Project
Illumina Body Map 2: averaged replicates
Navigation
Downloads
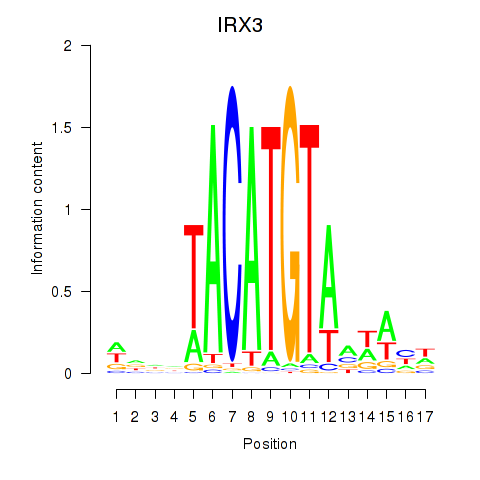
Results for IRX3
Z-value: 2.20
Transcription factors associated with IRX3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
IRX3
|
ENSG00000177508.11 | iroquois homeobox 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| IRX3 | hg19_v2_chr16_-_54320675_54320715 | 0.14 | 4.5e-01 | Click! |
Activity profile of IRX3 motif
Sorted Z-values of IRX3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_54320078 | 8.13 |
ENST00000231009.2
|
GZMK
|
granzyme K (granzyme 3; tryptase II) |
| chr15_-_22473353 | 5.97 |
ENST00000557788.2
|
IGHV4OR15-8
|
immunoglobulin heavy variable 4/OR15-8 (non-functional) |
| chr14_-_106453155 | 5.53 |
ENST00000390594.2
|
IGHV1-2
|
immunoglobulin heavy variable 1-2 |
| chr14_+_22386325 | 5.12 |
ENST00000390439.2
|
TRAV13-2
|
T cell receptor alpha variable 13-2 |
| chr1_+_26644441 | 5.11 |
ENST00000374213.2
|
CD52
|
CD52 molecule |
| chr22_+_23154239 | 5.08 |
ENST00000390315.2
|
IGLV3-10
|
immunoglobulin lambda variable 3-10 |
| chr13_-_46716969 | 4.92 |
ENST00000435666.2
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin) |
| chr14_-_107179265 | 4.81 |
ENST00000390634.2
|
IGHV2-70
|
immunoglobulin heavy variable 2-70 |
| chr1_+_158259558 | 4.78 |
ENST00000368170.3
|
CD1C
|
CD1c molecule |
| chr2_-_89545079 | 4.77 |
ENST00000468494.1
|
IGKV2-30
|
immunoglobulin kappa variable 2-30 |
| chr4_+_71091786 | 4.39 |
ENST00000317987.5
|
FDCSP
|
follicular dendritic cell secreted protein |
| chr14_-_106781017 | 4.08 |
ENST00000390612.2
|
IGHV4-28
|
immunoglobulin heavy variable 4-28 |
| chr22_+_23161491 | 3.87 |
ENST00000390316.2
|
IGLV3-9
|
immunoglobulin lambda variable 3-9 (gene/pseudogene) |
| chr3_+_151451707 | 3.81 |
ENST00000356517.3
|
AADACL2
|
arylacetamide deacetylase-like 2 |
| chr2_+_89975669 | 3.73 |
ENST00000474213.1
|
IGKV2D-30
|
immunoglobulin kappa variable 2D-30 |
| chr11_+_60223225 | 3.62 |
ENST00000524807.1
ENST00000345732.4 |
MS4A1
|
membrane-spanning 4-domains, subfamily A, member 1 |
| chr6_-_108145499 | 3.54 |
ENST00000369020.3
ENST00000369022.2 |
SCML4
|
sex comb on midleg-like 4 (Drosophila) |
| chr2_+_7865923 | 3.47 |
ENST00000417930.1
|
AC092580.4
|
AC092580.4 |
| chr4_+_156824840 | 3.36 |
ENST00000536354.2
|
TDO2
|
tryptophan 2,3-dioxygenase |
| chr14_+_22204418 | 3.35 |
ENST00000390426.2
|
TRAV4
|
T cell receptor alpha variable 4 |
| chr14_-_106494587 | 3.20 |
ENST00000390597.2
|
IGHV2-5
|
immunoglobulin heavy variable 2-5 |
| chr1_+_117297007 | 3.12 |
ENST00000369478.3
ENST00000369477.1 |
CD2
|
CD2 molecule |
| chr2_+_90139056 | 3.00 |
ENST00000492446.1
|
IGKV1D-16
|
immunoglobulin kappa variable 1D-16 |
| chr11_-_118213455 | 2.90 |
ENST00000300692.4
|
CD3D
|
CD3d molecule, delta (CD3-TCR complex) |
| chr14_-_106725723 | 2.86 |
ENST00000390609.2
|
IGHV3-23
|
immunoglobulin heavy variable 3-23 |
| chr14_-_106758101 | 2.83 |
ENST00000390611.2
|
IGHV2-26
|
immunoglobulin heavy variable 2-26 |
| chr14_-_106845789 | 2.82 |
ENST00000390617.2
|
IGHV3-35
|
immunoglobulin heavy variable 3-35 (non-functional) |
| chr14_-_106478603 | 2.79 |
ENST00000390596.2
|
IGHV4-4
|
immunoglobulin heavy variable 4-4 |
| chr5_+_35856951 | 2.75 |
ENST00000303115.3
ENST00000343305.4 ENST00000506850.1 ENST00000511982.1 |
IL7R
|
interleukin 7 receptor |
| chr2_+_90121477 | 2.73 |
ENST00000483379.1
|
IGKV1D-17
|
immunoglobulin kappa variable 1D-17 |
| chr11_-_118213360 | 2.70 |
ENST00000529594.1
|
CD3D
|
CD3d molecule, delta (CD3-TCR complex) |
| chr2_-_136875712 | 2.70 |
ENST00000241393.3
|
CXCR4
|
chemokine (C-X-C motif) receptor 4 |
| chr7_-_38289173 | 2.66 |
ENST00000436911.2
|
TRGC2
|
T cell receptor gamma constant 2 |
| chr14_-_106967788 | 2.63 |
ENST00000390622.2
|
IGHV1-46
|
immunoglobulin heavy variable 1-46 |
| chr14_-_106830057 | 2.62 |
ENST00000390616.2
|
IGHV4-34
|
immunoglobulin heavy variable 4-34 |
| chr12_+_5541267 | 2.62 |
ENST00000423158.3
|
NTF3
|
neurotrophin 3 |
| chr14_+_22433675 | 2.62 |
ENST00000390442.3
|
TRAV12-3
|
T cell receptor alpha variable 12-3 |
| chr14_-_106805716 | 2.58 |
ENST00000438142.2
|
IGHV4-31
|
immunoglobulin heavy variable 4-31 |
| chr10_-_96829246 | 2.49 |
ENST00000371270.3
ENST00000535898.1 ENST00000539050.1 |
CYP2C8
|
cytochrome P450, family 2, subfamily C, polypeptide 8 |
| chr6_-_49834209 | 2.45 |
ENST00000507853.1
|
CRISP1
|
cysteine-rich secretory protein 1 |
| chr14_-_107131560 | 2.37 |
ENST00000390632.2
|
IGHV3-66
|
immunoglobulin heavy variable 3-66 |
| chr11_+_60739115 | 2.33 |
ENST00000344028.5
ENST00000346437.4 |
CD6
|
CD6 molecule |
| chr2_-_89417335 | 2.29 |
ENST00000490686.1
|
IGKV1-17
|
immunoglobulin kappa variable 1-17 |
| chr22_+_23063100 | 2.27 |
ENST00000390309.2
|
IGLV3-19
|
immunoglobulin lambda variable 3-19 |
| chr5_-_156390230 | 2.25 |
ENST00000407087.3
ENST00000274532.2 |
TIMD4
|
T-cell immunoglobulin and mucin domain containing 4 |
| chr11_-_74408739 | 2.25 |
ENST00000529912.1
|
CHRDL2
|
chordin-like 2 |
| chr22_+_40297079 | 2.23 |
ENST00000344138.4
ENST00000543252.1 |
GRAP2
|
GRB2-related adaptor protein 2 |
| chr14_-_106692191 | 2.23 |
ENST00000390607.2
|
IGHV3-21
|
immunoglobulin heavy variable 3-21 |
| chr1_+_196788887 | 2.22 |
ENST00000320493.5
ENST00000367424.4 ENST00000367421.3 |
CFHR1
CFHR2
|
complement factor H-related 1 complement factor H-related 2 |
| chr6_-_49834240 | 2.22 |
ENST00000335847.4
|
CRISP1
|
cysteine-rich secretory protein 1 |
| chr18_-_56985776 | 2.22 |
ENST00000587244.1
|
CPLX4
|
complexin 4 |
| chr14_-_106926724 | 2.19 |
ENST00000434710.1
|
IGHV3-43
|
immunoglobulin heavy variable 3-43 |
| chr6_-_24799117 | 2.13 |
ENST00000565469.1
|
RP3-369A17.5
|
chromosome 6 open reading frame 229 |
| chr14_-_106963409 | 2.11 |
ENST00000390621.2
|
IGHV1-45
|
immunoglobulin heavy variable 1-45 |
| chr1_-_7913089 | 2.09 |
ENST00000361696.5
|
UTS2
|
urotensin 2 |
| chr12_-_10251603 | 2.07 |
ENST00000457018.2
|
CLEC1A
|
C-type lectin domain family 1, member A |
| chr11_-_118213331 | 2.05 |
ENST00000392884.2
|
CD3D
|
CD3d molecule, delta (CD3-TCR complex) |
| chr5_-_156362666 | 2.04 |
ENST00000406964.1
|
TIMD4
|
T-cell immunoglobulin and mucin domain containing 4 |
| chr6_+_71104588 | 2.02 |
ENST00000418403.1
|
RP11-462G2.1
|
RP11-462G2.1 |
| chr14_-_106610852 | 2.02 |
ENST00000390603.2
|
IGHV3-15
|
immunoglobulin heavy variable 3-15 |
| chr1_+_160709029 | 2.01 |
ENST00000444090.2
ENST00000441662.2 |
SLAMF7
|
SLAM family member 7 |
| chr11_+_60739249 | 1.98 |
ENST00000542157.1
ENST00000433107.2 ENST00000452451.2 ENST00000352009.5 |
CD6
|
CD6 molecule |
| chr2_+_90259593 | 1.98 |
ENST00000471857.1
|
IGKV1D-8
|
immunoglobulin kappa variable 1D-8 |
| chr2_-_89459813 | 1.98 |
ENST00000390256.2
|
IGKV6-21
|
immunoglobulin kappa variable 6-21 (non-functional) |
| chr2_-_89521942 | 1.98 |
ENST00000482769.1
|
IGKV2-28
|
immunoglobulin kappa variable 2-28 |
| chr4_+_69681710 | 1.97 |
ENST00000265403.7
ENST00000458688.2 |
UGT2B10
|
UDP glucuronosyltransferase 2 family, polypeptide B10 |
| chr1_+_160709055 | 1.97 |
ENST00000368043.3
ENST00000368042.3 ENST00000458602.2 ENST00000458104.2 |
SLAMF7
|
SLAM family member 7 |
| chr12_-_10542617 | 1.95 |
ENST00000240618.6
|
KLRK1
|
killer cell lectin-like receptor subfamily K, member 1 |
| chr2_-_158300556 | 1.95 |
ENST00000264192.3
|
CYTIP
|
cytohesin 1 interacting protein |
| chr2_+_119699864 | 1.95 |
ENST00000541757.1
ENST00000412481.1 |
MARCO
|
macrophage receptor with collagenous structure |
| chr2_+_114163945 | 1.92 |
ENST00000453673.3
|
IGKV1OR2-108
|
immunoglobulin kappa variable 1/OR2-108 (non-functional) |
| chr14_-_106791536 | 1.89 |
ENST00000390613.2
|
IGHV3-30
|
immunoglobulin heavy variable 3-30 |
| chr19_-_54974894 | 1.89 |
ENST00000333834.4
|
LENG9
|
leukocyte receptor cluster (LRC) member 9 |
| chr14_-_106471723 | 1.89 |
ENST00000390595.2
|
IGHV1-3
|
immunoglobulin heavy variable 1-3 |
| chr1_-_169733789 | 1.89 |
ENST00000454271.1
ENST00000609271.1 |
RP1-117P20.3
SELE
|
RP1-117P20.3 selectin E |
| chr4_+_40195262 | 1.87 |
ENST00000503941.1
|
RHOH
|
ras homolog family member H |
| chr12_+_101869096 | 1.86 |
ENST00000551346.1
|
SPIC
|
Spi-C transcription factor (Spi-1/PU.1 related) |
| chr1_+_81001398 | 1.85 |
ENST00000418041.1
ENST00000443104.1 |
RP5-887A10.1
|
RP5-887A10.1 |
| chrX_+_78426469 | 1.84 |
ENST00000276077.1
|
GPR174
|
G protein-coupled receptor 174 |
| chr1_+_81106951 | 1.81 |
ENST00000443565.1
|
RP5-887A10.1
|
RP5-887A10.1 |
| chr15_-_81616446 | 1.81 |
ENST00000302824.6
|
STARD5
|
StAR-related lipid transfer (START) domain containing 5 |
| chr4_-_38784592 | 1.80 |
ENST00000502321.1
|
TLR10
|
toll-like receptor 10 |
| chr15_+_81225699 | 1.79 |
ENST00000560027.1
|
KIAA1199
|
KIAA1199 |
| chr16_+_32063311 | 1.78 |
ENST00000426099.1
|
AC142381.1
|
AC142381.1 |
| chr12_-_9885888 | 1.75 |
ENST00000327839.3
|
CLECL1
|
C-type lectin-like 1 |
| chr1_+_160709076 | 1.75 |
ENST00000359331.4
ENST00000495334.1 |
SLAMF7
|
SLAM family member 7 |
| chr1_-_157522180 | 1.75 |
ENST00000356953.4
ENST00000368188.2 ENST00000368190.3 ENST00000368189.3 |
FCRL5
|
Fc receptor-like 5 |
| chr3_+_100354442 | 1.74 |
ENST00000475887.1
|
GPR128
|
G protein-coupled receptor 128 |
| chr14_-_106586656 | 1.69 |
ENST00000390602.2
|
IGHV3-13
|
immunoglobulin heavy variable 3-13 |
| chr11_+_60739140 | 1.68 |
ENST00000313421.7
|
CD6
|
CD6 molecule |
| chr4_-_69434245 | 1.63 |
ENST00000317746.2
|
UGT2B17
|
UDP glucuronosyltransferase 2 family, polypeptide B17 |
| chr17_+_34639793 | 1.62 |
ENST00000394465.2
ENST00000394463.2 ENST00000378342.4 |
CCL4L2
|
chemokine (C-C motif) ligand 4-like 2 |
| chr17_+_34430980 | 1.62 |
ENST00000250151.4
|
CCL4
|
chemokine (C-C motif) ligand 4 |
| chr1_+_196912902 | 1.62 |
ENST00000476712.2
ENST00000367415.5 |
CFHR2
|
complement factor H-related 2 |
| chr2_+_85921414 | 1.59 |
ENST00000263863.4
ENST00000524600.1 |
GNLY
|
granulysin |
| chr11_+_57365150 | 1.59 |
ENST00000457869.1
ENST00000340687.6 ENST00000378323.4 ENST00000378324.2 ENST00000403558.1 |
SERPING1
|
serpin peptidase inhibitor, clade G (C1 inhibitor), member 1 |
| chr17_+_38673270 | 1.58 |
ENST00000578280.1
|
RP5-1028K7.2
|
RP5-1028K7.2 |
| chr14_+_95047725 | 1.56 |
ENST00000554760.1
ENST00000554866.1 ENST00000329597.7 ENST00000556775.1 |
SERPINA5
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 5 |
| chr1_-_114414316 | 1.54 |
ENST00000528414.1
ENST00000538253.1 ENST00000460620.1 ENST00000420377.2 ENST00000525799.1 ENST00000359785.5 |
PTPN22
|
protein tyrosine phosphatase, non-receptor type 22 (lymphoid) |
| chr17_-_46657473 | 1.53 |
ENST00000332503.5
|
HOXB4
|
homeobox B4 |
| chr1_+_57320437 | 1.52 |
ENST00000361249.3
|
C8A
|
complement component 8, alpha polypeptide |
| chr10_+_96698406 | 1.52 |
ENST00000260682.6
|
CYP2C9
|
cytochrome P450, family 2, subfamily C, polypeptide 9 |
| chr2_-_88427568 | 1.51 |
ENST00000393750.3
ENST00000295834.3 |
FABP1
|
fatty acid binding protein 1, liver |
| chrX_-_55291163 | 1.51 |
ENST00000519203.1
ENST00000374951.1 |
PAGE3
|
P antigen family, member 3 (prostate associated) |
| chr9_-_4679419 | 1.51 |
ENST00000609131.1
ENST00000607997.1 |
RP11-6J24.6
|
RP11-6J24.6 |
| chr7_-_41742697 | 1.51 |
ENST00000242208.4
|
INHBA
|
inhibin, beta A |
| chr16_+_33605231 | 1.51 |
ENST00000570121.2
|
IGHV3OR16-12
|
immunoglobulin heavy variable 3/OR16-12 (non-functional) |
| chr2_+_89923550 | 1.51 |
ENST00000509129.1
|
IGKV1D-37
|
immunoglobulin kappa variable 1D-37 (non-functional) |
| chr16_-_33647696 | 1.51 |
ENST00000558425.1
ENST00000569103.2 |
RP11-812E19.9
|
Uncharacterized protein |
| chr2_+_143635222 | 1.50 |
ENST00000375773.2
ENST00000409512.1 ENST00000410015.2 |
KYNU
|
kynureninase |
| chr12_-_9268707 | 1.48 |
ENST00000318602.7
|
A2M
|
alpha-2-macroglobulin |
| chr2_-_88125471 | 1.48 |
ENST00000398146.3
|
RGPD2
|
RANBP2-like and GRIP domain containing 2 |
| chr2_+_217735493 | 1.47 |
ENST00000456163.1
|
AC007557.1
|
HCG1816075; Uncharacterized protein |
| chr3_+_177159695 | 1.47 |
ENST00000442937.1
|
LINC00578
|
long intergenic non-protein coding RNA 578 |
| chr8_-_86253888 | 1.47 |
ENST00000522389.1
ENST00000432364.2 ENST00000517618.1 |
CA1
|
carbonic anhydrase I |
| chr2_+_113885138 | 1.47 |
ENST00000409930.3
|
IL1RN
|
interleukin 1 receptor antagonist |
| chr12_+_9980113 | 1.46 |
ENST00000537723.1
|
KLRF1
|
killer cell lectin-like receptor subfamily F, member 1 |
| chr21_-_34185944 | 1.45 |
ENST00000479548.1
|
C21orf62
|
chromosome 21 open reading frame 62 |
| chr22_+_22516550 | 1.45 |
ENST00000390284.2
|
IGLV4-60
|
immunoglobulin lambda variable 4-60 |
| chr14_+_95047744 | 1.44 |
ENST00000553511.1
ENST00000554633.1 ENST00000555681.1 ENST00000554276.1 |
SERPINA5
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 5 |
| chr11_-_118083600 | 1.44 |
ENST00000524477.1
|
AMICA1
|
adhesion molecule, interacts with CXADR antigen 1 |
| chr1_+_171060018 | 1.43 |
ENST00000367755.4
ENST00000392085.2 ENST00000542847.1 ENST00000538429.1 ENST00000479749.1 |
FMO3
|
flavin containing monooxygenase 3 |
| chr17_+_18380051 | 1.42 |
ENST00000581545.1
ENST00000582333.1 ENST00000328114.6 ENST00000412421.2 ENST00000583322.1 ENST00000584941.1 |
LGALS9C
|
lectin, galactoside-binding, soluble, 9C |
| chr2_+_85921522 | 1.42 |
ENST00000409696.3
|
GNLY
|
granulysin |
| chr12_+_47473369 | 1.42 |
ENST00000546455.1
|
PCED1B
|
PC-esterase domain containing 1B |
| chr12_-_91398796 | 1.40 |
ENST00000261172.3
ENST00000551767.1 |
EPYC
|
epiphycan |
| chr11_+_114166536 | 1.39 |
ENST00000299964.3
|
NNMT
|
nicotinamide N-methyltransferase |
| chr22_+_40297105 | 1.39 |
ENST00000540310.1
|
GRAP2
|
GRB2-related adaptor protein 2 |
| chr5_+_159848807 | 1.38 |
ENST00000352433.5
|
PTTG1
|
pituitary tumor-transforming 1 |
| chr12_+_29542557 | 1.37 |
ENST00000550906.1
|
OVCH1-AS1
|
OVCH1 antisense RNA 1 |
| chr22_-_50970919 | 1.36 |
ENST00000329363.4
ENST00000437588.1 |
ODF3B
|
outer dense fiber of sperm tails 3B |
| chr1_+_207070775 | 1.36 |
ENST00000391929.3
ENST00000294984.2 ENST00000367093.3 |
IL24
|
interleukin 24 |
| chr11_-_5255861 | 1.36 |
ENST00000380299.3
|
HBD
|
hemoglobin, delta |
| chr12_-_68553512 | 1.36 |
ENST00000229135.3
|
IFNG
|
interferon, gamma |
| chr17_-_29641104 | 1.36 |
ENST00000577894.1
ENST00000330927.4 |
EVI2B
|
ecotropic viral integration site 2B |
| chr10_-_101841588 | 1.33 |
ENST00000370418.3
|
CPN1
|
carboxypeptidase N, polypeptide 1 |
| chr1_+_196857144 | 1.32 |
ENST00000367416.2
ENST00000251424.4 ENST00000367418.2 |
CFHR4
|
complement factor H-related 4 |
| chr16_+_30484054 | 1.32 |
ENST00000564118.1
ENST00000454514.2 ENST00000433423.2 |
ITGAL
|
integrin, alpha L (antigen CD11A (p180), lymphocyte function-associated antigen 1; alpha polypeptide) |
| chr3_+_122044084 | 1.31 |
ENST00000264474.3
ENST00000479204.1 |
CSTA
|
cystatin A (stefin A) |
| chr7_+_23210760 | 1.29 |
ENST00000366347.4
|
AC005082.1
|
Uncharacterized protein |
| chr10_-_90712520 | 1.27 |
ENST00000224784.6
|
ACTA2
|
actin, alpha 2, smooth muscle, aorta |
| chr1_+_242011485 | 1.27 |
ENST00000423131.1
ENST00000523590.1 |
EXO1
|
exonuclease 1 |
| chr4_-_70725856 | 1.26 |
ENST00000226444.3
|
SULT1E1
|
sulfotransferase family 1E, estrogen-preferring, member 1 |
| chrX_+_12809463 | 1.19 |
ENST00000380663.3
ENST00000380668.5 ENST00000398491.2 ENST00000489404.1 |
PRPS2
|
phosphoribosyl pyrophosphate synthetase 2 |
| chr1_-_89488510 | 1.19 |
ENST00000564665.1
ENST00000370481.4 |
GBP3
|
guanylate binding protein 3 |
| chr22_+_37309662 | 1.18 |
ENST00000403662.3
ENST00000262825.5 |
CSF2RB
|
colony stimulating factor 2 receptor, beta, low-affinity (granulocyte-macrophage) |
| chr5_-_13944652 | 1.17 |
ENST00000265104.4
|
DNAH5
|
dynein, axonemal, heavy chain 5 |
| chr5_-_64858944 | 1.17 |
ENST00000508421.1
ENST00000510693.1 ENST00000514814.1 ENST00000515497.1 ENST00000396679.1 |
CENPK
|
centromere protein K |
| chr14_+_22236722 | 1.17 |
ENST00000390428.3
|
TRAV6
|
T cell receptor alpha variable 6 |
| chr7_+_139529040 | 1.17 |
ENST00000455353.1
ENST00000458722.1 ENST00000411653.1 |
TBXAS1
|
thromboxane A synthase 1 (platelet) |
| chr2_+_231090471 | 1.17 |
ENST00000373645.3
|
SP140
|
SP140 nuclear body protein |
| chr3_+_111630451 | 1.16 |
ENST00000495180.1
|
PHLDB2
|
pleckstrin homology-like domain, family B, member 2 |
| chr13_+_113030625 | 1.15 |
ENST00000283550.3
|
SPACA7
|
sperm acrosome associated 7 |
| chr2_+_189839046 | 1.15 |
ENST00000304636.3
ENST00000317840.5 |
COL3A1
|
collagen, type III, alpha 1 |
| chr17_+_34431212 | 1.15 |
ENST00000394495.1
|
CCL4
|
chemokine (C-C motif) ligand 4 |
| chr4_-_84035905 | 1.15 |
ENST00000311507.4
|
PLAC8
|
placenta-specific 8 |
| chr12_-_21757774 | 1.14 |
ENST00000261195.2
|
GYS2
|
glycogen synthase 2 (liver) |
| chr5_+_110559603 | 1.14 |
ENST00000512453.1
|
CAMK4
|
calcium/calmodulin-dependent protein kinase IV |
| chr2_+_231090433 | 1.13 |
ENST00000486687.2
ENST00000350136.5 ENST00000392045.3 ENST00000417495.3 ENST00000343805.6 ENST00000420434.3 |
SP140
|
SP140 nuclear body protein |
| chr1_+_22964073 | 1.12 |
ENST00000402322.1
|
C1QA
|
complement component 1, q subcomponent, A chain |
| chr8_-_101724989 | 1.12 |
ENST00000517403.1
|
PABPC1
|
poly(A) binding protein, cytoplasmic 1 |
| chr21_-_34186006 | 1.12 |
ENST00000490358.1
|
C21orf62
|
chromosome 21 open reading frame 62 |
| chr7_+_139528952 | 1.11 |
ENST00000416849.2
ENST00000436047.2 ENST00000414508.2 ENST00000448866.1 |
TBXAS1
|
thromboxane A synthase 1 (platelet) |
| chr17_-_29641084 | 1.11 |
ENST00000544462.1
|
EVI2B
|
ecotropic viral integration site 2B |
| chr14_+_22554680 | 1.08 |
ENST00000390451.2
|
TRAV23DV6
|
T cell receptor alpha variable 23/delta variable 6 |
| chr14_-_105487381 | 1.07 |
ENST00000392590.3
ENST00000336219.3 |
CDCA4
|
cell division cycle associated 4 |
| chr3_-_191000172 | 1.06 |
ENST00000427544.2
|
UTS2B
|
urotensin 2B |
| chr21_-_47613673 | 1.06 |
ENST00000594486.1
|
AP001468.1
|
Protein LOC101060037 |
| chr12_+_69742121 | 1.06 |
ENST00000261267.2
ENST00000549690.1 ENST00000548839.1 |
LYZ
|
lysozyme |
| chr7_+_139529085 | 1.06 |
ENST00000539806.1
|
TBXAS1
|
thromboxane A synthase 1 (platelet) |
| chr3_+_177159744 | 1.04 |
ENST00000439009.1
|
LINC00578
|
long intergenic non-protein coding RNA 578 |
| chr16_+_31539183 | 1.04 |
ENST00000302312.4
|
AHSP
|
alpha hemoglobin stabilizing protein |
| chr2_-_231989808 | 1.04 |
ENST00000258400.3
|
HTR2B
|
5-hydroxytryptamine (serotonin) receptor 2B, G protein-coupled |
| chr17_-_54893250 | 1.04 |
ENST00000397862.2
|
C17orf67
|
chromosome 17 open reading frame 67 |
| chr4_+_69962185 | 1.03 |
ENST00000305231.7
|
UGT2B7
|
UDP glucuronosyltransferase 2 family, polypeptide B7 |
| chr14_+_100485712 | 1.03 |
ENST00000544450.2
|
EVL
|
Enah/Vasp-like |
| chr6_+_130742231 | 1.03 |
ENST00000545622.1
|
TMEM200A
|
transmembrane protein 200A |
| chr1_+_196743912 | 1.02 |
ENST00000367425.4
|
CFHR3
|
complement factor H-related 3 |
| chr3_+_186383741 | 1.02 |
ENST00000232003.4
|
HRG
|
histidine-rich glycoprotein |
| chr3_+_46395219 | 1.01 |
ENST00000445132.2
ENST00000292301.4 |
CCR2
|
chemokine (C-C motif) receptor 2 |
| chr5_+_118668846 | 1.01 |
ENST00000513374.1
|
TNFAIP8
|
tumor necrosis factor, alpha-induced protein 8 |
| chr5_+_162887556 | 1.01 |
ENST00000393915.4
ENST00000432118.2 ENST00000358715.3 |
HMMR
|
hyaluronan-mediated motility receptor (RHAMM) |
| chr15_+_38226827 | 1.01 |
ENST00000559502.1
ENST00000558148.1 ENST00000558158.1 |
TMCO5A
|
transmembrane and coiled-coil domains 5A |
| chr6_+_116782527 | 1.01 |
ENST00000368606.3
ENST00000368605.1 |
FAM26F
|
family with sequence similarity 26, member F |
| chr12_+_20963632 | 0.99 |
ENST00000540853.1
ENST00000261196.2 |
SLCO1B3
|
solute carrier organic anion transporter family, member 1B3 |
| chr6_-_32784687 | 0.98 |
ENST00000447394.1
ENST00000438763.2 |
HLA-DOB
|
major histocompatibility complex, class II, DO beta |
| chr2_-_69180012 | 0.97 |
ENST00000481498.1
|
GKN2
|
gastrokine 2 |
| chr11_-_104905840 | 0.97 |
ENST00000526568.1
ENST00000393136.4 ENST00000531166.1 ENST00000534497.1 ENST00000527979.1 ENST00000446369.1 ENST00000353247.5 ENST00000528974.1 ENST00000533400.1 ENST00000525825.1 ENST00000436863.3 |
CASP1
|
caspase 1, apoptosis-related cysteine peptidase |
| chr12_-_10605929 | 0.96 |
ENST00000347831.5
ENST00000359151.3 |
KLRC1
|
killer cell lectin-like receptor subfamily C, member 1 |
| chr15_-_31521567 | 0.96 |
ENST00000560812.1
ENST00000559853.1 ENST00000558109.1 |
RP11-16E12.2
|
RP11-16E12.2 |
| chr16_+_56995854 | 0.95 |
ENST00000566128.1
|
CETP
|
cholesteryl ester transfer protein, plasma |
| chr4_-_42154895 | 0.94 |
ENST00000502486.1
ENST00000504360.1 |
BEND4
|
BEN domain containing 4 |
| chr16_-_54882145 | 0.94 |
ENST00000561591.1
|
RP11-1136G4.1
|
RP11-1136G4.1 |
| chr11_+_827248 | 0.93 |
ENST00000527089.1
ENST00000530183.1 |
EFCAB4A
|
EF-hand calcium binding domain 4A |
| chrX_+_12924732 | 0.93 |
ENST00000218032.6
ENST00000311912.5 |
TLR8
|
toll-like receptor 8 |
| chr4_+_70146217 | 0.93 |
ENST00000335568.5
ENST00000511240.1 |
UGT2B28
|
UDP glucuronosyltransferase 2 family, polypeptide B28 |
| chr14_-_92247032 | 0.93 |
ENST00000556661.1
ENST00000553676.1 ENST00000554560.1 |
CATSPERB
|
catsper channel auxiliary subunit beta |
| chr1_+_109256067 | 0.93 |
ENST00000271311.2
|
FNDC7
|
fibronectin type III domain containing 7 |
| chr3_+_186330712 | 0.92 |
ENST00000411641.2
ENST00000273784.5 |
AHSG
|
alpha-2-HS-glycoprotein |
| chr6_-_26216872 | 0.92 |
ENST00000244601.3
|
HIST1H2BG
|
histone cluster 1, H2bg |
Network of associatons between targets according to the STRING database.
First level regulatory network of IRX3
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 4.9 | GO:0019442 | tryptophan catabolic process to acetyl-CoA(GO:0019442) |
| 1.0 | 3.1 | GO:0001868 | regulation of complement activation, lectin pathway(GO:0001868) negative regulation of complement activation, lectin pathway(GO:0001869) |
| 1.0 | 3.0 | GO:0002818 | intracellular defense response(GO:0002818) |
| 0.7 | 2.1 | GO:0046005 | positive regulation of circadian sleep/wake cycle, REM sleep(GO:0046005) |
| 0.7 | 2.1 | GO:0032829 | regulation of CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0032829) positive regulation of CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0032831) |
| 0.7 | 2.0 | GO:2000502 | negative regulation of natural killer cell chemotaxis(GO:2000502) |
| 0.6 | 1.9 | GO:0006711 | estrogen catabolic process(GO:0006711) |
| 0.5 | 3.6 | GO:2000501 | regulation of natural killer cell chemotaxis(GO:2000501) positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.5 | 1.5 | GO:0060279 | positive regulation of ovulation(GO:0060279) |
| 0.5 | 3.0 | GO:0061107 | seminal vesicle development(GO:0061107) |
| 0.5 | 7.7 | GO:0045059 | positive thymic T cell selection(GO:0045059) |
| 0.5 | 0.9 | GO:2001183 | negative regulation of interleukin-12 secretion(GO:2001183) |
| 0.4 | 1.7 | GO:0000912 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) |
| 0.4 | 4.7 | GO:1902715 | positive regulation of interferon-gamma secretion(GO:1902715) |
| 0.4 | 72.6 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.4 | 4.8 | GO:0048003 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.4 | 1.1 | GO:0002372 | myeloid dendritic cell cytokine production(GO:0002372) |
| 0.4 | 4.5 | GO:0097267 | omega-hydroxylase P450 pathway(GO:0097267) |
| 0.3 | 1.4 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.3 | 4.9 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.3 | 1.0 | GO:2001027 | negative regulation of endothelial cell chemotaxis(GO:2001027) |
| 0.3 | 1.0 | GO:2000451 | astrocyte chemotaxis(GO:0035700) positive regulation of immune complex clearance by monocytes and macrophages(GO:0090265) positive regulation of T cell extravasation(GO:2000409) positive regulation of CD8-positive, alpha-beta T cell extravasation(GO:2000451) regulation of astrocyte chemotaxis(GO:2000458) |
| 0.3 | 1.0 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) |
| 0.3 | 2.7 | GO:0044800 | fusion of virus membrane with host plasma membrane(GO:0019064) membrane fusion involved in viral entry into host cell(GO:0039663) multi-organism membrane fusion(GO:0044800) |
| 0.3 | 6.2 | GO:1900017 | positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.3 | 2.5 | GO:2000660 | negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.3 | 0.8 | GO:0043105 | regulation of GTP cyclohydrolase I activity(GO:0043095) negative regulation of GTP cyclohydrolase I activity(GO:0043105) |
| 0.3 | 1.3 | GO:0030070 | insulin processing(GO:0030070) |
| 0.3 | 0.8 | GO:0046167 | glycerol-3-phosphate biosynthetic process(GO:0046167) |
| 0.3 | 4.7 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.3 | 0.8 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.3 | 1.5 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.3 | 2.8 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.2 | 0.2 | GO:0046136 | positive regulation of vitamin metabolic process(GO:0046136) positive regulation of vitamin D biosynthetic process(GO:0060557) positive regulation of calcidiol 1-monooxygenase activity(GO:0060559) |
| 0.2 | 0.7 | GO:0050787 | antibiotic metabolic process(GO:0016999) detoxification of mercury ion(GO:0050787) |
| 0.2 | 1.2 | GO:0006015 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.2 | 1.2 | GO:0038043 | interleukin-5-mediated signaling pathway(GO:0038043) interleukin-3-mediated signaling pathway(GO:0038156) |
| 0.2 | 5.4 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.2 | 5.6 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.2 | 1.3 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.2 | 0.6 | GO:0001810 | regulation of type I hypersensitivity(GO:0001810) positive regulation of type I hypersensitivity(GO:0001812) type I hypersensitivity(GO:0016068) |
| 0.2 | 1.0 | GO:0010513 | positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) |
| 0.2 | 0.8 | GO:0045726 | positive regulation of integrin biosynthetic process(GO:0045726) |
| 0.2 | 3.0 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.2 | 0.6 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.2 | 0.8 | GO:0002740 | negative regulation of cytokine secretion involved in immune response(GO:0002740) |
| 0.2 | 0.6 | GO:1903925 | signal transduction involved in intra-S DNA damage checkpoint(GO:0072428) response to bisphenol A(GO:1903925) cellular response to bisphenol A(GO:1903926) |
| 0.2 | 0.6 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.2 | 4.0 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.2 | 1.6 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.2 | 1.1 | GO:1900623 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.2 | 0.3 | GO:0032765 | positive regulation of mast cell cytokine production(GO:0032765) |
| 0.2 | 0.9 | GO:0034196 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.2 | 0.6 | GO:0046379 | extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.1 | 0.4 | GO:1903414 | spleen trabecula formation(GO:0060345) iron cation export(GO:1903414) ferrous iron export(GO:1903988) |
| 0.1 | 0.7 | GO:0039516 | modulation by virus of host molecular function(GO:0039506) suppression by virus of host molecular function(GO:0039507) suppression by virus of host catalytic activity(GO:0039513) modulation by virus of host catalytic activity(GO:0039516) suppression by virus of host cysteine-type endopeptidase activity involved in apoptotic process(GO:0039650) negative regulation by symbiont of host catalytic activity(GO:0052053) negative regulation by symbiont of host molecular function(GO:0052056) modulation by symbiont of host catalytic activity(GO:0052148) |
| 0.1 | 1.0 | GO:0007079 | mitotic chromosome movement towards spindle pole(GO:0007079) |
| 0.1 | 5.1 | GO:0045730 | respiratory burst(GO:0045730) |
| 0.1 | 0.3 | GO:0002503 | peptide antigen assembly with MHC class II protein complex(GO:0002503) |
| 0.1 | 0.8 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.1 | 0.4 | GO:1901291 | negative regulation of double-strand break repair via single-strand annealing(GO:1901291) |
| 0.1 | 0.5 | GO:0035425 | autocrine signaling(GO:0035425) |
| 0.1 | 0.5 | GO:0060298 | positive regulation of sarcomere organization(GO:0060298) |
| 0.1 | 0.3 | GO:0021503 | neural fold bending(GO:0021503) |
| 0.1 | 0.7 | GO:0050916 | sensory perception of sweet taste(GO:0050916) |
| 0.1 | 0.5 | GO:0070171 | negative regulation of tooth mineralization(GO:0070171) |
| 0.1 | 2.6 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.1 | 0.5 | GO:0036509 | trimming of terminal mannose on B branch(GO:0036509) trimming of first mannose on A branch(GO:0036511) trimming of second mannose on A branch(GO:0036512) |
| 0.1 | 1.8 | GO:0002455 | humoral immune response mediated by circulating immunoglobulin(GO:0002455) |
| 0.1 | 1.8 | GO:0006662 | glycerol ether metabolic process(GO:0006662) |
| 0.1 | 1.0 | GO:0042904 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.1 | 0.4 | GO:0050883 | negative regulation of sodium:proton antiporter activity(GO:0032416) musculoskeletal movement, spinal reflex action(GO:0050883) |
| 0.1 | 0.4 | GO:1902161 | positive regulation of cyclic nucleotide-gated ion channel activity(GO:1902161) |
| 0.1 | 1.3 | GO:0002291 | T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:0002291) |
| 0.1 | 1.1 | GO:2000623 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.1 | 18.9 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.1 | 0.8 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.1 | 1.5 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.1 | 0.3 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.1 | 2.2 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.1 | 1.6 | GO:0034356 | NAD biosynthesis via nicotinamide riboside salvage pathway(GO:0034356) |
| 0.1 | 1.9 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 0.1 | 0.6 | GO:0035690 | cellular response to drug(GO:0035690) |
| 0.1 | 1.7 | GO:0020027 | hemoglobin metabolic process(GO:0020027) |
| 0.1 | 0.8 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.1 | 1.9 | GO:0070508 | sterol import(GO:0035376) cholesterol import(GO:0070508) |
| 0.1 | 0.6 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.1 | 0.7 | GO:0006880 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.1 | 0.4 | GO:0033686 | positive regulation of luteinizing hormone secretion(GO:0033686) |
| 0.1 | 0.1 | GO:0032886 | regulation of microtubule-based process(GO:0032886) |
| 0.1 | 0.4 | GO:0080154 | regulation of fertilization(GO:0080154) |
| 0.1 | 0.1 | GO:0031346 | positive regulation of cell projection organization(GO:0031346) |
| 0.1 | 0.7 | GO:0006939 | smooth muscle contraction(GO:0006939) |
| 0.1 | 0.9 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.1 | 0.5 | GO:0002767 | immune response-inhibiting cell surface receptor signaling pathway(GO:0002767) |
| 0.1 | 6.0 | GO:0042267 | natural killer cell mediated cytotoxicity(GO:0042267) |
| 0.1 | 0.6 | GO:2001206 | positive regulation of osteoclast development(GO:2001206) |
| 0.1 | 0.5 | GO:0048241 | epinephrine transport(GO:0048241) |
| 0.1 | 1.0 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.1 | 0.3 | GO:0016062 | adaptation of rhodopsin mediated signaling(GO:0016062) light adaption(GO:0036367) |
| 0.1 | 0.6 | GO:0070269 | pyroptosis(GO:0070269) |
| 0.1 | 0.9 | GO:0010886 | positive regulation of cholesterol storage(GO:0010886) |
| 0.1 | 0.5 | GO:0051712 | positive regulation of killing of cells of other organism(GO:0051712) |
| 0.1 | 0.2 | GO:0044537 | regulation of circulating fibrinogen levels(GO:0044537) |
| 0.1 | 0.7 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.1 | 0.2 | GO:0021965 | spinal cord ventral commissure morphogenesis(GO:0021965) |
| 0.1 | 0.5 | GO:0098706 | ferric iron import into cell(GO:0097461) ferric iron import across plasma membrane(GO:0098706) |
| 0.1 | 0.5 | GO:0043631 | mRNA polyadenylation(GO:0006378) RNA polyadenylation(GO:0043631) |
| 0.1 | 0.3 | GO:0010751 | negative regulation of nitric oxide mediated signal transduction(GO:0010751) |
| 0.1 | 0.7 | GO:0043950 | positive regulation of cAMP-mediated signaling(GO:0043950) |
| 0.1 | 1.1 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.1 | 0.2 | GO:0044205 | 'de novo' UMP biosynthetic process(GO:0044205) |
| 0.1 | 0.3 | GO:0010966 | regulation of phosphate transport(GO:0010966) |
| 0.0 | 0.1 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 0.0 | 0.7 | GO:0031584 | activation of phospholipase D activity(GO:0031584) |
| 0.0 | 0.9 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.0 | 0.2 | GO:0060467 | egg activation(GO:0007343) negative regulation of fertilization(GO:0060467) |
| 0.0 | 0.4 | GO:0003383 | apical constriction(GO:0003383) |
| 0.0 | 0.6 | GO:0042723 | thiamine-containing compound metabolic process(GO:0042723) |
| 0.0 | 0.2 | GO:0061358 | negative regulation of Wnt protein secretion(GO:0061358) |
| 0.0 | 2.2 | GO:0007194 | negative regulation of adenylate cyclase activity(GO:0007194) |
| 0.0 | 0.7 | GO:0015747 | urate transport(GO:0015747) |
| 0.0 | 0.6 | GO:0043383 | negative T cell selection(GO:0043383) |
| 0.0 | 0.2 | GO:0070173 | regulation of enamel mineralization(GO:0070173) |
| 0.0 | 1.0 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.0 | 1.0 | GO:0045724 | nonmotile primary cilium assembly(GO:0035058) positive regulation of cilium assembly(GO:0045724) regulation of nonmotile primary cilium assembly(GO:1902855) positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.0 | 0.3 | GO:0071550 | death-inducing signaling complex assembly(GO:0071550) |
| 0.0 | 0.6 | GO:0032494 | response to peptidoglycan(GO:0032494) |
| 0.0 | 0.5 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 2.0 | GO:0030155 | regulation of cell adhesion(GO:0030155) |
| 0.0 | 1.1 | GO:0010248 | establishment or maintenance of transmembrane electrochemical gradient(GO:0010248) |
| 0.0 | 0.6 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.0 | 0.4 | GO:0060372 | regulation of atrial cardiac muscle cell membrane repolarization(GO:0060372) |
| 0.0 | 0.8 | GO:0070129 | regulation of mitochondrial translation(GO:0070129) |
| 0.0 | 1.3 | GO:0030449 | regulation of complement activation(GO:0030449) |
| 0.0 | 0.7 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.0 | 0.0 | GO:0002921 | negative regulation of humoral immune response(GO:0002921) |
| 0.0 | 0.5 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.0 | 0.1 | GO:0018002 | N-terminal peptidyl-serine acetylation(GO:0017198) N-terminal peptidyl-glutamic acid acetylation(GO:0018002) peptidyl-serine acetylation(GO:0030920) |
| 0.0 | 1.6 | GO:0043029 | T cell homeostasis(GO:0043029) |
| 0.0 | 1.3 | GO:0000042 | protein targeting to Golgi(GO:0000042) |
| 0.0 | 1.1 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.4 | GO:0035986 | senescence-associated heterochromatin focus assembly(GO:0035986) |
| 0.0 | 0.4 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 0.2 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.0 | 0.6 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 0.0 | 0.3 | GO:0070887 | cellular response to chemical stimulus(GO:0070887) |
| 0.0 | 0.2 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 0.0 | 0.5 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.5 | GO:0015684 | ferrous iron transport(GO:0015684) ferrous iron transmembrane transport(GO:1903874) |
| 0.0 | 1.3 | GO:0034080 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.0 | 0.9 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.0 | 0.2 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.0 | 0.4 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 | 1.4 | GO:0071353 | cellular response to interleukin-4(GO:0071353) |
| 0.0 | 2.3 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.2 | GO:1990481 | mRNA pseudouridine synthesis(GO:1990481) |
| 0.0 | 0.9 | GO:0050908 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 | 2.4 | GO:0045576 | mast cell activation(GO:0045576) |
| 0.0 | 0.3 | GO:0060174 | limb bud formation(GO:0060174) |
| 0.0 | 0.6 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 0.0 | 0.7 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.0 | 0.3 | GO:0009117 | nucleoside phosphate metabolic process(GO:0006753) nucleotide metabolic process(GO:0009117) |
| 0.0 | 1.5 | GO:0002755 | MyD88-dependent toll-like receptor signaling pathway(GO:0002755) |
| 0.0 | 1.2 | GO:0051895 | negative regulation of focal adhesion assembly(GO:0051895) |
| 0.0 | 0.8 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.0 | 0.3 | GO:0071169 | establishment of protein localization to chromatin(GO:0071169) |
| 0.0 | 0.5 | GO:0045838 | positive regulation of membrane potential(GO:0045838) |
| 0.0 | 0.3 | GO:0045351 | type I interferon biosynthetic process(GO:0045351) |
| 0.0 | 0.3 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.0 | 1.8 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 0.0 | 0.4 | GO:0030889 | negative regulation of B cell proliferation(GO:0030889) |
| 0.0 | 0.4 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.0 | 0.2 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.0 | 0.2 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.0 | 0.5 | GO:0046602 | regulation of mitotic centrosome separation(GO:0046602) |
| 0.0 | 0.2 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) |
| 0.0 | 0.5 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.1 | GO:0045903 | positive regulation of translational fidelity(GO:0045903) |
| 0.0 | 0.9 | GO:0006907 | pinocytosis(GO:0006907) |
| 0.0 | 0.4 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.0 | 0.7 | GO:0014850 | response to muscle activity(GO:0014850) |
| 0.0 | 0.3 | GO:0022417 | protein maturation by protein folding(GO:0022417) |
| 0.0 | 0.2 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 0.0 | 1.2 | GO:0046676 | negative regulation of insulin secretion(GO:0046676) |
| 0.0 | 0.6 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.0 | 0.4 | GO:0031937 | positive regulation of chromatin silencing(GO:0031937) |
| 0.0 | 2.4 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.0 | 3.2 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 | 0.9 | GO:0051482 | positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G-protein coupled signaling pathway(GO:0051482) |
| 0.0 | 1.9 | GO:0001824 | blastocyst development(GO:0001824) |
| 0.0 | 0.8 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.5 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.0 | 0.3 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.0 | 0.5 | GO:0097186 | amelogenesis(GO:0097186) |
| 0.0 | 0.2 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.0 | 0.7 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.7 | GO:0051281 | positive regulation of release of sequestered calcium ion into cytosol(GO:0051281) |
| 0.0 | 0.8 | GO:0032402 | melanosome transport(GO:0032402) |
| 0.0 | 0.1 | GO:0048749 | compound eye development(GO:0048749) |
| 0.0 | 0.2 | GO:1904251 | regulation of bile acid biosynthetic process(GO:0070857) regulation of bile acid metabolic process(GO:1904251) |
| 0.0 | 0.2 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.5 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.6 | GO:0007398 | ectoderm development(GO:0007398) |
| 0.0 | 0.1 | GO:0030581 | intracellular transport of viral protein in host cell(GO:0019060) symbiont intracellular protein transport in host(GO:0030581) intracellular protein transport in other organism involved in symbiotic interaction(GO:0051708) |
| 0.0 | 0.3 | GO:0035640 | exploration behavior(GO:0035640) |
| 0.0 | 0.3 | GO:0051256 | mitotic spindle midzone assembly(GO:0051256) |
| 0.0 | 1.0 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.0 | 0.2 | GO:0002052 | positive regulation of neuroblast proliferation(GO:0002052) |
| 0.0 | 0.2 | GO:0034505 | tooth mineralization(GO:0034505) |
| 0.0 | 0.5 | GO:1903214 | regulation of protein targeting to mitochondrion(GO:1903214) positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.0 | 0.9 | GO:2000816 | negative regulation of mitotic sister chromatid separation(GO:2000816) |
| 0.0 | 0.3 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 0.1 | GO:0060694 | negative regulation of response to interferon-gamma(GO:0060331) negative regulation of interferon-gamma-mediated signaling pathway(GO:0060336) regulation of cholesterol transporter activity(GO:0060694) cellular response to vitamin E(GO:0071306) pancreatic stellate cell proliferation(GO:0072343) response to metformin(GO:1901558) regulation of pancreatic stellate cell proliferation(GO:2000229) negative regulation of pancreatic stellate cell proliferation(GO:2000230) |
| 0.0 | 0.2 | GO:0031629 | synaptic vesicle fusion to presynaptic active zone membrane(GO:0031629) vesicle fusion to plasma membrane(GO:0099500) |
| 0.0 | 1.1 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.0 | 0.4 | GO:0030947 | regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030947) |
| 0.0 | 0.2 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.0 | 1.5 | GO:0002250 | adaptive immune response(GO:0002250) |
| 0.0 | 0.3 | GO:0045824 | negative regulation of innate immune response(GO:0045824) |
| 0.0 | 0.2 | GO:0008217 | regulation of blood pressure(GO:0008217) |
| 0.0 | 0.6 | GO:0001701 | in utero embryonic development(GO:0001701) |
| 0.0 | 0.1 | GO:1902075 | cellular response to salt(GO:1902075) response to sodium arsenite(GO:1903935) cellular response to sodium arsenite(GO:1903936) |
| 0.0 | 0.6 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 0.6 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.0 | 0.5 | GO:0051156 | glucose 6-phosphate metabolic process(GO:0051156) |
| 0.0 | 0.4 | GO:0042113 | B cell activation(GO:0042113) |
| 0.0 | 0.1 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.0 | 0.1 | GO:2000078 | glandular epithelial cell maturation(GO:0002071) type B pancreatic cell maturation(GO:0072560) positive regulation of type B pancreatic cell development(GO:2000078) |
| 0.0 | 0.4 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 0.1 | GO:0033504 | floor plate development(GO:0033504) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.0 | GO:0036026 | protein C inhibitor-TMPRSS7 complex(GO:0036024) protein C inhibitor-TMPRSS11E complex(GO:0036025) protein C inhibitor-PLAT complex(GO:0036026) protein C inhibitor-PLAU complex(GO:0036027) protein C inhibitor-thrombin complex(GO:0036028) protein C inhibitor-KLK3 complex(GO:0036029) protein C inhibitor-plasma kallikrein complex(GO:0036030) serine protease inhibitor complex(GO:0097180) protein C inhibitor-coagulation factor V complex(GO:0097181) protein C inhibitor-coagulation factor Xa complex(GO:0097182) protein C inhibitor-coagulation factor XI complex(GO:0097183) |
| 0.6 | 7.7 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.6 | 29.7 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.5 | 1.5 | GO:0043511 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.5 | 1.4 | GO:0097179 | protease inhibitor complex(GO:0097179) |
| 0.5 | 1.4 | GO:0033150 | cytoskeletal calyx(GO:0033150) |
| 0.4 | 3.0 | GO:0097013 | phagocytic vesicle lumen(GO:0097013) |
| 0.4 | 1.2 | GO:0030526 | granulocyte macrophage colony-stimulating factor receptor complex(GO:0030526) |
| 0.3 | 0.9 | GO:0030895 | apolipoprotein B mRNA editing enzyme complex(GO:0030895) |
| 0.3 | 1.2 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.3 | 1.7 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 0.3 | 1.1 | GO:0005889 | hydrogen:potassium-exchanging ATPase complex(GO:0005889) |
| 0.2 | 6.3 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.2 | 1.1 | GO:0005602 | complement component C1 complex(GO:0005602) |
| 0.2 | 3.5 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.2 | 1.3 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.2 | 0.6 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.2 | 0.5 | GO:0097679 | other organism cytoplasm(GO:0097679) |
| 0.2 | 1.5 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.1 | 1.1 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.1 | 4.8 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.1 | 1.5 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.1 | 1.3 | GO:0034687 | integrin alphaL-beta2 complex(GO:0034687) |
| 0.1 | 0.6 | GO:0031905 | early endosome lumen(GO:0031905) |
| 0.1 | 0.5 | GO:0034680 | integrin alpha10-beta1 complex(GO:0034680) |
| 0.1 | 0.9 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.1 | 1.2 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.1 | 18.4 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.1 | 1.8 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.1 | 1.3 | GO:0045179 | apical cortex(GO:0045179) |
| 0.1 | 0.2 | GO:0000806 | Y chromosome(GO:0000806) |
| 0.1 | 0.4 | GO:0002081 | outer acrosomal membrane(GO:0002081) |
| 0.1 | 0.6 | GO:0072557 | IPAF inflammasome complex(GO:0072557) |
| 0.1 | 1.2 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.1 | 1.0 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.1 | 0.7 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.1 | 0.4 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.1 | 0.7 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.1 | 1.0 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.1 | 0.3 | GO:1990425 | ryanodine receptor complex(GO:1990425) |
| 0.1 | 0.8 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.1 | 0.5 | GO:0043196 | varicosity(GO:0043196) |
| 0.1 | 3.1 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.1 | 0.5 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.1 | 0.6 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.0 | 0.1 | GO:0005595 | collagen type XII trimer(GO:0005595) |
| 0.0 | 1.1 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.8 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.8 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.3 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.0 | 0.9 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 0.5 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.4 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.0 | 1.1 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.9 | GO:0036020 | endolysosome membrane(GO:0036020) |
| 0.0 | 0.2 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.0 | 2.0 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.8 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.0 | 0.2 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.7 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) ripoptosome(GO:0097342) |
| 0.0 | 1.0 | GO:0031105 | septin complex(GO:0031105) |
| 0.0 | 0.1 | GO:0036502 | Derlin-1-VIMP complex(GO:0036502) |
| 0.0 | 1.5 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 7.8 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 0.4 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 3.0 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.0 | 0.3 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.0 | 0.3 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.3 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.0 | 0.1 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 0.0 | 0.5 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 3.1 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 0.4 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.0 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.0 | 0.6 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.0 | 0.3 | GO:0060091 | kinocilium(GO:0060091) |
| 0.0 | 0.1 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.0 | 0.1 | GO:0031415 | NatA complex(GO:0031415) |
| 0.0 | 1.2 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 34.2 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 0.9 | GO:0097546 | ciliary base(GO:0097546) |
| 0.0 | 3.9 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.5 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.2 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.5 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 1.9 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
| 0.0 | 0.5 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 2.2 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.3 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 0.8 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 1.1 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.2 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 1.6 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 1.0 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.3 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.9 | GO:0043235 | receptor complex(GO:0043235) |
| 0.0 | 0.8 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 1.4 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 0.2 | GO:0043220 | Schmidt-Lanterman incisure(GO:0043220) |
| 0.0 | 0.1 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.0 | 0.6 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 0.1 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.0 | 0.2 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.0 | 0.2 | GO:0005771 | multivesicular body(GO:0005771) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.3 | GO:0036134 | thromboxane-A synthase activity(GO:0004796) 12-hydroxyheptadecatrienoic acid synthase activity(GO:0036134) |
| 0.8 | 4.0 | GO:0033695 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 0.7 | 2.8 | GO:0004917 | interleukin-7 receptor activity(GO:0004917) |
| 0.7 | 6.0 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.6 | 3.4 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.5 | 3.6 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.5 | 1.5 | GO:0005150 | interleukin-1, Type I receptor binding(GO:0005150) |
| 0.5 | 4.8 | GO:0030884 | lipid antigen binding(GO:0030882) endogenous lipid antigen binding(GO:0030883) exogenous lipid antigen binding(GO:0030884) |
| 0.5 | 29.7 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.4 | 1.3 | GO:0051908 | double-stranded DNA 5'-3' exodeoxyribonuclease activity(GO:0051908) |
| 0.4 | 2.6 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.3 | 1.4 | GO:0030760 | nicotinamide N-methyltransferase activity(GO:0008112) pyridine N-methyltransferase activity(GO:0030760) |
| 0.3 | 3.2 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.3 | 59.5 | GO:0003823 | antigen binding(GO:0003823) |
| 0.3 | 1.7 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.3 | 1.1 | GO:0008900 | hydrogen:potassium-exchanging ATPase activity(GO:0008900) |
| 0.3 | 0.8 | GO:0044549 | GTP cyclohydrolase binding(GO:0044549) |
| 0.3 | 0.8 | GO:0004370 | glycerol kinase activity(GO:0004370) |
| 0.3 | 1.8 | GO:0043120 | tumor necrosis factor binding(GO:0043120) |
| 0.3 | 1.5 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.3 | 1.5 | GO:0016822 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.2 | 0.7 | GO:0004947 | bradykinin receptor activity(GO:0004947) |
| 0.2 | 2.2 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.2 | 0.9 | GO:0017129 | triglyceride binding(GO:0017129) |
| 0.2 | 1.2 | GO:0004914 | interleukin-3 receptor activity(GO:0004912) interleukin-5 receptor activity(GO:0004914) |
| 0.2 | 1.1 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.2 | 1.5 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.2 | 1.0 | GO:0004910 | interleukin-1, Type II, blocking receptor activity(GO:0004910) |
| 0.2 | 1.5 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.2 | 2.7 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.2 | 1.3 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) steroid sulfotransferase activity(GO:0050294) |
| 0.2 | 0.7 | GO:0048030 | disaccharide binding(GO:0048030) |
| 0.2 | 0.7 | GO:0050309 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.2 | 3.0 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.2 | 2.0 | GO:0032393 | MHC class I receptor activity(GO:0032393) |
| 0.2 | 0.6 | GO:0010348 | lithium:proton antiporter activity(GO:0010348) |
| 0.1 | 0.4 | GO:0097689 | iron channel activity(GO:0097689) |
| 0.1 | 7.7 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 0.9 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.1 | 0.6 | GO:0035651 | AP-1 adaptor complex binding(GO:0035650) AP-3 adaptor complex binding(GO:0035651) |
| 0.1 | 0.7 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.1 | 0.8 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.1 | 0.4 | GO:0047017 | geranylgeranyl reductase activity(GO:0045550) prostaglandin-F synthase activity(GO:0047017) |
| 0.1 | 0.4 | GO:0034039 | 8-oxo-7,8-dihydroguanine DNA N-glycosylase activity(GO:0034039) |
| 0.1 | 1.2 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.1 | 1.8 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.1 | 0.6 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.1 | 0.5 | GO:0051431 | corticotropin-releasing hormone receptor 2 binding(GO:0051431) |
| 0.1 | 0.2 | GO:0017098 | sulfonylurea receptor binding(GO:0017098) |
| 0.1 | 0.5 | GO:0005277 | acetylcholine transmembrane transporter activity(GO:0005277) secondary active organic cation transmembrane transporter activity(GO:0008513) acetate ester transmembrane transporter activity(GO:1901375) |
| 0.1 | 1.3 | GO:0030369 | ICAM-3 receptor activity(GO:0030369) |
| 0.1 | 1.9 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.1 | 0.6 | GO:0019763 | immunoglobulin receptor activity(GO:0019763) |
| 0.1 | 0.7 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.1 | 0.8 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.1 | 0.5 | GO:0018685 | alkane 1-monooxygenase activity(GO:0018685) |
| 0.1 | 1.8 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.1 | 0.7 | GO:0016402 | pristanoyl-CoA oxidase activity(GO:0016402) |
| 0.1 | 0.4 | GO:0005260 | channel-conductance-controlling ATPase activity(GO:0005260) |
| 0.1 | 2.5 | GO:0045125 | bioactive lipid receptor activity(GO:0045125) |
| 0.1 | 0.3 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.1 | 0.5 | GO:0051916 | granulocyte colony-stimulating factor binding(GO:0051916) |
| 0.1 | 0.3 | GO:0019961 | interferon binding(GO:0019961) |
| 0.1 | 0.5 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.1 | 1.1 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.1 | 1.0 | GO:0043176 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.1 | 0.2 | GO:0070538 | oleic acid binding(GO:0070538) |
| 0.1 | 1.0 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.1 | 0.4 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.1 | 0.7 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.1 | 0.3 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 0.1 | 0.9 | GO:0043225 | anion transmembrane-transporting ATPase activity(GO:0043225) |
| 0.1 | 1.5 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.1 | 2.1 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.1 | 0.6 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.1 | 0.8 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 5.0 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.1 | 0.5 | GO:0008823 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.1 | 7.3 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.1 | 2.0 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.1 | 1.7 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.1 | 0.5 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.1 | 0.2 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.1 | 1.1 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.1 | 0.6 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.1 | 1.3 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.1 | 0.2 | GO:0015403 | thiamine uptake transmembrane transporter activity(GO:0015403) |
| 0.1 | 1.1 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.1 | 0.3 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.1 | 0.5 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.1 | 0.2 | GO:0051800 | phosphatidylinositol-3,4-bisphosphate 3-phosphatase activity(GO:0051800) |
| 0.1 | 0.5 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.1 | 0.5 | GO:0004873 | asialoglycoprotein receptor activity(GO:0004873) |
| 0.0 | 2.1 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 1.6 | GO:0008329 | signaling pattern recognition receptor activity(GO:0008329) pattern recognition receptor activity(GO:0038187) |
| 0.0 | 0.8 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.0 | 1.0 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 1.4 | GO:0001848 | complement binding(GO:0001848) |
| 0.0 | 0.2 | GO:0072545 | tyrosine binding(GO:0072545) |
| 0.0 | 0.6 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.0 | 0.1 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.1 | GO:0019862 | IgA binding(GO:0019862) |
| 0.0 | 0.2 | GO:0038047 | beta-endorphin receptor activity(GO:0004979) morphine receptor activity(GO:0038047) |
| 0.0 | 1.1 | GO:0008656 | cysteine-type endopeptidase activator activity involved in apoptotic process(GO:0008656) |
| 0.0 | 0.4 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.0 | 1.6 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.2 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.0 | 0.5 | GO:0033612 | receptor serine/threonine kinase binding(GO:0033612) |
| 0.0 | 0.7 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 0.2 | GO:0016635 | oxidoreductase activity, acting on the CH-CH group of donors, quinone or related compound as acceptor(GO:0016635) |
| 0.0 | 0.1 | GO:1990190 | peptide-glutamate-N-acetyltransferase activity(GO:1990190) |
| 0.0 | 0.5 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.6 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.4 | GO:0086007 | voltage-gated calcium channel activity involved in cardiac muscle cell action potential(GO:0086007) |
| 0.0 | 8.8 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 2.4 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.4 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.0 | 0.8 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.2 | GO:0017060 | 3-galactosyl-N-acetylglucosaminide 4-alpha-L-fucosyltransferase activity(GO:0017060) |
| 0.0 | 3.0 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.1 | GO:0061752 | telomeric repeat-containing RNA binding(GO:0061752) |
| 0.0 | 0.2 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.0 | 0.2 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.0 | 0.5 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.0 | 1.1 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.7 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.0 | 1.1 | GO:0004950 | G-protein coupled chemoattractant receptor activity(GO:0001637) chemokine receptor activity(GO:0004950) |
| 0.0 | 0.2 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 0.3 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 0.2 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.0 | 0.3 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.1 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.0 | 0.1 | GO:0005137 | interleukin-5 receptor binding(GO:0005137) |
| 0.0 | 2.9 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.3 | GO:0015377 | cation:chloride symporter activity(GO:0015377) |
| 0.0 | 0.7 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.4 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 0.4 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 1.2 | GO:0019825 | oxygen binding(GO:0019825) |
| 0.0 | 0.7 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.5 | GO:0015299 | solute:proton antiporter activity(GO:0015299) |
| 0.0 | 1.7 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 6.2 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 0.5 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.7 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 0.1 | GO:1990932 | 5.8S rRNA binding(GO:1990932) |
| 0.0 | 0.1 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.0 | 0.1 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.0 | 0.3 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.0 | 0.0 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.0 | 0.4 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 0.1 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 0.5 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.1 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.0 | 0.1 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 0.2 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.0 | 0.3 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.0 | 0.4 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.1 | GO:0030345 | extracellular matrix structural constituent conferring compression resistance(GO:0030021) structural constituent of tooth enamel(GO:0030345) |
| 0.0 | 0.2 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.1 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.0 | 1.0 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 2.6 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.8 | GO:0050840 | extracellular matrix binding(GO:0050840) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 9.0 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.1 | 3.4 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.1 | 1.3 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.1 | 1.2 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.1 | 4.4 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.1 | 0.4 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.0 | 1.6 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 1.2 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 1.4 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.5 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.0 | 1.5 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 2.2 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 2.3 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 2.2 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 2.0 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 2.6 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.0 | 1.3 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 10.2 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 2.4 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 0.2 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 1.8 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.7 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 1.7 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.1 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.0 | 6.3 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.8 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 0.3 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.4 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 0.6 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.8 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.0 | 1.0 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 0.9 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 1.0 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.0 | 0.7 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.0 | 0.6 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 1.8 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 1.0 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.2 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 0.6 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.6 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 0.2 | PID AURORA A PATHWAY | Aurora A signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.7 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.3 | 7.7 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.3 | 7.9 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.2 | 4.0 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.2 | 4.9 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.1 | 4.6 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.1 | 3.1 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.1 | 1.9 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.1 | 2.7 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.1 | 1.3 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.1 | 1.5 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.1 | 6.6 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.1 | 1.1 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.1 | 3.2 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.1 | 1.0 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.1 | 6.4 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.1 | 0.9 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.1 | 1.7 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.1 | 1.0 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.1 | 1.0 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.1 | 0.9 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.1 | 6.6 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.1 | 1.5 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.1 | 1.1 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 2.2 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.7 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.0 | 0.8 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.9 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 1.6 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.0 | 0.6 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.0 | 3.5 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 1.3 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 2.6 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 0.6 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 0.8 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.8 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 1.2 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.5 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 2.2 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.6 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.5 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.4 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.5 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.7 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 2.5 | REACTOME ACTIVATION OF THE MRNA UPON BINDING OF THE CAP BINDING COMPLEX AND EIFS AND SUBSEQUENT BINDING TO 43S | Genes involved in Activation of the mRNA upon binding of the cap-binding complex and eIFs, and subsequent binding to 43S |
| 0.0 | 1.0 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.7 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.2 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 0.4 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 0.5 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.3 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.0 | 1.8 | REACTOME 3 UTR MEDIATED TRANSLATIONAL REGULATION | Genes involved in 3' -UTR-mediated translational regulation |
| 0.0 | 0.3 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 1.2 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.0 | 0.3 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.9 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 0.8 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.3 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 1.4 | REACTOME REGULATION OF MITOTIC CELL CYCLE | Genes involved in Regulation of mitotic cell cycle |
| 0.0 | 0.3 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 1.8 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.3 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 0.5 | REACTOME POST NMDA RECEPTOR ACTIVATION EVENTS | Genes involved in Post NMDA receptor activation events |