Project
Illumina Body Map 2: averaged replicates
Navigation
Downloads
Results for IRX5
Z-value: 1.73
Transcription factors associated with IRX5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
IRX5
|
ENSG00000176842.10 | iroquois homeobox 5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| IRX5 | hg19_v2_chr16_+_54964740_54964789 | 0.09 | 6.2e-01 | Click! |
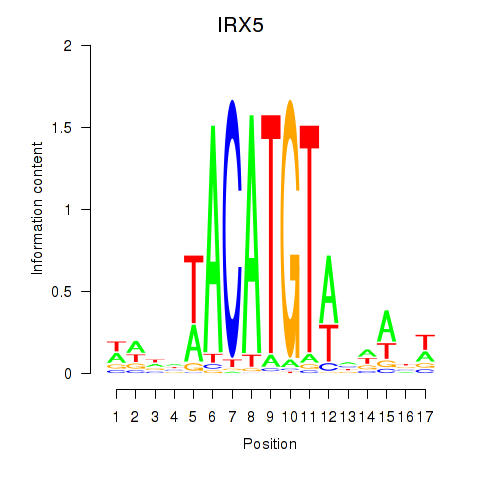
Activity profile of IRX5 motif
Sorted Z-values of IRX5 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_+_54901540 | 4.83 |
ENST00000539562.2
|
UNC13C
|
unc-13 homolog C (C. elegans) |
| chr3_+_35681728 | 4.07 |
ENST00000421492.1
ENST00000458225.1 ENST00000337271.5 |
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa |
| chr14_+_39944025 | 3.99 |
ENST00000554328.1
ENST00000556620.1 ENST00000557197.1 |
RP11-111A21.1
|
RP11-111A21.1 |
| chr7_+_123241908 | 3.88 |
ENST00000434204.1
ENST00000437535.1 ENST00000451215.1 |
ASB15
|
ankyrin repeat and SOCS box containing 15 |
| chr4_+_76995855 | 3.71 |
ENST00000355810.4
ENST00000349321.3 |
ART3
|
ADP-ribosyltransferase 3 |
| chr12_+_21679220 | 3.45 |
ENST00000256969.2
|
C12orf39
|
chromosome 12 open reading frame 39 |
| chr14_-_23876801 | 3.27 |
ENST00000356287.3
|
MYH6
|
myosin, heavy chain 6, cardiac muscle, alpha |
| chr14_-_23877474 | 2.94 |
ENST00000405093.3
|
MYH6
|
myosin, heavy chain 6, cardiac muscle, alpha |
| chr4_+_141264597 | 2.77 |
ENST00000338517.4
ENST00000394203.3 ENST00000506322.1 |
SCOC
|
short coiled-coil protein |
| chr7_+_54398390 | 2.68 |
ENST00000458615.1
ENST00000426205.1 |
RP11-436F9.1
|
RP11-436F9.1 |
| chr3_+_158787041 | 2.64 |
ENST00000471575.1
ENST00000476809.1 ENST00000485419.1 |
IQCJ-SCHIP1
|
IQCJ-SCHIP1 readthrough |
| chr1_-_169733789 | 2.62 |
ENST00000454271.1
ENST00000609271.1 |
RP1-117P20.3
SELE
|
RP1-117P20.3 selectin E |
| chr9_+_105757590 | 2.59 |
ENST00000374798.3
ENST00000487798.1 |
CYLC2
|
cylicin, basic protein of sperm head cytoskeleton 2 |
| chr1_+_248128535 | 2.55 |
ENST00000366480.3
|
OR2AK2
|
olfactory receptor, family 2, subfamily AK, member 2 |
| chr19_+_50691437 | 2.55 |
ENST00000598205.1
|
MYH14
|
myosin, heavy chain 14, non-muscle |
| chr14_-_47601041 | 2.53 |
ENST00000554762.1
|
MDGA2
|
MAM domain containing glycosylphosphatidylinositol anchor 2 |
| chr5_+_121187625 | 2.49 |
ENST00000321339.1
|
FTMT
|
ferritin mitochondrial |
| chr10_-_96829246 | 2.41 |
ENST00000371270.3
ENST00000535898.1 ENST00000539050.1 |
CYP2C8
|
cytochrome P450, family 2, subfamily C, polypeptide 8 |
| chr1_+_196857144 | 2.39 |
ENST00000367416.2
ENST00000251424.4 ENST00000367418.2 |
CFHR4
|
complement factor H-related 4 |
| chr20_+_10199566 | 2.20 |
ENST00000430336.1
|
SNAP25
|
synaptosomal-associated protein, 25kDa |
| chr12_+_126107042 | 2.20 |
ENST00000535886.1
|
TMEM132B
|
transmembrane protein 132B |
| chr3_+_156799587 | 2.17 |
ENST00000469196.1
|
RP11-6F2.5
|
RP11-6F2.5 |
| chr11_+_59807748 | 2.12 |
ENST00000278855.2
ENST00000532905.1 |
PLAC1L
|
oocyte secreted protein 2 |
| chr20_+_10199468 | 2.12 |
ENST00000254976.2
ENST00000304886.2 |
SNAP25
|
synaptosomal-associated protein, 25kDa |
| chr1_-_213020991 | 2.12 |
ENST00000332912.3
|
C1orf227
|
chromosome 1 open reading frame 227 |
| chr3_-_20053741 | 2.11 |
ENST00000389050.4
|
PP2D1
|
protein phosphatase 2C-like domain containing 1 |
| chr4_-_186392898 | 1.97 |
ENST00000510617.1
ENST00000307588.3 |
CCDC110
|
coiled-coil domain containing 110 |
| chr1_-_112106556 | 1.90 |
ENST00000443498.1
|
ADORA3
|
adenosine A3 receptor |
| chr7_-_99573640 | 1.90 |
ENST00000411734.1
|
AZGP1
|
alpha-2-glycoprotein 1, zinc-binding |
| chr12_+_24376201 | 1.86 |
ENST00000540733.1
ENST00000539583.1 |
RP11-778H2.1
|
RP11-778H2.1 |
| chr12_-_78934441 | 1.85 |
ENST00000546865.1
ENST00000547089.1 |
RP11-171L9.1
|
RP11-171L9.1 |
| chr1_-_112106578 | 1.84 |
ENST00000369717.4
|
ADORA3
|
adenosine A3 receptor |
| chr22_+_18721427 | 1.81 |
ENST00000342888.3
|
AC008132.1
|
Uncharacterized protein |
| chr6_-_49834240 | 1.79 |
ENST00000335847.4
|
CRISP1
|
cysteine-rich secretory protein 1 |
| chr19_-_50266580 | 1.78 |
ENST00000246801.3
|
TSKS
|
testis-specific serine kinase substrate |
| chr6_+_22569784 | 1.77 |
ENST00000510882.2
|
HDGFL1
|
hepatoma derived growth factor-like 1 |
| chr12_-_88423164 | 1.76 |
ENST00000298699.2
ENST00000550553.1 |
C12orf50
|
chromosome 12 open reading frame 50 |
| chrX_+_47863734 | 1.73 |
ENST00000304355.5
|
SPACA5
|
sperm acrosome associated 5 |
| chr7_-_99573677 | 1.72 |
ENST00000292401.4
|
AZGP1
|
alpha-2-glycoprotein 1, zinc-binding |
| chr6_-_49834209 | 1.71 |
ENST00000507853.1
|
CRISP1
|
cysteine-rich secretory protein 1 |
| chr4_+_114066764 | 1.67 |
ENST00000511380.1
|
ANK2
|
ankyrin 2, neuronal |
| chr3_+_186383741 | 1.65 |
ENST00000232003.4
|
HRG
|
histidine-rich glycoprotein |
| chr17_-_10450866 | 1.63 |
ENST00000578017.1
|
MYH2
|
myosin, heavy chain 2, skeletal muscle, adult |
| chr6_-_52705641 | 1.62 |
ENST00000370989.2
|
GSTA5
|
glutathione S-transferase alpha 5 |
| chr8_+_87878640 | 1.62 |
ENST00000518476.1
|
CNBD1
|
cyclic nucleotide binding domain containing 1 |
| chr6_-_50016364 | 1.61 |
ENST00000322246.4
|
DEFB112
|
defensin, beta 112 |
| chr8_-_17555164 | 1.60 |
ENST00000297488.6
|
MTUS1
|
microtubule associated tumor suppressor 1 |
| chr5_+_36608280 | 1.56 |
ENST00000513646.1
|
SLC1A3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3 |
| chr12_-_127174806 | 1.52 |
ENST00000545853.1
ENST00000537478.1 |
RP11-407A16.3
|
RP11-407A16.3 |
| chr6_+_22569678 | 1.49 |
ENST00000230012.3
|
HDGFL1
|
hepatoma derived growth factor-like 1 |
| chr6_+_29429217 | 1.48 |
ENST00000396792.2
|
OR2H1
|
olfactory receptor, family 2, subfamily H, member 1 |
| chr9_-_113100088 | 1.47 |
ENST00000374510.4
ENST00000423740.2 ENST00000374511.3 ENST00000374507.4 |
TXNDC8
|
thioredoxin domain containing 8 (spermatozoa) |
| chr7_+_129847688 | 1.46 |
ENST00000297819.3
|
SSMEM1
|
serine-rich single-pass membrane protein 1 |
| chrX_-_52258669 | 1.45 |
ENST00000441417.1
|
XAGE1A
|
X antigen family, member 1A |
| chr4_-_80329356 | 1.39 |
ENST00000358842.3
|
GK2
|
glycerol kinase 2 |
| chr16_-_54882145 | 1.39 |
ENST00000561591.1
|
RP11-1136G4.1
|
RP11-1136G4.1 |
| chr1_+_196912902 | 1.39 |
ENST00000476712.2
ENST00000367415.5 |
CFHR2
|
complement factor H-related 2 |
| chrX_+_52513455 | 1.38 |
ENST00000446098.1
|
XAGE1C
|
X antigen family, member 1C |
| chr8_-_74495065 | 1.30 |
ENST00000523533.1
|
STAU2
|
staufen double-stranded RNA binding protein 2 |
| chr3_+_108855558 | 1.30 |
ENST00000467240.1
ENST00000477643.1 ENST00000479039.1 ENST00000593799.1 |
RP11-59E19.1
|
RP11-59E19.1 |
| chr1_-_85870177 | 1.29 |
ENST00000542148.1
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1 |
| chr11_+_34999328 | 1.29 |
ENST00000526309.1
|
PDHX
|
pyruvate dehydrogenase complex, component X |
| chr17_-_67264947 | 1.27 |
ENST00000586811.1
|
ABCA5
|
ATP-binding cassette, sub-family A (ABC1), member 5 |
| chrX_-_15333736 | 1.25 |
ENST00000380470.3
|
ASB11
|
ankyrin repeat and SOCS box containing 11 |
| chr18_-_70305745 | 1.23 |
ENST00000581073.1
|
CBLN2
|
cerebellin 2 precursor |
| chr19_-_48059113 | 1.23 |
ENST00000391901.3
ENST00000314121.4 ENST00000448976.1 |
ZNF541
|
zinc finger protein 541 |
| chr4_+_185973026 | 1.22 |
ENST00000506338.1
ENST00000509017.1 |
RP11-386B13.3
|
RP11-386B13.3 |
| chr4_+_113739244 | 1.22 |
ENST00000503271.1
ENST00000503423.1 ENST00000506722.1 |
ANK2
|
ankyrin 2, neuronal |
| chr12_+_10365404 | 1.18 |
ENST00000266458.5
ENST00000421801.2 ENST00000544284.1 ENST00000545047.1 ENST00000543602.1 ENST00000545887.1 |
GABARAPL1
|
GABA(A) receptor-associated protein like 1 |
| chr3_+_186158134 | 1.17 |
ENST00000414102.1
|
RP11-78H24.1
|
RP11-78H24.1 |
| chr1_+_63989004 | 1.17 |
ENST00000371088.4
|
EFCAB7
|
EF-hand calcium binding domain 7 |
| chr4_+_41361616 | 1.15 |
ENST00000513024.1
|
LIMCH1
|
LIM and calponin homology domains 1 |
| chr1_-_186365908 | 1.15 |
ENST00000598663.1
|
AL596220.1
|
Uncharacterized protein |
| chr20_+_44330651 | 1.15 |
ENST00000305479.2
|
WFDC13
|
WAP four-disulfide core domain 13 |
| chrX_-_120066151 | 1.14 |
ENST00000419982.2
ENST00000416816.1 |
CT47A12
|
cancer/testis antigen family 47, member A12 |
| chrX_-_138790348 | 1.14 |
ENST00000414978.1
ENST00000519895.1 |
MCF2
|
MCF.2 cell line derived transforming sequence |
| chr1_-_152086556 | 1.13 |
ENST00000368804.1
|
TCHH
|
trichohyalin |
| chr12_+_2912363 | 1.13 |
ENST00000544366.1
|
FKBP4
|
FK506 binding protein 4, 59kDa |
| chr5_+_169758393 | 1.12 |
ENST00000521471.1
ENST00000518357.1 ENST00000436248.3 |
CTB-114C7.3
|
CTB-114C7.3 |
| chr20_+_38633006 | 1.11 |
ENST00000432633.1
ENST00000445606.1 |
RP11-101E14.2
|
RP11-101E14.2 |
| chr4_+_159131596 | 1.11 |
ENST00000512481.1
|
TMEM144
|
transmembrane protein 144 |
| chr11_-_35547572 | 1.10 |
ENST00000378880.2
|
PAMR1
|
peptidase domain containing associated with muscle regeneration 1 |
| chr5_+_128300810 | 1.10 |
ENST00000262462.4
|
SLC27A6
|
solute carrier family 27 (fatty acid transporter), member 6 |
| chr2_-_105372203 | 1.09 |
ENST00000424321.1
|
AC068057.2
|
long intergenic non-protein coding RNA 1114 |
| chr2_-_88427568 | 1.08 |
ENST00000393750.3
ENST00000295834.3 |
FABP1
|
fatty acid binding protein 1, liver |
| chrX_+_47867194 | 1.07 |
ENST00000376940.3
|
SPACA5
|
sperm acrosome associated 5 |
| chr4_+_144312659 | 1.07 |
ENST00000509992.1
|
GAB1
|
GRB2-associated binding protein 1 |
| chr1_+_240408560 | 1.05 |
ENST00000441342.1
ENST00000545751.1 |
FMN2
|
formin 2 |
| chr1_-_116383738 | 1.04 |
ENST00000320238.3
|
NHLH2
|
nescient helix loop helix 2 |
| chrX_-_120071012 | 1.04 |
ENST00000457020.2
|
CT47A11
|
cancer/testis antigen family 47, member A11 |
| chr5_-_36001108 | 1.03 |
ENST00000333811.4
|
UGT3A1
|
UDP glycosyltransferase 3 family, polypeptide A1 |
| chr6_+_53948221 | 1.03 |
ENST00000460844.2
|
MLIP
|
muscular LMNA-interacting protein |
| chr11_+_63057412 | 1.02 |
ENST00000544661.1
|
SLC22A10
|
solute carrier family 22, member 10 |
| chr12_-_120315074 | 1.02 |
ENST00000261833.7
ENST00000392521.2 |
CIT
|
citron (rho-interacting, serine/threonine kinase 21) |
| chr2_+_210517895 | 1.01 |
ENST00000447185.1
|
MAP2
|
microtubule-associated protein 2 |
| chr9_-_21166659 | 1.01 |
ENST00000380225.1
|
IFNA21
|
interferon, alpha 21 |
| chr1_-_85156417 | 1.01 |
ENST00000422026.1
|
SSX2IP
|
synovial sarcoma, X breakpoint 2 interacting protein |
| chr11_-_89653576 | 1.01 |
ENST00000420869.1
|
TRIM49D1
|
tripartite motif containing 49D1 |
| chr10_-_91403625 | 1.01 |
ENST00000322191.6
ENST00000342512.3 ENST00000371774.2 |
PANK1
|
pantothenate kinase 1 |
| chr3_+_68053359 | 1.00 |
ENST00000478136.1
|
FAM19A1
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A1 |
| chrY_+_23698778 | 1.00 |
ENST00000303902.5
|
RBMY1A1
|
RNA binding motif protein, Y-linked, family 1, member A1 |
| chr6_-_117747015 | 1.00 |
ENST00000368508.3
ENST00000368507.3 |
ROS1
|
c-ros oncogene 1 , receptor tyrosine kinase |
| chrX_-_33229636 | 0.99 |
ENST00000357033.4
|
DMD
|
dystrophin |
| chr12_+_8309630 | 0.99 |
ENST00000396570.3
|
ZNF705A
|
zinc finger protein 705A |
| chr20_-_3762087 | 0.97 |
ENST00000379756.3
|
SPEF1
|
sperm flagellar 1 |
| chr8_+_9009296 | 0.96 |
ENST00000521718.1
|
RP11-10A14.4
|
Uncharacterized protein |
| chr17_-_3301704 | 0.95 |
ENST00000322608.2
|
OR1E1
|
olfactory receptor, family 1, subfamily E, member 1 |
| chr5_+_140566 | 0.95 |
ENST00000502646.1
|
PLEKHG4B
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 4B |
| chr14_-_23623577 | 0.93 |
ENST00000422941.2
ENST00000453702.1 |
SLC7A8
|
solute carrier family 7 (amino acid transporter light chain, L system), member 8 |
| chrX_+_9935392 | 0.93 |
ENST00000445307.2
|
AC002365.1
|
Homo sapiens uncharacterized LOC100288814 (LOC100288814), mRNA. |
| chr5_+_175490540 | 0.92 |
ENST00000515817.1
|
FAM153B
|
family with sequence similarity 153, member B |
| chr14_+_56127960 | 0.92 |
ENST00000553624.1
|
KTN1
|
kinectin 1 (kinesin receptor) |
| chrX_+_52240504 | 0.92 |
ENST00000399805.2
|
XAGE1B
|
X antigen family, member 1B |
| chr12_+_58335360 | 0.91 |
ENST00000300145.3
|
XRCC6BP1
|
XRCC6 binding protein 1 |
| chr2_+_228735763 | 0.91 |
ENST00000373666.2
|
DAW1
|
dynein assembly factor with WDR repeat domains 1 |
| chr2_+_5866581 | 0.90 |
ENST00000413960.1
|
AC010729.2
|
AC010729.2 |
| chr15_+_66585555 | 0.90 |
ENST00000319194.5
ENST00000525134.2 ENST00000441424.2 |
DIS3L
|
DIS3 mitotic control homolog (S. cerevisiae)-like |
| chr12_-_58329819 | 0.90 |
ENST00000551421.1
|
RP11-620J15.3
|
RP11-620J15.3 |
| chr10_-_97175444 | 0.90 |
ENST00000486141.2
|
SORBS1
|
sorbin and SH3 domain containing 1 |
| chr9_+_96846740 | 0.89 |
ENST00000288976.3
|
PTPDC1
|
protein tyrosine phosphatase domain containing 1 |
| chr10_+_96698406 | 0.89 |
ENST00000260682.6
|
CYP2C9
|
cytochrome P450, family 2, subfamily C, polypeptide 9 |
| chr1_+_87012922 | 0.89 |
ENST00000263723.5
|
CLCA4
|
chloride channel accessory 4 |
| chr17_+_22022437 | 0.89 |
ENST00000540040.1
|
MTRNR2L1
|
MT-RNR2-like 1 |
| chr11_+_124134723 | 0.88 |
ENST00000524943.2
|
OR8G5
|
olfactory receptor, family 8, subfamily G, member 5 |
| chr18_-_46784778 | 0.88 |
ENST00000582399.1
|
DYM
|
dymeclin |
| chr11_-_1619524 | 0.87 |
ENST00000412090.1
|
KRTAP5-2
|
keratin associated protein 5-2 |
| chr15_-_80634126 | 0.87 |
ENST00000558244.1
|
LINC00927
|
long intergenic non-protein coding RNA 927 |
| chr2_-_39456673 | 0.87 |
ENST00000378803.1
ENST00000395035.3 |
CDKL4
|
cyclin-dependent kinase-like 4 |
| chrX_+_47990039 | 0.86 |
ENST00000304270.5
|
SPACA5B
|
sperm acrosome associated 5B |
| chr5_-_145895753 | 0.86 |
ENST00000311104.2
|
GPR151
|
G protein-coupled receptor 151 |
| chr1_-_21377383 | 0.86 |
ENST00000374935.3
|
EIF4G3
|
eukaryotic translation initiation factor 4 gamma, 3 |
| chr7_+_871559 | 0.85 |
ENST00000421580.1
|
SUN1
|
Sad1 and UNC84 domain containing 1 |
| chr3_+_173116225 | 0.84 |
ENST00000457714.1
|
NLGN1
|
neuroligin 1 |
| chr8_+_30496078 | 0.84 |
ENST00000517349.1
|
SMIM18
|
small integral membrane protein 18 |
| chr15_-_80634104 | 0.83 |
ENST00000561432.1
ENST00000558913.1 ENST00000558578.1 |
LINC00927
|
long intergenic non-protein coding RNA 927 |
| chr14_-_23624511 | 0.83 |
ENST00000529705.2
|
SLC7A8
|
solute carrier family 7 (amino acid transporter light chain, L system), member 8 |
| chr16_+_87636474 | 0.82 |
ENST00000284262.2
|
JPH3
|
junctophilin 3 |
| chr2_+_78143006 | 0.82 |
ENST00000443419.1
|
AC073628.1
|
AC073628.1 |
| chr10_-_23528745 | 0.82 |
ENST00000376501.5
|
C10orf115
|
chromosome 10 open reading frame 115 |
| chr17_+_58641908 | 0.81 |
ENST00000592917.1
|
RP11-15E18.4
|
Uncharacterized protein |
| chr4_+_159131630 | 0.81 |
ENST00000504569.1
ENST00000509278.1 ENST00000514558.1 ENST00000503200.1 |
TMEM144
|
transmembrane protein 144 |
| chr15_+_21145765 | 0.81 |
ENST00000553416.1
|
CT60
|
cancer/testis antigen 60 (non-protein coding) |
| chr2_+_35056422 | 0.81 |
ENST00000592523.1
ENST00000588944.1 ENST00000585391.1 ENST00000591221.1 ENST00000586769.1 ENST00000588650.1 |
AC012593.1
|
AC012593.1 |
| chr8_+_12803146 | 0.80 |
ENST00000447063.2
|
KIAA1456
|
KIAA1456 |
| chr11_+_55029628 | 0.79 |
ENST00000417545.2
|
TRIM48
|
tripartite motif containing 48 |
| chr15_-_64665911 | 0.79 |
ENST00000606793.1
ENST00000561349.1 ENST00000560278.1 |
CTD-2116N17.1
|
Uncharacterized protein |
| chr5_+_66675200 | 0.78 |
ENST00000503106.1
|
RP11-434D9.1
|
RP11-434D9.1 |
| chr17_+_58018269 | 0.78 |
ENST00000591035.1
|
RP11-178C3.1
|
Uncharacterized protein |
| chr3_+_81043000 | 0.78 |
ENST00000482617.1
|
RP11-6B4.1
|
RP11-6B4.1 |
| chr13_+_76362974 | 0.77 |
