Project
Illumina Body Map 2: averaged replicates
Navigation
Downloads
Results for LHX6
Z-value: 1.31
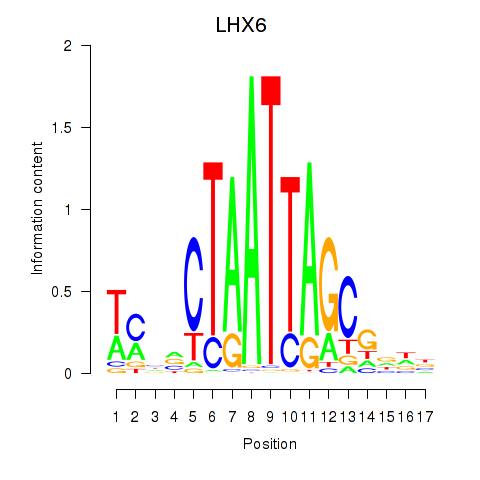
Transcription factors associated with LHX6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
LHX6
|
ENSG00000106852.11 | LIM homeobox 6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| LHX6 | hg19_v2_chr9_-_124976154_124976177 | 0.11 | 5.5e-01 | Click! |
Activity profile of LHX6 motif
Sorted Z-values of LHX6 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_37723336 | 1.64 |
ENST00000450180.1
|
GPR141
|
G protein-coupled receptor 141 |
| chr6_-_62996066 | 1.41 |
ENST00000281156.4
|
KHDRBS2
|
KH domain containing, RNA binding, signal transduction associated 2 |
| chr15_+_51669513 | 1.38 |
ENST00000558426.1
|
GLDN
|
gliomedin |
| chr11_-_102651343 | 1.34 |
ENST00000279441.4
ENST00000539681.1 |
MMP10
|
matrix metallopeptidase 10 (stromelysin 2) |
| chr5_-_35938674 | 1.33 |
ENST00000397366.1
ENST00000513623.1 ENST00000514524.1 ENST00000397367.2 |
CAPSL
|
calcyphosine-like |
| chr12_+_8666126 | 1.32 |
ENST00000299665.2
|
CLEC4D
|
C-type lectin domain family 4, member D |
| chr12_-_45269251 | 1.21 |
ENST00000553120.1
|
NELL2
|
NEL-like 2 (chicken) |
| chr12_-_45269430 | 1.16 |
ENST00000395487.2
|
NELL2
|
NEL-like 2 (chicken) |
| chr16_-_55866997 | 1.07 |
ENST00000360526.3
ENST00000361503.4 |
CES1
|
carboxylesterase 1 |
| chr21_-_22175341 | 1.07 |
ENST00000416768.1
ENST00000452561.1 ENST00000419299.1 ENST00000437238.1 |
LINC00320
|
long intergenic non-protein coding RNA 320 |
| chr18_+_61143994 | 0.97 |
ENST00000382771.4
|
SERPINB5
|
serpin peptidase inhibitor, clade B (ovalbumin), member 5 |
| chr6_+_30457244 | 0.96 |
ENST00000376630.4
|
HLA-E
|
major histocompatibility complex, class I, E |
| chr12_-_45270077 | 0.94 |
ENST00000551601.1
ENST00000549027.1 ENST00000452445.2 |
NELL2
|
NEL-like 2 (chicken) |
| chr17_+_11757333 | 0.94 |
ENST00000579703.1
|
DNAH9
|
dynein, axonemal, heavy chain 9 |
| chr9_-_71161505 | 0.94 |
ENST00000446290.1
|
RP11-274B18.2
|
RP11-274B18.2 |
| chr9_-_38424443 | 0.92 |
ENST00000377694.1
|
IGFBPL1
|
insulin-like growth factor binding protein-like 1 |
| chr1_-_21620877 | 0.89 |
ENST00000527991.1
|
ECE1
|
endothelin converting enzyme 1 |
| chr6_-_167040693 | 0.89 |
ENST00000366863.2
|
RPS6KA2
|
ribosomal protein S6 kinase, 90kDa, polypeptide 2 |
| chr12_-_772901 | 0.88 |
ENST00000305108.4
|
NINJ2
|
ninjurin 2 |
| chr12_-_45270151 | 0.87 |
ENST00000429094.2
|
NELL2
|
NEL-like 2 (chicken) |
| chr14_+_60386853 | 0.87 |
ENST00000570145.1
|
LRRC9
|
leucine rich repeat containing 9 |
| chr18_-_44181442 | 0.82 |
ENST00000398722.4
|
LOXHD1
|
lipoxygenase homology domains 1 |
| chr18_-_67624160 | 0.82 |
ENST00000581982.1
ENST00000280200.4 |
CD226
|
CD226 molecule |
| chr7_-_14029515 | 0.78 |
ENST00000430479.1
ENST00000405218.2 ENST00000343495.5 |
ETV1
|
ets variant 1 |
| chr17_-_73937028 | 0.78 |
ENST00000586631.2
|
FBF1
|
Fas (TNFRSF6) binding factor 1 |
| chr7_-_36764142 | 0.78 |
ENST00000258749.5
ENST00000535891.1 |
AOAH
|
acyloxyacyl hydrolase (neutrophil) |
| chr9_+_1050331 | 0.76 |
ENST00000382255.3
ENST00000382251.3 ENST00000412350.2 |
DMRT2
|
doublesex and mab-3 related transcription factor 2 |
| chr12_-_45269769 | 0.76 |
ENST00000548826.1
|
NELL2
|
NEL-like 2 (chicken) |
| chr5_-_171881362 | 0.73 |
ENST00000519643.1
|
SH3PXD2B
|
SH3 and PX domains 2B |
| chr10_+_71562180 | 0.72 |
ENST00000517713.1
ENST00000522165.1 ENST00000520133.1 |
COL13A1
|
collagen, type XIII, alpha 1 |
| chr19_+_29374337 | 0.72 |
ENST00000591285.1
|
AC011524.1
|
AC011524.1 |
| chr2_+_90043607 | 0.72 |
ENST00000462693.1
|
IGKV2D-24
|
immunoglobulin kappa variable 2D-24 (non-functional) |
| chr10_+_71561630 | 0.70 |
ENST00000398974.3
ENST00000398971.3 ENST00000398968.3 ENST00000398966.3 ENST00000398964.3 ENST00000398969.3 ENST00000356340.3 ENST00000398972.3 ENST00000398973.3 |
COL13A1
|
collagen, type XIII, alpha 1 |
| chr10_+_71561649 | 0.70 |
ENST00000398978.3
ENST00000354547.3 ENST00000357811.3 |
COL13A1
|
collagen, type XIII, alpha 1 |
| chr19_-_42916499 | 0.70 |
ENST00000601189.1
ENST00000599211.1 |
LIPE
|
lipase, hormone-sensitive |
| chr5_-_82969405 | 0.68 |
ENST00000510978.1
|
HAPLN1
|
hyaluronan and proteoglycan link protein 1 |
| chr4_+_71296204 | 0.68 |
ENST00000413702.1
|
MUC7
|
mucin 7, secreted |
| chr5_+_142149932 | 0.68 |
ENST00000274498.4
|
ARHGAP26
|
Rho GTPase activating protein 26 |
| chr7_-_111424506 | 0.68 |
ENST00000450156.1
ENST00000494651.2 |
DOCK4
|
dedicator of cytokinesis 4 |
| chr7_+_37723450 | 0.68 |
ENST00000447769.1
|
GPR141
|
G protein-coupled receptor 141 |
| chr21_-_43916433 | 0.67 |
ENST00000291536.3
|
RSPH1
|
radial spoke head 1 homolog (Chlamydomonas) |
| chr6_+_122931366 | 0.67 |
ENST00000368452.2
ENST00000368448.1 ENST00000392490.1 |
PKIB
|
protein kinase (cAMP-dependent, catalytic) inhibitor beta |
| chr17_-_62208169 | 0.65 |
ENST00000606895.1
|
ERN1
|
endoplasmic reticulum to nucleus signaling 1 |
| chr19_-_14889349 | 0.65 |
ENST00000315576.3
ENST00000392967.2 ENST00000346057.1 ENST00000353876.1 ENST00000353005.1 |
EMR2
|
egf-like module containing, mucin-like, hormone receptor-like 2 |
| chr15_-_64673630 | 0.64 |
ENST00000558008.1
ENST00000559519.1 ENST00000380258.2 |
KIAA0101
|
KIAA0101 |
| chr19_-_52307357 | 0.63 |
ENST00000594900.1
|
FPR1
|
formyl peptide receptor 1 |
| chr9_+_12695702 | 0.63 |
ENST00000381136.2
|
TYRP1
|
tyrosinase-related protein 1 |
| chr12_+_16500599 | 0.62 |
ENST00000535309.1
ENST00000540056.1 ENST00000396209.1 ENST00000540126.1 |
MGST1
|
microsomal glutathione S-transferase 1 |
| chr3_+_85008089 | 0.62 |
ENST00000383699.3
|
CADM2
|
cell adhesion molecule 2 |
| chr6_-_167040731 | 0.62 |
ENST00000265678.4
|
RPS6KA2
|
ribosomal protein S6 kinase, 90kDa, polypeptide 2 |
| chr2_-_89476644 | 0.61 |
ENST00000484817.1
|
IGKV2-24
|
immunoglobulin kappa variable 2-24 |
| chr7_-_64467031 | 0.61 |
ENST00000394323.2
|
ERV3-1
|
endogenous retrovirus group 3, member 1 |
| chr11_-_118023594 | 0.61 |
ENST00000529878.1
|
SCN4B
|
sodium channel, voltage-gated, type IV, beta subunit |
| chr13_+_37006421 | 0.61 |
ENST00000255465.4
|
CCNA1
|
cyclin A1 |
| chr10_+_103986085 | 0.60 |
ENST00000370005.3
|
ELOVL3
|
ELOVL fatty acid elongase 3 |
| chr14_-_100842588 | 0.60 |
ENST00000556645.1
ENST00000556209.1 ENST00000556504.1 ENST00000556435.1 ENST00000554772.1 ENST00000553581.1 ENST00000553769.2 ENST00000554605.1 ENST00000557722.1 ENST00000553413.1 ENST00000553524.1 ENST00000358655.4 |
WARS
|
tryptophanyl-tRNA synthetase |
| chr6_+_148663729 | 0.60 |
ENST00000367467.3
|
SASH1
|
SAM and SH3 domain containing 1 |
| chr21_-_43916296 | 0.60 |
ENST00000398352.3
|
RSPH1
|
radial spoke head 1 homolog (Chlamydomonas) |
| chr20_+_19867150 | 0.59 |
ENST00000255006.6
|
RIN2
|
Ras and Rab interactor 2 |
| chr6_+_148593425 | 0.59 |
ENST00000367469.1
|
SASH1
|
SAM and SH3 domain containing 1 |
| chr7_+_70597109 | 0.59 |
ENST00000333538.5
|
WBSCR17
|
Williams-Beuren syndrome chromosome region 17 |
| chr13_+_37005967 | 0.58 |
ENST00000440264.1
ENST00000449823.1 |
CCNA1
|
cyclin A1 |
| chr4_-_174255400 | 0.58 |
ENST00000506267.1
|
HMGB2
|
high mobility group box 2 |
| chr4_+_144312659 | 0.58 |
ENST00000509992.1
|
GAB1
|
GRB2-associated binding protein 1 |
| chr4_-_103749205 | 0.58 |
ENST00000508249.1
|
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr3_+_130162328 | 0.57 |
ENST00000512482.1
|
COL6A5
|
collagen, type VI, alpha 5 |
| chr12_-_91546926 | 0.56 |
ENST00000550758.1
|
DCN
|
decorin |
| chrX_+_37639302 | 0.56 |
ENST00000545017.1
ENST00000536160.1 |
CYBB
|
cytochrome b-245, beta polypeptide |
| chr7_-_36764062 | 0.55 |
ENST00000435386.1
|
AOAH
|
acyloxyacyl hydrolase (neutrophil) |
| chr3_-_157221128 | 0.55 |
ENST00000392833.2
ENST00000362010.2 |
VEPH1
|
ventricular zone expressed PH domain-containing 1 |
| chr16_-_55867146 | 0.54 |
ENST00000422046.2
|
CES1
|
carboxylesterase 1 |
| chr5_+_66300464 | 0.54 |
ENST00000436277.1
|
MAST4
|
microtubule associated serine/threonine kinase family member 4 |
| chr11_-_35287243 | 0.53 |
ENST00000464522.2
|
SLC1A2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
| chr1_-_232598163 | 0.53 |
ENST00000308942.4
|
SIPA1L2
|
signal-induced proliferation-associated 1 like 2 |
| chr7_-_111424462 | 0.52 |
ENST00000437129.1
|
DOCK4
|
dedicator of cytokinesis 4 |
| chr4_+_158493642 | 0.52 |
ENST00000507108.1
ENST00000455598.1 ENST00000509450.1 |
RP11-364P22.1
|
RP11-364P22.1 |
| chr3_-_149095652 | 0.52 |
ENST00000305366.3
|
TM4SF1
|
transmembrane 4 L six family member 1 |
| chr4_+_144354644 | 0.51 |
ENST00000512843.1
|
GAB1
|
GRB2-associated binding protein 1 |
| chr18_-_67624412 | 0.49 |
ENST00000580335.1
|
CD226
|
CD226 molecule |
| chr13_+_37006398 | 0.49 |
ENST00000418263.1
|
CCNA1
|
cyclin A1 |
| chr2_-_152118276 | 0.49 |
ENST00000409092.1
|
RBM43
|
RNA binding motif protein 43 |
| chr18_+_60190682 | 0.49 |
ENST00000588676.1
|
ZCCHC2
|
zinc finger, CCHC domain containing 2 |
| chr12_-_9760482 | 0.48 |
ENST00000229402.3
|
KLRB1
|
killer cell lectin-like receptor subfamily B, member 1 |
| chr12_+_7014126 | 0.48 |
ENST00000415834.1
ENST00000436789.1 |
LRRC23
|
leucine rich repeat containing 23 |
| chr12_+_16500037 | 0.48 |
ENST00000536371.1
ENST00000010404.2 |
MGST1
|
microsomal glutathione S-transferase 1 |
| chr11_-_102668879 | 0.47 |
ENST00000315274.6
|
MMP1
|
matrix metallopeptidase 1 (interstitial collagenase) |
| chr4_-_174255536 | 0.46 |
ENST00000446922.2
|
HMGB2
|
high mobility group box 2 |
| chr8_-_13134045 | 0.46 |
ENST00000512044.2
|
DLC1
|
deleted in liver cancer 1 |
| chr18_+_44526744 | 0.45 |
ENST00000585469.1
|
KATNAL2
|
katanin p60 subunit A-like 2 |
| chr19_+_12902289 | 0.45 |
ENST00000302754.4
|
JUNB
|
jun B proto-oncogene |
| chr1_+_40713573 | 0.45 |
ENST00000372766.3
|
TMCO2
|
transmembrane and coiled-coil domains 2 |
| chr5_+_66300446 | 0.45 |
ENST00000261569.7
|
MAST4
|
microtubule associated serine/threonine kinase family member 4 |
| chr6_+_29691198 | 0.44 |
ENST00000440587.2
ENST00000434407.2 |
HLA-F
|
major histocompatibility complex, class I, F |
| chr11_+_59856130 | 0.44 |
ENST00000278888.3
|
MS4A2
|
membrane-spanning 4-domains, subfamily A, member 2 |
| chr11_+_76156045 | 0.43 |
ENST00000533988.1
ENST00000524490.1 ENST00000334736.3 ENST00000343878.3 ENST00000533972.1 |
C11orf30
|
chromosome 11 open reading frame 30 |
| chr12_+_16500571 | 0.42 |
ENST00000543076.1
ENST00000396210.3 |
MGST1
|
microsomal glutathione S-transferase 1 |
| chr15_-_77712477 | 0.42 |
ENST00000560626.2
|
PEAK1
|
pseudopodium-enriched atypical kinase 1 |
| chr1_-_204380919 | 0.41 |
ENST00000367188.4
|
PPP1R15B
|
protein phosphatase 1, regulatory subunit 15B |
| chr3_-_157221380 | 0.41 |
ENST00000468233.1
|
VEPH1
|
ventricular zone expressed PH domain-containing 1 |
| chr5_-_110848189 | 0.41 |
ENST00000296632.3
ENST00000512160.1 ENST00000509887.1 |
STARD4
|
StAR-related lipid transfer (START) domain containing 4 |
| chr11_-_76155700 | 0.40 |
ENST00000572035.1
|
RP11-111M22.3
|
RP11-111M22.3 |
| chr5_+_142149955 | 0.40 |
ENST00000378004.3
|
ARHGAP26
|
Rho GTPase activating protein 26 |
| chr18_-_67623906 | 0.40 |
ENST00000583955.1
|
CD226
|
CD226 molecule |
| chr1_-_48866517 | 0.39 |
ENST00000371841.1
|
SPATA6
|
spermatogenesis associated 6 |
| chr16_+_31885079 | 0.39 |
ENST00000300870.10
ENST00000394846.3 |
ZNF267
|
zinc finger protein 267 |
| chr19_-_10121144 | 0.39 |
ENST00000264828.3
|
COL5A3
|
collagen, type V, alpha 3 |
| chr3_-_157221357 | 0.39 |
ENST00000494677.1
|
VEPH1
|
ventricular zone expressed PH domain-containing 1 |
| chr7_-_151330218 | 0.39 |
ENST00000476632.1
|
PRKAG2
|
protein kinase, AMP-activated, gamma 2 non-catalytic subunit |
| chr10_+_71561704 | 0.39 |
ENST00000520267.1
|
COL13A1
|
collagen, type XIII, alpha 1 |
| chr2_-_192711968 | 0.38 |
ENST00000304141.4
|
SDPR
|
serum deprivation response |
| chr15_-_77712429 | 0.38 |
ENST00000564328.1
ENST00000558305.1 |
PEAK1
|
pseudopodium-enriched atypical kinase 1 |
| chr11_-_33913708 | 0.38 |
ENST00000257818.2
|
LMO2
|
LIM domain only 2 (rhombotin-like 1) |
| chr9_+_12775011 | 0.38 |
ENST00000319264.3
|
LURAP1L
|
leucine rich adaptor protein 1-like |
| chr17_+_66511540 | 0.38 |
ENST00000588188.2
|
PRKAR1A
|
protein kinase, cAMP-dependent, regulatory, type I, alpha |
| chr19_-_12833361 | 0.37 |
ENST00000592287.1
|
TNPO2
|
transportin 2 |
| chr7_+_21582638 | 0.37 |
ENST00000409508.3
ENST00000328843.6 |
DNAH11
|
dynein, axonemal, heavy chain 11 |
| chr2_+_29341037 | 0.37 |
ENST00000449202.1
|
CLIP4
|
CAP-GLY domain containing linker protein family, member 4 |
| chr5_-_110848253 | 0.37 |
ENST00000505803.1
ENST00000502322.1 |
STARD4
|
StAR-related lipid transfer (START) domain containing 4 |
| chr12_-_53045948 | 0.37 |
ENST00000309680.3
|
KRT2
|
keratin 2 |
| chr6_+_29691056 | 0.36 |
ENST00000414333.1
ENST00000334668.4 ENST00000259951.7 |
HLA-F
|
major histocompatibility complex, class I, F |
| chr2_-_55276320 | 0.36 |
ENST00000357376.3
|
RTN4
|
reticulon 4 |
| chr6_+_34204642 | 0.36 |
ENST00000347617.6
ENST00000401473.3 ENST00000311487.5 ENST00000447654.1 ENST00000395004.3 |
HMGA1
|
high mobility group AT-hook 1 |
| chr1_-_152386732 | 0.35 |
ENST00000271835.3
|
CRNN
|
cornulin |
| chr2_+_65454926 | 0.35 |
ENST00000542850.1
ENST00000377982.4 |
ACTR2
|
ARP2 actin-related protein 2 homolog (yeast) |
| chr18_+_616672 | 0.35 |
ENST00000338387.7
|
CLUL1
|
clusterin-like 1 (retinal) |
| chr15_+_77713222 | 0.35 |
ENST00000558176.1
|
HMG20A
|
high mobility group 20A |
| chr16_-_66583994 | 0.35 |
ENST00000564917.1
|
TK2
|
thymidine kinase 2, mitochondrial |
| chr6_-_27880174 | 0.34 |
ENST00000303324.2
|
OR2B2
|
olfactory receptor, family 2, subfamily B, member 2 |
| chr12_+_7014064 | 0.34 |
ENST00000443597.2
|
LRRC23
|
leucine rich repeat containing 23 |
| chrY_+_14813160 | 0.33 |
ENST00000338981.3
|
USP9Y
|
ubiquitin specific peptidase 9, Y-linked |
| chr6_-_138833630 | 0.33 |
ENST00000533765.1
|
NHSL1
|
NHS-like 1 |
| chr3_-_112693759 | 0.33 |
ENST00000440122.2
ENST00000490004.1 |
CD200R1
|
CD200 receptor 1 |
| chr13_+_24825824 | 0.33 |
ENST00000434675.1
ENST00000494772.1 |
SPATA13
|
spermatogenesis associated 13 |
| chr17_+_74261413 | 0.32 |
ENST00000587913.1
|
UBALD2
|
UBA-like domain containing 2 |
| chr7_-_86849836 | 0.31 |
ENST00000455575.1
|
TMEM243
|
transmembrane protein 243, mitochondrial |
| chr6_-_49681235 | 0.31 |
ENST00000339139.4
|
CRISP2
|
cysteine-rich secretory protein 2 |
| chr2_-_74618907 | 0.31 |
ENST00000421392.1
ENST00000437375.1 |
DCTN1
|
dynactin 1 |
| chrX_+_113818545 | 0.31 |
ENST00000371951.1
ENST00000276198.1 ENST00000371950.3 |
HTR2C
|
5-hydroxytryptamine (serotonin) receptor 2C, G protein-coupled |
| chr21_-_22175450 | 0.30 |
ENST00000435279.2
|
LINC00320
|
long intergenic non-protein coding RNA 320 |
| chr2_-_201936302 | 0.30 |
ENST00000453765.1
ENST00000452799.1 ENST00000446678.1 ENST00000418596.3 |
FAM126B
|
family with sequence similarity 126, member B |
| chr12_+_7013897 | 0.30 |
ENST00000007969.8
ENST00000323702.5 |
LRRC23
|
leucine rich repeat containing 23 |
| chr5_+_115358088 | 0.29 |
ENST00000600981.3
|
CTD-2287O16.3
|
CTD-2287O16.3 |
| chr19_-_12833164 | 0.29 |
ENST00000356861.5
|
TNPO2
|
transportin 2 |
| chr2_-_101925055 | 0.29 |
ENST00000295317.3
|
RNF149
|
ring finger protein 149 |
| chr4_-_66536196 | 0.29 |
ENST00000511294.1
|
EPHA5
|
EPH receptor A5 |
| chr14_+_57671888 | 0.29 |
ENST00000391612.1
|
AL391152.1
|
AL391152.1 |
| chr20_+_30467600 | 0.29 |
ENST00000375934.4
ENST00000375922.4 |
TTLL9
|
tubulin tyrosine ligase-like family, member 9 |
| chr3_+_172468472 | 0.28 |
ENST00000232458.5
ENST00000392692.3 |
ECT2
|
epithelial cell transforming sequence 2 oncogene |
| chr19_+_41869894 | 0.28 |
ENST00000413014.2
|
TMEM91
|
transmembrane protein 91 |
| chr2_+_65454863 | 0.28 |
ENST00000260641.5
|
ACTR2
|
ARP2 actin-related protein 2 homolog (yeast) |
| chr8_+_42010464 | 0.28 |
ENST00000518421.1
ENST00000174653.3 ENST00000396926.3 ENST00000521280.1 ENST00000522288.1 |
AP3M2
|
adaptor-related protein complex 3, mu 2 subunit |
| chr12_+_66696322 | 0.27 |
ENST00000247815.4
|
HELB
|
helicase (DNA) B |
| chr16_-_66584059 | 0.27 |
ENST00000417693.3
ENST00000544898.1 ENST00000569718.1 ENST00000527284.1 ENST00000299697.7 ENST00000451102.2 |
TK2
|
thymidine kinase 2, mitochondrial |
| chrX_-_24690771 | 0.27 |
ENST00000379145.1
|
PCYT1B
|
phosphate cytidylyltransferase 1, choline, beta |
| chr6_+_26402517 | 0.27 |
ENST00000414912.2
|
BTN3A1
|
butyrophilin, subfamily 3, member A1 |
| chr12_+_21654714 | 0.26 |
ENST00000542038.1
ENST00000540141.1 ENST00000229314.5 |
GOLT1B
|
golgi transport 1B |
| chr17_+_59489112 | 0.26 |
ENST00000335108.2
|
C17orf82
|
chromosome 17 open reading frame 82 |
| chr11_-_102714534 | 0.26 |
ENST00000299855.5
|
MMP3
|
matrix metallopeptidase 3 (stromelysin 1, progelatinase) |
| chr17_-_42906965 | 0.26 |
ENST00000586267.1
|
GJC1
|
gap junction protein, gamma 1, 45kDa |
| chr11_-_76155618 | 0.25 |
ENST00000530759.1
|
RP11-111M22.3
|
RP11-111M22.3 |
| chr1_+_81771806 | 0.25 |
ENST00000370721.1
ENST00000370727.1 ENST00000370725.1 ENST00000370723.1 ENST00000370728.1 ENST00000370730.1 |
LPHN2
|
latrophilin 2 |
| chr15_-_64673665 | 0.25 |
ENST00000300035.4
|
KIAA0101
|
KIAA0101 |
| chr4_-_103749179 | 0.25 |
ENST00000502690.1
|
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr12_-_110883346 | 0.25 |
ENST00000547365.1
|
ARPC3
|
actin related protein 2/3 complex, subunit 3, 21kDa |
| chr2_+_113763031 | 0.25 |
ENST00000259211.6
|
IL36A
|
interleukin 36, alpha |
| chr6_+_26402465 | 0.25 |
ENST00000476549.2
ENST00000289361.6 ENST00000450085.2 ENST00000425234.2 ENST00000427334.1 ENST00000506698.1 |
BTN3A1
|
butyrophilin, subfamily 3, member A1 |
| chr17_-_38210644 | 0.24 |
ENST00000394128.2
ENST00000394127.2 ENST00000356271.3 ENST00000535071.2 ENST00000580885.1 ENST00000543759.2 ENST00000537674.2 ENST00000580517.1 ENST00000578161.1 |
MED24
|
mediator complex subunit 24 |
| chr13_+_98086445 | 0.24 |
ENST00000245304.4
|
RAP2A
|
RAP2A, member of RAS oncogene family |
| chr9_+_71944241 | 0.24 |
ENST00000257515.8
|
FAM189A2
|
family with sequence similarity 189, member A2 |
| chr4_-_46126093 | 0.24 |
ENST00000295452.4
|
GABRG1
|
gamma-aminobutyric acid (GABA) A receptor, gamma 1 |
| chr4_-_121843985 | 0.24 |
ENST00000264808.3
ENST00000428209.2 ENST00000515109.1 ENST00000394435.2 |
PRDM5
|
PR domain containing 5 |
| chrX_-_47509994 | 0.24 |
ENST00000343894.4
|
ELK1
|
ELK1, member of ETS oncogene family |
| chr12_+_64798826 | 0.24 |
ENST00000540203.1
|
XPOT
|
exportin, tRNA |
| chr10_-_99447024 | 0.24 |
ENST00000370626.3
|
AVPI1
|
arginine vasopressin-induced 1 |
| chrX_+_77166172 | 0.23 |
ENST00000343533.5
ENST00000350425.4 ENST00000341514.6 |
ATP7A
|
ATPase, Cu++ transporting, alpha polypeptide |
| chr18_-_21017817 | 0.23 |
ENST00000542162.1
ENST00000383233.3 ENST00000582336.1 ENST00000450466.2 ENST00000578520.1 ENST00000399707.1 |
TMEM241
|
transmembrane protein 241 |
| chr4_+_165675197 | 0.23 |
ENST00000515485.1
|
RP11-294O2.2
|
RP11-294O2.2 |
| chr11_+_5710919 | 0.23 |
ENST00000379965.3
ENST00000425490.1 |
TRIM22
|
tripartite motif containing 22 |
| chr7_-_86849883 | 0.23 |
ENST00000433078.1
|
TMEM243
|
transmembrane protein 243, mitochondrial |
| chr11_-_121986923 | 0.23 |
ENST00000560104.1
|
BLID
|
BH3-like motif containing, cell death inducer |
| chr13_+_19756173 | 0.23 |
ENST00000382988.2
|
RP11-408E5.4
|
RP11-408E5.4 |
| chr3_+_129207033 | 0.23 |
ENST00000507221.1
|
IFT122
|
intraflagellar transport 122 homolog (Chlamydomonas) |
| chr19_+_17858509 | 0.23 |
ENST00000594202.1
ENST00000252771.7 ENST00000389133.4 |
FCHO1
|
FCH domain only 1 |
| chr4_+_66536248 | 0.23 |
ENST00000514260.1
ENST00000507117.1 |
RP11-807H7.1
|
RP11-807H7.1 |
| chrX_+_15525426 | 0.23 |
ENST00000342014.6
|
BMX
|
BMX non-receptor tyrosine kinase |
| chr6_-_33385854 | 0.23 |
ENST00000488478.1
|
CUTA
|
cutA divalent cation tolerance homolog (E. coli) |
| chr5_-_147286065 | 0.22 |
ENST00000318315.4
ENST00000515291.1 |
C5orf46
|
chromosome 5 open reading frame 46 |
| chr12_-_21654479 | 0.22 |
ENST00000421138.2
ENST00000444129.2 ENST00000539672.1 ENST00000542432.1 ENST00000536964.1 ENST00000536240.1 ENST00000396093.3 ENST00000314748.6 |
RECQL
|
RecQ protein-like (DNA helicase Q1-like) |
| chr5_-_82969363 | 0.22 |
ENST00000503117.1
|
HAPLN1
|
hyaluronan and proteoglycan link protein 1 |
| chr17_-_74733404 | 0.22 |
ENST00000508921.3
ENST00000583836.1 ENST00000358156.6 ENST00000392485.2 ENST00000359995.5 |
SRSF2
|
serine/arginine-rich splicing factor 2 |
| chr22_-_42343117 | 0.22 |
ENST00000407253.3
ENST00000215980.5 |
CENPM
|
centromere protein M |
| chr5_+_161277603 | 0.22 |
ENST00000519621.1
|
GABRA1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1 |
| chr19_-_51071302 | 0.22 |
ENST00000389201.3
ENST00000600381.1 |
LRRC4B
|
leucine rich repeat containing 4B |
| chr3_-_112693865 | 0.22 |
ENST00000471858.1
ENST00000295863.4 ENST00000308611.3 |
CD200R1
|
CD200 receptor 1 |
| chr7_+_150782945 | 0.22 |
ENST00000463381.1
|
AGAP3
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 3 |
| chr13_-_76111945 | 0.22 |
ENST00000355801.4
ENST00000406936.3 |
COMMD6
|
COMM domain containing 6 |
| chr8_+_133975231 | 0.21 |
ENST00000518058.1
|
TG
|
thyroglobulin |
| chr11_+_77532233 | 0.21 |
ENST00000525409.1
|
AAMDC
|
adipogenesis associated, Mth938 domain containing |
| chr1_+_145438469 | 0.20 |
ENST00000369317.4
|
TXNIP
|
thioredoxin interacting protein |
| chr3_-_160823040 | 0.20 |
ENST00000484127.1
ENST00000492353.1 ENST00000473142.1 ENST00000468268.1 ENST00000460353.1 ENST00000320474.4 ENST00000392781.2 |
B3GALNT1
|
beta-1,3-N-acetylgalactosaminyltransferase 1 (globoside blood group) |
Network of associatons between targets according to the STRING database.
First level regulatory network of LHX6
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.5 | GO:0071449 | cellular response to lipid hydroperoxide(GO:0071449) |
| 0.4 | 1.6 | GO:0090119 | vesicle-mediated cholesterol transport(GO:0090119) |
| 0.3 | 1.0 | GO:0042270 | antigen processing and presentation of endogenous peptide antigen via MHC class Ib(GO:0002476) protection from natural killer cell mediated cytotoxicity(GO:0042270) |
| 0.3 | 1.7 | GO:0002729 | positive regulation of natural killer cell cytokine production(GO:0002729) |
| 0.3 | 4.9 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.3 | 0.8 | GO:0010877 | lipid transport involved in lipid storage(GO:0010877) |
| 0.2 | 0.6 | GO:0043438 | acetoacetic acid metabolic process(GO:0043438) |
| 0.2 | 1.2 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.1 | 1.2 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.1 | 0.9 | GO:0034959 | substance P catabolic process(GO:0010814) calcitonin catabolic process(GO:0010816) endothelin maturation(GO:0034959) |
| 0.1 | 0.7 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 0.1 | 0.7 | GO:0046092 | deoxycytidine metabolic process(GO:0046092) |
| 0.1 | 0.6 | GO:1904844 | response to L-glutamine(GO:1904844) cellular response to L-glutamine(GO:1904845) |
| 0.1 | 0.8 | GO:0061055 | myotome development(GO:0061055) |
| 0.1 | 0.6 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.1 | 0.6 | GO:0016344 | meiotic chromosome movement towards spindle pole(GO:0016344) |
| 0.1 | 0.8 | GO:0002480 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-independent(GO:0002480) |
| 0.1 | 0.2 | GO:2000646 | positive regulation of receptor catabolic process(GO:2000646) |
| 0.1 | 1.0 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) |
| 0.1 | 0.3 | GO:0043397 | corticotropin-releasing hormone secretion(GO:0043396) regulation of corticotropin-releasing hormone secretion(GO:0043397) |
| 0.1 | 0.4 | GO:0090402 | oncogene-induced cell senescence(GO:0090402) |
| 0.1 | 0.6 | GO:0090063 | positive regulation of microtubule nucleation(GO:0090063) |
| 0.1 | 1.4 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.1 | 0.7 | GO:0036289 | mRNA splicing via endonucleolytic cleavage and ligation involved in unfolded protein response(GO:0030969) peptidyl-serine autophosphorylation(GO:0036289) mRNA splicing, via endonucleolytic cleavage and ligation(GO:0070054) mRNA endonucleolytic cleavage involved in unfolded protein response(GO:0070055) |
| 0.1 | 0.2 | GO:0042245 | RNA repair(GO:0042245) |
| 0.1 | 0.2 | GO:0034759 | regulation of iron ion transport(GO:0034756) regulation of iron ion transmembrane transport(GO:0034759) cellular response to lead ion(GO:0071284) |
| 0.1 | 0.2 | GO:0060830 | ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) |
| 0.1 | 0.5 | GO:0060136 | embryonic process involved in female pregnancy(GO:0060136) |
| 0.1 | 0.7 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.1 | 0.2 | GO:0090340 | positive regulation of high-density lipoprotein particle assembly(GO:0090108) positive regulation of secretion of lysosomal enzymes(GO:0090340) |
| 0.1 | 0.4 | GO:1903898 | negative regulation of PERK-mediated unfolded protein response(GO:1903898) |
| 0.0 | 0.5 | GO:0070779 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.0 | 0.1 | GO:0071140 | resolution of recombination intermediates(GO:0071139) resolution of mitotic recombination intermediates(GO:0071140) |
| 0.0 | 1.0 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 5.5 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.0 | 0.1 | GO:0032877 | positive regulation of DNA endoreduplication(GO:0032877) negative regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071930) |
| 0.0 | 0.2 | GO:0036079 | GDP-fucose transport(GO:0015783) purine nucleotide-sugar transport(GO:0036079) |
| 0.0 | 0.3 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.0 | 0.3 | GO:0086021 | SA node cell to atrial cardiac muscle cell communication by electrical coupling(GO:0086021) |
| 0.0 | 0.2 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.0 | 0.6 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.0 | 0.4 | GO:0019368 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.7 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 1.4 | GO:0007141 | male meiosis I(GO:0007141) |
| 0.0 | 0.1 | GO:0046901 | tetrahydrofolylpolyglutamate biosynthetic process(GO:0046901) |
| 0.0 | 0.6 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.0 | 0.4 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.0 | 0.9 | GO:0003341 | cilium movement(GO:0003341) |
| 0.0 | 0.6 | GO:0086016 | AV node cell action potential(GO:0086016) AV node cell to bundle of His cell signaling(GO:0086027) |
| 0.0 | 0.2 | GO:0044828 | negative regulation by host of viral genome replication(GO:0044828) |
| 0.0 | 0.5 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.0 | 0.9 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.0 | 0.3 | GO:0097475 | motor neuron migration(GO:0097475) |
| 0.0 | 0.9 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.0 | 0.1 | GO:0072356 | chromosome passenger complex localization to kinetochore(GO:0072356) |
| 0.0 | 0.4 | GO:0003356 | regulation of cilium beat frequency(GO:0003356) |
| 0.0 | 0.1 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.0 | 0.3 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.0 | 0.2 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.0 | 1.1 | GO:0035728 | response to hepatocyte growth factor(GO:0035728) |
| 0.0 | 0.2 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.0 | 0.4 | GO:0051546 | keratinocyte migration(GO:0051546) |
| 0.0 | 0.5 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.0 | 0.2 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.0 | 0.1 | GO:1904933 | regulation of cell proliferation in midbrain(GO:1904933) |
| 0.0 | 0.1 | GO:1900377 | negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) |
| 0.0 | 0.0 | GO:0014717 | regulation of satellite cell activation involved in skeletal muscle regeneration(GO:0014717) satellite cell activation involved in skeletal muscle regeneration(GO:0014901) |
| 0.0 | 0.7 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.4 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.0 | 0.2 | GO:0071394 | cellular response to testosterone stimulus(GO:0071394) |
| 0.0 | 0.4 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.0 | 1.2 | GO:0035082 | axoneme assembly(GO:0035082) |
| 0.0 | 0.3 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 0.5 | GO:0072643 | interferon-gamma secretion(GO:0072643) |
| 0.0 | 0.2 | GO:0000733 | DNA strand renaturation(GO:0000733) |
| 0.0 | 0.1 | GO:0010166 | wax biosynthetic process(GO:0010025) wax metabolic process(GO:0010166) |
| 0.0 | 0.4 | GO:0006853 | carnitine shuttle(GO:0006853) |
| 0.0 | 0.2 | GO:0015705 | iodide transport(GO:0015705) |
| 0.0 | 0.3 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.0 | 0.3 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.0 | 0.2 | GO:0045719 | negative regulation of glycogen biosynthetic process(GO:0045719) |
| 0.0 | 0.8 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.2 | GO:1900246 | positive regulation of RIG-I signaling pathway(GO:1900246) |
| 0.0 | 0.2 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 0.0 | 0.2 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.0 | 0.1 | GO:0030174 | regulation of DNA-dependent DNA replication initiation(GO:0030174) |
| 0.0 | 0.2 | GO:1990440 | positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990440) |
| 0.0 | 0.8 | GO:0050982 | detection of mechanical stimulus(GO:0050982) |
| 0.0 | 0.2 | GO:0002347 | response to tumor cell(GO:0002347) |
| 0.0 | 1.6 | GO:0048024 | regulation of mRNA splicing, via spliceosome(GO:0048024) |
| 0.0 | 0.3 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.0 | 0.2 | GO:0051256 | mitotic spindle midzone assembly(GO:0051256) |
| 0.0 | 1.3 | GO:0002292 | T cell differentiation involved in immune response(GO:0002292) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.5 | GO:0030936 | collagen type XIII trimer(GO:0005600) transmembrane collagen trimer(GO:0030936) |
| 0.3 | 1.7 | GO:0097123 | cyclin A1-CDK2 complex(GO:0097123) |
| 0.2 | 0.7 | GO:0097679 | other organism cytoplasm(GO:0097679) |
| 0.2 | 1.8 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.1 | 0.9 | GO:0031302 | intrinsic component of endosome membrane(GO:0031302) |
| 0.1 | 0.7 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.1 | 0.4 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.1 | 0.6 | GO:0036338 | viral envelope(GO:0019031) viral membrane(GO:0036338) |
| 0.1 | 0.4 | GO:0097224 | sperm connecting piece(GO:0097224) |
| 0.1 | 0.3 | GO:0032997 | Fc receptor complex(GO:0032997) Fc-epsilon receptor I complex(GO:0032998) |
| 0.1 | 1.3 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.1 | 0.5 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.1 | 0.2 | GO:0002944 | cyclin K-CDK12 complex(GO:0002944) cyclin K-CDK13 complex(GO:0002945) |
| 0.0 | 0.7 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.0 | 0.3 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 0.0 | 0.6 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.3 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 0.6 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.6 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.2 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.0 | 0.4 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.0 | 0.8 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 2.0 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 0.6 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.3 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 0.6 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 0.4 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 2.2 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.0 | 0.1 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.0 | 0.2 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.0 | 0.2 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.0 | 1.2 | GO:0030286 | dynein complex(GO:0030286) |
| 0.0 | 0.4 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 1.3 | GO:0101003 | ficolin-1-rich granule membrane(GO:0101003) |
| 0.0 | 1.8 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 2.3 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 0.2 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.0 | 0.3 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.2 | GO:0031010 | ISWI-type complex(GO:0031010) |
| 0.0 | 0.3 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.7 | GO:0002102 | podosome(GO:0002102) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.3 | GO:0050528 | acyloxyacyl hydrolase activity(GO:0050528) |
| 0.3 | 1.6 | GO:0047374 | methylumbelliferyl-acetate deacetylase activity(GO:0047374) |
| 0.2 | 0.7 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.2 | 1.0 | GO:0044378 | non-sequence-specific DNA binding, bending(GO:0044378) |
| 0.2 | 0.6 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.1 | 0.8 | GO:0046979 | TAP2 binding(GO:0046979) |
| 0.1 | 0.7 | GO:0004797 | deoxycytidine kinase activity(GO:0004137) thymidine kinase activity(GO:0004797) |
| 0.1 | 0.6 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 0.1 | 0.3 | GO:0017116 | single-stranded DNA-dependent ATP-dependent DNA helicase activity(GO:0017116) |
| 0.1 | 0.3 | GO:0047273 | galactosylgalactosylglucosylceramide beta-D-acetylgalactosaminyltransferase activity(GO:0047273) |
| 0.1 | 0.6 | GO:0005124 | N-formyl peptide receptor activity(GO:0004982) scavenger receptor binding(GO:0005124) |
| 0.1 | 1.0 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.1 | 1.5 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.1 | 0.6 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.1 | 0.2 | GO:0016532 | superoxide dismutase copper chaperone activity(GO:0016532) copper-dependent protein binding(GO:0032767) |
| 0.1 | 0.3 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.1 | 0.5 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.1 | 0.2 | GO:0001133 | RNA polymerase II transcription factor activity, sequence-specific transcription regulatory region DNA binding(GO:0001133) |
| 0.0 | 1.4 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.0 | 0.5 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.2 | GO:0035514 | DNA demethylase activity(GO:0035514) |
| 0.0 | 0.2 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.0 | 0.2 | GO:0036080 | GDP-fucose transmembrane transporter activity(GO:0005457) purine nucleotide-sugar transmembrane transporter activity(GO:0036080) |
| 0.0 | 1.4 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 1.2 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 0.3 | GO:0086020 | gap junction channel activity involved in SA node cell-atrial cardiac muscle cell electrical coupling(GO:0086020) |
| 0.0 | 0.7 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 1.3 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.0 | 0.2 | GO:0004346 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.0 | 0.5 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.4 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 1.4 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.1 | GO:0008841 | tetrahydrofolylpolyglutamate synthase activity(GO:0004326) dihydrofolate synthase activity(GO:0008841) |
| 0.0 | 0.6 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.0 | 0.2 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.0 | 0.3 | GO:0019863 | IgE binding(GO:0019863) |
| 0.0 | 0.4 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.0 | 0.9 | GO:0017127 | cholesterol transporter activity(GO:0017127) |
| 0.0 | 0.1 | GO:0050252 | retinol O-fatty-acyltransferase activity(GO:0050252) |
| 0.0 | 0.4 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.3 | GO:0043176 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.0 | 0.1 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.0 | 0.2 | GO:0099529 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.0 | 0.9 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.7 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.0 | 1.3 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.4 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.3 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 2.9 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.1 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.0 | 0.7 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 1.6 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 0.6 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.8 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.1 | GO:0000285 | 1-phosphatidylinositol-3-phosphate 5-kinase activity(GO:0000285) |
| 0.0 | 0.2 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.0 | 0.1 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.0 | 0.5 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 1.1 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.6 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 0.2 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.5 | GO:0042169 | SH2 domain binding(GO:0042169) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.6 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.1 | 1.7 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 3.5 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 2.6 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.0 | 1.5 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 6.1 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.8 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 0.2 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.0 | 1.3 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 0.8 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 1.0 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.9 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 0.7 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.7 | NABA CORE MATRISOME | Ensemble of genes encoding core extracellular matrix including ECM glycoproteins, collagens and proteoglycans |
| 0.0 | 1.2 | PID RAC1 PATHWAY | RAC1 signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.1 | 2.4 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.0 | 1.0 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 2.0 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.4 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 1.7 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 2.9 | REACTOME EXTRACELLULAR MATRIX ORGANIZATION | Genes involved in Extracellular matrix organization |
| 0.0 | 1.5 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 1.1 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 0.8 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.3 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 2.3 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.7 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.6 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.4 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.8 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.0 | 0.6 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.3 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.5 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.6 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.0 | 0.4 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |