Project
Illumina Body Map 2: averaged replicates
Navigation
Downloads
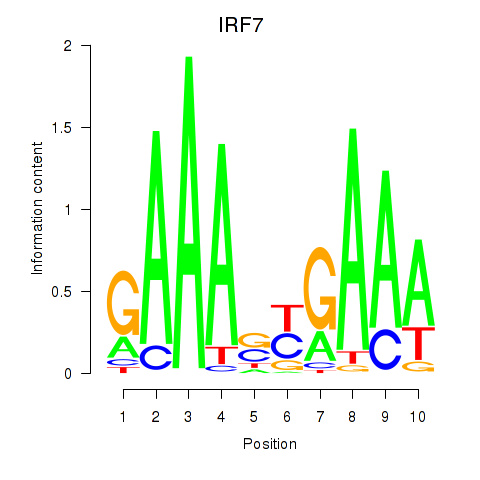
Results for IRF7
Z-value: 2.72
Transcription factors associated with IRF7
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
IRF7
|
ENSG00000185507.15 | interferon regulatory factor 7 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| IRF7 | hg19_v2_chr11_-_615570_615728 | -0.54 | 1.6e-03 | Click! |
Activity profile of IRF7 motif
Sorted Z-values of IRF7 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_91574142 | 9.56 |
ENST00000547937.1
|
DCN
|
decorin |
| chr2_-_151344172 | 6.87 |
ENST00000375734.2
ENST00000263895.4 ENST00000454202.1 |
RND3
|
Rho family GTPase 3 |
| chr6_-_46889694 | 6.47 |
ENST00000283296.7
ENST00000362015.4 ENST00000456426.2 |
GPR116
|
G protein-coupled receptor 116 |
| chr4_+_41614909 | 6.02 |
ENST00000509454.1
ENST00000396595.3 ENST00000381753.4 |
LIMCH1
|
LIM and calponin homology domains 1 |
| chr11_-_2160180 | 5.85 |
ENST00000381406.4
|
IGF2
|
insulin-like growth factor 2 (somatomedin A) |
| chr12_+_19358228 | 5.68 |
ENST00000424268.1
ENST00000543806.1 |
PLEKHA5
|
pleckstrin homology domain containing, family A member 5 |
| chr1_-_85870177 | 5.64 |
ENST00000542148.1
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1 |
| chr11_-_111794446 | 5.44 |
ENST00000527950.1
|
CRYAB
|
crystallin, alpha B |
| chr2_-_216240386 | 5.42 |
ENST00000438981.1
|
FN1
|
fibronectin 1 |
| chr3_+_119316721 | 5.40 |
ENST00000488919.1
ENST00000495992.1 |
PLA1A
|
phospholipase A1 member A |
| chr4_-_186732048 | 5.34 |
ENST00000448662.2
ENST00000439049.1 ENST00000420158.1 ENST00000431808.1 ENST00000319471.9 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr1_+_162602244 | 5.17 |
ENST00000367922.3
ENST00000367921.3 |
DDR2
|
discoidin domain receptor tyrosine kinase 2 |
| chr4_-_152147579 | 5.03 |
ENST00000304527.4
ENST00000455740.1 ENST00000424281.1 ENST00000409598.4 |
SH3D19
|
SH3 domain containing 19 |
| chr14_+_94577074 | 4.96 |
ENST00000444961.1
ENST00000448882.1 ENST00000557098.1 ENST00000554800.1 ENST00000556544.1 ENST00000298902.5 ENST00000555819.1 ENST00000557634.1 ENST00000555744.1 |
IFI27
|
interferon, alpha-inducible protein 27 |
| chr13_+_110958124 | 4.94 |
ENST00000400163.2
|
COL4A2
|
collagen, type IV, alpha 2 |
| chr5_+_140734570 | 4.85 |
ENST00000571252.1
|
PCDHGA4
|
protocadherin gamma subfamily A, 4 |
| chr3_+_119316689 | 4.71 |
ENST00000273371.4
|
PLA1A
|
phospholipase A1 member A |
| chr4_-_87281196 | 4.66 |
ENST00000359221.3
|
MAPK10
|
mitogen-activated protein kinase 10 |
| chr7_-_122526499 | 4.29 |
ENST00000412584.2
|
CADPS2
|
Ca++-dependent secretion activator 2 |
| chr11_+_27062272 | 4.28 |
ENST00000529202.1
ENST00000533566.1 |
BBOX1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1 |
| chr2_+_239756671 | 4.26 |
ENST00000448943.2
|
TWIST2
|
twist family bHLH transcription factor 2 |
| chr12_-_91572278 | 4.23 |
ENST00000425043.1
ENST00000420120.2 ENST00000441303.2 ENST00000456569.2 |
DCN
|
decorin |
| chrX_-_34675391 | 4.19 |
ENST00000275954.3
|
TMEM47
|
transmembrane protein 47 |
| chr4_+_41614720 | 4.14 |
ENST00000509277.1
|
LIMCH1
|
LIM and calponin homology domains 1 |
| chr9_+_6215799 | 4.12 |
ENST00000417746.2
ENST00000456383.2 |
IL33
|
interleukin 33 |
| chr10_+_695888 | 4.06 |
ENST00000441152.2
|
PRR26
|
proline rich 26 |
| chr15_+_96875657 | 4.06 |
ENST00000559679.1
ENST00000394171.2 |
NR2F2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr5_+_32710736 | 4.05 |
ENST00000415685.2
|
NPR3
|
natriuretic peptide receptor C/guanylate cyclase C (atrionatriuretic peptide receptor C) |
| chr4_-_138453606 | 4.03 |
ENST00000412923.2
ENST00000344876.4 ENST00000507846.1 ENST00000510305.1 |
PCDH18
|
protocadherin 18 |
| chr8_+_39770803 | 4.01 |
ENST00000518237.1
|
IDO1
|
indoleamine 2,3-dioxygenase 1 |
| chr4_-_87281224 | 4.00 |
ENST00000395169.3
ENST00000395161.2 |
MAPK10
|
mitogen-activated protein kinase 10 |
| chr12_-_22063787 | 3.98 |
ENST00000544039.1
|
ABCC9
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 9 |
| chr4_+_113739244 | 3.93 |
ENST00000503271.1
ENST00000503423.1 ENST00000506722.1 |
ANK2
|
ankyrin 2, neuronal |
| chr4_-_164253738 | 3.91 |
ENST00000509586.1
ENST00000504391.1 ENST00000512819.1 |
NPY1R
|
neuropeptide Y receptor Y1 |
| chr9_-_95186739 | 3.84 |
ENST00000375550.4
|
OMD
|
osteomodulin |
| chr3_+_186435065 | 3.84 |
ENST00000287611.2
ENST00000265023.4 |
KNG1
|
kininogen 1 |
| chr11_+_34643600 | 3.83 |
ENST00000530286.1
ENST00000533754.1 |
EHF
|
ets homologous factor |
| chr9_+_74764340 | 3.83 |
ENST00000376986.1
ENST00000358399.3 |
GDA
|
guanine deaminase |
| chr11_+_19799327 | 3.81 |
ENST00000540292.1
|
NAV2
|
neuron navigator 2 |
| chr7_-_16921601 | 3.80 |
ENST00000402239.3
ENST00000310398.2 ENST00000414935.1 |
AGR3
|
anterior gradient 3 |
| chr17_-_67057114 | 3.80 |
ENST00000370732.2
|
ABCA9
|
ATP-binding cassette, sub-family A (ABC1), member 9 |
| chr10_+_123923205 | 3.63 |
ENST00000369004.3
ENST00000260733.3 |
TACC2
|
transforming, acidic coiled-coil containing protein 2 |
| chr11_+_77774897 | 3.60 |
ENST00000281030.2
|
THRSP
|
thyroid hormone responsive |
| chr4_+_89300158 | 3.51 |
ENST00000502870.1
|
HERC6
|
HECT and RLD domain containing E3 ubiquitin protein ligase family member 6 |
| chr17_-_67057203 | 3.48 |
ENST00000340001.4
|
ABCA9
|
ATP-binding cassette, sub-family A (ABC1), member 9 |
| chr8_+_97597148 | 3.42 |
ENST00000521590.1
|
SDC2
|
syndecan 2 |
| chr1_-_207119738 | 3.40 |
ENST00000356495.4
|
PIGR
|
polymeric immunoglobulin receptor |
| chr2_+_234526272 | 3.39 |
ENST00000373450.4
|
UGT1A1
|
UDP glucuronosyltransferase 1 family, polypeptide A8 |
| chr10_-_71169031 | 3.38 |
ENST00000373307.1
|
TACR2
|
tachykinin receptor 2 |
| chr20_+_56136136 | 3.36 |
ENST00000319441.4
ENST00000543666.1 |
PCK1
|
phosphoenolpyruvate carboxykinase 1 (soluble) |
| chr9_-_95298314 | 3.30 |
ENST00000344604.5
ENST00000375540.1 |
ECM2
|
extracellular matrix protein 2, female organ and adipocyte specific |
| chr17_-_67057047 | 3.30 |
ENST00000495634.1
ENST00000453985.2 ENST00000585714.1 |
ABCA9
|
ATP-binding cassette, sub-family A (ABC1), member 9 |
| chr5_+_140625147 | 3.29 |
ENST00000231173.3
|
PCDHB15
|
protocadherin beta 15 |
| chr2_-_217560248 | 3.26 |
ENST00000233813.4
|
IGFBP5
|
insulin-like growth factor binding protein 5 |
| chr12_-_91576750 | 3.25 |
ENST00000228329.5
ENST00000303320.3 ENST00000052754.5 |
DCN
|
decorin |
| chr12_-_91573249 | 3.24 |
ENST00000550099.1
ENST00000546391.1 ENST00000551354.1 |
DCN
|
decorin |
| chr12_+_41831485 | 3.21 |
ENST00000539469.2
ENST00000298919.7 |
PDZRN4
|
PDZ domain containing ring finger 4 |
| chr7_-_122526799 | 3.19 |
ENST00000334010.7
ENST00000313070.7 |
CADPS2
|
Ca++-dependent secretion activator 2 |
| chr6_+_118869452 | 3.16 |
ENST00000357525.5
|
PLN
|
phospholamban |
| chr15_+_80733570 | 3.14 |
ENST00000533983.1
ENST00000527771.1 ENST00000525103.1 |
ARNT2
|
aryl-hydrocarbon receptor nuclear translocator 2 |
| chr1_+_82266053 | 3.11 |
ENST00000370715.1
ENST00000370713.1 ENST00000319517.6 ENST00000370717.2 ENST00000394879.1 ENST00000271029.4 ENST00000335786.5 |
LPHN2
|
latrophilin 2 |
| chr9_-_119162885 | 3.08 |
ENST00000445861.2
|
PAPPA-AS1
|
PAPPA antisense RNA 1 |
| chr3_+_186435137 | 3.08 |
ENST00000447445.1
|
KNG1
|
kininogen 1 |
| chr2_+_169658928 | 3.07 |
ENST00000317647.7
ENST00000445023.2 |
NOSTRIN
|
nitric oxide synthase trafficking |
| chr10_+_24498060 | 3.07 |
ENST00000376454.3
ENST00000376452.3 |
KIAA1217
|
KIAA1217 |
| chr3_-_112329110 | 3.05 |
ENST00000479368.1
|
CCDC80
|
coiled-coil domain containing 80 |
| chr9_-_21187598 | 2.99 |
ENST00000421715.1
|
IFNA4
|
interferon, alpha 4 |
| chr5_+_38845960 | 2.98 |
ENST00000502536.1
|
OSMR
|
oncostatin M receptor |
| chr9_-_95298254 | 2.96 |
ENST00000444490.2
|
ECM2
|
extracellular matrix protein 2, female organ and adipocyte specific |
| chr3_-_168865522 | 2.95 |
ENST00000464456.1
|
MECOM
|
MDS1 and EVI1 complex locus |
| chr5_+_140749803 | 2.93 |
ENST00000576222.1
|
PCDHGB3
|
protocadherin gamma subfamily B, 3 |
| chr1_-_114430169 | 2.92 |
ENST00000393316.3
|
BCL2L15
|
BCL2-like 15 |
| chr10_+_24497704 | 2.92 |
ENST00000376456.4
ENST00000458595.1 |
KIAA1217
|
KIAA1217 |
| chr12_+_7167980 | 2.91 |
ENST00000360817.5
ENST00000402681.3 |
C1S
|
complement component 1, s subcomponent |
| chr11_+_111789580 | 2.89 |
ENST00000278601.5
|
C11orf52
|
chromosome 11 open reading frame 52 |
| chr5_+_140501581 | 2.89 |
ENST00000194152.1
|
PCDHB4
|
protocadherin beta 4 |
| chr2_-_152118352 | 2.88 |
ENST00000331426.5
|
RBM43
|
RNA binding motif protein 43 |
| chr4_-_138453559 | 2.88 |
ENST00000511115.1
|
PCDH18
|
protocadherin 18 |
| chr12_-_91576561 | 2.87 |
ENST00000547568.2
ENST00000552962.1 |
DCN
|
decorin |
| chrX_+_48620147 | 2.81 |
ENST00000303227.6
|
GLOD5
|
glyoxalase domain containing 5 |
| chrX_+_99899180 | 2.78 |
ENST00000373004.3
|
SRPX2
|
sushi-repeat containing protein, X-linked 2 |
| chr3_+_29322437 | 2.78 |
ENST00000434693.2
|
RBMS3
|
RNA binding motif, single stranded interacting protein 3 |
| chr1_-_147232669 | 2.76 |
ENST00000369237.1
|
GJA5
|
gap junction protein, alpha 5, 40kDa |
| chr15_+_96876340 | 2.76 |
ENST00000453270.2
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr12_-_91573316 | 2.71 |
ENST00000393155.1
|
DCN
|
decorin |
| chr7_-_122526411 | 2.71 |
ENST00000449022.2
|
CADPS2
|
Ca++-dependent secretion activator 2 |
| chr2_+_36923901 | 2.69 |
ENST00000457137.2
|
VIT
|
vitrin |
| chr12_-_59314246 | 2.65 |
ENST00000320743.3
|
LRIG3
|
leucine-rich repeats and immunoglobulin-like domains 3 |
| chr11_-_119293872 | 2.62 |
ENST00000524970.1
|
THY1
|
Thy-1 cell surface antigen |
| chr2_+_234580525 | 2.61 |
ENST00000609637.1
|
UGT1A1
|
UDP glucuronosyltransferase 1 family, polypeptide A8 |
| chr3_-_100565249 | 2.61 |
ENST00000495591.1
ENST00000383691.4 ENST00000466947.1 |
ABI3BP
|
ABI family, member 3 (NESH) binding protein |
| chr12_-_91573132 | 2.61 |
ENST00000550563.1
ENST00000546370.1 |
DCN
|
decorin |
| chr11_+_69455855 | 2.61 |
ENST00000227507.2
ENST00000536559.1 |
CCND1
|
cyclin D1 |
| chr12_+_19358192 | 2.60 |
ENST00000538305.1
|
PLEKHA5
|
pleckstrin homology domain containing, family A member 5 |
| chr7_+_55177416 | 2.56 |
ENST00000450046.1
ENST00000454757.2 |
EGFR
|
epidermal growth factor receptor |
| chr8_+_85618155 | 2.55 |
ENST00000523850.1
ENST00000521376.1 |
RALYL
|
RALY RNA binding protein-like |
| chr9_-_13175823 | 2.53 |
ENST00000545857.1
|
MPDZ
|
multiple PDZ domain protein |
| chr16_-_71598823 | 2.51 |
ENST00000566202.1
|
ZNF19
|
zinc finger protein 19 |
| chr1_-_190446759 | 2.50 |
ENST00000367462.3
|
BRINP3
|
bone morphogenetic protein/retinoic acid inducible neural-specific 3 |
| chr6_-_137366096 | 2.48 |
ENST00000316649.5
ENST00000367746.3 |
IL20RA
|
interleukin 20 receptor, alpha |
| chr17_+_28443819 | 2.46 |
ENST00000479218.2
|
NSRP1
|
nuclear speckle splicing regulatory protein 1 |
| chr5_+_68788594 | 2.45 |
ENST00000396442.2
ENST00000380766.2 |
OCLN
|
occludin |
| chr7_+_134576151 | 2.44 |
ENST00000393118.2
|
CALD1
|
caldesmon 1 |
| chr9_+_74764278 | 2.44 |
ENST00000238018.4
ENST00000376989.3 |
GDA
|
guanine deaminase |
| chr5_+_92919043 | 2.44 |
ENST00000327111.3
|
NR2F1
|
nuclear receptor subfamily 2, group F, member 1 |
| chr4_-_187112626 | 2.41 |
ENST00000596414.1
|
AC110771.1
|
Uncharacterized protein |
| chr7_-_111424462 | 2.37 |
ENST00000437129.1
|
DOCK4
|
dedicator of cytokinesis 4 |
| chr12_-_10324716 | 2.36 |
ENST00000545927.1
ENST00000432556.2 ENST00000309539.3 ENST00000544577.1 |
OLR1
|
oxidized low density lipoprotein (lectin-like) receptor 1 |
| chrX_+_102883620 | 2.35 |
ENST00000372626.3
|
TCEAL1
|
transcription elongation factor A (SII)-like 1 |
| chr2_-_188419078 | 2.35 |
ENST00000437725.1
ENST00000409676.1 ENST00000339091.4 ENST00000420747.1 |
TFPI
|
tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor) |
| chr6_-_137365402 | 2.34 |
ENST00000541547.1
|
IL20RA
|
interleukin 20 receptor, alpha |
| chr14_-_35183755 | 2.33 |
ENST00000555765.1
|
CFL2
|
cofilin 2 (muscle) |
| chr4_-_89951028 | 2.33 |
ENST00000506913.1
|
FAM13A
|
family with sequence similarity 13, member A |
| chr12_-_7245125 | 2.31 |
ENST00000542285.1
ENST00000540610.1 |
C1R
|
complement component 1, r subcomponent |
| chr10_+_18629628 | 2.31 |
ENST00000377329.4
|
CACNB2
|
calcium channel, voltage-dependent, beta 2 subunit |
| chr9_+_71986182 | 2.30 |
ENST00000303068.7
|
FAM189A2
|
family with sequence similarity 189, member A2 |
| chr5_+_38846101 | 2.29 |
ENST00000274276.3
|
OSMR
|
oncostatin M receptor |
| chr10_-_115423792 | 2.29 |
ENST00000369360.3
ENST00000360478.3 ENST00000359988.3 ENST00000369358.4 |
NRAP
|
nebulin-related anchoring protein |
| chr10_-_61495760 | 2.28 |
ENST00000395347.1
|
SLC16A9
|
solute carrier family 16, member 9 |
| chr6_-_53530474 | 2.28 |
ENST00000370905.3
|
KLHL31
|
kelch-like family member 31 |
| chr11_+_46402583 | 2.27 |
ENST00000359803.3
|
MDK
|
midkine (neurite growth-promoting factor 2) |
| chr17_+_68100989 | 2.27 |
ENST00000585558.1
ENST00000392670.1 |
KCNJ16
|
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr2_+_234580499 | 2.27 |
ENST00000354728.4
|
UGT1A9
|
UDP glucuronosyltransferase 1 family, polypeptide A9 |
| chr3_+_29322803 | 2.26 |
ENST00000396583.3
ENST00000383767.2 |
RBMS3
|
RNA binding motif, single stranded interacting protein 3 |
| chr6_-_94129244 | 2.25 |
ENST00000369303.4
ENST00000369297.1 |
EPHA7
|
EPH receptor A7 |
| chr17_-_67138015 | 2.24 |
ENST00000284425.2
ENST00000590645.1 |
ABCA6
|
ATP-binding cassette, sub-family A (ABC1), member 6 |
| chr8_+_19171128 | 2.24 |
ENST00000265807.3
|
SH2D4A
|
SH2 domain containing 4A |
| chr17_+_53343577 | 2.23 |
ENST00000573945.1
|
HLF
|
hepatic leukemia factor |
| chr3_-_149388682 | 2.22 |
ENST00000475579.1
|
WWTR1
|
WW domain containing transcription regulator 1 |
| chr3_-_178865747 | 2.21 |
ENST00000435560.1
|
RP11-360P21.2
|
RP11-360P21.2 |
| chrX_-_99891796 | 2.21 |
ENST00000373020.4
|
TSPAN6
|
tetraspanin 6 |
| chr12_+_12878829 | 2.20 |
ENST00000326765.6
|
APOLD1
|
apolipoprotein L domain containing 1 |
| chr2_+_48796120 | 2.20 |
ENST00000394754.1
|
STON1-GTF2A1L
|
STON1-GTF2A1L readthrough |
| chr11_-_104769141 | 2.19 |
ENST00000508062.1
ENST00000422698.2 |
CASP12
|
caspase 12 (gene/pseudogene) |
| chr6_-_137366163 | 2.19 |
ENST00000367748.1
|
IL20RA
|
interleukin 20 receptor, alpha |
| chr15_-_37393406 | 2.18 |
ENST00000338564.5
ENST00000558313.1 ENST00000340545.5 |
MEIS2
|
Meis homeobox 2 |
| chr2_-_188419200 | 2.18 |
ENST00000233156.3
ENST00000426055.1 ENST00000453013.1 ENST00000417013.1 |
TFPI
|
tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor) |
| chr6_+_160693591 | 2.17 |
ENST00000419196.1
|
RP1-276N6.2
|
RP1-276N6.2 |
| chr7_-_14029515 | 2.16 |
ENST00000430479.1
ENST00000405218.2 ENST00000343495.5 |
ETV1
|
ets variant 1 |
| chr2_+_205410723 | 2.15 |
ENST00000358768.2
ENST00000351153.1 ENST00000349953.3 |
PARD3B
|
par-3 family cell polarity regulator beta |
| chr13_-_33780133 | 2.14 |
ENST00000399365.3
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13 |
| chr7_-_93519471 | 2.14 |
ENST00000451238.1
|
TFPI2
|
tissue factor pathway inhibitor 2 |
| chr12_-_71148413 | 2.13 |
ENST00000440835.2
ENST00000549308.1 ENST00000550661.1 |
PTPRR
|
protein tyrosine phosphatase, receptor type, R |
| chr3_+_29322851 | 2.13 |
ENST00000445033.1
|
RBMS3
|
RNA binding motif, single stranded interacting protein 3 |
| chr8_+_17434689 | 2.13 |
ENST00000398074.3
|
PDGFRL
|
platelet-derived growth factor receptor-like |
| chr14_-_54420133 | 2.13 |
ENST00000559501.1
ENST00000558984.1 |
BMP4
|
bone morphogenetic protein 4 |
| chr7_-_16505440 | 2.13 |
ENST00000307068.4
|
SOSTDC1
|
sclerostin domain containing 1 |
| chr17_+_53343171 | 2.09 |
ENST00000430986.2
|
HLF
|
hepatic leukemia factor |
| chr14_-_35182994 | 2.08 |
ENST00000341223.3
|
CFL2
|
cofilin 2 (muscle) |
| chr9_+_27109440 | 2.06 |
ENST00000519080.1
|
TEK
|
TEK tyrosine kinase, endothelial |
| chr5_-_41261540 | 2.06 |
ENST00000263413.3
|
C6
|
complement component 6 |
| chr12_-_7245152 | 2.06 |
ENST00000542220.2
|
C1R
|
complement component 1, r subcomponent |
| chr3_-_47950745 | 2.05 |
ENST00000429422.1
|
MAP4
|
microtubule-associated protein 4 |
| chr10_-_97321112 | 2.04 |
ENST00000607232.1
ENST00000371227.4 ENST00000371249.2 ENST00000371247.2 ENST00000371246.2 ENST00000393949.1 ENST00000353505.5 ENST00000347291.4 |
SORBS1
|
sorbin and SH3 domain containing 1 |
| chr17_+_36584662 | 2.04 |
ENST00000431231.2
ENST00000437668.3 |
ARHGAP23
|
Rho GTPase activating protein 23 |
| chr12_+_8850277 | 2.04 |
ENST00000539923.1
ENST00000537189.1 |
RIMKLB
|
ribosomal modification protein rimK-like family member B |
| chr17_-_56492989 | 2.04 |
ENST00000583753.1
|
RNF43
|
ring finger protein 43 |
| chr12_-_28124903 | 2.03 |
ENST00000395872.1
ENST00000354417.3 ENST00000201015.4 |
PTHLH
|
parathyroid hormone-like hormone |
| chr5_+_140593509 | 2.02 |
ENST00000341948.4
|
PCDHB13
|
protocadherin beta 13 |
| chr3_-_151176497 | 2.02 |
ENST00000282466.3
|
IGSF10
|
immunoglobulin superfamily, member 10 |
| chr10_-_75401500 | 2.01 |
ENST00000359322.4
|
MYOZ1
|
myozenin 1 |
| chr5_-_131630931 | 2.01 |
ENST00000431054.1
|
P4HA2
|
prolyl 4-hydroxylase, alpha polypeptide II |
| chr3_-_116163830 | 2.01 |
ENST00000333617.4
|
LSAMP
|
limbic system-associated membrane protein |
| chr3_-_114035026 | 1.99 |
ENST00000570269.1
|
RP11-553L6.5
|
RP11-553L6.5 |
| chr3_-_28390298 | 1.98 |
ENST00000457172.1
|
AZI2
|
5-azacytidine induced 2 |
| chr21_+_17791648 | 1.98 |
ENST00000602892.1
ENST00000418813.2 ENST00000435697.1 |
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chrX_-_13835398 | 1.97 |
ENST00000475307.1
|
GPM6B
|
glycoprotein M6B |
| chr1_+_110453109 | 1.96 |
ENST00000525659.1
|
CSF1
|
colony stimulating factor 1 (macrophage) |
| chr2_+_207804278 | 1.95 |
ENST00000272852.3
|
CPO
|
carboxypeptidase O |
| chr8_-_81787006 | 1.94 |
ENST00000327835.3
|
ZNF704
|
zinc finger protein 704 |
| chr9_+_27109392 | 1.93 |
ENST00000406359.4
|
TEK
|
TEK tyrosine kinase, endothelial |
| chr3_-_28390120 | 1.91 |
ENST00000334100.6
|
AZI2
|
5-azacytidine induced 2 |
| chr16_-_21289627 | 1.91 |
ENST00000396023.2
ENST00000415987.2 |
CRYM
|
crystallin, mu |
| chr15_+_71228826 | 1.91 |
ENST00000558456.1
ENST00000560158.2 ENST00000558808.1 ENST00000559806.1 ENST00000559069.1 |
LRRC49
|
leucine rich repeat containing 49 |
| chr2_+_210444748 | 1.91 |
ENST00000392194.1
|
MAP2
|
microtubule-associated protein 2 |
| chr1_-_227505826 | 1.90 |
ENST00000334218.5
ENST00000366766.2 ENST00000366764.2 |
CDC42BPA
|
CDC42 binding protein kinase alpha (DMPK-like) |
| chr11_+_46402297 | 1.90 |
ENST00000405308.2
|
MDK
|
midkine (neurite growth-promoting factor 2) |
| chrX_-_31285042 | 1.89 |
ENST00000378680.2
ENST00000378723.3 |
DMD
|
dystrophin |
| chr3_+_100120441 | 1.89 |
ENST00000489752.1
|
LNP1
|
leukemia NUP98 fusion partner 1 |
| chrX_-_13835461 | 1.88 |
ENST00000316715.4
ENST00000356942.5 |
GPM6B
|
glycoprotein M6B |
| chr12_-_71148357 | 1.88 |
ENST00000378778.1
|
PTPRR
|
protein tyrosine phosphatase, receptor type, R |
| chr20_+_5892037 | 1.88 |
ENST00000378961.4
|
CHGB
|
chromogranin B (secretogranin 1) |
| chr6_-_105585022 | 1.88 |
ENST00000314641.5
|
BVES
|
blood vessel epicardial substance |
| chr18_+_47088401 | 1.87 |
ENST00000261292.4
ENST00000427224.2 ENST00000580036.1 |
LIPG
|
lipase, endothelial |
| chr15_-_99789736 | 1.85 |
ENST00000560235.1
ENST00000394132.2 ENST00000560860.1 ENST00000558078.1 ENST00000394136.1 ENST00000262074.4 ENST00000558613.1 ENST00000394130.1 ENST00000560772.1 |
TTC23
|
tetratricopeptide repeat domain 23 |
| chr2_+_153191706 | 1.85 |
ENST00000288670.9
|
FMNL2
|
formin-like 2 |
| chr2_+_205410516 | 1.85 |
ENST00000406610.2
ENST00000462231.1 |
PARD3B
|
par-3 family cell polarity regulator beta |
| chr19_-_58485895 | 1.84 |
ENST00000314391.3
|
C19orf18
|
chromosome 19 open reading frame 18 |
| chr13_+_102104980 | 1.83 |
ENST00000545560.2
|
ITGBL1
|
integrin, beta-like 1 (with EGF-like repeat domains) |
| chr4_-_186732241 | 1.82 |
ENST00000421639.1
|
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr12_-_7245018 | 1.81 |
ENST00000543835.1
ENST00000535233.2 |
C1R
|
complement component 1, r subcomponent |
| chr18_-_10787140 | 1.81 |
ENST00000383408.2
|
PIEZO2
|
piezo-type mechanosensitive ion channel component 2 |
| chr17_-_76975925 | 1.80 |
ENST00000591274.1
ENST00000589906.1 ENST00000591778.1 ENST00000589775.2 ENST00000585407.1 ENST00000262776.3 |
LGALS3BP
|
lectin, galactoside-binding, soluble, 3 binding protein |
| chr1_+_176432298 | 1.80 |
ENST00000367661.3
ENST00000367662.3 |
PAPPA2
|
pappalysin 2 |
| chr4_+_15376165 | 1.79 |
ENST00000382383.3
ENST00000429690.1 |
C1QTNF7
|
C1q and tumor necrosis factor related protein 7 |
| chr9_+_131684562 | 1.77 |
ENST00000421063.2
|
PHYHD1
|
phytanoyl-CoA dioxygenase domain containing 1 |
| chr17_+_28443799 | 1.76 |
ENST00000584423.1
ENST00000247026.5 |
NSRP1
|
nuclear speckle splicing regulatory protein 1 |
| chr9_-_13279563 | 1.76 |
ENST00000541718.1
|
MPDZ
|
multiple PDZ domain protein |
| chr12_+_54519842 | 1.75 |
ENST00000508564.1
|
RP11-834C11.4
|
RP11-834C11.4 |
| chr4_-_129491686 | 1.74 |
ENST00000514265.1
|
RP11-184M15.1
|
RP11-184M15.1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of IRF7
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 6.3 | GO:0006147 | guanine catabolic process(GO:0006147) |
| 1.9 | 28.5 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 1.8 | 5.4 | GO:1904235 | regulation of substrate-dependent cell migration, cell attachment to substrate(GO:1904235) positive regulation of substrate-dependent cell migration, cell attachment to substrate(GO:1904237) |
| 1.7 | 6.8 | GO:0009956 | radial pattern formation(GO:0009956) |
| 1.3 | 6.4 | GO:0060611 | mammary gland fat development(GO:0060611) positive regulation of macrophage colony-stimulating factor signaling pathway(GO:1902228) positive regulation of response to macrophage colony-stimulating factor(GO:1903971) positive regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903974) positive regulation of microglial cell migration(GO:1904141) |
| 1.2 | 6.0 | GO:0070980 | biphenyl catabolic process(GO:0070980) |
| 1.1 | 6.7 | GO:0030421 | defecation(GO:0030421) |
| 1.1 | 5.4 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 1.1 | 3.3 | GO:1904204 | regulation of skeletal muscle hypertrophy(GO:1904204) |
| 1.1 | 3.2 | GO:1901877 | regulation of calcium ion binding(GO:1901876) negative regulation of calcium ion binding(GO:1901877) regulation of calcium ion import into sarcoplasmic reticulum(GO:1902080) negative regulation of calcium ion import into sarcoplasmic reticulum(GO:1902081) |
| 1.0 | 10.2 | GO:1990504 | dense core granule exocytosis(GO:1990504) |
| 1.0 | 4.0 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.9 | 2.8 | GO:1990029 | atrial ventricular junction remodeling(GO:0003294) atrial cardiac muscle cell to AV node cell communication by electrical coupling(GO:0086044) bundle of His cell to Purkinje myocyte communication by electrical coupling(GO:0086054) Purkinje myocyte to ventricular cardiac muscle cell communication by electrical coupling(GO:0086055) regulation of Purkinje myocyte action potential(GO:0098906) vasomotion(GO:1990029) |
| 0.9 | 2.7 | GO:0071314 | cellular response to cocaine(GO:0071314) |
| 0.9 | 2.6 | GO:0061033 | secretion by lung epithelial cell involved in lung growth(GO:0061033) |
| 0.8 | 3.4 | GO:0006114 | glycerol biosynthetic process(GO:0006114) |
| 0.8 | 2.5 | GO:0002590 | regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002589) negative regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002590) |
| 0.8 | 2.4 | GO:0031448 | regulation of fast-twitch skeletal muscle fiber contraction(GO:0031446) positive regulation of fast-twitch skeletal muscle fiber contraction(GO:0031448) |
| 0.8 | 6.5 | GO:0044565 | dendritic cell proliferation(GO:0044565) |
| 0.8 | 3.1 | GO:1905123 | regulation of endosome organization(GO:1904978) regulation of glucosylceramidase activity(GO:1905123) |
| 0.7 | 2.9 | GO:0010983 | positive regulation of high-density lipoprotein particle clearance(GO:0010983) |
| 0.7 | 1.4 | GO:0035990 | tendon cell differentiation(GO:0035990) tendon formation(GO:0035992) |
| 0.7 | 5.7 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.7 | 6.3 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.7 | 5.5 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.7 | 3.4 | GO:0002415 | immunoglobulin transcytosis in epithelial cells mediated by polymeric immunoglobulin receptor(GO:0002415) |
| 0.7 | 3.4 | GO:0035106 | negative regulation of luteinizing hormone secretion(GO:0033685) operant conditioning(GO:0035106) |
| 0.7 | 5.3 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.7 | 2.6 | GO:2000298 | regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000298) |
| 0.7 | 3.3 | GO:0001834 | trophectodermal cell proliferation(GO:0001834) regulation of trophectodermal cell proliferation(GO:1904073) positive regulation of trophectodermal cell proliferation(GO:1904075) |
| 0.6 | 3.9 | GO:0003134 | BMP signaling pathway involved in heart induction(GO:0003130) endodermal-mesodermal cell signaling(GO:0003133) endodermal-mesodermal cell signaling involved in heart induction(GO:0003134) |
| 0.6 | 2.6 | GO:0043006 | activation of phospholipase A2 activity by calcium-mediated signaling(GO:0043006) |
| 0.6 | 3.8 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.6 | 5.0 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.6 | 1.9 | GO:0060931 | sinoatrial node cell development(GO:0060931) |
| 0.6 | 8.7 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.6 | 1.8 | GO:1901254 | modulation by host of viral RNA genome replication(GO:0044830) positive regulation of intracellular transport of viral material(GO:1901254) |
| 0.6 | 1.8 | GO:0061537 | glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 0.6 | 4.8 | GO:0051025 | negative regulation of immunoglobulin secretion(GO:0051025) |
| 0.6 | 1.8 | GO:0060981 | cell migration involved in coronary angiogenesis(GO:0060981) |
| 0.6 | 3.5 | GO:0051552 | flavone metabolic process(GO:0051552) |
| 0.6 | 1.7 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.6 | 1.7 | GO:0060829 | regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060827) negative regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060829) |
| 0.6 | 4.0 | GO:1903385 | regulation of homophilic cell adhesion(GO:1903385) |
| 0.5 | 5.9 | GO:2000467 | positive regulation of glycogen (starch) synthase activity(GO:2000467) |
| 0.5 | 2.1 | GO:0060648 | mammary gland bud morphogenesis(GO:0060648) |
| 0.5 | 2.7 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.5 | 1.5 | GO:0007493 | endodermal cell fate determination(GO:0007493) |
| 0.5 | 3.4 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.5 | 0.9 | GO:0006533 | aspartate catabolic process(GO:0006533) |
| 0.5 | 10.4 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.4 | 4.9 | GO:0051611 | negative regulation of neurotransmitter uptake(GO:0051581) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.4 | 1.8 | GO:0060392 | negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 0.4 | 5.6 | GO:1900038 | negative regulation of cellular response to hypoxia(GO:1900038) |
| 0.4 | 1.7 | GO:1903045 | neural crest cell migration involved in sympathetic nervous system development(GO:1903045) |
| 0.4 | 1.3 | GO:1901252 | regulation of intracellular transport of viral material(GO:1901252) |
| 0.4 | 1.7 | GO:0034343 | type III interferon production(GO:0034343) regulation of type III interferon production(GO:0034344) |
| 0.4 | 2.1 | GO:0001970 | positive regulation of activation of membrane attack complex(GO:0001970) |
| 0.4 | 2.0 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.4 | 2.4 | GO:0061107 | seminal vesicle development(GO:0061107) |
| 0.4 | 0.8 | GO:0002541 | activation of plasma proteins involved in acute inflammatory response(GO:0002541) |
| 0.4 | 1.6 | GO:0072615 | nitrogen catabolite regulation of transcription from RNA polymerase II promoter(GO:0001079) nitrogen catabolite activation of transcription from RNA polymerase II promoter(GO:0001080) regulation of urea metabolic process(GO:0034255) intracellular bile acid receptor signaling pathway(GO:0038185) interleukin-17 secretion(GO:0072615) nitrogen catabolite regulation of transcription(GO:0090293) nitrogen catabolite activation of transcription(GO:0090294) regulation of nitrogen cycle metabolic process(GO:1903314) positive regulation of glutamate metabolic process(GO:2000213) regulation of ammonia assimilation cycle(GO:2001248) positive regulation of ammonia assimilation cycle(GO:2001250) |
| 0.4 | 2.4 | GO:0060316 | positive regulation of ryanodine-sensitive calcium-release channel activity(GO:0060316) |
| 0.4 | 0.4 | GO:0072011 | glomerular endothelium development(GO:0072011) |
| 0.4 | 10.1 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.4 | 1.5 | GO:0018197 | peptidyl-aspartic acid modification(GO:0018197) peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.4 | 1.5 | GO:0099640 | axo-dendritic protein transport(GO:0099640) |
| 0.4 | 1.5 | GO:1901846 | positive regulation of cell communication by electrical coupling involved in cardiac conduction(GO:1901846) |
| 0.4 | 1.1 | GO:0046416 | D-amino acid metabolic process(GO:0046416) |
| 0.4 | 1.1 | GO:0061075 | regulation of transcription from RNA polymerase II promoter involved in forebrain neuron fate commitment(GO:0021882) cerebral cortex GABAergic interneuron fate commitment(GO:0021893) positive regulation of neural retina development(GO:0061075) positive regulation of retina development in camera-type eye(GO:1902868) positive regulation of amacrine cell differentiation(GO:1902871) |
| 0.4 | 4.6 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.3 | 1.4 | GO:0002426 | immunoglobulin production in mucosal tissue(GO:0002426) |
| 0.3 | 4.3 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.3 | 1.2 | GO:0019056 | modulation by virus of host transcription(GO:0019056) positive regulation of sprouting of injured axon(GO:0048687) positive regulation of axon extension involved in regeneration(GO:0048691) modulation by symbiont of host transcription(GO:0052026) |
| 0.3 | 1.2 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
| 0.3 | 2.0 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.3 | 1.5 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 0.3 | 2.3 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.3 | 2.2 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.3 | 4.2 | GO:0033601 | positive regulation of mammary gland epithelial cell proliferation(GO:0033601) |
| 0.3 | 2.5 | GO:0072734 | response to staurosporine(GO:0072733) cellular response to staurosporine(GO:0072734) |
| 0.3 | 4.4 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.3 | 1.1 | GO:0001905 | activation of membrane attack complex(GO:0001905) regulation of activation of membrane attack complex(GO:0001969) negative regulation of activation of membrane attack complex(GO:0001971) |
| 0.3 | 0.8 | GO:0050720 | interleukin-1 beta biosynthetic process(GO:0050720) |
| 0.3 | 1.1 | GO:0042109 | lymphotoxin A production(GO:0032641) lymphotoxin A biosynthetic process(GO:0042109) |
| 0.3 | 1.6 | GO:0015692 | lead ion transport(GO:0015692) |
| 0.3 | 0.8 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 0.3 | 0.8 | GO:1902362 | melanocyte apoptotic process(GO:1902362) |
| 0.3 | 3.3 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.2 | 2.2 | GO:0048630 | skeletal muscle tissue growth(GO:0048630) |
| 0.2 | 1.0 | GO:0046909 | intermembrane transport(GO:0046909) protein transport from ciliary membrane to plasma membrane(GO:1903445) |
| 0.2 | 1.7 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.2 | 8.6 | GO:0036150 | phosphatidylserine acyl-chain remodeling(GO:0036150) |
| 0.2 | 0.7 | GO:1902548 | negative regulation of cellular response to vascular endothelial growth factor stimulus(GO:1902548) |
| 0.2 | 0.9 | GO:1990926 | short-term synaptic potentiation(GO:1990926) |
| 0.2 | 8.5 | GO:0003298 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.2 | 4.3 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.2 | 2.9 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.2 | 3.8 | GO:0097475 | motor neuron migration(GO:0097475) |
| 0.2 | 0.2 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.2 | 0.7 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 0.2 | 1.3 | GO:0045196 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.2 | 1.1 | GO:0010513 | positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) |
| 0.2 | 5.4 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.2 | 1.5 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.2 | 1.3 | GO:2000661 | positive regulation of interleukin-1-mediated signaling pathway(GO:2000661) |
| 0.2 | 1.2 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.2 | 0.6 | GO:0006425 | glutaminyl-tRNA aminoacylation(GO:0006425) |
| 0.2 | 2.2 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.2 | 1.3 | GO:0022614 | membrane to membrane docking(GO:0022614) |
| 0.2 | 4.7 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.2 | 1.5 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.2 | 2.0 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.2 | 0.7 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.2 | 1.3 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.2 | 0.5 | GO:0072579 | molybdenum incorporation into molybdenum-molybdopterin complex(GO:0018315) metal incorporation into metallo-molybdopterin complex(GO:0042040) glycine receptor clustering(GO:0072579) |
| 0.2 | 0.9 | GO:1900039 | positive regulation of cellular response to hypoxia(GO:1900039) |
| 0.2 | 1.3 | GO:0035995 | skeletal muscle myosin thick filament assembly(GO:0030241) detection of muscle stretch(GO:0035995) |
| 0.2 | 0.4 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.2 | 0.7 | GO:0002408 | myeloid dendritic cell chemotaxis(GO:0002408) |
| 0.2 | 0.2 | GO:0060664 | epithelial cell proliferation involved in salivary gland morphogenesis(GO:0060664) |
| 0.2 | 0.7 | GO:0097156 | fasciculation of motor neuron axon(GO:0097156) |
| 0.2 | 1.0 | GO:0071603 | endothelial cell-cell adhesion(GO:0071603) |
| 0.2 | 1.7 | GO:0070560 | protein secretion by platelet(GO:0070560) |
| 0.2 | 0.8 | GO:0060830 | ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) |
| 0.2 | 4.0 | GO:0030157 | pancreatic juice secretion(GO:0030157) |
| 0.2 | 0.8 | GO:0009257 | 10-formyltetrahydrofolate biosynthetic process(GO:0009257) |
| 0.2 | 3.8 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.2 | 2.5 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.2 | 2.8 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.2 | 1.0 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.2 | 0.8 | GO:0071874 | response to norepinephrine(GO:0071873) cellular response to norepinephrine stimulus(GO:0071874) |
| 0.2 | 0.8 | GO:0033590 | response to cobalamin(GO:0033590) |
| 0.2 | 0.8 | GO:0006045 | N-acetylglucosamine biosynthetic process(GO:0006045) glucosamine-containing compound biosynthetic process(GO:1901073) |
| 0.2 | 0.6 | GO:0018352 | protein-pyridoxal-5-phosphate linkage(GO:0018352) |
| 0.2 | 2.0 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 0.2 | 3.6 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.1 | 2.5 | GO:1904896 | ESCRT complex disassembly(GO:1904896) ESCRT III complex disassembly(GO:1904903) |
| 0.1 | 1.0 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.1 | 3.6 | GO:0006853 | carnitine shuttle(GO:0006853) |
| 0.1 | 0.4 | GO:0060300 | regulation of cytokine activity(GO:0060300) |
| 0.1 | 0.1 | GO:0032240 | negative regulation of nucleobase-containing compound transport(GO:0032240) negative regulation of RNA export from nucleus(GO:0046832) |
| 0.1 | 0.9 | GO:0006001 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 0.1 | 2.0 | GO:0015705 | iodide transport(GO:0015705) |
| 0.1 | 1.0 | GO:1902731 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) negative regulation of chondrocyte proliferation(GO:1902731) |
| 0.1 | 2.3 | GO:0072178 | nephric duct morphogenesis(GO:0072178) |
| 0.1 | 1.5 | GO:0070842 | aggresome assembly(GO:0070842) |
| 0.1 | 1.6 | GO:1903944 | regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.1 | 1.9 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.1 | 1.6 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.1 | 1.2 | GO:0031666 | positive regulation of lipopolysaccharide-mediated signaling pathway(GO:0031666) |
| 0.1 | 0.8 | GO:0033489 | cholesterol biosynthetic process via desmosterol(GO:0033489) cholesterol biosynthetic process via lathosterol(GO:0033490) |
| 0.1 | 2.4 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.1 | 6.7 | GO:0045124 | regulation of bone resorption(GO:0045124) |
| 0.1 | 1.6 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.1 | 0.4 | GO:0006624 | vacuolar protein processing(GO:0006624) |
| 0.1 | 2.2 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.1 | 0.4 | GO:0050955 | thermoception(GO:0050955) |
| 0.1 | 2.6 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.1 | 2.0 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.1 | 1.2 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.1 | 0.9 | GO:0003065 | positive regulation of heart rate by epinephrine(GO:0003065) |
| 0.1 | 0.5 | GO:0044376 | RNA polymerase II complex import to nucleus(GO:0044376) RNA polymerase III complex localization to nucleus(GO:1990022) |
| 0.1 | 0.4 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.1 | 0.5 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.1 | 5.2 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.1 | 2.3 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.1 | 1.7 | GO:0070673 | response to interleukin-18(GO:0070673) |
| 0.1 | 6.7 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.1 | 0.3 | GO:0046022 | positive regulation of transcription from RNA polymerase II promoter during mitosis(GO:0046022) |
| 0.1 | 0.5 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.1 | 0.8 | GO:0006777 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) |
| 0.1 | 0.8 | GO:0071104 | response to interleukin-9(GO:0071104) |
| 0.1 | 1.7 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.1 | 0.8 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.1 | 2.0 | GO:0045725 | positive regulation of glycogen biosynthetic process(GO:0045725) |
| 0.1 | 1.2 | GO:0046628 | positive regulation of insulin receptor signaling pathway(GO:0046628) |
| 0.1 | 2.1 | GO:0035791 | platelet-derived growth factor receptor-beta signaling pathway(GO:0035791) |
| 0.1 | 0.3 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.1 | 1.5 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.1 | 0.8 | GO:1900004 | negative regulation of serine-type endopeptidase activity(GO:1900004) negative regulation of serine-type peptidase activity(GO:1902572) |
| 0.1 | 17.3 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.1 | 0.6 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.1 | 0.5 | GO:0048680 | positive regulation of axon regeneration(GO:0048680) |
| 0.1 | 1.0 | GO:0021678 | third ventricle development(GO:0021678) |
| 0.1 | 2.0 | GO:0019511 | peptidyl-proline hydroxylation(GO:0019511) |
| 0.1 | 0.8 | GO:0051901 | positive regulation of mitochondrial depolarization(GO:0051901) |
| 0.1 | 1.0 | GO:0035986 | senescence-associated heterochromatin focus assembly(GO:0035986) |
| 0.1 | 0.3 | GO:0055099 | detection of endogenous stimulus(GO:0009726) response to high density lipoprotein particle(GO:0055099) |
| 0.1 | 0.2 | GO:0046102 | adenosine catabolic process(GO:0006154) inosine metabolic process(GO:0046102) inosine biosynthetic process(GO:0046103) |
| 0.1 | 1.4 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.1 | 2.0 | GO:0032331 | negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.1 | 0.8 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.1 | 1.7 | GO:1904355 | positive regulation of telomere capping(GO:1904355) |
| 0.1 | 0.5 | GO:0046541 | saliva secretion(GO:0046541) |
| 0.1 | 0.5 | GO:1903772 | virus maturation(GO:0019075) regulation of viral budding via host ESCRT complex(GO:1903772) |
| 0.1 | 1.0 | GO:2001054 | negative regulation of mesenchymal cell apoptotic process(GO:2001054) |
| 0.1 | 1.6 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.1 | 0.6 | GO:0034627 | 'de novo' NAD biosynthetic process(GO:0034627) |
| 0.1 | 0.9 | GO:1903671 | negative regulation of sprouting angiogenesis(GO:1903671) |
| 0.1 | 2.4 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.1 | 1.3 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.1 | 2.4 | GO:0035635 | entry of bacterium into host cell(GO:0035635) |
| 0.1 | 0.9 | GO:0010962 | regulation of glycogen biosynthetic process(GO:0005979) regulation of glucan biosynthetic process(GO:0010962) |
| 0.1 | 0.5 | GO:0070445 | oligodendrocyte progenitor proliferation(GO:0070444) regulation of oligodendrocyte progenitor proliferation(GO:0070445) |
| 0.1 | 2.3 | GO:0071498 | cellular response to fluid shear stress(GO:0071498) |
| 0.1 | 3.6 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.1 | 0.7 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.1 | 1.5 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.1 | 1.0 | GO:0060065 | uterus development(GO:0060065) |
| 0.1 | 0.4 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.1 | 0.5 | GO:0021800 | cerebral cortex tangential migration(GO:0021800) |
| 0.1 | 1.3 | GO:2000651 | positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.1 | 0.6 | GO:0010727 | negative regulation of hydrogen peroxide metabolic process(GO:0010727) |
| 0.1 | 3.1 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.1 | 3.6 | GO:0046825 | regulation of protein export from nucleus(GO:0046825) |
| 0.1 | 2.2 | GO:0045601 | regulation of endothelial cell differentiation(GO:0045601) |
| 0.1 | 3.1 | GO:0050999 | regulation of nitric-oxide synthase activity(GO:0050999) |
| 0.1 | 1.6 | GO:0010801 | negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 0.1 | 0.2 | GO:0060151 | peroxisome localization(GO:0060151) microtubule-based peroxisome localization(GO:0060152) |
| 0.1 | 0.6 | GO:0044413 | evasion or tolerance of host defenses by virus(GO:0019049) avoidance of host defenses(GO:0044413) evasion or tolerance of host defenses(GO:0044415) avoidance of defenses of other organism involved in symbiotic interaction(GO:0051832) evasion or tolerance of defenses of other organism involved in symbiotic interaction(GO:0051834) |
| 0.1 | 3.6 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.1 | 0.3 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.1 | 0.6 | GO:0032233 | positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.1 | 0.4 | GO:0048549 | endosome localization(GO:0032439) positive regulation of pinocytosis(GO:0048549) |
| 0.1 | 1.7 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 0.1 | 0.8 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.1 | 0.3 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.1 | 0.3 | GO:2000676 | positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.1 | 0.2 | GO:0045726 | positive regulation of integrin biosynthetic process(GO:0045726) |
| 0.1 | 0.5 | GO:0072189 | ureter development(GO:0072189) |
| 0.1 | 1.0 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.1 | 2.1 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.1 | 1.5 | GO:0042359 | vitamin D metabolic process(GO:0042359) |
| 0.1 | 0.2 | GO:0048294 | negative regulation of isotype switching to IgE isotypes(GO:0048294) |
| 0.1 | 0.7 | GO:0036152 | phosphatidylethanolamine acyl-chain remodeling(GO:0036152) |
| 0.1 | 0.6 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.1 | 0.7 | GO:0045745 | positive regulation of G-protein coupled receptor protein signaling pathway(GO:0045745) |
| 0.0 | 1.1 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.0 | 2.4 | GO:2001222 | regulation of neuron migration(GO:2001222) |
| 0.0 | 0.2 | GO:0043335 | protein unfolding(GO:0043335) |
| 0.0 | 0.2 | GO:1902037 | negative regulation of hematopoietic stem cell differentiation(GO:1902037) |
| 0.0 | 2.8 | GO:0031529 | ruffle organization(GO:0031529) |
| 0.0 | 0.7 | GO:0060044 | negative regulation of cardiac muscle tissue growth(GO:0055022) negative regulation of cardiac muscle cell proliferation(GO:0060044) negative regulation of heart growth(GO:0061117) |
| 0.0 | 9.0 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 | 0.6 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.0 | 0.4 | GO:0050435 | beta-amyloid metabolic process(GO:0050435) |
| 0.0 | 0.5 | GO:0015747 | urate transport(GO:0015747) |
| 0.0 | 1.3 | GO:0001783 | B cell apoptotic process(GO:0001783) |
| 0.0 | 0.6 | GO:0008627 | intrinsic apoptotic signaling pathway in response to osmotic stress(GO:0008627) |
| 0.0 | 0.1 | GO:0051342 | regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051342) negative regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051344) |
| 0.0 | 3.9 | GO:0003151 | outflow tract morphogenesis(GO:0003151) |
| 0.0 | 9.1 | GO:0045727 | positive regulation of translation(GO:0045727) |
| 0.0 | 1.9 | GO:0019835 | cytolysis(GO:0019835) |
| 0.0 | 3.6 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.0 | 2.3 | GO:0046329 | negative regulation of JNK cascade(GO:0046329) |
| 0.0 | 11.2 | GO:0031032 | actomyosin structure organization(GO:0031032) |
| 0.0 | 0.3 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
| 0.0 | 0.7 | GO:0032148 | activation of protein kinase B activity(GO:0032148) |
| 0.0 | 0.6 | GO:0051386 | regulation of neurotrophin TRK receptor signaling pathway(GO:0051386) |
| 0.0 | 0.3 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.0 | 5.1 | GO:0030010 | establishment of cell polarity(GO:0030010) |
| 0.0 | 0.3 | GO:0061737 | leukotriene signaling pathway(GO:0061737) |
| 0.0 | 1.3 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 1.5 | GO:0022400 | regulation of rhodopsin mediated signaling pathway(GO:0022400) |
| 0.0 | 0.3 | GO:0060012 | synaptic transmission, glycinergic(GO:0060012) |
| 0.0 | 0.3 | GO:0055091 | phospholipid homeostasis(GO:0055091) |
| 0.0 | 2.5 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.0 | 0.1 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.0 | 0.6 | GO:0015866 | ADP transport(GO:0015866) |
| 0.0 | 0.2 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 0.0 | 1.1 | GO:1901687 | glutathione derivative metabolic process(GO:1901685) glutathione derivative biosynthetic process(GO:1901687) |
| 0.0 | 0.3 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.0 | 0.6 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 1.1 | GO:0006836 | neurotransmitter transport(GO:0006836) |
| 0.0 | 1.3 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.0 | 2.2 | GO:0008542 | visual learning(GO:0008542) |
| 0.0 | 0.5 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.0 | 1.9 | GO:0014047 | glutamate secretion(GO:0014047) |
| 0.0 | 0.5 | GO:0061088 | regulation of sequestering of zinc ion(GO:0061088) |
| 0.0 | 2.3 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 1.1 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.0 | 4.5 | GO:0048706 | embryonic skeletal system development(GO:0048706) |
| 0.0 | 1.7 | GO:0006699 | bile acid biosynthetic process(GO:0006699) |
| 0.0 | 0.5 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 1.0 | GO:0014904 | myotube cell development(GO:0014904) |
| 0.0 | 0.9 | GO:0009081 | branched-chain amino acid metabolic process(GO:0009081) branched-chain amino acid catabolic process(GO:0009083) |
| 0.0 | 0.8 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.0 | 0.3 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.0 | 1.4 | GO:0045599 | negative regulation of fat cell differentiation(GO:0045599) |
| 0.0 | 0.6 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.0 | 3.8 | GO:0008652 | cellular amino acid biosynthetic process(GO:0008652) |
| 0.0 | 0.1 | GO:1904383 | response to sodium phosphate(GO:1904383) |
| 0.0 | 1.0 | GO:0043268 | positive regulation of potassium ion transport(GO:0043268) |
| 0.0 | 0.3 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.0 | 0.7 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.2 | GO:0050771 | negative regulation of axonogenesis(GO:0050771) |
| 0.0 | 0.7 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.0 | 0.1 | GO:0043686 | co-translational protein modification(GO:0043686) |
| 0.0 | 0.5 | GO:0042481 | regulation of odontogenesis(GO:0042481) |
| 0.0 | 0.2 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.0 | 0.0 | GO:0072716 | response to actinomycin D(GO:0072716) |
| 0.0 | 0.2 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.0 | 0.5 | GO:0070863 | positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.0 | 0.2 | GO:0010944 | negative regulation of transcription by competitive promoter binding(GO:0010944) |
| 0.0 | 0.5 | GO:0070884 | regulation of calcineurin-NFAT signaling cascade(GO:0070884) |
| 0.0 | 1.3 | GO:0034219 | carbohydrate transmembrane transport(GO:0034219) |
| 0.0 | 8.0 | GO:0006869 | lipid transport(GO:0006869) |
| 0.0 | 0.2 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.0 | 0.6 | GO:0006590 | thyroid hormone generation(GO:0006590) |
| 0.0 | 0.1 | GO:0070632 | spindle pole body duplication(GO:0030474) spindle pole body organization(GO:0051300) spindle pole body localization(GO:0070631) establishment of spindle pole body localization(GO:0070632) spindle pole body localization to nuclear envelope(GO:0071789) establishment of spindle pole body localization to nuclear envelope(GO:0071790) |
| 0.0 | 0.3 | GO:0070535 | histone H2A K63-linked ubiquitination(GO:0070535) |
| 0.0 | 1.4 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.0 | 0.1 | GO:1904381 | Golgi apparatus mannose trimming(GO:1904381) |
| 0.0 | 0.2 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.0 | 0.2 | GO:0098719 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.1 | GO:0051083 | 'de novo' cotranslational protein folding(GO:0051083) |
| 0.0 | 0.5 | GO:0045668 | negative regulation of osteoblast differentiation(GO:0045668) |
| 0.0 | 0.1 | GO:1904428 | negative regulation of tubulin deacetylation(GO:1904428) |
| 0.0 | 0.3 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.0 | 0.4 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.0 | 0.4 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.0 | 1.6 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 0.7 | GO:0060765 | regulation of androgen receptor signaling pathway(GO:0060765) |
| 0.0 | 0.9 | GO:0000281 | mitotic cytokinesis(GO:0000281) |
| 0.0 | 0.4 | GO:0008535 | respiratory chain complex IV assembly(GO:0008535) |
| 0.0 | 0.1 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.0 | 0.5 | GO:0045920 | negative regulation of exocytosis(GO:0045920) |
| 0.0 | 0.6 | GO:0019228 | neuronal action potential(GO:0019228) |
| 0.0 | 1.8 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.0 | 1.8 | GO:1903955 | positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.0 | 0.2 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.0 | 1.5 | GO:0030449 | regulation of complement activation(GO:0030449) regulation of protein activation cascade(GO:2000257) |
| 0.0 | 0.4 | GO:0006536 | glutamate metabolic process(GO:0006536) |
| 0.0 | 0.1 | GO:0040018 | positive regulation of multicellular organism growth(GO:0040018) |
| 0.0 | 0.3 | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
| 0.0 | 1.1 | GO:0043647 | inositol phosphate metabolic process(GO:0043647) |
| 0.0 | 1.0 | GO:0030879 | mammary gland development(GO:0030879) |
| 0.0 | 0.9 | GO:0030819 | positive regulation of cAMP biosynthetic process(GO:0030819) |
| 0.0 | 0.5 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.0 | 0.5 | GO:0006356 | regulation of transcription from RNA polymerase I promoter(GO:0006356) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 28.5 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 1.8 | 5.3 | GO:0005900 | oncostatin-M receptor complex(GO:0005900) |
| 0.9 | 6.4 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 0.9 | 2.6 | GO:0097489 | multivesicular body, internal vesicle lumen(GO:0097489) |
| 0.7 | 22.6 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.7 | 4.0 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.6 | 2.4 | GO:0036027 | protein C inhibitor-TMPRSS7 complex(GO:0036024) protein C inhibitor-TMPRSS11E complex(GO:0036025) protein C inhibitor-PLAT complex(GO:0036026) protein C inhibitor-PLAU complex(GO:0036027) protein C inhibitor-thrombin complex(GO:0036028) protein C inhibitor-KLK3 complex(GO:0036029) protein C inhibitor-plasma kallikrein complex(GO:0036030) serine protease inhibitor complex(GO:0097180) protein C inhibitor-coagulation factor V complex(GO:0097181) protein C inhibitor-coagulation factor Xa complex(GO:0097182) protein C inhibitor-coagulation factor XI complex(GO:0097183) |
| 0.6 | 1.8 | GO:1902737 | dendritic filopodium(GO:1902737) |
| 0.6 | 6.5 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.6 | 1.7 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.5 | 6.0 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.5 | 1.6 | GO:0030895 | apolipoprotein B mRNA editing enzyme complex(GO:0030895) |
| 0.5 | 4.0 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.4 | 1.3 | GO:0071065 | alpha9-beta1 integrin-vascular cell adhesion molecule-1 complex(GO:0071065) |
| 0.4 | 4.0 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.4 | 5.1 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.4 | 2.3 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.3 | 4.5 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.3 | 2.0 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.3 | 4.0 | GO:0030478 | actin cap(GO:0030478) |
| 0.3 | 10.1 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.3 | 1.6 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.3 | 1.5 | GO:0031673 | H zone(GO:0031673) |
| 0.3 | 6.0 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.2 | 4.4 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.2 | 2.4 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.2 | 1.7 | GO:1990357 | terminal web(GO:1990357) |
| 0.2 | 3.8 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.2 | 1.0 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 0.2 | 0.8 | GO:0070931 | Golgi-associated vesicle lumen(GO:0070931) |
| 0.2 | 5.0 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.2 | 2.3 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.2 | 6.7 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.1 | 4.2 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.1 | 1.3 | GO:0043196 | varicosity(GO:0043196) |
| 0.1 | 3.7 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.1 | 2.4 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.1 | 5.4 | GO:0043034 | costamere(GO:0043034) |
| 0.1 | 2.3 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.1 | 5.8 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.1 | 0.5 | GO:0072534 | perineuronal net(GO:0072534) |
| 0.1 | 0.8 | GO:1990812 | growth cone filopodium(GO:1990812) |
| 0.1 | 0.9 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.1 | 0.9 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 0.1 | 1.0 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.1 | 7.0 | GO:0032420 | stereocilium(GO:0032420) |
| 0.1 | 1.8 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.1 | 1.6 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.1 | 2.4 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.1 | 3.2 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.1 | 2.8 | GO:0005922 | connexon complex(GO:0005922) |
| 0.1 | 4.4 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.1 | 0.9 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.1 | 0.9 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.1 | 1.8 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.1 | 1.0 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.1 | 2.7 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.1 | 17.3 | GO:0030018 | Z disc(GO:0030018) |
| 0.1 | 9.7 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.1 | 2.1 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.1 | 3.0 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.1 | 1.3 | GO:0036038 | MKS complex(GO:0036038) |
| 0.1 | 1.2 | GO:0008091 | spectrin(GO:0008091) |
| 0.1 | 1.0 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.1 | 1.5 | GO:0043203 | axon hillock(GO:0043203) |
| 0.1 | 1.0 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.1 | 2.1 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.1 | 4.6 | GO:0016235 | aggresome(GO:0016235) |
| 0.1 | 0.8 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.1 | 1.0 | GO:0097433 | dense body(GO:0097433) |
| 0.1 | 8.0 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.1 | 0.6 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.1 | 8.2 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.1 | 10.4 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.1 | 2.1 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 0.6 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.0 | 0.4 | GO:0044354 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.0 | 1.0 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.0 | 0.8 | GO:0032059 | bleb(GO:0032059) |
| 0.0 | 5.2 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 3.7 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 4.5 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 1.3 | GO:0071437 | invadopodium(GO:0071437) |
| 0.0 | 1.8 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.0 | 1.6 | GO:0032590 | dendrite membrane(GO:0032590) |
| 0.0 | 3.7 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 3.0 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 4.5 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 3.0 | GO:0098857 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 1.8 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.7 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.3 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.0 | 0.4 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.0 | 0.7 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 0.0 | 1.2 | GO:0035267 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.0 | 0.7 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 1.1 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.5 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.3 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.0 | 0.6 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.0 | 4.3 | GO:0030665 | clathrin-coated vesicle membrane(GO:0030665) |
| 0.0 | 0.2 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
| 0.0 | 1.3 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 0.8 | GO:0030666 | endocytic vesicle membrane(GO:0030666) |
| 0.0 | 1.7 | GO:0098793 | presynapse(GO:0098793) |
| 0.0 | 4.2 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 0.2 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.0 | 2.2 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 1.2 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.0 | 0.1 | GO:0070762 | nuclear pore transmembrane ring(GO:0070762) |
| 0.0 | 0.3 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 0.8 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 0.5 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.2 | GO:0072588 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 0.0 | 0.1 | GO:0005846 | nuclear cap binding complex(GO:0005846) |
| 0.0 | 0.2 | GO:0071682 | endocytic vesicle lumen(GO:0071682) |
| 0.0 | 0.1 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 1.1 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.7 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 1.3 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.0 | 0.2 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.0 | 6.1 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 0.1 | GO:0031595 | proteasome regulatory particle, lid subcomplex(GO:0008541) nuclear proteasome complex(GO:0031595) |
| 0.0 | 1.3 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.4 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.0 | 0.6 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 0.5 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.1 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.0 | 1.0 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 0.1 | GO:0042405 | nuclear inclusion body(GO:0042405) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 6.3 | GO:0008892 | guanine deaminase activity(GO:0008892) |
| 1.4 | 10.1 | GO:0052739 | phosphatidylserine 1-acylhydrolase activity(GO:0052739) |
| 1.4 | 8.5 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 1.4 | 4.2 | GO:0072591 | citrate-L-glutamate ligase activity(GO:0072591) |
| 1.3 | 6.3 | GO:0070052 | collagen V binding(GO:0070052) |
| 1.2 | 3.6 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 1.1 | 3.3 | GO:0004613 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 1.1 | 4.3 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 1.0 | 4.0 | GO:0033754 | indoleamine 2,3-dioxygenase activity(GO:0033754) |
| 1.0 | 4.0 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 1.0 | 3.9 | GO:0005019 | platelet-derived growth factor beta-receptor activity(GO:0005019) |
| 1.0 | 3.9 | GO:0001601 | peptide YY receptor activity(GO:0001601) |
| 0.9 | 2.8 | GO:0086078 | gap junction channel activity involved in atrial cardiac muscle cell-AV node cell electrical coupling(GO:0086076) gap junction channel activity involved in bundle of His cell-Purkinje myocyte electrical coupling(GO:0086078) gap junction channel activity involved in Purkinje myocyte-ventricular cardiac muscle cell electrical coupling(GO:0086079) |
| 0.9 | 5.3 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.8 | 5.6 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.8 | 6.4 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.7 | 8.7 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.7 | 9.9 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.7 | 3.4 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.5 | 2.6 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.5 | 4.0 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.5 | 1.5 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.5 | 1.9 | GO:0016639 | oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 0.5 | 2.3 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 0.5 | 1.4 | GO:0047225 | acetylgalactosaminyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0047225) |
| 0.4 | 14.2 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.4 | 3.0 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.4 | 1.3 | GO:0098640 | integrin binding involved in cell-matrix adhesion(GO:0098640) |
| 0.4 | 8.3 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.4 | 1.6 | GO:1902122 | chenodeoxycholic acid binding(GO:1902122) |
| 0.4 | 1.6 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.4 | 3.4 | GO:0019763 | immunoglobulin receptor activity(GO:0019763) |
| 0.4 | 18.8 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.4 | 1.5 | GO:0008426 | protein kinase C inhibitor activity(GO:0008426) |
| 0.4 | 1.1 | GO:0030626 | U12 snRNA binding(GO:0030626) |
| 0.3 | 5.2 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.3 | 2.0 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.3 | 5.7 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.3 | 26.1 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.3 | 2.1 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.3 | 1.6 | GO:0015086 | cadmium ion transmembrane transporter activity(GO:0015086) cobalt ion transmembrane transporter activity(GO:0015087) lead ion transmembrane transporter activity(GO:0015094) ferrous iron uptake transmembrane transporter activity(GO:0015639) |
| 0.3 | 4.2 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.3 | 1.8 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.3 | 3.9 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.3 | 2.3 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.3 | 3.3 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.2 | 2.2 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.2 | 2.0 | GO:0015111 | iodide transmembrane transporter activity(GO:0015111) |
| 0.2 | 2.6 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.2 | 6.3 | GO:0097153 | cysteine-type endopeptidase activity involved in apoptotic process(GO:0097153) |
| 0.2 | 2.0 | GO:0019798 | procollagen-proline 4-dioxygenase activity(GO:0004656) procollagen-proline dioxygenase activity(GO:0019798) |
| 0.2 | 1.1 | GO:0050115 | myosin-light-chain-phosphatase activity(GO:0050115) |
| 0.2 | 1.5 | GO:0042954 | lipoprotein transporter activity(GO:0042954) |
| 0.2 | 0.6 | GO:0004819 | glutamine-tRNA ligase activity(GO:0004819) |
| 0.2 | 0.8 | GO:0008761 | UDP-N-acetylglucosamine 2-epimerase activity(GO:0008761) N-acylmannosamine kinase activity(GO:0009384) |
| 0.2 | 12.8 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.2 | 8.0 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.2 | 0.5 | GO:0061599 | molybdopterin adenylyltransferase activity(GO:0061598) molybdopterin molybdotransferase activity(GO:0061599) |
| 0.2 | 0.5 | GO:0046848 | hydroxyapatite binding(GO:0046848) |
| 0.2 | 2.9 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.2 | 1.3 | GO:0004909 | interleukin-1, Type I, activating receptor activity(GO:0004909) |
| 0.2 | 3.3 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.2 | 0.5 | GO:0070336 | flap-structured DNA binding(GO:0070336) |
| 0.2 | 0.8 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.2 | 1.3 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.2 | 2.1 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.2 | 4.8 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.2 | 0.8 | GO:0030151 | molybdenum ion binding(GO:0030151) |
| 0.2 | 0.8 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 0.1 | 0.4 | GO:0016768 | spermine synthase activity(GO:0016768) |
| 0.1 | 1.3 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.1 | 2.8 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.1 | 1.1 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.1 | 1.7 | GO:1990459 | transferrin receptor binding(GO:1990459) |
| 0.1 | 2.0 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.1 | 3.1 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.1 | 1.4 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.1 | 1.2 | GO:0043426 | MRF binding(GO:0043426) |
| 0.1 | 1.8 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.1 | 1.2 | GO:1904929 | coreceptor activity involved in Wnt signaling pathway, planar cell polarity pathway(GO:1904929) |
| 0.1 | 1.5 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.1 | 3.9 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.1 | 1.9 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.1 | 4.0 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.1 | 7.7 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.1 | 1.3 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.1 | 4.5 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.1 | 0.8 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.1 | 1.0 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.1 | 0.3 | GO:0034041 | sterol-transporting ATPase activity(GO:0034041) |
| 0.1 | 4.5 | GO:0005521 | lamin binding(GO:0005521) |
| 0.1 | 25.6 | GO:0008201 | heparin binding(GO:0008201) |
| 0.1 | 1.7 | GO:0019871 | potassium channel inhibitor activity(GO:0019870) sodium channel inhibitor activity(GO:0019871) |
| 0.1 | 0.3 | GO:0070506 | high-density lipoprotein particle receptor activity(GO:0070506) |
| 0.1 | 1.2 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.1 | 2.4 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.1 | 1.2 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 1.9 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) intronic transcription regulatory region DNA binding(GO:0044213) |
| 0.1 | 1.5 | GO:0005221 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.1 | 0.6 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.1 | 0.8 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.1 | 0.9 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.1 | 0.2 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.1 | 1.3 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.1 | 14.2 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.1 | 2.0 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.1 | 5.7 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.1 | 0.7 | GO:0060698 | endoribonuclease inhibitor activity(GO:0060698) |
| 0.1 | 1.6 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.1 | 1.1 | GO:0016641 | oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor(GO:0016641) |
| 0.1 | 1.1 | GO:0051378 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.1 | 0.9 | GO:0102338 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.1 | 0.6 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.1 | 0.6 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.1 | 1.5 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.1 | 2.3 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.1 | 1.3 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.1 | 1.7 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.1 | 0.3 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.1 | 7.9 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 0.6 | GO:0070573 | metallodipeptidase activity(GO:0070573) |
| 0.1 | 0.5 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.1 | 0.4 | GO:0047820 | D-glutamate cyclase activity(GO:0047820) |
| 0.1 | 5.1 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.1 | 0.5 | GO:0016167 | glial cell-derived neurotrophic factor receptor activity(GO:0016167) |
| 0.1 | 1.5 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) acetylcholine binding(GO:0042166) |
| 0.1 | 2.6 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.1 | 3.5 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 1.1 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 2.4 | GO:0017046 | peptide hormone binding(GO:0017046) |
| 0.0 | 2.0 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 1.7 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 0.7 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 1.0 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 7.3 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 1.5 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 1.7 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 0.2 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.0 | 0.3 | GO:0016933 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.0 | 0.3 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 0.9 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) potassium ion leak channel activity(GO:0022841) |
| 0.0 | 0.8 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 1.1 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 6.5 | GO:0042626 | ATPase activity, coupled to transmembrane movement of substances(GO:0042626) |
| 0.0 | 1.9 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.5 | GO:0019966 | interleukin-1 binding(GO:0019966) |
| 0.0 | 0.6 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) |
| 0.0 | 0.8 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.1 | GO:0046538 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.0 | 0.6 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.0 | 0.1 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 0.0 | 0.2 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.0 | 2.7 | GO:0005539 | glycosaminoglycan binding(GO:0005539) |
| 0.0 | 1.3 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 0.8 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.0 | 1.7 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 0.2 | GO:0016427 | tRNA (cytosine) methyltransferase activity(GO:0016427) |
| 0.0 | 0.5 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 0.3 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.0 | 1.0 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 1.7 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 1.9 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 0.7 | GO:0030553 | cGMP binding(GO:0030553) |
| 0.0 | 0.2 | GO:0004882 | androgen receptor activity(GO:0004882) |
| 0.0 | 1.5 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 1.2 | GO:0071814 | lipoprotein particle binding(GO:0071813) protein-lipid complex binding(GO:0071814) |
| 0.0 | 1.1 | GO:0001848 | complement binding(GO:0001848) |
| 0.0 | 0.3 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.0 | 0.6 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.2 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.0 | 0.7 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.2 | GO:0016434 | rRNA (cytosine) methyltransferase activity(GO:0016434) |
| 0.0 | 4.1 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.4 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.4 | GO:0042805 | actinin binding(GO:0042805) |
| 0.0 | 3.2 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.8 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.0 | 1.9 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.0 | 0.3 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 3.0 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.5 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.0 | 0.1 | GO:0003696 | satellite DNA binding(GO:0003696) |
| 0.0 | 0.3 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) G-quadruplex DNA binding(GO:0051880) |
| 0.0 | 0.7 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 1.6 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.0 | 0.7 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 0.2 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.0 | 0.2 | GO:0022821 | potassium:proton antiporter activity(GO:0015386) potassium ion antiporter activity(GO:0022821) |
| 0.0 | 11.3 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.0 | 1.1 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.2 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.0 | 20.9 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 2.0 | GO:0001158 | enhancer sequence-specific DNA binding(GO:0001158) |
| 0.0 | 0.9 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 0.8 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 1.4 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 1.7 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.0 | 1.6 | GO:0035257 | nuclear hormone receptor binding(GO:0035257) |
| 0.0 | 1.0 | GO:0015144 | carbohydrate transmembrane transporter activity(GO:0015144) carbohydrate transporter activity(GO:1901476) |
| 0.0 | 0.6 | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups(GO:0016765) |
| 0.0 | 0.9 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 1.0 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 0.2 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.0 | 0.3 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.0 | 0.8 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.5 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.4 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.6 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.3 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.1 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 1.4 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.0 | 0.3 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 2.2 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.3 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 0.5 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.0 | GO:0004556 | alpha-amylase activity(GO:0004556) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 32.0 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.2 | 16.5 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.2 | 9.8 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.2 | 8.6 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.1 | 3.6 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.1 | 2.9 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.1 | 1.0 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.1 | 9.8 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.1 | 2.9 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.1 | 7.1 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.1 | 1.3 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.1 | 3.2 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.1 | 4.1 | PID TCPTP PATHWAY | Signaling events mediated by TCPTP |
| 0.1 | 1.9 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.1 | 7.5 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 5.3 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.1 | 3.2 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.1 | 6.0 | PID BMP PATHWAY | BMP receptor signaling |
| 0.1 | 6.4 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.1 | 23.0 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 2.5 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.1 | 6.2 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.1 | 4.4 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.1 | 0.8 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.1 | 4.2 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.1 | 1.8 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.1 | 3.6 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 1.8 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.0 | 1.6 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 4.5 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 3.4 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.7 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 1.3 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 1.1 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.0 | 2.0 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 1.3 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.0 | 0.1 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 1.5 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 1.4 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.0 | 0.8 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 1.0 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 2.2 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 1.6 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.9 | PID INTEGRIN1 PATHWAY | Beta1 integrin cell surface interactions |
| 0.0 | 2.0 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.6 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 1.1 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.0 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.0 | 0.9 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 0.2 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.0 | 1.0 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.3 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
| 0.0 | 0.6 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 0.8 | PID FANCONI PATHWAY | Fanconi anemia pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 28.3 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.4 | 12.9 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.4 | 10.9 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.3 | 6.1 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.3 | 8.2 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.3 | 8.3 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.2 | 6.0 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.2 | 2.6 | REACTOME SHC1 EVENTS IN EGFR SIGNALING | Genes involved in SHC1 events in EGFR signaling |
| 0.2 | 3.3 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.2 | 3.8 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.2 | 2.9 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.2 | 6.9 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.2 | 5.4 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.2 | 3.9 | REACTOME FORMATION OF FIBRIN CLOT CLOTTING CASCADE | Genes involved in Formation of Fibrin Clot (Clotting Cascade) |
| 0.2 | 1.2 | REACTOME MRNA PROCESSING | Genes involved in mRNA Processing |
| 0.1 | 4.0 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.1 | 5.2 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.1 | 3.4 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.1 | 8.6 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 2.8 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.1 | 2.5 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.1 | 2.6 | REACTOME ACTIVATED POINT MUTANTS OF FGFR2 | Genes involved in Activated point mutants of FGFR2 |
| 0.1 | 3.3 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.1 | 1.3 | REACTOME OPSINS | Genes involved in Opsins |
| 0.1 | 6.5 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 2.0 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.1 | 2.4 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.1 | 2.0 | REACTOME TRAF6 MEDIATED INDUCTION OF TAK1 COMPLEX | Genes involved in TRAF6 mediated induction of TAK1 complex |
| 0.1 | 1.5 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.1 | 2.8 | REACTOME SIGNALING BY PDGF | Genes involved in Signaling by PDGF |
| 0.1 | 1.7 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.1 | 4.0 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 2.3 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.1 | 2.4 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.1 | 2.2 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.1 | 1.1 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.1 | 1.7 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.1 | 1.8 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.1 | 2.9 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.1 | 2.2 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.1 | 1.2 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.1 | 2.4 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.1 | 1.1 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 2.6 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.9 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.6 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 0.6 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 1.4 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 4.0 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 2.2 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.0 | 1.7 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 1.2 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.9 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 2.8 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 2.1 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.8 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.0 | 0.7 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 1.1 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 1.2 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.0 | 1.1 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 2.9 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.8 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 1.1 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.3 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.3 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 0.6 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 3.6 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.2 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 2.0 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.4 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 1.4 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 1.7 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 1.3 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 4.9 | REACTOME GENERIC TRANSCRIPTION PATHWAY | Genes involved in Generic Transcription Pathway |
| 0.0 | 0.8 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.3 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 2.3 | REACTOME G ALPHA Q SIGNALLING EVENTS | Genes involved in G alpha (q) signalling events |