ENST00000497947.2
|
LMO7
|
LIM domain 7 |
| chr10_-_51958906 | 0.77 |
ENST00000489640.1
|
ASAH2
|
N-acylsphingosine amidohydrolase (non-lysosomal ceramidase) 2 |
| chr12_+_11187087 | 0.77 |
ENST00000601123.1
|
AC018630.1
|
Uncharacterized protein |
| chr3_+_155838337 | 0.76 |
ENST00000490337.1
ENST00000389636.5 |
KCNAB1
|
potassium voltage-gated channel, shaker-related subfamily, beta member 1 |
| chr1_-_89641680 | 0.76 |
ENST00000294671.2
|
GBP7
|
guanylate binding protein 7 |
| chr1_+_10509971 | 0.76 |
ENST00000320498.4
|
CORT
|
cortistatin |
| chr1_-_179457805 | 0.76 |
ENST00000600581.1
|
AL160286.1
|
Uncharacterized protein |
| chrY_-_24038660 | 0.76 |
ENST00000382677.3
|
RBMY1D
|
RNA binding motif protein, Y-linked, family 1, member D |
| chr2_+_38177620 | 0.76 |
ENST00000402091.3
|
RMDN2
|
regulator of microtubule dynamics 2 |
| chr2_-_183106641 | 0.74 |
ENST00000346717.4
|
PDE1A
|
phosphodiesterase 1A, calmodulin-dependent |
| chr8_+_120885949 | 0.74 |
ENST00000523492.1
ENST00000286234.5 |
DEPTOR
|
DEP domain containing MTOR-interacting protein |
| chr12_-_21556577 | 0.74 |
ENST00000450590.1
ENST00000453443.1 |
SLCO1A2
|
solute carrier organic anion transporter family, member 1A2 |
| chr7_+_6121296 | 0.73 |
ENST00000428901.1
|
AC004895.4
|
AC004895.4 |
| chr18_+_616672 | 0.73 |
ENST00000338387.7
|
CLUL1
|
clusterin-like 1 (retinal) |
| chrX_+_79675965 | 0.73 |
ENST00000308293.5
|
FAM46D
|
family with sequence similarity 46, member D |
| chr1_+_15668240 | 0.72 |
ENST00000444385.1
|
FHAD1
|
forkhead-associated (FHA) phosphopeptide binding domain 1 |
| chr3_-_139199565 | 0.72 |
ENST00000511956.1
|
RBP2
|
retinol binding protein 2, cellular |
| chr1_-_241683001 | 0.72 |
ENST00000366560.3
|
FH
|
fumarate hydratase |
| chr4_+_122722466 | 0.71 |
ENST00000243498.5
ENST00000379663.3 ENST00000509800.1 |
EXOSC9
|
exosome component 9 |
| chr4_+_69962185 | 0.69 |
ENST00000305231.7
|
UGT2B7
|
UDP glucuronosyltransferase 2 family, polypeptide B7 |
| chr9_+_117904097 | 0.69 |
ENST00000374016.1
|
DEC1
|
deleted in esophageal cancer 1 |
| chr5_-_177207634 | 0.69 |
ENST00000513554.1
ENST00000440605.3 |
FAM153A
|
family with sequence similarity 153, member A |
| chr2_+_34935472 | 0.68 |
ENST00000604250.1
|
AC073218.2
|
AC073218.2 |
| chr3_+_52245458 | 0.68 |
ENST00000459884.1
|
ALAS1
|
aminolevulinate, delta-, synthase 1 |
| chr5_+_96079240 | 0.68 |
ENST00000515663.1
|
CAST
|
calpastatin |
| chr14_+_56127989 | 0.67 |
ENST00000555573.1
|
KTN1
|
kinectin 1 (kinesin receptor) |
| chr1_-_201438282 | 0.67 |
ENST00000367311.3
ENST00000367309.1 |
PHLDA3
|
pleckstrin homology-like domain, family A, member 3 |
| chr15_+_66585879 | 0.66 |
ENST00000319212.4
|
DIS3L
|
DIS3 mitotic control homolog (S. cerevisiae)-like |
| chr13_-_34250861 | 0.65 |
ENST00000445227.1
ENST00000454681.2 |
RP11-141M1.3
|
RP11-141M1.3 |
| chr13_-_41768654 | 0.64 |
ENST00000379483.3
|
KBTBD7
|
kelch repeat and BTB (POZ) domain containing 7 |
| chr3_-_72897545 | 0.64 |
ENST00000325599.8
|
SHQ1
|
SHQ1, H/ACA ribonucleoprotein assembly factor |
| chr14_+_35747825 | 0.63 |
ENST00000540871.1
|
PSMA6
|
proteasome (prosome, macropain) subunit, alpha type, 6 |
| chr3_+_180586536 | 0.63 |
ENST00000465551.1
|
FXR1
|
fragile X mental retardation, autosomal homolog 1 |
| chr1_-_21377447 | 0.63 |
ENST00000374937.3
ENST00000264211.8 |
EIF4G3
|
eukaryotic translation initiation factor 4 gamma, 3 |
| chr9_-_125330838 | 0.63 |
ENST00000304865.2
|
OR1L8
|
olfactory receptor, family 1, subfamily L, member 8 |
| chr18_+_6729725 | 0.63 |
ENST00000400091.2
ENST00000583410.1 ENST00000584387.1 |
ARHGAP28
|
Rho GTPase activating protein 28 |
| chr2_+_113299990 | 0.63 |
ENST00000537335.1
ENST00000417433.2 |
POLR1B
|
polymerase (RNA) I polypeptide B, 128kDa |
| chr16_+_21689835 | 0.62 |
ENST00000286149.4
ENST00000388958.3 |
OTOA
|
otoancorin |
| chr4_+_69962212 | 0.61 |
ENST00000508661.1
|
UGT2B7
|
UDP glucuronosyltransferase 2 family, polypeptide B7 |
| chr6_+_116850174 | 0.61 |
ENST00000416171.2
ENST00000368597.2 ENST00000452373.1 ENST00000405399.1 |
FAM26D
|
family with sequence similarity 26, member D |
| chr3_-_139199589 | 0.59 |
ENST00000506825.1
|
RBP2
|
retinol binding protein 2, cellular |
| chr3_-_146323019 | 0.59 |
ENST00000492200.1
ENST00000482567.1 |
PLSCR5
|
phospholipid scramblase family, member 5 |
| chr5_+_169010638 | 0.59 |
ENST00000265295.4
ENST00000506574.1 ENST00000515224.1 ENST00000508247.1 ENST00000513941.1 |
SPDL1
|
spindle apparatus coiled-coil protein 1 |
| chr8_+_9009202 | 0.59 |
ENST00000518496.1
|
RP11-10A14.4
|
Uncharacterized protein |
| chr12_+_71833550 | 0.59 |
ENST00000266674.5
|
LGR5
|
leucine-rich repeat containing G protein-coupled receptor 5 |
| chr10_-_15130767 | 0.58 |
ENST00000356189.5
|
ACBD7
|
acyl-CoA binding domain containing 7 |
| chr18_-_69449517 | 0.57 |
ENST00000584810.1
|
RP11-723G8.2
|
Uncharacterized protein |
| chr12_+_10365082 | 0.57 |
ENST00000545859.1
|
GABARAPL1
|
GABA(A) receptor-associated protein like 1 |
| chr4_-_22444733 | 0.57 |
ENST00000508133.1
|
GPR125
|
G protein-coupled receptor 125 |
| chr1_-_62190793 | 0.56 |
ENST00000371177.2
ENST00000606498.1 |
TM2D1
|
TM2 domain containing 1 |
| chr11_+_22688150 | 0.55 |
ENST00000454584.2
|
GAS2
|
growth arrest-specific 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of IRX5
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 4.3 | GO:1990926 | short-term synaptic potentiation(GO:1990926) |
| 0.6 | 2.9 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.5 | 0.5 | GO:0006533 | aspartate catabolic process(GO:0006533) |
| 0.5 | 4.8 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.5 | 1.4 | GO:0046167 | glycerol-3-phosphate biosynthetic process(GO:0046167) |
| 0.4 | 1.7 | GO:2001027 | negative regulation of endothelial cell chemotaxis(GO:2001027) |
| 0.4 | 6.2 | GO:0055009 | atrial cardiac muscle tissue development(GO:0003228) atrial cardiac muscle tissue morphogenesis(GO:0055009) |
| 0.4 | 1.1 | GO:0051758 | homologous chromosome movement towards spindle pole involved in homologous chromosome segregation(GO:0051758) |
| 0.3 | 2.4 | GO:0002933 | lipid hydroxylation(GO:0002933) |
| 0.3 | 0.8 | GO:0060733 | regulation of eIF2 alpha phosphorylation by amino acid starvation(GO:0060733) regulation of translational initiation in response to starvation(GO:0071262) positive regulation of translational initiation in response to starvation(GO:0071264) |
| 0.2 | 2.2 | GO:0097577 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.2 | 0.7 | GO:0006106 | fumarate metabolic process(GO:0006106) |
| 0.2 | 0.8 | GO:0048789 | cytoskeletal matrix organization at active zone(GO:0048789) neurexin clustering involved in presynaptic membrane assembly(GO:0097115) retrograde trans-synaptic signaling by trans-synaptic protein complex(GO:0098942) |
| 0.2 | 1.0 | GO:0010966 | regulation of phosphate transport(GO:0010966) |
| 0.2 | 0.9 | GO:0016098 | monoterpenoid metabolic process(GO:0016098) |
| 0.2 | 0.7 | GO:0034476 | U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) |
| 0.2 | 3.5 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.2 | 2.8 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.2 | 2.8 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.2 | 0.6 | GO:0090071 | negative regulation of ribosome biogenesis(GO:0090071) |
| 0.2 | 0.6 | GO:1904425 | negative regulation of GTP binding(GO:1904425) |
| 0.1 | 3.7 | GO:0001973 | adenosine receptor signaling pathway(GO:0001973) |
| 0.1 | 0.4 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.1 | 1.6 | GO:0070779 | gamma-aminobutyric acid biosynthetic process(GO:0009449) D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.1 | 0.5 | GO:0032053 | ciliary basal body organization(GO:0032053) |
| 0.1 | 1.3 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.1 | 0.2 | GO:0002879 | positive regulation of acute inflammatory response to non-antigenic stimulus(GO:0002879) |
| 0.1 | 1.6 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.1 | 0.4 | GO:0086046 | membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.1 | 2.6 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 0.1 | 0.4 | GO:0006428 | isoleucyl-tRNA aminoacylation(GO:0006428) |
| 0.1 | 1.5 | GO:0006662 | glycerol ether metabolic process(GO:0006662) |
| 0.1 | 0.4 | GO:0070458 | cellular detoxification of nitrogen compound(GO:0070458) |
| 0.1 | 0.9 | GO:0044821 | meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.1 | 1.3 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.1 | 1.0 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.1 | 1.7 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.1 | 3.2 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.1 | 1.6 | GO:0016075 | rRNA catabolic process(GO:0016075) |
| 0.1 | 0.4 | GO:0072560 | glandular epithelial cell maturation(GO:0002071) type B pancreatic cell maturation(GO:0072560) |
| 0.1 | 1.1 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.1 | 2.5 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.1 | 3.7 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.1 | 1.3 | GO:0006527 | arginine catabolic process(GO:0006527) negative regulation of cellular response to hypoxia(GO:1900038) |
| 0.1 | 1.1 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.1 | 0.2 | GO:0035397 | helper T cell enhancement of adaptive immune response(GO:0035397) |
| 0.1 | 1.0 | GO:0015747 | urate transport(GO:0015747) |
| 0.1 | 0.4 | GO:0032571 | response to vitamin K(GO:0032571) regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904207) positive regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904209) bone regeneration(GO:1990523) |
| 0.1 | 1.1 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.1 | 0.3 | GO:1900425 | negative regulation of defense response to bacterium(GO:1900425) |
| 0.1 | 1.3 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 0.1 | 0.6 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.1 | 1.3 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.1 | 0.3 | GO:0016095 | polyprenol catabolic process(GO:0016095) |
| 0.1 | 2.5 | GO:0021522 | spinal cord motor neuron differentiation(GO:0021522) |
| 0.0 | 0.4 | GO:0051597 | response to methylmercury(GO:0051597) |
| 0.0 | 1.8 | GO:0015695 | organic cation transport(GO:0015695) |
| 0.0 | 1.0 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.0 | 0.7 | GO:0006782 | protoporphyrinogen IX biosynthetic process(GO:0006782) |
| 0.0 | 0.6 | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.0 | 1.0 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.2 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.0 | 0.3 | GO:0060423 | foregut regionalization(GO:0060423) lung field specification(GO:0060424) lung induction(GO:0060492) |
| 0.0 | 1.1 | GO:0010745 | negative regulation of macrophage derived foam cell differentiation(GO:0010745) |
| 0.0 | 0.7 | GO:0045792 | negative regulation of cell size(GO:0045792) |
| 0.0 | 0.8 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.0 | 0.8 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
| 0.0 | 0.7 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.0 | 0.3 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.0 | 0.7 | GO:2000675 | negative regulation of type B pancreatic cell apoptotic process(GO:2000675) |
| 0.0 | 1.1 | GO:0031115 | copper ion transport(GO:0006825) negative regulation of microtubule polymerization(GO:0031115) |
| 0.0 | 0.9 | GO:0045725 | positive regulation of glycogen biosynthetic process(GO:0045725) |
| 0.0 | 1.9 | GO:0034219 | carbohydrate transmembrane transport(GO:0034219) |
| 0.0 | 0.2 | GO:0007079 | mitotic chromosome movement towards spindle pole(GO:0007079) |
| 0.0 | 0.3 | GO:0097475 | motor neuron migration(GO:0097475) |
| 0.0 | 0.4 | GO:0032119 | sequestering of zinc ion(GO:0032119) regulation of sequestering of zinc ion(GO:0061088) |
| 0.0 | 1.0 | GO:0044705 | mating behavior(GO:0007617) multi-organism reproductive behavior(GO:0044705) |
| 0.0 | 0.4 | GO:0032886 | regulation of microtubule-based process(GO:0032886) |
| 0.0 | 0.4 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.0 | 0.3 | GO:0061470 | T follicular helper cell differentiation(GO:0061470) |
| 0.0 | 0.2 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.0 | 0.3 | GO:0042695 | thelarche(GO:0042695) mammary gland branching involved in thelarche(GO:0060744) |
| 0.0 | 6.6 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.2 | GO:2001206 | positive regulation of osteoclast development(GO:2001206) |
| 0.0 | 0.6 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 0.6 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.4 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.0 | 1.1 | GO:0035728 | response to hepatocyte growth factor(GO:0035728) |
| 0.0 | 1.1 | GO:0000038 | very long-chain fatty acid metabolic process(GO:0000038) |
| 0.0 | 1.3 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.4 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 0.7 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.8 | GO:0035640 | exploration behavior(GO:0035640) |
| 0.0 | 0.3 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.0 | 4.1 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.0 | 0.5 | GO:0043518 | negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) |
| 0.0 | 0.2 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.0 | 0.4 | GO:0051531 | NFAT protein import into nucleus(GO:0051531) |
| 0.0 | 1.1 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 | 0.3 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.0 | 0.6 | GO:2000637 | positive regulation of gene silencing by miRNA(GO:2000637) |
| 0.0 | 0.3 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 1.8 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 1.5 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 0.5 | GO:0051156 | glucose 6-phosphate metabolic process(GO:0051156) |
| 0.0 | 0.5 | GO:0000042 | protein targeting to Golgi(GO:0000042) |
| 0.0 | 0.3 | GO:2000269 | regulation of fibroblast apoptotic process(GO:2000269) |
| 0.0 | 0.3 | GO:1903959 | regulation of anion transmembrane transport(GO:1903959) |
| 0.0 | 1.5 | GO:0006446 | regulation of translational initiation(GO:0006446) |
| 0.0 | 0.4 | GO:0001502 | cartilage condensation(GO:0001502) |
| 0.0 | 0.5 | GO:0040036 | regulation of fibroblast growth factor receptor signaling pathway(GO:0040036) |
| 0.0 | 0.5 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.0 | 1.0 | GO:0021954 | central nervous system neuron development(GO:0021954) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.6 | GO:0033150 | cytoskeletal calyx(GO:0033150) |
| 0.5 | 4.3 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 0.5 | 4.8 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.4 | 2.5 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.3 | 7.8 | GO:0032982 | myosin filament(GO:0032982) |
| 0.3 | 0.8 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 0.1 | 1.0 | GO:0043541 | UDP-N-acetylglucosamine transferase complex(GO:0043541) |
| 0.1 | 0.9 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.1 | 1.3 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.1 | 2.3 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.1 | 0.8 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.1 | 1.0 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.1 | 1.0 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.1 | 1.3 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.1 | 3.5 | GO:0043034 | costamere(GO:0043034) |
| 0.1 | 0.8 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.1 | 1.1 | GO:0045179 | apical cortex(GO:0045179) |
| 0.1 | 1.5 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.1 | 0.9 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.0 | 0.6 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.3 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.0 | 1.7 | GO:0036019 | endolysosome(GO:0036019) |
| 0.0 | 1.8 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.0 | 0.4 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.0 | 0.7 | GO:0045239 | tricarboxylic acid cycle enzyme complex(GO:0045239) |
| 0.0 | 0.4 | GO:0000801 | central element(GO:0000801) |
| 0.0 | 0.4 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.0 | 0.8 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 0.9 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.4 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.0 | 0.4 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 1.1 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.6 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.4 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 2.6 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 1.0 | GO:0030118 | clathrin coat(GO:0030118) |
| 0.0 | 2.9 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 0.7 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.4 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 0.2 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.0 | 0.4 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 0.2 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.8 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 3.3 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 3.7 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.3 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 0.1 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 6.2 | GO:0017018 | myosin phosphatase activity(GO:0017018) |
| 0.7 | 3.3 | GO:0034875 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 0.5 | 1.4 | GO:0004370 | glycerol kinase activity(GO:0004370) |
| 0.4 | 3.7 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.3 | 1.1 | GO:0032767 | copper-dependent protein binding(GO:0032767) |
| 0.2 | 3.7 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.2 | 0.7 | GO:0003870 | 5-aminolevulinate synthase activity(GO:0003870) N-succinyltransferase activity(GO:0016749) |
| 0.2 | 2.5 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.2 | 0.7 | GO:0051800 | phosphatidylinositol-3,4-bisphosphate 3-phosphatase activity(GO:0051800) |
| 0.2 | 1.3 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.2 | 3.6 | GO:0008320 | protein transmembrane transporter activity(GO:0008320) |
| 0.2 | 1.6 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.2 | 2.8 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.2 | 2.6 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.2 | 1.1 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.1 | 1.0 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.1 | 1.3 | GO:0034603 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 0.1 | 4.8 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.1 | 1.8 | GO:0019534 | toxin transporter activity(GO:0019534) |
| 0.1 | 0.5 | GO:0050309 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.1 | 1.3 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.1 | 1.1 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.1 | 2.5 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.1 | 0.7 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.1 | 1.5 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.1 | 0.4 | GO:0004822 | isoleucine-tRNA ligase activity(GO:0004822) |
| 0.1 | 1.7 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.1 | 0.8 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.1 | 4.3 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.1 | 1.1 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 0.1 | 1.6 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.1 | 1.0 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.1 | 1.0 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.1 | 0.3 | GO:0047751 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) steroid dehydrogenase activity, acting on the CH-CH group of donors(GO:0033765) cholestenone 5-alpha-reductase activity(GO:0047751) |
| 0.1 | 0.7 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.1 | 0.2 | GO:0010348 | lithium:proton antiporter activity(GO:0010348) |
| 0.1 | 1.7 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.1 | 0.5 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.6 | GO:0033592 | RNA strand annealing activity(GO:0033592) |
| 0.0 | 2.2 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 2.5 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 1.2 | GO:0016918 | retinal binding(GO:0016918) |
| 0.0 | 2.9 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.6 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 3.2 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 1.0 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.0 | 0.3 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.0 | 0.9 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 1.5 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.0 | 0.2 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 0.0 | 1.4 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.8 | GO:0099604 | calcium-release channel activity(GO:0015278) ligand-gated calcium channel activity(GO:0099604) |
| 0.0 | 0.7 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 1.8 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.0 | 0.4 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.0 | 1.8 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.4 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.0 | 0.2 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 0.8 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.4 | GO:0086007 | voltage-gated calcium channel activity involved in cardiac muscle cell action potential(GO:0086007) |
| 0.0 | 0.8 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.0 | 0.6 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.0 | 0.9 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.6 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.0 | 0.7 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.0 | 1.9 | GO:1901476 | carbohydrate transmembrane transporter activity(GO:0015144) carbohydrate transporter activity(GO:1901476) |
| 0.0 | 1.6 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 1.8 | GO:0016303 | 1-phosphatidylinositol-3-kinase activity(GO:0016303) |
| 0.0 | 0.8 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 4.4 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 0.6 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 2.5 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.7 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.3 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 0.9 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 4.5 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 0.4 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.9 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.3 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.5 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 0.7 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.3 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.0 | 0.7 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.0 | 1.0 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.0 | 1.0 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.0 | 2.6 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.1 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 0.0 | 0.9 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 1.5 | PID ERBB1 RECEPTOR PROXIMAL PATHWAY | EGF receptor (ErbB1) signaling pathway |
| 0.0 | 2.6 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.0 | 1.6 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.9 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.8 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 1.0 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 1.0 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 0.6 | PID INSULIN PATHWAY | Insulin Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 4.3 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.2 | 3.3 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.1 | 3.7 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.1 | 7.2 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 1.3 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.1 | 0.7 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.1 | 2.5 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 1.4 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 1.0 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.7 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 3.3 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 1.5 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 1.0 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.0 | 1.5 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.5 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.0 | 6.4 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.0 | 0.7 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.7 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 1.1 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.3 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.4 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.0 | 0.8 | REACTOME MUSCLE CONTRACTION | Genes involved in Muscle contraction |
| 0.0 | 0.8 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.0 | 0.6 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.3 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.3 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.0 | 0.6 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 5.7 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.7 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.6 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 1.5 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 0.4 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.5 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.8 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.4 | REACTOME TRAF6 MEDIATED NFKB ACTIVATION | Genes involved in TRAF6 mediated NF-kB activation |
| 0.0 | 0.8 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.3 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |