positive regulation of urine volume(GO:0035810) |
| 0.0 | 0.2 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.0 | 0.4 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.0 | 0.0 | GO:1990167 | protein K27-linked deubiquitination(GO:1990167) protein K33-linked deubiquitination(GO:1990168) |
| 0.0 | 0.1 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.0 | 0.0 | GO:0051350 | negative regulation of lyase activity(GO:0051350) |
| 0.0 | 0.2 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.4 | GO:0043046 | DNA methylation involved in gamete generation(GO:0043046) |
| 0.0 | 0.3 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.0 | 0.1 | GO:0006868 | glutamine transport(GO:0006868) L-cystine transport(GO:0015811) |
| 0.0 | 0.1 | GO:0060965 | negative regulation of gene silencing by miRNA(GO:0060965) |
| 0.0 | 0.2 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.0 | 0.2 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.5 | GO:0042026 | protein refolding(GO:0042026) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0019008 | molybdopterin synthase complex(GO:0019008) |
| 0.2 | 0.6 | GO:1990716 | axonemal central apparatus(GO:1990716) |
| 0.2 | 1.1 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.1 | 1.3 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.1 | 0.3 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 0.1 | 0.5 | GO:0071149 | TEAD-2-YAP complex(GO:0071149) |
| 0.1 | 0.8 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 0.1 | 0.3 | GO:0036457 | keratohyalin granule(GO:0036457) |
| 0.1 | 1.5 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.1 | 0.7 | GO:0043541 | UDP-N-acetylglucosamine transferase complex(GO:0043541) |
| 0.1 | 1.8 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.1 | 1.5 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 0.1 | 0.7 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.1 | 1.7 | GO:0032982 | myosin filament(GO:0032982) |
| 0.1 | 0.4 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.1 | 0.7 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.1 | 0.2 | GO:1990723 | cytoplasmic periphery of the nuclear pore complex(GO:1990723) |
| 0.0 | 0.2 | GO:0034271 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.0 | 0.3 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.0 | 0.2 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 0.0 | 0.2 | GO:0045293 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.0 | 1.3 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 0.8 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.0 | 0.4 | GO:0071547 | piP-body(GO:0071547) |
| 0.0 | 0.5 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.0 | 0.3 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.0 | 0.6 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.1 | GO:0070939 | Dsl1p complex(GO:0070939) |
| 0.0 | 0.7 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.2 | GO:0045272 | plasma membrane respiratory chain complex I(GO:0045272) |
| 0.0 | 0.1 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.0 | 0.1 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
| 0.0 | 0.3 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.4 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.1 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.0 | 0.2 | GO:0030905 | retromer, tubulation complex(GO:0030905) |
| 0.0 | 0.9 | GO:0036019 | endolysosome(GO:0036019) |
| 0.0 | 0.8 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.0 | 1.0 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.0 | 0.4 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.0 | 1.1 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.2 | GO:0030678 | mitochondrial ribonuclease P complex(GO:0030678) |
| 0.0 | 0.3 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.0 | 0.2 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.0 | 0.1 | GO:0031592 | centrosomal corona(GO:0031592) |
| 0.0 | 0.2 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.0 | 0.5 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.0 | 0.2 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.0 | 0.4 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 0.3 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.1 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.0 | 1.8 | GO:0031970 | organelle envelope lumen(GO:0031970) |
| 0.0 | 0.5 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.1 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.0 | 0.2 | GO:0034709 | methylosome(GO:0034709) |
| 0.0 | 0.3 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 0.0 | GO:0005873 | plus-end kinesin complex(GO:0005873) |
| 0.0 | 0.9 | GO:0030118 | clathrin coat(GO:0030118) |
| 0.0 | 0.0 | GO:0031371 | ubiquitin conjugating enzyme complex(GO:0031371) |
| 0.0 | 0.2 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 0.2 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.8 | GO:0015030 | Cajal body(GO:0015030) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.5 | GO:0015068 | amidinotransferase activity(GO:0015067) glycine amidinotransferase activity(GO:0015068) |
| 0.5 | 1.5 | GO:0102007 | lactonohydrolase activity(GO:0046573) acyl-L-homoserine-lactone lactonohydrolase activity(GO:0102007) |
| 0.2 | 0.7 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.2 | 0.9 | GO:1902122 | chenodeoxycholic acid binding(GO:1902122) |
| 0.2 | 0.8 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.2 | 1.3 | GO:0018636 | phenanthrene 9,10-monooxygenase activity(GO:0018636) ketosteroid monooxygenase activity(GO:0047086) |
| 0.2 | 1.1 | GO:0004447 | iodide peroxidase activity(GO:0004447) |
| 0.1 | 0.6 | GO:0004031 | aldehyde oxidase activity(GO:0004031) |
| 0.1 | 1.2 | GO:0004305 | ethanolamine kinase activity(GO:0004305) |
| 0.1 | 0.6 | GO:0033265 | acetylcholinesterase activity(GO:0003990) choline binding(GO:0033265) |
| 0.1 | 0.4 | GO:0030366 | molybdopterin synthase activity(GO:0030366) |
| 0.1 | 0.9 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.1 | 0.3 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 0.1 | 0.7 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.1 | 0.8 | GO:0005124 | scavenger receptor binding(GO:0005124) |
| 0.1 | 0.9 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) |
| 0.1 | 0.3 | GO:0015403 | thiamine uptake transmembrane transporter activity(GO:0015403) |
| 0.1 | 0.6 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.1 | 0.7 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.1 | 1.5 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.1 | 0.2 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 0.1 | 1.9 | GO:0031432 | titin binding(GO:0031432) |
| 0.1 | 0.4 | GO:0010861 | thyroid hormone receptor activator activity(GO:0010861) thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.1 | 0.4 | GO:0052740 | 1-acyl-2-lysophosphatidylserine acylhydrolase activity(GO:0052740) |
| 0.1 | 0.6 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.1 | 0.7 | GO:0005497 | androgen binding(GO:0005497) |
| 0.1 | 0.4 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.1 | 0.8 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.1 | 0.4 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.1 | 0.2 | GO:0004315 | 3-oxoacyl-[acyl-carrier-protein] synthase activity(GO:0004315) |
| 0.1 | 0.4 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.1 | 0.2 | GO:0046848 | hydroxyapatite binding(GO:0046848) |
| 0.1 | 0.5 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.1 | 0.7 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.1 | 0.2 | GO:0004947 | bradykinin receptor activity(GO:0004947) |
| 0.1 | 0.3 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.1 | 0.4 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.1 | 0.2 | GO:0034602 | oxoglutarate dehydrogenase (NAD+) activity(GO:0034602) |
| 0.1 | 0.5 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.1 | 1.3 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.1 | 0.7 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.1 | 0.4 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.1 | 0.3 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.1 | 0.8 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.1 | 1.3 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.1 | 0.8 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.1 | 0.7 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.1 | 0.2 | GO:0003955 | NAD(P)H dehydrogenase (quinone) activity(GO:0003955) 3-oxoacyl-[acyl-carrier-protein] reductase (NADH) activity(GO:0047025) |
| 0.1 | 0.6 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 1.9 | GO:0019865 | immunoglobulin binding(GO:0019865) |
| 0.0 | 0.1 | GO:0010309 | acireductone dioxygenase [iron(II)-requiring] activity(GO:0010309) |
| 0.0 | 0.6 | GO:0009374 | biotin carboxylase activity(GO:0004075) biotin binding(GO:0009374) |
| 0.0 | 0.3 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.0 | 0.3 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.0 | 0.6 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.0 | 0.2 | GO:0004775 | succinate-CoA ligase (ADP-forming) activity(GO:0004775) |
| 0.0 | 1.1 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.2 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.0 | 1.4 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 0.1 | GO:0004507 | steroid 11-beta-monooxygenase activity(GO:0004507) corticosterone 18-monooxygenase activity(GO:0047783) |
| 0.0 | 0.1 | GO:0016422 | mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 0.0 | 0.9 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 1.0 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.1 | GO:0008193 | tRNA guanylyltransferase activity(GO:0008193) |
| 0.0 | 0.4 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.0 | 0.1 | GO:0022865 | transmembrane electron transfer carrier(GO:0022865) |
| 0.0 | 0.3 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 0.0 | 0.6 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.0 | 0.6 | GO:0008391 | arachidonic acid monooxygenase activity(GO:0008391) arachidonic acid epoxygenase activity(GO:0008392) |
| 0.0 | 0.4 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 0.0 | 0.2 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.0 | 0.7 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.1 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.0 | 0.1 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 0.0 | 0.3 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.0 | 0.4 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 0.3 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.0 | 0.1 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.0 | 0.2 | GO:0034604 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 0.0 | 0.1 | GO:0016429 | tRNA (adenine) methyltransferase activity(GO:0016426) tRNA (adenine-N1-)-methyltransferase activity(GO:0016429) |
| 0.0 | 0.3 | GO:0043176 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.0 | 1.3 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.0 | 0.1 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.7 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.0 | 0.2 | GO:0008523 | sodium-dependent multivitamin transmembrane transporter activity(GO:0008523) |
| 0.0 | 0.2 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 1.3 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 2.2 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.3 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.7 | GO:0016646 | oxidoreductase activity, acting on the CH-NH group of donors, NAD or NADP as acceptor(GO:0016646) |
| 0.0 | 2.9 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.2 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.0 | 0.6 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 0.7 | GO:0019200 | carbohydrate kinase activity(GO:0019200) |
| 0.0 | 0.3 | GO:0030548 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.0 | 0.1 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.3 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.0 | 0.3 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 1.5 | GO:0051213 | dioxygenase activity(GO:0051213) |
| 0.0 | 0.3 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.0 | 0.5 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.9 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.0 | 0.5 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.8 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.7 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.8 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.2 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.0 | 0.0 | GO:0008969 | phosphohistidine phosphatase activity(GO:0008969) |
| 0.0 | 0.1 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.0 | 0.1 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.0 | 0.1 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.0 | 0.3 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.4 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 0.2 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 0.0 | GO:1904455 | ubiquitin-specific protease activity involved in negative regulation of ERAD pathway(GO:1904455) |
| 0.0 | 0.1 | GO:0015186 | L-cystine transmembrane transporter activity(GO:0015184) L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.0 | 0.2 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.2 | GO:0008035 | high-density lipoprotein particle binding(GO:0008035) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.1 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.9 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 1.2 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.0 | 0.9 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.3 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 0.6 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.6 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.1 | 1.3 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.0 | 1.3 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.9 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.6 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.0 | 2.6 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.6 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.6 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 1.0 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 1.2 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 1.3 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.0 | 0.3 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 0.5 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.3 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 0.2 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.3 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.8 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.0 | 0.8 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 1.0 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.6 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.0 | 0.4 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 1.1 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 0.6 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.2 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 0.4 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.6 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.3 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 0.0 | REACTOME CDK MEDIATED PHOSPHORYLATION AND REMOVAL OF CDC6 | Genes involved in CDK-mediated phosphorylation and removal of Cdc6 |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.2 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.2 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 3.4 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.3 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.0 | 0.2 | REACTOME ACTIVATION OF CHAPERONES BY ATF6 ALPHA | Genes involved in Activation of Chaperones by ATF6-alpha |