Project
Illumina Body Map 2: averaged replicates
Navigation
Downloads
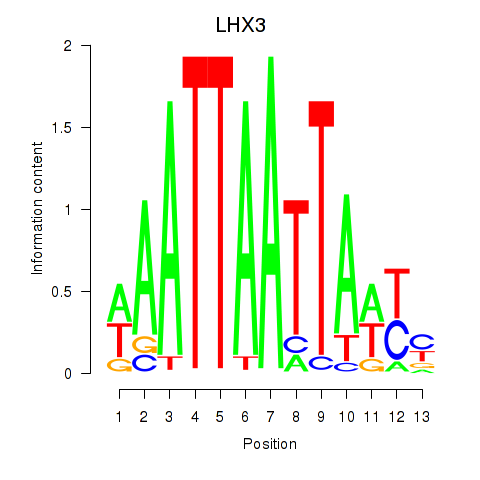
Results for LHX3
Z-value: 1.57
Transcription factors associated with LHX3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
LHX3
|
ENSG00000107187.11 | LIM homeobox 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| LHX3 | hg19_v2_chr9_-_139094988_139095012 | -0.41 | 1.9e-02 | Click! |
Activity profile of LHX3 motif
Sorted Z-values of LHX3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_155484103 | 4.94 |
ENST00000302068.4
|
FGB
|
fibrinogen beta chain |
| chr4_+_155484155 | 4.68 |
ENST00000509493.1
|
FGB
|
fibrinogen beta chain |
| chr3_+_151591422 | 4.30 |
ENST00000362032.5
|
SUCNR1
|
succinate receptor 1 |
| chr5_-_39274617 | 3.47 |
ENST00000510188.1
|
FYB
|
FYN binding protein |
| chr8_-_66750978 | 3.25 |
ENST00000523253.1
|
PDE7A
|
phosphodiesterase 7A |
| chr2_+_218994002 | 2.82 |
ENST00000428565.1
|
CXCR2
|
chemokine (C-X-C motif) receptor 2 |
| chr12_-_10282681 | 2.65 |
ENST00000533022.1
|
CLEC7A
|
C-type lectin domain family 7, member A |
| chr7_-_36764062 | 2.61 |
ENST00000435386.1
|
AOAH
|
acyloxyacyl hydrolase (neutrophil) |
| chr19_+_14693888 | 2.60 |
ENST00000547437.1
ENST00000397439.2 ENST00000417570.1 |
CLEC17A
|
C-type lectin domain family 17, member A |
| chr2_+_135596180 | 2.60 |
ENST00000283054.4
ENST00000392928.1 |
ACMSD
|
aminocarboxymuconate semialdehyde decarboxylase |
| chr12_-_10282742 | 2.55 |
ENST00000298523.5
ENST00000396484.2 ENST00000310002.4 |
CLEC7A
|
C-type lectin domain family 7, member A |
| chr7_-_36764142 | 2.53 |
ENST00000258749.5
ENST00000535891.1 |
AOAH
|
acyloxyacyl hydrolase (neutrophil) |
| chr2_-_89160117 | 2.51 |
ENST00000390238.2
|
IGKJ5
|
immunoglobulin kappa joining 5 |
| chr19_-_51920952 | 2.43 |
ENST00000356298.5
ENST00000339313.5 ENST00000529627.1 ENST00000439889.2 ENST00000353836.5 ENST00000432469.2 |
SIGLEC10
|
sialic acid binding Ig-like lectin 10 |
| chr12_-_10282836 | 2.29 |
ENST00000304084.8
ENST00000353231.5 ENST00000525605.1 |
CLEC7A
|
C-type lectin domain family 7, member A |
| chr3_+_186692745 | 2.22 |
ENST00000438590.1
|
ST6GAL1
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 |
| chr20_-_7238861 | 2.14 |
ENST00000428954.1
|
RP11-19D2.1
|
RP11-19D2.1 |
| chr2_+_135596106 | 2.10 |
ENST00000356140.5
|
ACMSD
|
aminocarboxymuconate semialdehyde decarboxylase |
| chr11_+_59824060 | 2.08 |
ENST00000395032.2
ENST00000358152.2 |
MS4A3
|
membrane-spanning 4-domains, subfamily A, member 3 (hematopoietic cell-specific) |
| chr3_+_63953415 | 2.07 |
ENST00000484332.1
|
ATXN7
|
ataxin 7 |
| chr1_-_92952433 | 1.97 |
ENST00000294702.5
|
GFI1
|
growth factor independent 1 transcription repressor |
| chr7_-_115799942 | 1.97 |
ENST00000484212.1
|
TFEC
|
transcription factor EC |
| chr1_-_160549235 | 1.85 |
ENST00000368054.3
ENST00000368048.3 ENST00000311224.4 ENST00000368051.3 ENST00000534968.1 |
CD84
|
CD84 molecule |
| chr17_-_39093672 | 1.84 |
ENST00000209718.3
ENST00000436344.3 ENST00000485751.1 |
KRT23
|
keratin 23 (histone deacetylase inducible) |
| chr20_+_5987890 | 1.84 |
ENST00000378868.4
|
CRLS1
|
cardiolipin synthase 1 |
| chr1_-_150944411 | 1.81 |
ENST00000368949.4
|
CERS2
|
ceramide synthase 2 |
| chr7_-_87342564 | 1.79 |
ENST00000265724.3
ENST00000416177.1 |
ABCB1
|
ATP-binding cassette, sub-family B (MDR/TAP), member 1 |
| chr11_+_59824127 | 1.79 |
ENST00000278865.3
|
MS4A3
|
membrane-spanning 4-domains, subfamily A, member 3 (hematopoietic cell-specific) |
| chrX_+_107288280 | 1.78 |
ENST00000458383.1
|
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chr5_-_88120083 | 1.74 |
ENST00000509373.1
|
MEF2C
|
myocyte enhancer factor 2C |
| chr6_-_159466136 | 1.68 |
ENST00000367066.3
ENST00000326965.6 |
TAGAP
|
T-cell activation RhoGTPase activating protein |
| chr16_-_28634874 | 1.67 |
ENST00000395609.1
ENST00000350842.4 |
SULT1A1
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 1 |
| chr6_-_66417107 | 1.50 |
ENST00000370621.3
ENST00000370618.3 ENST00000503581.1 ENST00000393380.2 |
EYS
|
eyes shut homolog (Drosophila) |
| chr1_+_244214577 | 1.47 |
ENST00000358704.4
|
ZBTB18
|
zinc finger and BTB domain containing 18 |
| chr5_-_88120151 | 1.47 |
ENST00000506716.1
|
MEF2C
|
myocyte enhancer factor 2C |
| chr14_+_22959438 | 1.46 |
ENST00000390489.1
|
TRAJ48
|
T cell receptor alpha joining 48 |
| chr2_-_145278475 | 1.45 |
ENST00000558170.2
|
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr4_-_36245561 | 1.43 |
ENST00000506189.1
|
ARAP2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
| chr19_+_50016411 | 1.32 |
ENST00000426395.3
ENST00000600273.1 ENST00000599988.1 |
FCGRT
|
Fc fragment of IgG, receptor, transporter, alpha |
| chr5_-_173217916 | 1.31 |
ENST00000523617.1
|
CTB-43E15.4
|
CTB-43E15.4 |
| chrX_+_9431324 | 1.30 |
ENST00000407597.2
ENST00000424279.1 ENST00000536365.1 ENST00000441088.1 ENST00000380961.1 ENST00000415293.1 |
TBL1X
|
transducin (beta)-like 1X-linked |
| chr14_+_22963806 | 1.28 |
ENST00000390493.1
|
TRAJ44
|
T cell receptor alpha joining 44 |
| chr9_-_128246769 | 1.26 |
ENST00000444226.1
|
MAPKAP1
|
mitogen-activated protein kinase associated protein 1 |
| chr6_+_130339710 | 1.24 |
ENST00000526087.1
ENST00000533560.1 ENST00000361794.2 |
L3MBTL3
|
l(3)mbt-like 3 (Drosophila) |
| chr12_+_59989918 | 1.19 |
ENST00000547379.1
ENST00000549465.1 |
SLC16A7
|
solute carrier family 16 (monocarboxylate transporter), member 7 |
| chr12_-_118628315 | 1.17 |
ENST00000540561.1
|
TAOK3
|
TAO kinase 3 |
| chr17_-_27418537 | 1.16 |
ENST00000408971.2
|
TIAF1
|
TGFB1-induced anti-apoptotic factor 1 |
| chr14_-_52436247 | 1.15 |
ENST00000597846.1
|
AL358333.1
|
HCG2013195; Uncharacterized protein |
| chr14_+_62164340 | 1.15 |
ENST00000557538.1
ENST00000539097.1 |
HIF1A
|
hypoxia inducible factor 1, alpha subunit (basic helix-loop-helix transcription factor) |
| chr8_-_57123815 | 1.14 |
ENST00000316981.3
ENST00000423799.2 ENST00000429357.2 |
PLAG1
|
pleiomorphic adenoma gene 1 |
| chr14_+_22465771 | 1.13 |
ENST00000390445.2
|
TRAV17
|
T cell receptor alpha variable 17 |
| chr1_-_150208320 | 1.12 |
ENST00000534220.1
|
ANP32E
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chr3_+_41236325 | 1.12 |
ENST00000426215.1
ENST00000405570.1 |
CTNNB1
|
catenin (cadherin-associated protein), beta 1, 88kDa |
| chr17_-_64225508 | 1.11 |
ENST00000205948.6
|
APOH
|
apolipoprotein H (beta-2-glycoprotein I) |
| chr12_-_118628350 | 1.11 |
ENST00000537952.1
ENST00000537822.1 |
TAOK3
|
TAO kinase 3 |
| chr10_+_104263743 | 1.08 |
ENST00000369902.3
ENST00000369899.2 ENST00000423559.2 |
SUFU
|
suppressor of fused homolog (Drosophila) |
| chr3_+_141106643 | 1.07 |
ENST00000514251.1
|
ZBTB38
|
zinc finger and BTB domain containing 38 |
| chr6_-_9933500 | 1.06 |
ENST00000492169.1
|
OFCC1
|
orofacial cleft 1 candidate 1 |
| chr19_+_48949030 | 1.04 |
ENST00000253237.5
|
GRWD1
|
glutamate-rich WD repeat containing 1 |
| chr1_-_150208412 | 1.03 |
ENST00000532744.1
ENST00000369114.5 ENST00000369115.2 ENST00000369116.4 |
ANP32E
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chr1_+_66820058 | 1.02 |
ENST00000480109.2
|
PDE4B
|
phosphodiesterase 4B, cAMP-specific |
| chr5_-_173217931 | 1.01 |
ENST00000522731.1
|
CTB-43E15.4
|
CTB-43E15.4 |
| chr12_-_78934441 | 0.99 |
ENST00000546865.1
ENST00000547089.1 |
RP11-171L9.1
|
RP11-171L9.1 |
| chr17_-_60142609 | 0.99 |
ENST00000397786.2
|
MED13
|
mediator complex subunit 13 |
| chrX_+_107288197 | 0.99 |
ENST00000415430.3
|
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chr5_-_88119580 | 0.98 |
ENST00000539796.1
|
MEF2C
|
myocyte enhancer factor 2C |
| chr1_-_150208363 | 0.96 |
ENST00000436748.2
|
ANP32E
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chr4_-_41884620 | 0.91 |
ENST00000504870.1
|
LINC00682
|
long intergenic non-protein coding RNA 682 |
| chr3_-_141747950 | 0.91 |
ENST00000497579.1
|
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr1_-_150208291 | 0.90 |
ENST00000533654.1
|
ANP32E
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chr10_-_74283694 | 0.89 |
ENST00000398763.4
ENST00000418483.2 ENST00000489666.2 |
MICU1
|
mitochondrial calcium uptake 1 |
| chr19_+_50016610 | 0.87 |
ENST00000596975.1
|
FCGRT
|
Fc fragment of IgG, receptor, transporter, alpha |
| chr7_+_39017504 | 0.87 |
ENST00000403058.1
|
POU6F2
|
POU class 6 homeobox 2 |
| chr19_+_48949087 | 0.86 |
ENST00000598711.1
|
GRWD1
|
glutamate-rich WD repeat containing 1 |
| chrM_-_14670 | 0.86 |
ENST00000361681.2
|
MT-ND6
|
mitochondrially encoded NADH dehydrogenase 6 |
| chr2_-_8715616 | 0.85 |
ENST00000418358.1
|
AC011747.3
|
AC011747.3 |
| chr17_-_38956205 | 0.84 |
ENST00000306658.7
|
KRT28
|
keratin 28 |
| chr12_-_7656357 | 0.84 |
ENST00000396620.3
ENST00000432237.2 ENST00000359156.4 |
CD163
|
CD163 molecule |
| chr10_-_115904361 | 0.83 |
ENST00000428953.1
ENST00000543782.1 |
C10orf118
|
chromosome 10 open reading frame 118 |
| chr11_-_67981046 | 0.83 |
ENST00000402789.1
ENST00000402185.2 ENST00000458496.1 |
SUV420H1
|
suppressor of variegation 4-20 homolog 1 (Drosophila) |
| chrX_-_110655306 | 0.82 |
ENST00000371993.2
|
DCX
|
doublecortin |
| chr15_+_64680003 | 0.80 |
ENST00000261884.3
|
TRIP4
|
thyroid hormone receptor interactor 4 |
| chr3_+_121902511 | 0.79 |
ENST00000490131.1
|
CASR
|
calcium-sensing receptor |
| chr4_+_95128748 | 0.79 |
ENST00000359052.4
|
SMARCAD1
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 |
| chr3_+_68055366 | 0.78 |
ENST00000496687.1
|
FAM19A1
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A1 |
| chr2_-_99279928 | 0.77 |
ENST00000414521.2
|
MGAT4A
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme A |
| chr12_-_42631529 | 0.76 |
ENST00000548917.1
|
YAF2
|
YY1 associated factor 2 |
| chr5_+_67586465 | 0.75 |
ENST00000336483.5
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
| chr6_+_114178512 | 0.74 |
ENST00000368635.4
|
MARCKS
|
myristoylated alanine-rich protein kinase C substrate |
| chr7_-_105332084 | 0.74 |
ENST00000472195.1
|
ATXN7L1
|
ataxin 7-like 1 |
| chr1_-_24126023 | 0.73 |
ENST00000429356.1
|
GALE
|
UDP-galactose-4-epimerase |
| chr4_-_120243545 | 0.73 |
ENST00000274024.3
|
FABP2
|
fatty acid binding protein 2, intestinal |
| chr6_+_26104104 | 0.73 |
ENST00000377803.2
|
HIST1H4C
|
histone cluster 1, H4c |
| chr7_+_107224364 | 0.72 |
ENST00000491150.1
|
BCAP29
|
B-cell receptor-associated protein 29 |
| chr4_-_23891693 | 0.72 |
ENST00000264867.2
|
PPARGC1A
|
peroxisome proliferator-activated receptor gamma, coactivator 1 alpha |
| chr12_+_49740700 | 0.72 |
ENST00000549441.2
ENST00000395069.3 |
DNAJC22
|
DnaJ (Hsp40) homolog, subfamily C, member 22 |
| chr4_-_23891658 | 0.71 |
ENST00000507380.1
|
PPARGC1A
|
peroxisome proliferator-activated receptor gamma, coactivator 1 alpha |
| chrX_+_107288239 | 0.69 |
ENST00000217957.5
|
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chr4_+_113568207 | 0.69 |
ENST00000511529.1
|
LARP7
|
La ribonucleoprotein domain family, member 7 |
| chr11_-_67980744 | 0.69 |
ENST00000401547.2
ENST00000453170.1 ENST00000304363.4 |
SUV420H1
|
suppressor of variegation 4-20 homolog 1 (Drosophila) |
| chrX_-_39186610 | 0.66 |
ENST00000429281.1
ENST00000448597.1 |
RP11-265P11.2
|
RP11-265P11.2 |
| chr15_+_48483736 | 0.66 |
ENST00000417307.2
ENST00000559641.1 |
CTXN2
SLC12A1
|
cortexin 2 solute carrier family 12 (sodium/potassium/chloride transporter), member 1 |
| chr1_-_24126051 | 0.63 |
ENST00000445705.1
|
GALE
|
UDP-galactose-4-epimerase |
| chr9_-_215744 | 0.63 |
ENST00000382387.2
|
C9orf66
|
chromosome 9 open reading frame 66 |
| chr17_-_71223839 | 0.63 |
ENST00000579872.1
ENST00000580032.1 |
FAM104A
|
family with sequence similarity 104, member A |
| chr3_-_47517302 | 0.62 |
ENST00000441517.2
ENST00000545718.1 |
SCAP
|
SREBF chaperone |
| chr4_+_41937131 | 0.61 |
ENST00000504986.1
ENST00000508448.1 ENST00000513702.1 ENST00000325094.5 |
TMEM33
|
transmembrane protein 33 |
| chr14_-_104408032 | 0.61 |
ENST00000455348.2
|
RD3L
|
retinal degeneration 3-like |
| chr19_-_43969796 | 0.59 |
ENST00000244333.3
|
LYPD3
|
LY6/PLAUR domain containing 3 |
| chrX_+_108779004 | 0.58 |
ENST00000218004.1
|
NXT2
|
nuclear transport factor 2-like export factor 2 |
| chr20_+_11008408 | 0.57 |
ENST00000378252.1
|
C20orf187
|
chromosome 20 open reading frame 187 |
| chrX_-_106243294 | 0.56 |
ENST00000255495.7
|
MORC4
|
MORC family CW-type zinc finger 4 |
| chr14_+_55494323 | 0.56 |
ENST00000339298.2
|
SOCS4
|
suppressor of cytokine signaling 4 |
| chr13_-_81801115 | 0.55 |
ENST00000567258.1
|
LINC00564
|
long intergenic non-protein coding RNA 564 |
| chr1_+_115572415 | 0.54 |
ENST00000256592.1
|
TSHB
|
thyroid stimulating hormone, beta |
| chr10_-_90611566 | 0.52 |
ENST00000371930.4
|
ANKRD22
|
ankyrin repeat domain 22 |
| chr17_+_35851570 | 0.52 |
ENST00000394386.1
|
DUSP14
|
dual specificity phosphatase 14 |
| chr1_+_168250194 | 0.50 |
ENST00000367821.3
|
TBX19
|
T-box 19 |
| chr5_+_179135246 | 0.49 |
ENST00000508787.1
|
CANX
|
calnexin |
| chr16_-_1538765 | 0.49 |
ENST00000447419.2
ENST00000440447.2 |
PTX4
|
pentraxin 4, long |
| chr3_+_69812877 | 0.49 |
ENST00000457080.1
ENST00000328528.6 |
MITF
|
microphthalmia-associated transcription factor |
| chr5_+_126984710 | 0.49 |
ENST00000379445.3
|
CTXN3
|
cortexin 3 |
| chr19_-_11457162 | 0.48 |
ENST00000590482.1
|
TMEM205
|
transmembrane protein 205 |
| chr11_-_559377 | 0.48 |
ENST00000486629.1
|
C11orf35
|
chromosome 11 open reading frame 35 |
| chr3_+_138340067 | 0.47 |
ENST00000479848.1
|
FAIM
|
Fas apoptotic inhibitory molecule |
| chr19_+_11457175 | 0.46 |
ENST00000458408.1
ENST00000586451.1 ENST00000588592.1 |
CCDC159
|
coiled-coil domain containing 159 |
| chr18_+_72201664 | 0.46 |
ENST00000358821.3
|
CNDP1
|
carnosine dipeptidase 1 (metallopeptidase M20 family) |
| chr10_-_27529486 | 0.44 |
ENST00000375888.1
|
ACBD5
|
acyl-CoA binding domain containing 5 |
| chr2_+_185463093 | 0.44 |
ENST00000302277.6
|
ZNF804A
|
zinc finger protein 804A |
| chr14_+_55493920 | 0.44 |
ENST00000395472.2
ENST00000555846.1 |
SOCS4
|
suppressor of cytokine signaling 4 |
| chr6_-_138833630 | 0.44 |
ENST00000533765.1
|
NHSL1
|
NHS-like 1 |
| chr6_-_47445214 | 0.43 |
ENST00000604014.1
|
RP11-385F7.1
|
RP11-385F7.1 |
| chr8_-_25281747 | 0.42 |
ENST00000421054.2
|
GNRH1
|
gonadotropin-releasing hormone 1 (luteinizing-releasing hormone) |
| chrX_-_106243451 | 0.42 |
ENST00000355610.4
ENST00000535534.1 |
MORC4
|
MORC family CW-type zinc finger 4 |
| chr17_-_10372875 | 0.40 |
ENST00000255381.2
|
MYH4
|
myosin, heavy chain 4, skeletal muscle |
| chr8_+_119294456 | 0.39 |
ENST00000366457.2
|
AC023590.1
|
Uncharacterized protein |
| chr2_+_234826016 | 0.38 |
ENST00000324695.4
ENST00000433712.2 |
TRPM8
|
transient receptor potential cation channel, subfamily M, member 8 |
| chr14_-_107049312 | 0.38 |
ENST00000390627.2
|
IGHV3-53
|
immunoglobulin heavy variable 3-53 |
| chr10_+_106937525 | 0.37 |
ENST00000369699.4
|
SORCS3
|
sortilin-related VPS10 domain containing receptor 3 |
| chr14_+_39583427 | 0.36 |
ENST00000308317.6
ENST00000396249.2 ENST00000250379.8 ENST00000534684.2 ENST00000527381.1 |
GEMIN2
|
gem (nuclear organelle) associated protein 2 |
| chr12_+_40787194 | 0.35 |
ENST00000425730.2
ENST00000454784.4 |
MUC19
|
mucin 19, oligomeric |
| chr5_-_78809950 | 0.35 |
ENST00000334082.6
|
HOMER1
|
homer homolog 1 (Drosophila) |
| chr4_+_158141899 | 0.34 |
ENST00000264426.9
ENST00000506284.1 |
GRIA2
|
glutamate receptor, ionotropic, AMPA 2 |
| chr3_+_138340049 | 0.34 |
ENST00000464668.1
|
FAIM
|
Fas apoptotic inhibitory molecule |
| chrM_+_10464 | 0.33 |
ENST00000361335.1
|
MT-ND4L
|
mitochondrially encoded NADH dehydrogenase 4L |
| chr19_+_11457232 | 0.33 |
ENST00000587531.1
|
CCDC159
|
coiled-coil domain containing 159 |
| chrX_+_84258832 | 0.31 |
ENST00000373173.2
|
APOOL
|
apolipoprotein O-like |
| chr18_+_28956740 | 0.30 |
ENST00000308128.4
ENST00000359747.4 |
DSG4
|
desmoglein 4 |
| chr14_-_106668095 | 0.30 |
ENST00000390606.2
|
IGHV3-20
|
immunoglobulin heavy variable 3-20 |
| chr15_-_20193370 | 0.29 |
ENST00000558565.2
|
IGHV3OR15-7
|
immunoglobulin heavy variable 3/OR15-7 (pseudogene) |
| chr15_-_75748115 | 0.29 |
ENST00000360439.4
|
SIN3A
|
SIN3 transcription regulator family member A |
| chr19_+_9296279 | 0.28 |
ENST00000344248.2
|
OR7D2
|
olfactory receptor, family 7, subfamily D, member 2 |
| chr7_-_122840015 | 0.28 |
ENST00000194130.2
|
SLC13A1
|
solute carrier family 13 (sodium/sulfate symporter), member 1 |
| chr16_+_28565230 | 0.28 |
ENST00000317058.3
|
CCDC101
|
coiled-coil domain containing 101 |
| chr8_+_91233750 | 0.27 |
ENST00000523406.1
|
LINC00534
|
long intergenic non-protein coding RNA 534 |
| chr12_+_4385230 | 0.27 |
ENST00000536537.1
|
CCND2
|
cyclin D2 |
| chr13_+_113301358 | 0.27 |
ENST00000356049.1
|
C13orf35
|
chromosome 13 open reading frame 35 |
| chr1_-_45988542 | 0.26 |
ENST00000424390.1
|
PRDX1
|
peroxiredoxin 1 |
| chr7_-_15014398 | 0.26 |
ENST00000437998.1
|
DGKB
|
diacylglycerol kinase, beta 90kDa |
| chr16_+_15489629 | 0.26 |
ENST00000396385.3
|
MPV17L
|
MPV17 mitochondrial membrane protein-like |
| chr16_+_72459838 | 0.26 |
ENST00000564508.1
|
AC004158.3
|
AC004158.3 |
| chr16_+_89334512 | 0.25 |
ENST00000602042.1
|
AC137932.1
|
AC137932.1 |
| chr4_+_74301880 | 0.25 |
ENST00000395792.2
ENST00000226359.2 |
AFP
|
alpha-fetoprotein |
| chr20_-_656823 | 0.25 |
ENST00000246104.6
|
SCRT2
|
scratch family zinc finger 2 |
| chr11_-_5799897 | 0.24 |
ENST00000317093.2
|
OR52N5
|
olfactory receptor, family 52, subfamily N, member 5 |
| chr19_-_6279932 | 0.24 |
ENST00000252674.7
|
MLLT1
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 1 |
| chr4_+_158142750 | 0.23 |
ENST00000505888.1
ENST00000449365.1 |
GRIA2
|
glutamate receptor, ionotropic, AMPA 2 |
| chr6_+_23337918 | 0.23 |
ENST00000431001.1
|
RP11-439H9.1
|
RP11-439H9.1 |
| chr1_-_182640988 | 0.22 |
ENST00000367556.1
|
RGS8
|
regulator of G-protein signaling 8 |
| chr11_-_327537 | 0.22 |
ENST00000602735.1
|
IFITM3
|
interferon induced transmembrane protein 3 |
| chr6_-_49712091 | 0.21 |
ENST00000371159.4
|
CRISP3
|
cysteine-rich secretory protein 3 |
| chr18_+_72201829 | 0.21 |
ENST00000582365.1
|
CNDP1
|
carnosine dipeptidase 1 (metallopeptidase M20 family) |
| chr12_-_53994805 | 0.21 |
ENST00000328463.7
|
ATF7
|
activating transcription factor 7 |
| chrX_+_24167746 | 0.20 |
ENST00000428571.1
ENST00000539115.1 |
ZFX
|
zinc finger protein, X-linked |
| chr18_+_6774000 | 0.20 |
ENST00000532723.1
|
ARHGAP28
|
Rho GTPase activating protein 28 |
| chr5_+_55149150 | 0.20 |
ENST00000297015.3
|
IL31RA
|
interleukin 31 receptor A |
| chr5_-_59783882 | 0.20 |
ENST00000505507.2
ENST00000502484.2 |
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chr12_-_11287243 | 0.19 |
ENST00000539585.1
|
TAS2R30
|
taste receptor, type 2, member 30 |
| chr14_-_57197224 | 0.15 |
ENST00000554597.1
ENST00000556696.1 |
RP11-1085N6.3
|
Uncharacterized protein |
| chr2_-_14541060 | 0.15 |
ENST00000418420.1
ENST00000417751.1 |
LINC00276
|
long intergenic non-protein coding RNA 276 |
| chr2_-_231860596 | 0.15 |
ENST00000441063.1
ENST00000434094.1 ENST00000418330.1 ENST00000457803.1 ENST00000414876.1 ENST00000446741.1 ENST00000426904.1 |
AC105344.2
|
SPATA3 antisense RNA 1 (head to head) |
| chr4_+_71384257 | 0.13 |
ENST00000339336.4
|
AMTN
|
amelotin |
| chr5_+_59783941 | 0.12 |
ENST00000506884.1
ENST00000504876.2 |
PART1
|
prostate androgen-regulated transcript 1 (non-protein coding) |
| chr4_+_71384300 | 0.12 |
ENST00000504451.1
|
AMTN
|
amelotin |
| chr12_-_52761262 | 0.11 |
ENST00000257901.3
|
KRT85
|
keratin 85 |
| chr1_-_182641037 | 0.09 |
ENST00000483095.2
|
RGS8
|
regulator of G-protein signaling 8 |
| chr1_-_57285038 | 0.09 |
ENST00000343433.6
|
C1orf168
|
chromosome 1 open reading frame 168 |
| chr4_-_68829226 | 0.09 |
ENST00000396188.2
|
TMPRSS11A
|
transmembrane protease, serine 11A |
| chr15_-_75748143 | 0.08 |
ENST00000568431.1
ENST00000568309.1 ENST00000568190.1 ENST00000570115.1 ENST00000564778.1 |
SIN3A
|
SIN3 transcription regulator family member A |
| chr15_+_86686953 | 0.08 |
ENST00000421325.2
|
AGBL1
|
ATP/GTP binding protein-like 1 |
| chr2_+_78143006 | 0.07 |
ENST00000443419.1
|
AC073628.1
|
AC073628.1 |
| chr16_-_52061283 | 0.07 |
ENST00000566314.1
|
C16orf97
|
chromosome 16 open reading frame 97 |
| chr4_-_41749724 | 0.06 |
ENST00000510424.1
|
PHOX2B
|
paired-like homeobox 2b |
| chr4_-_103940791 | 0.06 |
ENST00000510559.1
ENST00000394789.3 ENST00000296422.7 |
SLC9B1
|
solute carrier family 9, subfamily B (NHA1, cation proton antiporter 1), member 1 |
| chr11_+_5009424 | 0.06 |
ENST00000300762.1
|
MMP26
|
matrix metallopeptidase 26 |
| chr18_-_5197501 | 0.05 |
ENST00000580650.1
|
C18orf42
|
chromosome 18 open reading frame 42 |
| chr6_-_49712147 | 0.05 |
ENST00000433368.2
ENST00000354620.4 |
CRISP3
|
cysteine-rich secretory protein 3 |
| chr4_-_68829144 | 0.04 |
ENST00000508048.1
|
TMPRSS11A
|
transmembrane protease, serine 11A |
| chr15_-_99057551 | 0.04 |
ENST00000558256.1
|
FAM169B
|
family with sequence similarity 169, member B |
| chrM_+_14741 | 0.04 |
ENST00000361789.2
|
MT-CYB
|
mitochondrially encoded cytochrome b |
Network of associatons between targets according to the STRING database.
First level regulatory network of LHX3
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.2 | GO:0002416 | IgG immunoglobulin transcytosis in epithelial cells mediated by FcRn immunoglobulin receptor(GO:0002416) |
| 0.7 | 9.6 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.7 | 4.7 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.7 | 2.0 | GO:0070105 | positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 0.6 | 4.2 | GO:0003172 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.5 | 1.4 | GO:1901860 | positive regulation of mitochondrial DNA metabolic process(GO:1901860) |
| 0.5 | 2.8 | GO:0038112 | interleukin-8-mediated signaling pathway(GO:0038112) |
| 0.5 | 1.8 | GO:0014011 | Schwann cell proliferation involved in axon regeneration(GO:0014011) negative regulation of Schwann cell migration(GO:1900148) regulation of Schwann cell proliferation involved in axon regeneration(GO:1905044) negative regulation of Schwann cell proliferation involved in axon regeneration(GO:1905045) |
| 0.4 | 1.8 | GO:0032661 | regulation of interleukin-18 production(GO:0032661) |
| 0.4 | 1.1 | GO:0021775 | smoothened signaling pathway involved in ventral spinal cord interneuron specification(GO:0021775) smoothened signaling pathway involved in spinal cord motor neuron cell fate specification(GO:0021776) |
| 0.3 | 7.5 | GO:0009756 | carbohydrate mediated signaling(GO:0009756) |
| 0.3 | 1.1 | GO:0007403 | glial cell fate determination(GO:0007403) canonical Wnt signaling pathway involved in positive regulation of cardiac outflow tract cell proliferation(GO:0061324) regulation of chromatin-mediated maintenance of transcription(GO:1904499) positive regulation of chromatin-mediated maintenance of transcription(GO:1904501) regulation of euchromatin binding(GO:1904793) |
| 0.2 | 1.1 | GO:0070101 | epithelial cell differentiation involved in mammary gland alveolus development(GO:0061030) positive regulation of chemokine-mediated signaling pathway(GO:0070101) |
| 0.2 | 1.8 | GO:1901529 | positive regulation of anion channel activity(GO:1901529) |
| 0.2 | 0.6 | GO:1903371 | regulation of endoplasmic reticulum tubular network organization(GO:1903371) |
| 0.2 | 1.8 | GO:0097068 | response to thyroxine(GO:0097068) response to L-phenylalanine derivative(GO:1904386) |
| 0.2 | 2.1 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.2 | 1.3 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.2 | 0.9 | GO:0007402 | ganglion mother cell fate determination(GO:0007402) |
| 0.2 | 1.4 | GO:0021767 | mammillary body development(GO:0021767) mammillary axonal complex development(GO:0061373) positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.1 | 1.2 | GO:0006848 | pyruvate transport(GO:0006848) pyruvate transmembrane transport(GO:1901475) |
| 0.1 | 3.5 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.1 | 0.4 | GO:1990637 | response to prolactin(GO:1990637) |
| 0.1 | 1.1 | GO:0034392 | negative regulation of smooth muscle cell apoptotic process(GO:0034392) |
| 0.1 | 0.9 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.1 | 0.4 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.1 | 0.3 | GO:0070175 | positive regulation of enamel mineralization(GO:0070175) |
| 0.1 | 2.2 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.1 | 1.2 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.1 | 3.2 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.1 | 0.6 | GO:0090206 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.1 | 1.2 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.1 | 1.4 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.1 | 0.4 | GO:1901675 | negative regulation of histone H3-K27 acetylation(GO:1901675) |
| 0.1 | 3.5 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.1 | 0.7 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.1 | 2.3 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.1 | 4.0 | GO:0043486 | histone exchange(GO:0043486) |
| 0.1 | 1.9 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.1 | 1.1 | GO:0060252 | positive regulation of glial cell proliferation(GO:0060252) |
| 0.1 | 1.7 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.1 | 0.4 | GO:0050955 | thermoception(GO:0050955) |
| 0.0 | 0.2 | GO:1904425 | negative regulation of GTP binding(GO:1904425) |
| 0.0 | 0.4 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.2 | GO:0010730 | negative regulation of hydrogen peroxide biosynthetic process(GO:0010730) |
| 0.0 | 1.5 | GO:0050962 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 | 1.0 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 0.8 | GO:0051531 | NFAT protein import into nucleus(GO:0051531) |
| 0.0 | 0.8 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.8 | GO:0070933 | histone H4 deacetylation(GO:0070933) |
| 0.0 | 1.0 | GO:2000272 | negative regulation of receptor activity(GO:2000272) |
| 0.0 | 0.4 | GO:0097475 | motor neuron migration(GO:0097475) |
| 0.0 | 0.5 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.1 | GO:0021934 | medulla oblongata development(GO:0021550) hindbrain tangential cell migration(GO:0021934) lateral line system development(GO:0048925) |
| 0.0 | 0.4 | GO:1900452 | regulation of long term synaptic depression(GO:1900452) |
| 0.0 | 0.9 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 0.8 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.7 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.0 | 3.0 | GO:0070268 | cornification(GO:0070268) |
| 0.0 | 0.5 | GO:0033189 | response to vitamin A(GO:0033189) |
| 0.0 | 0.3 | GO:0071481 | cellular response to X-ray(GO:0071481) |
| 0.0 | 0.8 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 0.7 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.0 | 0.5 | GO:0021983 | pituitary gland development(GO:0021983) |
| 0.0 | 0.2 | GO:0001542 | ovulation from ovarian follicle(GO:0001542) |
| 0.0 | 0.3 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.0 | 0.8 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.0 | 0.9 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.5 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 9.6 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.2 | 0.7 | GO:0042585 | germinal vesicle(GO:0042585) |
| 0.2 | 4.0 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.1 | 0.9 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.1 | 1.1 | GO:0070369 | Scrib-APC-beta-catenin complex(GO:0034750) beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.1 | 0.8 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.1 | 0.4 | GO:0060076 | postsynaptic density(GO:0014069) excitatory synapse(GO:0060076) postsynaptic specialization(GO:0099572) |
| 0.1 | 2.8 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.1 | 1.1 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.0 | 1.3 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.4 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.0 | 1.4 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.3 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.3 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.0 | 0.4 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.0 | 1.0 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 0.5 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.0 | 1.1 | GO:0097546 | ciliary base(GO:0097546) |
| 0.0 | 3.7 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.1 | GO:0098843 | postsynaptic endocytic zone(GO:0098843) |
| 0.0 | 3.9 | GO:0035579 | specific granule membrane(GO:0035579) |
| 0.0 | 1.5 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 1.0 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.8 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 0.4 | GO:0032982 | myosin filament(GO:0032982) |
| 0.0 | 2.7 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.6 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.6 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 1.5 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.3 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 6.5 | GO:0016607 | nuclear speck(GO:0016607) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 5.1 | GO:0050528 | acyloxyacyl hydrolase activity(GO:0050528) |
| 0.6 | 1.8 | GO:0008808 | cardiolipin synthase activity(GO:0008808) phosphatidyltransferase activity(GO:0030572) CDP-diacylglycerol-phosphatidylglycerol phosphatidyltransferase activity(GO:0043337) |
| 0.5 | 2.8 | GO:0004918 | interleukin-8 receptor activity(GO:0004918) |
| 0.4 | 2.2 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.4 | 2.6 | GO:0042806 | fucose binding(GO:0042806) |
| 0.3 | 1.4 | GO:0003978 | UDP-N-acetylglucosamine 4-epimerase activity(GO:0003974) UDP-glucose 4-epimerase activity(GO:0003978) |
| 0.3 | 1.8 | GO:0090554 | phosphatidylcholine-translocating ATPase activity(GO:0090554) |
| 0.3 | 1.2 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.2 | 1.7 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) steroid sulfotransferase activity(GO:0050294) |
| 0.2 | 2.2 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.2 | 0.7 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 0.2 | 7.5 | GO:0038187 | signaling pattern recognition receptor activity(GO:0008329) pattern recognition receptor activity(GO:0038187) |
| 0.2 | 4.2 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.2 | 1.1 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.1 | 1.8 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.1 | 4.5 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.1 | 1.4 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.1 | 1.9 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.1 | 4.7 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.1 | 9.6 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.1 | 2.3 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 1.0 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.0 | 4.0 | GO:0019212 | phosphatase inhibitor activity(GO:0019212) |
| 0.0 | 0.8 | GO:0043559 | insulin binding(GO:0043559) |
| 0.0 | 1.1 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 2.0 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 1.3 | GO:0070300 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) phosphatidic acid binding(GO:0070300) |
| 0.0 | 0.3 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.5 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 1.1 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 1.4 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.5 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.0 | 0.8 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 0.6 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.4 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.0 | 0.7 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.0 | 2.4 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.4 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 0.7 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 0.9 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.6 | GO:0043236 | laminin binding(GO:0043236) |
| 0.0 | 2.1 | GO:0036459 | thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) |
| 0.0 | 0.8 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.4 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.0 | 0.7 | GO:0005080 | protein kinase C binding(GO:0005080) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 10.4 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.1 | 6.1 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.1 | 2.8 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.1 | 2.3 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 1.9 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 1.1 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.0 | 1.3 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.0 | 3.5 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 5.8 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.4 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.0 | 0.8 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.6 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 2.3 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 1.8 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 2.0 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 1.8 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.3 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 1.1 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 0.5 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 9.6 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.2 | 4.7 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.1 | 1.8 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.1 | 1.6 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.1 | 3.5 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.1 | 4.0 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.1 | 1.5 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 2.7 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.1 | 1.0 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.1 | 2.6 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.1 | 0.7 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.1 | 1.7 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 1.1 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 2.8 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 2.0 | REACTOME CD28 DEPENDENT PI3K AKT SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.0 | 1.8 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.8 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 1.1 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.5 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 1.2 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.7 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 3.3 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 2.1 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |