Project
Illumina Body Map 2: averaged replicates
Navigation
Downloads
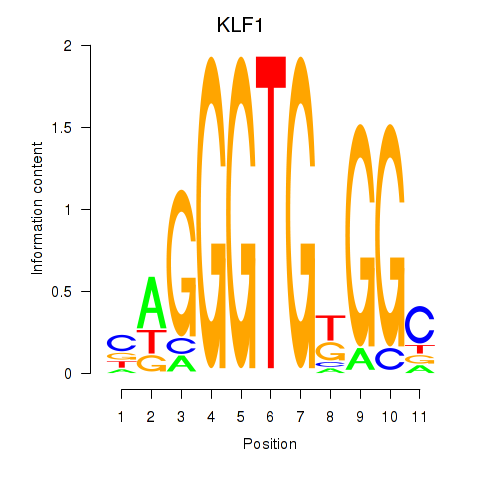
Results for KLF1
Z-value: 3.55
Transcription factors associated with KLF1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
KLF1
|
ENSG00000105610.4 | Kruppel like factor 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| KLF1 | hg19_v2_chr19_-_12997995_12998021 | 0.15 | 4.0e-01 | Click! |
Activity profile of KLF1 motif
Sorted Z-values of KLF1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_5248294 | 9.13 |
ENST00000335295.4
|
HBB
|
hemoglobin, beta |
| chr15_+_43885252 | 8.87 |
ENST00000453782.1
ENST00000300283.6 ENST00000437924.1 ENST00000450086.2 |
CKMT1B
|
creatine kinase, mitochondrial 1B |
| chr15_+_43985084 | 8.17 |
ENST00000434505.1
ENST00000411750.1 |
CKMT1A
|
creatine kinase, mitochondrial 1A |
| chr8_+_80523962 | 5.87 |
ENST00000518491.1
|
STMN2
|
stathmin-like 2 |
| chr13_+_53602894 | 4.68 |
ENST00000219022.2
|
OLFM4
|
olfactomedin 4 |
| chr11_-_62689046 | 4.27 |
ENST00000306960.3
ENST00000543973.1 |
CHRM1
|
cholinergic receptor, muscarinic 1 |
| chr20_-_52687059 | 4.21 |
ENST00000371435.2
ENST00000395961.3 |
BCAS1
|
breast carcinoma amplified sequence 1 |
| chr5_-_83680603 | 4.20 |
ENST00000296591.5
|
EDIL3
|
EGF-like repeats and discoidin I-like domains 3 |
| chr19_+_35607166 | 4.17 |
ENST00000604255.1
ENST00000346446.5 ENST00000344013.6 ENST00000603449.1 ENST00000406988.1 ENST00000605550.1 ENST00000604804.1 ENST00000605552.1 |
FXYD3
|
FXYD domain containing ion transport regulator 3 |
| chr7_-_80548493 | 4.12 |
ENST00000536800.1
|
SEMA3C
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3C |
| chr19_+_35606777 | 4.04 |
ENST00000604404.1
ENST00000435734.2 ENST00000603181.1 |
FXYD3
|
FXYD domain containing ion transport regulator 3 |
| chr8_-_75233563 | 3.85 |
ENST00000342232.4
|
JPH1
|
junctophilin 1 |
| chr11_+_1074870 | 3.70 |
ENST00000441003.2
ENST00000359061.5 |
MUC2
|
mucin 2, oligomeric mucus/gel-forming |
| chr17_+_38171681 | 3.53 |
ENST00000225474.2
ENST00000331769.2 ENST00000394148.3 ENST00000577675.1 |
CSF3
|
colony stimulating factor 3 (granulocyte) |
| chr12_+_57943781 | 3.47 |
ENST00000455537.2
ENST00000286452.5 |
KIF5A
|
kinesin family member 5A |
| chr8_-_27115903 | 3.41 |
ENST00000350889.4
ENST00000519997.1 ENST00000519614.1 ENST00000522908.1 ENST00000265770.7 |
STMN4
|
stathmin-like 4 |
| chr8_-_143867946 | 3.40 |
ENST00000301263.4
|
LY6D
|
lymphocyte antigen 6 complex, locus D |
| chr1_+_27189631 | 3.20 |
ENST00000339276.4
|
SFN
|
stratifin |
| chr1_+_47264711 | 3.19 |
ENST00000371923.4
ENST00000271153.4 ENST00000371919.4 |
CYP4B1
|
cytochrome P450, family 4, subfamily B, polypeptide 1 |
| chr19_+_35606692 | 3.19 |
ENST00000406242.3
ENST00000454903.2 |
FXYD3
|
FXYD domain containing ion transport regulator 3 |
| chr7_-_80548667 | 3.16 |
ENST00000265361.3
|
SEMA3C
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3C |
| chr4_-_90756769 | 3.15 |
ENST00000345009.4
ENST00000505199.1 ENST00000502987.1 |
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor) |
| chr17_+_38171614 | 3.08 |
ENST00000583218.1
ENST00000394149.3 |
CSF3
|
colony stimulating factor 3 (granulocyte) |
| chr8_-_27115931 | 3.07 |
ENST00000523048.1
|
STMN4
|
stathmin-like 4 |
| chr2_-_220174166 | 3.06 |
ENST00000409251.3
ENST00000451506.1 ENST00000295718.2 ENST00000446182.1 |
PTPRN
|
protein tyrosine phosphatase, receptor type, N |
| chr9_+_74764340 | 3.01 |
ENST00000376986.1
ENST00000358399.3 |
GDA
|
guanine deaminase |
| chr17_-_74023474 | 3.01 |
ENST00000301607.3
|
EVPL
|
envoplakin |
| chr2_+_17721937 | 2.99 |
ENST00000451533.1
|
VSNL1
|
visinin-like 1 |
| chr7_+_22766766 | 2.87 |
ENST00000426291.1
ENST00000401651.1 ENST00000258743.5 ENST00000420258.2 ENST00000407492.1 ENST00000401630.3 ENST00000406575.1 |
IL6
|
interleukin 6 (interferon, beta 2) |
| chrX_-_153151586 | 2.85 |
ENST00000370060.1
ENST00000370055.1 ENST00000420165.1 |
L1CAM
|
L1 cell adhesion molecule |
| chr1_-_39395165 | 2.85 |
ENST00000372985.3
|
RHBDL2
|
rhomboid, veinlet-like 2 (Drosophila) |
| chr3_+_238456 | 2.84 |
ENST00000427688.1
|
CHL1
|
cell adhesion molecule L1-like |
| chr12_-_8815404 | 2.74 |
ENST00000359478.2
ENST00000396549.2 |
MFAP5
|
microfibrillar associated protein 5 |
| chr3_+_181429704 | 2.73 |
ENST00000431565.2
ENST00000325404.1 |
SOX2
|
SRY (sex determining region Y)-box 2 |
| chr6_+_30850697 | 2.72 |
ENST00000509639.1
ENST00000412274.2 ENST00000507901.1 ENST00000507046.1 ENST00000437124.2 ENST00000454612.2 ENST00000396342.2 |
DDR1
|
discoidin domain receptor tyrosine kinase 1 |
| chr1_+_6845384 | 2.69 |
ENST00000303635.7
|
CAMTA1
|
calmodulin binding transcription activator 1 |
| chr10_+_105036909 | 2.68 |
ENST00000369849.4
|
INA
|
internexin neuronal intermediate filament protein, alpha |
| chr17_+_43972010 | 2.68 |
ENST00000334239.8
ENST00000446361.3 |
MAPT
|
microtubule-associated protein tau |
| chr12_+_56477093 | 2.68 |
ENST00000549672.1
ENST00000415288.2 |
ERBB3
|
v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 3 |
| chr5_-_142077569 | 2.63 |
ENST00000407758.1
ENST00000441680.2 ENST00000419524.2 |
FGF1
|
fibroblast growth factor 1 (acidic) |
| chr12_-_85306562 | 2.62 |
ENST00000551612.1
ENST00000450363.3 ENST00000552192.1 |
SLC6A15
|
solute carrier family 6 (neutral amino acid transporter), member 15 |
| chr11_+_394196 | 2.56 |
ENST00000331563.2
ENST00000531857.1 |
PKP3
|
plakophilin 3 |
| chr6_+_74405501 | 2.50 |
ENST00000437994.2
ENST00000422508.2 |
CD109
|
CD109 molecule |
| chr20_+_17207636 | 2.50 |
ENST00000262545.2
|
PCSK2
|
proprotein convertase subtilisin/kexin type 2 |
| chr6_-_3227877 | 2.49 |
ENST00000259818.7
|
TUBB2B
|
tubulin, beta 2B class IIb |
| chr14_+_42077552 | 2.46 |
ENST00000554120.1
|
LRFN5
|
leucine rich repeat and fibronectin type III domain containing 5 |
| chr17_-_74023291 | 2.45 |
ENST00000586740.1
|
EVPL
|
envoplakin |
| chr6_+_30850862 | 2.44 |
ENST00000504651.1
ENST00000512694.1 ENST00000515233.1 |
DDR1
|
discoidin domain receptor tyrosine kinase 1 |
| chr20_-_52687030 | 2.38 |
ENST00000411563.1
|
BCAS1
|
breast carcinoma amplified sequence 1 |
| chr15_-_76005170 | 2.36 |
ENST00000308508.5
|
CSPG4
|
chondroitin sulfate proteoglycan 4 |
| chr8_-_21988558 | 2.34 |
ENST00000312841.8
|
HR
|
hair growth associated |
| chr6_+_74405804 | 2.32 |
ENST00000287097.5
|
CD109
|
CD109 molecule |
| chr17_-_34122596 | 2.32 |
ENST00000250144.8
|
MMP28
|
matrix metallopeptidase 28 |
| chr11_+_62475130 | 2.31 |
ENST00000294117.5
|
GNG3
|
guanine nucleotide binding protein (G protein), gamma 3 |
| chr12_-_85306594 | 2.28 |
ENST00000266682.5
|
SLC6A15
|
solute carrier family 6 (neutral amino acid transporter), member 15 |
| chr6_+_150464155 | 2.25 |
ENST00000361131.4
|
PPP1R14C
|
protein phosphatase 1, regulatory (inhibitor) subunit 14C |
| chr13_-_45010939 | 2.23 |
ENST00000261489.2
|
TSC22D1
|
TSC22 domain family, member 1 |
| chr19_-_42916499 | 2.21 |
ENST00000601189.1
ENST00000599211.1 |
LIPE
|
lipase, hormone-sensitive |
| chr2_+_17721920 | 2.20 |
ENST00000295156.4
|
VSNL1
|
visinin-like 1 |
| chr3_+_238427 | 2.20 |
ENST00000397491.2
|
CHL1
|
cell adhesion molecule L1-like |
| chr19_+_35630022 | 2.19 |
ENST00000589209.1
|
FXYD1
|
FXYD domain containing ion transport regulator 1 |
| chr12_-_100378006 | 2.18 |
ENST00000547776.2
ENST00000329257.7 ENST00000547010.1 |
ANKS1B
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr20_+_44035200 | 2.15 |
ENST00000372717.1
ENST00000360981.4 |
DBNDD2
|
dysbindin (dystrobrevin binding protein 1) domain containing 2 |
| chr16_+_56225248 | 2.12 |
ENST00000262493.6
|
GNAO1
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide O |
| chr16_-_31147020 | 2.11 |
ENST00000568261.1
ENST00000567797.1 ENST00000317508.6 |
PRSS8
|
protease, serine, 8 |
| chr6_-_114664180 | 2.10 |
ENST00000312719.5
|
HS3ST5
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 5 |
| chr1_+_18434240 | 2.10 |
ENST00000251296.1
|
IGSF21
|
immunoglobin superfamily, member 21 |
| chr3_+_8775466 | 2.07 |
ENST00000343849.2
ENST00000397368.2 |
CAV3
|
caveolin 3 |
| chr2_-_220173685 | 2.06 |
ENST00000423636.2
ENST00000442029.1 ENST00000412847.1 |
PTPRN
|
protein tyrosine phosphatase, receptor type, N |
| chr8_+_21911054 | 2.05 |
ENST00000519850.1
ENST00000381470.3 |
DMTN
|
dematin actin binding protein |
| chr6_+_44238203 | 2.04 |
ENST00000451188.2
|
TMEM151B
|
transmembrane protein 151B |
| chr14_+_23846210 | 2.04 |
ENST00000339180.4
ENST00000342473.4 ENST00000397227.3 ENST00000555731.1 |
CMTM5
|
CKLF-like MARVEL transmembrane domain containing 5 |
| chr5_+_140501581 | 2.03 |
ENST00000194152.1
|
PCDHB4
|
protocadherin beta 4 |
| chr3_+_115342349 | 2.02 |
ENST00000393780.3
|
GAP43
|
growth associated protein 43 |
| chr10_+_71211212 | 2.01 |
ENST00000373290.2
|
TSPAN15
|
tetraspanin 15 |
| chr11_-_75379612 | 2.00 |
ENST00000526740.1
|
MAP6
|
microtubule-associated protein 6 |
| chr20_+_17207665 | 1.99 |
ENST00000536609.1
|
PCSK2
|
proprotein convertase subtilisin/kexin type 2 |
| chr12_-_8815215 | 1.97 |
ENST00000544889.1
ENST00000543369.1 |
MFAP5
|
microfibrillar associated protein 5 |
| chr5_-_172756506 | 1.96 |
ENST00000265087.4
|
STC2
|
stanniocalcin 2 |
| chr11_+_71903169 | 1.95 |
ENST00000393676.3
|
FOLR1
|
folate receptor 1 (adult) |
| chr2_+_219824357 | 1.91 |
ENST00000302625.4
|
CDK5R2
|
cyclin-dependent kinase 5, regulatory subunit 2 (p39) |
| chr12_-_57443886 | 1.90 |
ENST00000300119.3
|
MYO1A
|
myosin IA |
| chr16_-_67427389 | 1.89 |
ENST00000562206.1
ENST00000290942.5 ENST00000393957.2 |
TPPP3
|
tubulin polymerization-promoting protein family member 3 |
| chr11_-_75380165 | 1.84 |
ENST00000304771.3
|
MAP6
|
microtubule-associated protein 6 |
| chr14_+_24540761 | 1.82 |
ENST00000559207.1
|
CPNE6
|
copine VI (neuronal) |
| chr7_+_73245193 | 1.81 |
ENST00000340958.2
|
CLDN4
|
claudin 4 |
| chr16_+_89989687 | 1.81 |
ENST00000315491.7
ENST00000555576.1 ENST00000554336.1 ENST00000553967.1 |
TUBB3
|
Tubulin beta-3 chain |
| chr14_+_23845995 | 1.79 |
ENST00000359320.3
|
CMTM5
|
CKLF-like MARVEL transmembrane domain containing 5 |
| chr17_-_39661849 | 1.78 |
ENST00000246635.3
ENST00000336861.3 ENST00000587544.1 ENST00000587435.1 |
KRT13
|
keratin 13 |
| chr8_+_21912328 | 1.78 |
ENST00000432128.1
ENST00000443491.2 ENST00000517600.1 ENST00000523782.2 |
DMTN
|
dematin actin binding protein |
| chr2_+_220306745 | 1.78 |
ENST00000431523.1
ENST00000396698.1 ENST00000396695.2 |
SPEG
|
SPEG complex locus |
| chr12_-_8815477 | 1.76 |
ENST00000433590.2
|
MFAP5
|
microfibrillar associated protein 5 |
| chr12_-_8815299 | 1.76 |
ENST00000535336.1
|
MFAP5
|
microfibrillar associated protein 5 |
| chr17_-_39661947 | 1.74 |
ENST00000590425.1
|
KRT13
|
keratin 13 |
| chr14_+_21569245 | 1.73 |
ENST00000556585.2
|
TMEM253
|
transmembrane protein 253 |
| chr12_+_26274917 | 1.72 |
ENST00000538142.1
|
SSPN
|
sarcospan |
| chr3_-_168864315 | 1.70 |
ENST00000475754.1
ENST00000484519.1 |
MECOM
|
MDS1 and EVI1 complex locus |
| chr2_+_172950227 | 1.69 |
ENST00000341900.6
|
DLX1
|
distal-less homeobox 1 |
| chr17_-_27949911 | 1.69 |
ENST00000492276.2
ENST00000345068.5 ENST00000584602.1 |
CORO6
|
coronin 6 |
| chr22_+_38071615 | 1.68 |
ENST00000215909.5
|
LGALS1
|
lectin, galactoside-binding, soluble, 1 |
| chr17_+_33914424 | 1.66 |
ENST00000590432.1
|
AP2B1
|
adaptor-related protein complex 2, beta 1 subunit |
| chr10_-_47173994 | 1.66 |
ENST00000414655.2
ENST00000545298.1 ENST00000359178.4 ENST00000358140.4 ENST00000503031.1 |
ANXA8L1
LINC00842
|
annexin A8-like 1 long intergenic non-protein coding RNA 842 |
| chr16_+_30078811 | 1.65 |
ENST00000564688.1
|
ALDOA
|
aldolase A, fructose-bisphosphate |
| chr1_-_117210290 | 1.64 |
ENST00000369483.1
ENST00000369486.3 |
IGSF3
|
immunoglobulin superfamily, member 3 |
| chr12_-_49581152 | 1.61 |
ENST00000550811.1
|
TUBA1A
|
tubulin, alpha 1a |
| chr11_-_1606513 | 1.61 |
ENST00000382171.2
|
KRTAP5-1
|
keratin associated protein 5-1 |
| chr9_+_74764278 | 1.60 |
ENST00000238018.4
ENST00000376989.3 |
GDA
|
guanine deaminase |
| chr3_-_124560257 | 1.59 |
ENST00000496703.1
|
ITGB5
|
integrin, beta 5 |
| chr8_-_42065187 | 1.58 |
ENST00000270189.6
ENST00000352041.3 ENST00000220809.4 |
PLAT
|
plasminogen activator, tissue |
| chr1_-_31712401 | 1.58 |
ENST00000373736.2
|
NKAIN1
|
Na+/K+ transporting ATPase interacting 1 |
| chr3_-_168864427 | 1.58 |
ENST00000468789.1
|
MECOM
|
MDS1 and EVI1 complex locus |
| chr5_+_102595119 | 1.58 |
ENST00000510890.1
|
C5orf30
|
chromosome 5 open reading frame 30 |
| chr16_+_66400533 | 1.57 |
ENST00000341529.3
|
CDH5
|
cadherin 5, type 2 (vascular endothelium) |
| chr12_-_100378392 | 1.57 |
ENST00000549866.1
|
ANKS1B
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr5_+_71403061 | 1.57 |
ENST00000512974.1
ENST00000296755.7 |
MAP1B
|
microtubule-associated protein 1B |
| chr1_+_35225339 | 1.55 |
ENST00000339480.1
|
GJB4
|
gap junction protein, beta 4, 30.3kDa |
| chr2_-_51259641 | 1.55 |
ENST00000406316.2
ENST00000405581.1 |
NRXN1
|
neurexin 1 |
| chr9_+_112887772 | 1.53 |
ENST00000259318.7
|
AKAP2
|
A kinase (PRKA) anchor protein 2 |
| chr10_+_31610064 | 1.53 |
ENST00000446923.2
ENST00000559476.1 |
ZEB1
|
zinc finger E-box binding homeobox 1 |
| chr22_-_44258280 | 1.52 |
ENST00000540422.1
|
SULT4A1
|
sulfotransferase family 4A, member 1 |
| chr1_-_27709793 | 1.51 |
ENST00000374027.3
ENST00000374025.3 |
CD164L2
|
CD164 sialomucin-like 2 |
| chr1_-_145076068 | 1.51 |
ENST00000369345.4
|
PDE4DIP
|
phosphodiesterase 4D interacting protein |
| chr2_-_51259528 | 1.50 |
ENST00000404971.1
|
NRXN1
|
neurexin 1 |
| chr1_+_11866207 | 1.47 |
ENST00000312413.6
ENST00000346436.6 |
CLCN6
|
chloride channel, voltage-sensitive 6 |
| chr3_-_195538728 | 1.46 |
ENST00000349607.4
ENST00000346145.4 |
MUC4
|
mucin 4, cell surface associated |
| chr20_+_44034804 | 1.45 |
ENST00000357275.2
ENST00000372720.3 |
DBNDD2
|
dysbindin (dystrobrevin binding protein 1) domain containing 2 |
| chr18_-_5540471 | 1.45 |
ENST00000581833.1
ENST00000544123.1 ENST00000342933.3 ENST00000400111.3 ENST00000585142.1 |
EPB41L3
|
erythrocyte membrane protein band 4.1-like 3 |
| chr17_-_40575535 | 1.45 |
ENST00000357037.5
|
PTRF
|
polymerase I and transcript release factor |
| chr12_-_125398602 | 1.44 |
ENST00000541272.1
ENST00000535131.1 |
UBC
|
ubiquitin C |
| chr6_-_78173490 | 1.43 |
ENST00000369947.2
|
HTR1B
|
5-hydroxytryptamine (serotonin) receptor 1B, G protein-coupled |
| chr12_+_116997186 | 1.39 |
ENST00000306985.4
|
MAP1LC3B2
|
microtubule-associated protein 1 light chain 3 beta 2 |
| chr21_-_31311818 | 1.39 |
ENST00000535441.1
ENST00000309434.7 ENST00000327783.4 ENST00000389124.2 ENST00000389125.3 ENST00000399913.1 |
GRIK1
|
glutamate receptor, ionotropic, kainate 1 |
| chr19_-_36523529 | 1.34 |
ENST00000593074.1
|
CLIP3
|
CAP-GLY domain containing linker protein 3 |
| chr10_+_47746929 | 1.34 |
ENST00000340243.6
ENST00000374277.5 ENST00000449464.2 ENST00000538825.1 ENST00000335083.5 |
ANXA8L2
AL603965.1
|
annexin A8-like 2 Protein LOC100996760 |
| chr7_-_944631 | 1.33 |
ENST00000453175.2
|
ADAP1
|
ArfGAP with dual PH domains 1 |
| chr11_-_6440624 | 1.33 |
ENST00000311051.3
|
APBB1
|
amyloid beta (A4) precursor protein-binding, family B, member 1 (Fe65) |
| chr8_-_22089533 | 1.32 |
ENST00000321613.3
|
PHYHIP
|
phytanoyl-CoA 2-hydroxylase interacting protein |
| chr2_-_172750733 | 1.32 |
ENST00000392592.4
ENST00000422440.2 |
SLC25A12
|
solute carrier family 25 (aspartate/glutamate carrier), member 12 |
| chr3_-_48130314 | 1.31 |
ENST00000439356.1
ENST00000395734.3 ENST00000426837.2 |
MAP4
|
microtubule-associated protein 4 |
| chr8_-_22549856 | 1.28 |
ENST00000522910.1
|
EGR3
|
early growth response 3 |
| chr13_+_73632897 | 1.28 |
ENST00000377687.4
|
KLF5
|
Kruppel-like factor 5 (intestinal) |
| chr11_-_62313090 | 1.27 |
ENST00000528508.1
ENST00000533365.1 |
AHNAK
|
AHNAK nucleoprotein |
| chr8_+_26435359 | 1.27 |
ENST00000311151.5
|
DPYSL2
|
dihydropyrimidinase-like 2 |
| chr7_+_30960915 | 1.27 |
ENST00000441328.2
ENST00000409899.1 ENST00000409611.1 |
AQP1
|
aquaporin 1 (Colton blood group) |
| chr5_-_115910091 | 1.26 |
ENST00000257414.8
|
SEMA6A
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A |
| chr3_-_195538760 | 1.26 |
ENST00000475231.1
|
MUC4
|
mucin 4, cell surface associated |
| chr12_-_110939870 | 1.24 |
ENST00000447578.2
ENST00000546588.1 ENST00000360579.7 ENST00000549970.1 ENST00000549578.1 |
VPS29
|
vacuolar protein sorting 29 homolog (S. cerevisiae) |
| chr1_-_98510843 | 1.24 |
ENST00000413670.2
ENST00000538428.1 |
MIR137HG
|
MIR137 host gene (non-protein coding) |
| chr5_-_16617162 | 1.23 |
ENST00000306320.9
|
FAM134B
|
family with sequence similarity 134, member B |
| chr11_+_1860832 | 1.22 |
ENST00000252898.7
|
TNNI2
|
troponin I type 2 (skeletal, fast) |
| chr3_-_149293990 | 1.22 |
ENST00000472417.1
|
WWTR1
|
WW domain containing transcription regulator 1 |
| chr2_+_28974489 | 1.22 |
ENST00000455580.1
|
PPP1CB
|
protein phosphatase 1, catalytic subunit, beta isozyme |
| chr11_-_6426635 | 1.21 |
ENST00000608645.1
ENST00000608394.1 ENST00000529519.1 |
APBB1
|
amyloid beta (A4) precursor protein-binding, family B, member 1 (Fe65) |
| chr16_-_29910365 | 1.21 |
ENST00000346932.5
ENST00000350527.3 ENST00000537485.1 ENST00000568380.1 |
SEZ6L2
|
seizure related 6 homolog (mouse)-like 2 |
| chr17_+_39845134 | 1.20 |
ENST00000591776.1
ENST00000469257.1 |
EIF1
|
eukaryotic translation initiation factor 1 |
| chr22_-_39639021 | 1.20 |
ENST00000455790.1
|
PDGFB
|
platelet-derived growth factor beta polypeptide |
| chr19_+_13135790 | 1.19 |
ENST00000358552.3
|
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor) |
| chr1_+_209602156 | 1.19 |
ENST00000429156.1
ENST00000366437.3 ENST00000603283.1 ENST00000431096.1 |
MIR205HG
|
MIR205 host gene (non-protein coding) |
| chr15_+_78556809 | 1.19 |
ENST00000343789.3
ENST00000394852.3 |
DNAJA4
|
DnaJ (Hsp40) homolog, subfamily A, member 4 |
| chr1_-_94147385 | 1.17 |
ENST00000260502.6
|
BCAR3
|
breast cancer anti-estrogen resistance 3 |
| chr22_+_31477296 | 1.16 |
ENST00000426927.1
ENST00000440425.1 ENST00000358743.1 ENST00000347557.2 ENST00000333137.7 |
SMTN
|
smoothelin |
| chr10_-_104178857 | 1.16 |
ENST00000020673.5
|
PSD
|
pleckstrin and Sec7 domain containing |
| chr15_-_42386752 | 1.15 |
ENST00000290472.3
|
PLA2G4D
|
phospholipase A2, group IVD (cytosolic) |
| chr17_+_46125685 | 1.14 |
ENST00000579889.1
|
NFE2L1
|
nuclear factor, erythroid 2-like 1 |
| chr7_+_142982023 | 1.14 |
ENST00000359333.3
ENST00000409244.1 ENST00000409541.1 ENST00000410004.1 |
TMEM139
|
transmembrane protein 139 |
| chr1_-_59249732 | 1.13 |
ENST00000371222.2
|
JUN
|
jun proto-oncogene |
| chr2_+_45878790 | 1.13 |
ENST00000306156.3
|
PRKCE
|
protein kinase C, epsilon |
| chr12_+_48516463 | 1.12 |
ENST00000546465.1
|
PFKM
|
phosphofructokinase, muscle |
| chr2_-_26205340 | 1.12 |
ENST00000264712.3
|
KIF3C
|
kinesin family member 3C |
| chr15_+_90931450 | 1.11 |
ENST00000268182.5
ENST00000560738.1 ENST00000560418.1 |
IQGAP1
|
IQ motif containing GTPase activating protein 1 |
| chr12_+_7037461 | 1.11 |
ENST00000396684.2
|
ATN1
|
atrophin 1 |
| chr19_+_38880695 | 1.11 |
ENST00000587947.1
ENST00000338502.4 |
SPRED3
|
sprouty-related, EVH1 domain containing 3 |
| chr19_+_35629702 | 1.10 |
ENST00000351325.4
|
FXYD1
|
FXYD domain containing ion transport regulator 1 |
| chr1_-_27709816 | 1.09 |
ENST00000374030.1
|
CD164L2
|
CD164 sialomucin-like 2 |
| chr12_+_48516357 | 1.09 |
ENST00000549022.1
ENST00000547587.1 ENST00000312352.7 |
PFKM
|
phosphofructokinase, muscle |
| chr19_+_13135731 | 1.09 |
ENST00000587260.1
|
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor) |
| chr8_+_38614754 | 1.08 |
ENST00000521642.1
|
TACC1
|
transforming, acidic coiled-coil containing protein 1 |
| chr1_+_11866270 | 1.08 |
ENST00000376497.3
ENST00000376487.3 ENST00000376496.3 |
CLCN6
|
chloride channel, voltage-sensitive 6 |
| chr20_+_44035847 | 1.06 |
ENST00000372712.2
|
DBNDD2
|
dysbindin (dystrobrevin binding protein 1) domain containing 2 |
| chr9_-_33264557 | 1.06 |
ENST00000473781.1
ENST00000488499.1 |
BAG1
|
BCL2-associated athanogene |
| chr6_+_73331520 | 1.06 |
ENST00000342056.2
ENST00000355194.4 |
KCNQ5
|
potassium voltage-gated channel, KQT-like subfamily, member 5 |
| chrX_+_47077632 | 1.06 |
ENST00000457458.2
|
CDK16
|
cyclin-dependent kinase 16 |
| chr5_-_11903337 | 1.06 |
ENST00000502551.1
|
CTNND2
|
catenin (cadherin-associated protein), delta 2 |
| chr5_+_169931009 | 1.05 |
ENST00000328939.4
ENST00000390656.4 |
KCNIP1
|
Kv channel interacting protein 1 |
| chr12_+_72667203 | 1.05 |
ENST00000547300.1
|
TRHDE
|
thyrotropin-releasing hormone degrading enzyme |
| chr20_-_48099182 | 1.05 |
ENST00000371741.4
|
KCNB1
|
potassium voltage-gated channel, Shab-related subfamily, member 1 |
| chr17_-_7493390 | 1.04 |
ENST00000538513.2
ENST00000570788.1 ENST00000250055.2 |
SOX15
|
SRY (sex determining region Y)-box 15 |
| chr11_-_71781096 | 1.04 |
ENST00000535087.1
ENST00000535838.1 |
NUMA1
|
nuclear mitotic apparatus protein 1 |
| chr6_-_131291572 | 1.03 |
ENST00000529208.1
|
EPB41L2
|
erythrocyte membrane protein band 4.1-like 2 |
| chr2_-_219151984 | 1.03 |
ENST00000444000.1
ENST00000418569.1 |
TMBIM1
|
transmembrane BAX inhibitor motif containing 1 |
| chr7_+_86274145 | 1.03 |
ENST00000439827.1
ENST00000394720.2 ENST00000421579.1 |
GRM3
|
glutamate receptor, metabotropic 3 |
| chr5_-_87980753 | 1.02 |
ENST00000511014.2
|
LINC00461
|
long intergenic non-protein coding RNA 461 |
| chr3_+_183770835 | 1.02 |
ENST00000318351.1
|
HTR3C
|
5-hydroxytryptamine (serotonin) receptor 3C, ionotropic |
| chr3_-_186080012 | 1.01 |
ENST00000544847.1
ENST00000265022.3 |
DGKG
|
diacylglycerol kinase, gamma 90kDa |
| chr2_+_154728426 | 1.01 |
ENST00000392825.3
ENST00000434213.1 |
GALNT13
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 13 (GalNAc-T13) |
| chr17_-_46716647 | 1.01 |
ENST00000608940.1
|
RP11-357H14.17
|
RP11-357H14.17 |
| chr18_-_5540384 | 1.00 |
ENST00000584670.1
ENST00000582703.1 ENST00000580179.1 |
EPB41L3
|
erythrocyte membrane protein band 4.1-like 3 |
| chr2_-_55276320 | 1.00 |
ENST00000357376.3
|
RTN4
|
reticulon 4 |
| chr12_+_56498312 | 1.00 |
ENST00000552766.1
|
PA2G4
|
proliferation-associated 2G4, 38kDa |
| chr5_+_140480083 | 1.00 |
ENST00000231130.2
|
PCDHB3
|
protocadherin beta 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of KLF1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.6 | 10.4 | GO:0030185 | nitric oxide transport(GO:0030185) |
| 1.5 | 4.6 | GO:0006147 | guanine catabolic process(GO:0006147) |
| 1.5 | 5.9 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
| 1.3 | 5.1 | GO:1990502 | dense core granule maturation(GO:1990502) |
| 1.1 | 3.4 | GO:0050760 | negative regulation of thymidylate synthase biosynthetic process(GO:0050760) |
| 0.9 | 4.5 | GO:0030070 | insulin processing(GO:0030070) |
| 0.9 | 3.5 | GO:0099640 | axo-dendritic protein transport(GO:0099640) |
| 0.7 | 2.9 | GO:0002384 | hepatic immune response(GO:0002384) response to prolactin(GO:1990637) |
| 0.7 | 7.3 | GO:0003350 | pulmonary myocardium development(GO:0003350) |
| 0.6 | 1.9 | GO:0003147 | neural crest cell migration involved in heart formation(GO:0003147) anterior neural tube closure(GO:0061713) |
| 0.6 | 3.2 | GO:0010482 | epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 0.6 | 3.1 | GO:0051585 | negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of glutathione peroxidase activity(GO:1903282) positive regulation of glutathione peroxidase activity(GO:1903284) positive regulation of hydrogen peroxide catabolic process(GO:1903285) positive regulation of peroxidase activity(GO:2000470) |
| 0.6 | 1.7 | GO:0021893 | regulation of transcription from RNA polymerase II promoter involved in forebrain neuron fate commitment(GO:0021882) cerebral cortex GABAergic interneuron fate commitment(GO:0021893) positive regulation of neural retina development(GO:0061075) positive regulation of retina development in camera-type eye(GO:1902868) positive regulation of amacrine cell differentiation(GO:1902871) |
| 0.5 | 3.7 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.5 | 4.3 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 0.5 | 4.9 | GO:0015820 | leucine transport(GO:0015820) |
| 0.5 | 8.2 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.5 | 4.7 | GO:2000389 | regulation of neutrophil extravasation(GO:2000389) |
| 0.5 | 3.7 | GO:0072675 | osteoclast fusion(GO:0072675) |
| 0.4 | 1.3 | GO:0035508 | positive regulation of myosin-light-chain-phosphatase activity(GO:0035508) |
| 0.4 | 2.7 | GO:0090258 | negative regulation of mitochondrial fission(GO:0090258) |
| 0.4 | 2.2 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 0.4 | 6.6 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 0.4 | 0.4 | GO:0061196 | fungiform papilla development(GO:0061196) |
| 0.4 | 2.9 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.4 | 3.3 | GO:0010734 | protein glutathionylation(GO:0010731) regulation of protein glutathionylation(GO:0010732) negative regulation of protein glutathionylation(GO:0010734) |
| 0.4 | 2.3 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 0.4 | 3.8 | GO:0070560 | protein secretion by platelet(GO:0070560) |
| 0.4 | 1.1 | GO:0021722 | superior olivary nucleus development(GO:0021718) superior olivary nucleus maturation(GO:0021722) |
| 0.4 | 2.2 | GO:0093001 | glycolysis from storage polysaccharide through glucose-1-phosphate(GO:0093001) |
| 0.4 | 3.2 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.4 | 3.2 | GO:0018879 | biphenyl metabolic process(GO:0018879) |
| 0.4 | 2.1 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.3 | 1.0 | GO:0048627 | myoblast development(GO:0048627) |
| 0.3 | 3.1 | GO:0097116 | gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 0.3 | 2.0 | GO:1902045 | negative regulation of Fas signaling pathway(GO:1902045) |
| 0.3 | 1.3 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.3 | 0.9 | GO:0033214 | iron assimilation(GO:0033212) iron assimilation by chelation and transport(GO:0033214) positive regulation of bone mineralization involved in bone maturation(GO:1900159) negative regulation of tumor necrosis factor (ligand) superfamily member 11 production(GO:2000308) |
| 0.3 | 2.2 | GO:0010609 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902162) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.3 | 1.2 | GO:1905176 | metanephric glomerular mesangial cell development(GO:0072255) reversible differentiation(GO:0090677) cell dedifferentiation involved in phenotypic switching(GO:0090678) positive regulation of phenotypic switching(GO:1900241) regulation of vascular smooth muscle cell dedifferentiation(GO:1905174) positive regulation of vascular smooth muscle cell dedifferentiation(GO:1905176) vascular smooth muscle cell dedifferentiation(GO:1990936) |
| 0.3 | 1.4 | GO:0007198 | adenylate cyclase-inhibiting serotonin receptor signaling pathway(GO:0007198) |
| 0.3 | 1.7 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.3 | 1.1 | GO:0035669 | TRAM-dependent toll-like receptor signaling pathway(GO:0035668) TRAM-dependent toll-like receptor 4 signaling pathway(GO:0035669) |
| 0.3 | 1.7 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 0.3 | 1.1 | GO:0097156 | fasciculation of motor neuron axon(GO:0097156) |
| 0.3 | 2.5 | GO:1902669 | positive regulation of axon guidance(GO:1902669) |
| 0.3 | 5.2 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.3 | 1.0 | GO:0010701 | positive regulation of norepinephrine secretion(GO:0010701) |
| 0.3 | 3.6 | GO:0035290 | trunk segmentation(GO:0035290) trunk neural crest cell migration(GO:0036484) ventral trunk neural crest cell migration(GO:0036486) |
| 0.2 | 0.7 | GO:2000657 | regulation of apolipoprotein binding(GO:2000656) negative regulation of apolipoprotein binding(GO:2000657) |
| 0.2 | 2.7 | GO:0001714 | endodermal cell fate specification(GO:0001714) |
| 0.2 | 1.0 | GO:0000412 | histone peptidyl-prolyl isomerization(GO:0000412) |
| 0.2 | 1.5 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 0.2 | 3.4 | GO:0035634 | response to stilbenoid(GO:0035634) |
| 0.2 | 0.9 | GO:0060262 | regulation of N-terminal protein palmitoylation(GO:0060254) negative regulation of N-terminal protein palmitoylation(GO:0060262) negative regulation of protein lipidation(GO:1903060) |
| 0.2 | 2.3 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.2 | 0.7 | GO:0070093 | negative regulation of glucagon secretion(GO:0070093) |
| 0.2 | 1.8 | GO:0021592 | fourth ventricle development(GO:0021592) |
| 0.2 | 2.7 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.2 | 1.3 | GO:0016476 | regulation of embryonic cell shape(GO:0016476) |
| 0.2 | 6.4 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.2 | 0.6 | GO:0006990 | positive regulation of transcription from RNA polymerase II promoter involved in unfolded protein response(GO:0006990) |
| 0.2 | 3.8 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.2 | 0.8 | GO:0048687 | modulation by virus of host transcription(GO:0019056) positive regulation of sprouting of injured axon(GO:0048687) positive regulation of axon extension involved in regeneration(GO:0048691) modulation by symbiont of host transcription(GO:0052026) |
| 0.2 | 1.1 | GO:0035026 | leading edge cell differentiation(GO:0035026) |
| 0.2 | 2.5 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.2 | 1.3 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.2 | 2.6 | GO:0035766 | cell chemotaxis to fibroblast growth factor(GO:0035766) endothelial cell chemotaxis to fibroblast growth factor(GO:0035768) branch elongation involved in ureteric bud branching(GO:0060681) regulation of cell chemotaxis to fibroblast growth factor(GO:1904847) regulation of endothelial cell chemotaxis to fibroblast growth factor(GO:2000544) |
| 0.2 | 1.6 | GO:0045905 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.2 | 8.7 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.2 | 1.5 | GO:0097577 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.2 | 1.8 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.2 | 0.8 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.2 | 1.5 | GO:0045602 | negative regulation of endothelial cell differentiation(GO:0045602) |
| 0.2 | 0.8 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.2 | 2.1 | GO:0043587 | tongue morphogenesis(GO:0043587) |
| 0.1 | 1.8 | GO:1903142 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.1 | 1.9 | GO:0052805 | histidine catabolic process(GO:0006548) imidazole-containing compound catabolic process(GO:0052805) |
| 0.1 | 2.8 | GO:0010765 | positive regulation of sodium ion transport(GO:0010765) |
| 0.1 | 2.0 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 0.1 | 1.7 | GO:0009048 | dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.1 | 0.9 | GO:0097021 | lymphocyte migration into lymphoid organs(GO:0097021) |
| 0.1 | 1.3 | GO:0045586 | regulation of gamma-delta T cell differentiation(GO:0045586) |
| 0.1 | 0.7 | GO:0002934 | desmosome organization(GO:0002934) |
| 0.1 | 2.0 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.1 | 1.1 | GO:1900086 | positive regulation of peptidyl-tyrosine autophosphorylation(GO:1900086) |
| 0.1 | 2.6 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.1 | 1.6 | GO:0032808 | lacrimal gland development(GO:0032808) |
| 0.1 | 0.7 | GO:0042492 | gamma-delta T cell differentiation(GO:0042492) |
| 0.1 | 2.4 | GO:1900016 | negative regulation of fibroblast migration(GO:0010764) negative regulation of cytokine production involved in inflammatory response(GO:1900016) |
| 0.1 | 1.3 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.1 | 0.5 | GO:0086073 | bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 0.1 | 0.7 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.1 | 0.9 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 0.1 | 5.0 | GO:0035640 | exploration behavior(GO:0035640) |
| 0.1 | 0.6 | GO:2000563 | positive regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000563) |
| 0.1 | 0.3 | GO:1903179 | negative regulation of late endosome to lysosome transport(GO:1902823) regulation of dopamine biosynthetic process(GO:1903179) positive regulation of dopamine biosynthetic process(GO:1903181) negative regulation of protein catabolic process in the vacuole(GO:1904351) negative regulation of lysosomal protein catabolic process(GO:1905166) |
| 0.1 | 0.9 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.1 | 0.9 | GO:0061589 | calcium activated phosphatidylserine scrambling(GO:0061589) |
| 0.1 | 3.8 | GO:0060314 | regulation of ryanodine-sensitive calcium-release channel activity(GO:0060314) |
| 0.1 | 3.8 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.1 | 3.0 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.1 | 1.0 | GO:0032888 | regulation of spindle elongation(GO:0032887) regulation of mitotic spindle elongation(GO:0032888) anastral spindle assembly(GO:0055048) protein localization to spindle pole body(GO:0071988) regulation of protein localization to spindle pole body(GO:1902363) positive regulation of protein localization to spindle pole body(GO:1902365) positive regulation of mitotic spindle elongation(GO:1902846) |
| 0.1 | 0.3 | GO:0048203 | vesicle targeting, trans-Golgi to endosome(GO:0048203) |
| 0.1 | 1.3 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.1 | 1.7 | GO:1904903 | ESCRT complex disassembly(GO:1904896) ESCRT III complex disassembly(GO:1904903) |
| 0.1 | 1.7 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.1 | 4.1 | GO:0000717 | nucleotide-excision repair, DNA duplex unwinding(GO:0000717) |
| 0.1 | 0.7 | GO:0071926 | endocannabinoid signaling pathway(GO:0071926) regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.1 | 1.6 | GO:0042048 | olfactory behavior(GO:0042048) |
| 0.1 | 6.5 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.1 | 0.6 | GO:1902462 | regulation of mesenchymal stem cell proliferation(GO:1902460) positive regulation of mesenchymal stem cell proliferation(GO:1902462) |
| 0.1 | 5.2 | GO:0035774 | positive regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0035774) |
| 0.1 | 0.9 | GO:1903764 | regulation of potassium ion export across plasma membrane(GO:1903764) |
| 0.1 | 3.7 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
| 0.1 | 2.7 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.1 | 1.8 | GO:0061436 | establishment of skin barrier(GO:0061436) |
| 0.1 | 0.4 | GO:0097010 | eukaryotic translation initiation factor 4F complex assembly(GO:0097010) |
| 0.1 | 2.7 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.1 | 2.2 | GO:0019372 | lipoxygenase pathway(GO:0019372) |
| 0.1 | 1.1 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.1 | 1.6 | GO:0061339 | establishment of monopolar cell polarity(GO:0061162) establishment or maintenance of monopolar cell polarity(GO:0061339) |
| 0.1 | 8.7 | GO:2000649 | regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.1 | 0.3 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.1 | 0.7 | GO:0010025 | wax biosynthetic process(GO:0010025) wax metabolic process(GO:0010166) |
| 0.1 | 0.8 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.1 | 1.3 | GO:2000675 | negative regulation of type B pancreatic cell apoptotic process(GO:2000675) |
| 0.1 | 3.7 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.1 | 0.8 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 0.1 | 0.5 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.1 | 0.8 | GO:1902572 | negative regulation of serine-type endopeptidase activity(GO:1900004) negative regulation of serine-type peptidase activity(GO:1902572) |
| 0.1 | 1.2 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.1 | 0.3 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.1 | 0.5 | GO:1902031 | regulation of NADP metabolic process(GO:1902031) |
| 0.1 | 3.2 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.1 | 1.2 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.1 | 1.5 | GO:0030033 | microvillus assembly(GO:0030033) |
| 0.1 | 0.3 | GO:0006272 | leading strand elongation(GO:0006272) |
| 0.1 | 2.0 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.1 | 0.7 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 0.1 | 5.2 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.1 | 0.3 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.1 | 1.4 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.1 | 3.7 | GO:0050885 | neuromuscular process controlling balance(GO:0050885) |
| 0.1 | 0.5 | GO:0001878 | response to yeast(GO:0001878) |
| 0.1 | 1.4 | GO:0036148 | phosphatidylglycerol acyl-chain remodeling(GO:0036148) |
| 0.1 | 3.8 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.1 | 1.4 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.1 | 1.0 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.1 | 2.8 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.1 | 1.2 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.1 | 0.7 | GO:0015824 | proline transport(GO:0015824) |
| 0.1 | 0.6 | GO:0071847 | TNFSF11-mediated signaling pathway(GO:0071847) |
| 0.1 | 0.9 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.1 | 1.6 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.1 | 1.4 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.1 | 0.3 | GO:2000674 | regulation of type B pancreatic cell apoptotic process(GO:2000674) |
| 0.1 | 1.2 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.1 | 1.0 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.1 | 0.4 | GO:0008626 | granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 0.1 | 1.2 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 3.0 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.0 | 2.2 | GO:1903959 | regulation of anion transmembrane transport(GO:1903959) |
| 0.0 | 0.8 | GO:0035878 | nail development(GO:0035878) |
| 0.0 | 0.6 | GO:0019368 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.3 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 | 1.7 | GO:0002028 | regulation of sodium ion transport(GO:0002028) |
| 0.0 | 1.2 | GO:0031639 | plasminogen activation(GO:0031639) |
| 0.0 | 0.1 | GO:0010637 | negative regulation of mitochondrial fusion(GO:0010637) |
| 0.0 | 0.7 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.0 | 0.9 | GO:0021794 | thalamus development(GO:0021794) |
| 0.0 | 1.9 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
| 0.0 | 1.8 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.0 | 1.2 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.6 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.0 | 0.2 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.0 | 0.2 | GO:0032480 | negative regulation of type I interferon production(GO:0032480) |
| 0.0 | 1.0 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.0 | 0.2 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.0 | 0.3 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.0 | 1.2 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.0 | 1.5 | GO:0034035 | purine ribonucleoside bisphosphate metabolic process(GO:0034035) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) |
| 0.0 | 0.1 | GO:0090149 | mitochondrial membrane fission(GO:0090149) |
| 0.0 | 0.1 | GO:0060266 | positive regulation of respiratory burst involved in inflammatory response(GO:0060265) negative regulation of respiratory burst involved in inflammatory response(GO:0060266) negative regulation of respiratory burst(GO:0060268) |
| 0.0 | 2.1 | GO:0007223 | Wnt signaling pathway, calcium modulating pathway(GO:0007223) |
| 0.0 | 0.6 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 2.0 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.0 | 0.5 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.0 | 1.2 | GO:0030252 | growth hormone secretion(GO:0030252) |
| 0.0 | 1.9 | GO:0051123 | RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) |
| 0.0 | 0.2 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.0 | 5.6 | GO:0048813 | dendrite morphogenesis(GO:0048813) |
| 0.0 | 0.3 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.0 | 1.5 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.0 | 0.7 | GO:0035162 | embryonic hemopoiesis(GO:0035162) |
| 0.0 | 0.4 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.4 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.0 | 1.0 | GO:0021591 | ventricular system development(GO:0021591) |
| 0.0 | 1.3 | GO:0030516 | regulation of axon extension(GO:0030516) |
| 0.0 | 0.4 | GO:1900271 | regulation of long-term synaptic potentiation(GO:1900271) |
| 0.0 | 0.2 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.0 | 1.0 | GO:1901685 | glutathione derivative metabolic process(GO:1901685) glutathione derivative biosynthetic process(GO:1901687) |
| 0.0 | 0.5 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 0.0 | 0.3 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 1.9 | GO:0046785 | microtubule polymerization(GO:0046785) |
| 0.0 | 0.3 | GO:0070294 | renal sodium ion transport(GO:0003096) renal sodium ion absorption(GO:0070294) |
| 0.0 | 0.9 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 0.7 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 2.0 | GO:0008593 | regulation of Notch signaling pathway(GO:0008593) |
| 0.0 | 0.2 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.0 | 1.0 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 1.3 | GO:0046580 | negative regulation of Ras protein signal transduction(GO:0046580) |
| 0.0 | 1.1 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.0 | 0.9 | GO:0006260 | DNA replication(GO:0006260) |
| 0.0 | 1.0 | GO:0045742 | positive regulation of epidermal growth factor receptor signaling pathway(GO:0045742) |
| 0.0 | 0.1 | GO:1902463 | protein localization to cell leading edge(GO:1902463) |
| 0.0 | 0.3 | GO:0034244 | negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.0 | 0.5 | GO:0050771 | negative regulation of axonogenesis(GO:0050771) |
| 0.0 | 0.3 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.0 | 0.9 | GO:0033119 | negative regulation of RNA splicing(GO:0033119) |
| 0.0 | 0.8 | GO:0071378 | growth hormone receptor signaling pathway(GO:0060396) cellular response to growth hormone stimulus(GO:0071378) |
| 0.0 | 0.0 | GO:0060988 | lipid tube assembly(GO:0060988) |
| 0.0 | 2.9 | GO:0000187 | activation of MAPK activity(GO:0000187) |
| 0.0 | 0.2 | GO:0015812 | gamma-aminobutyric acid transport(GO:0015812) |
| 0.0 | 0.1 | GO:1901315 | negative regulation of histone ubiquitination(GO:0033183) histone H2A K63-linked ubiquitination(GO:0070535) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) |
| 0.0 | 1.3 | GO:0021987 | cerebral cortex development(GO:0021987) |
| 0.0 | 1.1 | GO:0006939 | smooth muscle contraction(GO:0006939) |
| 0.0 | 0.4 | GO:0014047 | glutamate secretion(GO:0014047) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 9.1 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 1.2 | 3.7 | GO:0070702 | inner mucus layer(GO:0070702) outer mucus layer(GO:0070703) |
| 0.6 | 2.9 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.4 | 1.3 | GO:0020003 | symbiont-containing vacuole(GO:0020003) symbiont-containing vacuole membrane(GO:0020005) |
| 0.4 | 1.6 | GO:0034684 | integrin alphav-beta5 complex(GO:0034684) |
| 0.4 | 2.7 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.4 | 1.5 | GO:0008043 | intracellular ferritin complex(GO:0008043) ferritin complex(GO:0070288) |
| 0.3 | 3.4 | GO:1990812 | growth cone filopodium(GO:1990812) |
| 0.3 | 1.6 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
| 0.3 | 8.2 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.3 | 1.8 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.3 | 3.8 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.3 | 3.5 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.3 | 1.1 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.3 | 2.2 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.2 | 4.0 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.2 | 0.9 | GO:0097232 | lamellar body membrane(GO:0097232) alveolar lamellar body membrane(GO:0097233) |
| 0.2 | 0.8 | GO:1902912 | pyruvate kinase complex(GO:1902912) |
| 0.2 | 0.8 | GO:0034676 | integrin alpha6-beta4 complex(GO:0034676) |
| 0.2 | 2.0 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.2 | 7.7 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.2 | 2.7 | GO:0005883 | neurofilament(GO:0005883) |
| 0.2 | 0.8 | GO:0071942 | XPC complex(GO:0071942) |
| 0.1 | 6.9 | GO:0030057 | desmosome(GO:0030057) |
| 0.1 | 2.0 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.1 | 0.9 | GO:0097013 | phagocytic vesicle lumen(GO:0097013) |
| 0.1 | 14.6 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.1 | 2.2 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.1 | 0.8 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.1 | 3.0 | GO:0005861 | troponin complex(GO:0005861) |
| 0.1 | 1.1 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.1 | 3.2 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.1 | 3.4 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.1 | 0.7 | GO:0002169 | 3-methylcrotonyl-CoA carboxylase complex, mitochondrial(GO:0002169) methylcrotonoyl-CoA carboxylase complex(GO:1905202) |
| 0.1 | 0.6 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 0.1 | 1.5 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.1 | 4.2 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.1 | 4.0 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.1 | 8.5 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.1 | 2.9 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.1 | 10.5 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.1 | 1.9 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.1 | 2.5 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.1 | 0.8 | GO:0016342 | catenin complex(GO:0016342) |
| 0.1 | 0.9 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.1 | 2.9 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.1 | 1.4 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.1 | 1.3 | GO:0008091 | spectrin(GO:0008091) |
| 0.1 | 3.3 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.1 | 0.3 | GO:0045293 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.1 | 1.6 | GO:0005922 | connexon complex(GO:0005922) |
| 0.1 | 2.7 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.1 | 11.6 | GO:0043204 | perikaryon(GO:0043204) |
| 0.1 | 1.0 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 1.0 | GO:0097227 | sperm annulus(GO:0097227) |
| 0.1 | 1.9 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 1.3 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 4.3 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.7 | GO:0030130 | clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.0 | 0.8 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.0 | 0.6 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.0 | 1.1 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.3 | GO:0032021 | NELF complex(GO:0032021) |
| 0.0 | 0.3 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 3.3 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 7.7 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 2.6 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 4.3 | GO:0043679 | axon terminus(GO:0043679) |
| 0.0 | 1.2 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 1.0 | GO:0046930 | pore complex(GO:0046930) |
| 0.0 | 4.1 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 3.3 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 0.2 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.0 | 4.6 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 0.8 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 1.3 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.4 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 2.6 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.7 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 3.5 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 0.6 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 4.6 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.1 | GO:0031417 | NatC complex(GO:0031417) |
| 0.0 | 0.7 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.1 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.0 | 5.1 | GO:0005875 | microtubule associated complex(GO:0005875) |
| 0.0 | 1.9 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 2.2 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.0 | 0.7 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.0 | 0.8 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.0 | 8.0 | GO:0005874 | microtubule(GO:0005874) |
| 0.0 | 3.7 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 7.7 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 0.3 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 0.7 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 1.1 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 2.0 | GO:0035579 | specific granule membrane(GO:0035579) |
| 0.0 | 0.8 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 2.6 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 0.3 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 1.7 | GO:0034399 | nuclear periphery(GO:0034399) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 4.9 | GO:0005298 | proline:sodium symporter activity(GO:0005298) |
| 1.5 | 4.6 | GO:0008892 | guanine deaminase activity(GO:0008892) |
| 1.5 | 9.1 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.8 | 8.0 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.8 | 3.2 | GO:0008426 | protein kinase C inhibitor activity(GO:0008426) |
| 0.7 | 2.2 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.7 | 2.7 | GO:0099609 | microtubule lateral binding(GO:0099609) |
| 0.6 | 3.1 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.6 | 1.9 | GO:0036440 | citrate (Si)-synthase activity(GO:0004108) citrate synthase activity(GO:0036440) |
| 0.6 | 1.7 | GO:0030395 | lactose binding(GO:0030395) |
| 0.5 | 4.3 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.5 | 2.6 | GO:0050115 | myosin-light-chain-phosphatase activity(GO:0050115) |
| 0.5 | 2.1 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 0.5 | 1.9 | GO:0061714 | folic acid receptor activity(GO:0061714) |
| 0.5 | 2.7 | GO:0030197 | extracellular matrix constituent, lubricant activity(GO:0030197) |
| 0.4 | 1.3 | GO:0035379 | carbon dioxide transmembrane transporter activity(GO:0035379) |
| 0.4 | 1.2 | GO:0045155 | electron transporter, transferring electrons from CoQH2-cytochrome c reductase complex and cytochrome c oxidase complex activity(GO:0045155) |
| 0.4 | 1.1 | GO:0016534 | cyclin-dependent protein kinase 5 activator activity(GO:0016534) |
| 0.4 | 3.1 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.4 | 8.3 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.3 | 5.2 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.3 | 5.4 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.3 | 3.8 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.3 | 2.9 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.3 | 1.7 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 0.3 | 1.3 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.2 | 3.5 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.2 | 2.1 | GO:0050656 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) 3'-phosphoadenosine 5'-phosphosulfate binding(GO:0050656) |
| 0.2 | 19.0 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.2 | 1.3 | GO:0097001 | ceramide binding(GO:0097001) |
| 0.2 | 2.2 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.2 | 3.0 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.2 | 1.4 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.2 | 3.4 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.2 | 0.6 | GO:0019948 | SUMO activating enzyme activity(GO:0019948) |
| 0.2 | 2.4 | GO:0015288 | porin activity(GO:0015288) |
| 0.2 | 2.0 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.2 | 3.1 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.2 | 3.6 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.2 | 1.0 | GO:0001641 | group II metabotropic glutamate receptor activity(GO:0001641) |
| 0.2 | 4.5 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.2 | 1.3 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.2 | 0.8 | GO:0010465 | nerve growth factor receptor activity(GO:0010465) |
| 0.2 | 2.1 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.2 | 1.1 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.2 | 1.1 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.1 | 2.1 | GO:0051430 | corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.1 | 8.2 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.1 | 0.9 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.1 | 0.8 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.1 | 3.2 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.1 | 2.2 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.1 | 0.3 | GO:0016422 | mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 0.1 | 1.7 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.1 | 0.7 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.1 | 1.5 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 0.9 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.1 | 1.4 | GO:0043176 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.1 | 1.4 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.1 | 1.3 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.1 | 1.5 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.1 | 2.3 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.1 | 0.7 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.1 | 0.7 | GO:0004485 | methylcrotonoyl-CoA carboxylase activity(GO:0004485) |
| 0.1 | 3.2 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.1 | 1.9 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.1 | 3.5 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.1 | 0.3 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 0.1 | 2.3 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.1 | 0.6 | GO:0051022 | GDP-dissociation inhibitor binding(GO:0051021) Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.1 | 0.4 | GO:0034057 | RNA strand-exchange activity(GO:0034057) |
| 0.1 | 2.6 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.1 | 1.6 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.1 | 1.0 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.1 | 0.5 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 0.1 | 0.9 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.1 | 0.8 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.1 | 1.6 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.1 | 0.6 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.1 | 0.4 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.1 | 2.0 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.1 | 0.5 | GO:0004471 | malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.1 | 0.9 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.1 | 1.4 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.1 | 0.9 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.1 | 0.4 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.1 | 0.5 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.1 | 1.2 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.1 | 6.0 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.1 | 0.5 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.1 | 2.7 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.1 | 7.2 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 1.8 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.1 | 11.6 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 2.1 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.1 | 1.5 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 0.3 | GO:0031748 | D1 dopamine receptor binding(GO:0031748) |
| 0.0 | 2.8 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 3.6 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 0.6 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 2.0 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.0 | 1.1 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.9 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 1.0 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.3 | GO:0086089 | voltage-gated potassium channel activity involved in atrial cardiac muscle cell action potential repolarization(GO:0086089) |
| 0.0 | 0.7 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 1.6 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 1.0 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.6 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 1.1 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.9 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 1.0 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 3.0 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.3 | GO:0050694 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.0 | 0.6 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.0 | 1.2 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 6.5 | GO:0002020 | protease binding(GO:0002020) |
| 0.0 | 5.3 | GO:0070851 | growth factor receptor binding(GO:0070851) |
| 0.0 | 1.0 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 1.1 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.9 | GO:0030955 | potassium ion binding(GO:0030955) |
| 0.0 | 0.8 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.0 | 3.6 | GO:0051219 | phosphoprotein binding(GO:0051219) |
| 0.0 | 1.6 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.0 | 1.3 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 0.1 | GO:0004052 | arachidonate 12-lipoxygenase activity(GO:0004052) |
| 0.0 | 14.5 | GO:0015631 | tubulin binding(GO:0015631) |
| 0.0 | 0.3 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.2 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 4.0 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.9 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.0 | 0.2 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.0 | 0.4 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 2.1 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.0 | 1.8 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 1.5 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.3 | GO:1901611 | phosphatidylglycerol binding(GO:1901611) |
| 0.0 | 2.8 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.7 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 2.3 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 2.2 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.0 | 0.7 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.4 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.0 | 0.4 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.7 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.2 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.0 | 2.3 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.9 | GO:0005272 | sodium channel activity(GO:0005272) |
| 0.0 | 1.1 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 1.4 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 1.0 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 0.1 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 0.2 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.0 | 0.5 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.0 | 0.8 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 0.1 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 1.2 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 1.2 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.0 | 1.4 | GO:0008565 | protein transporter activity(GO:0008565) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.6 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.1 | 4.2 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.1 | 2.7 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.1 | 2.7 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.1 | 1.9 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.1 | 24.2 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 2.5 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.1 | 5.1 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.1 | 3.0 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.1 | 7.5 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.1 | 4.0 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.1 | 5.9 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 5.1 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.1 | 2.0 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.1 | 3.0 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.0 | 3.1 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 0.9 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 2.6 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 2.0 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 0.8 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.0 | 1.7 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 2.0 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 1.1 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 1.6 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 3.7 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 1.2 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 1.6 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 3.2 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 0.7 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 1.3 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 1.9 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.7 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 1.9 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 1.7 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 6.5 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.7 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 0.4 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 0.6 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.7 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 1.5 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 1.3 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.4 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 1.2 | PID E2F PATHWAY | E2F transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 4.6 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.2 | 5.7 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.2 | 5.7 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.2 | 2.6 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.1 | 5.9 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.1 | 6.4 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.1 | 5.5 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.1 | 2.9 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.1 | 3.2 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.1 | 5.8 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.1 | 4.9 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 6.2 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 3.0 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.1 | 4.5 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.1 | 1.3 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.1 | 3.2 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.1 | 1.3 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.1 | 3.0 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.1 | 2.9 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.1 | 0.6 | REACTOME G BETA GAMMA SIGNALLING THROUGH PI3KGAMMA | Genes involved in G beta:gamma signalling through PI3Kgamma |
| 0.1 | 1.9 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.1 | 0.7 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.1 | 2.8 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.1 | 2.2 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.1 | 1.3 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.1 | 1.9 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.1 | 1.6 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.1 | 2.6 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.1 | 1.1 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.1 | 2.5 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.1 | 18.2 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.1 | 4.1 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 1.5 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 2.0 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 1.7 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 8.2 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 2.1 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 3.2 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 2.1 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 4.8 | REACTOME MUSCLE CONTRACTION | Genes involved in Muscle contraction |
| 0.0 | 1.7 | REACTOME REGULATION OF INSULIN SECRETION BY GLUCAGON LIKE PEPTIDE1 | Genes involved in Regulation of Insulin Secretion by Glucagon-like Peptide-1 |
| 0.0 | 1.0 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 1.0 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.5 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.9 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 1.4 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 2.2 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.0 | 0.8 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.0 | 2.1 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 2.0 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.8 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 1.0 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 1.0 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.0 | 0.3 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.0 | 1.0 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.0 | 0.8 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 0.6 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 1.2 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 0.7 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.0 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.0 | 0.3 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |