Project
Illumina Body Map 2: averaged replicates
Navigation
Downloads
Results for MEF2C
Z-value: 6.41
Transcription factors associated with MEF2C
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
MEF2C
|
ENSG00000081189.9 | myocyte enhancer factor 2C |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| MEF2C | hg19_v2_chr5_-_88120151_88120177 | 0.37 | 3.7e-02 | Click! |
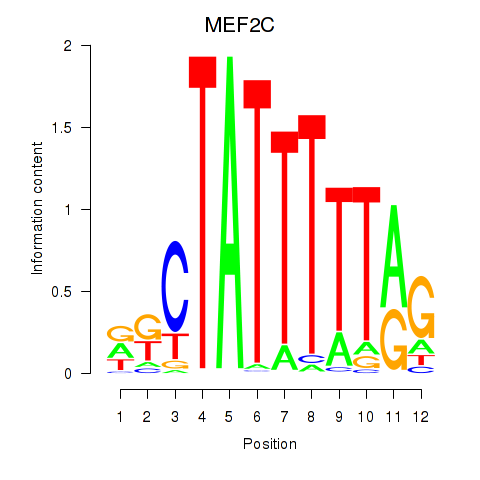
Activity profile of MEF2C motif
Sorted Z-values of MEF2C motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_19223523 | 55.51 |
ENST00000265968.3
|
CSRP3
|
cysteine and glycine-rich protein 3 (cardiac LIM protein) |
| chr12_-_111358372 | 53.49 |
ENST00000548438.1
ENST00000228841.8 |
MYL2
|
myosin, light chain 2, regulatory, cardiac, slow |
| chr3_-_39234074 | 48.18 |
ENST00000340369.3
ENST00000421646.1 ENST00000396251.1 |
XIRP1
|
xin actin-binding repeat containing 1 |
| chr1_-_201391149 | 47.32 |
ENST00000555948.1
ENST00000556362.1 |
TNNI1
|
troponin I type 1 (skeletal, slow) |
| chr5_+_191592 | 45.79 |
ENST00000328278.3
|
LRRC14B
|
leucine rich repeat containing 14B |
| chr17_+_37821593 | 45.79 |
ENST00000578283.1
|
TCAP
|
titin-cap |
| chr6_-_123958111 | 44.31 |
ENST00000542443.1
|
TRDN
|
triadin |
| chr6_-_123957942 | 42.66 |
ENST00000398178.3
|
TRDN
|
triadin |
| chr7_-_113559104 | 42.38 |
ENST00000284601.3
|
PPP1R3A
|
protein phosphatase 1, regulatory subunit 3A |
| chr22_-_36013368 | 41.06 |
ENST00000442617.1
ENST00000397326.2 ENST00000397328.1 ENST00000451685.1 |
MB
|
myoglobin |
| chr6_-_123958051 | 40.91 |
ENST00000546248.1
|
TRDN
|
triadin |
| chr2_+_168043793 | 37.22 |
ENST00000409273.1
ENST00000409605.1 |
XIRP2
|
xin actin-binding repeat containing 2 |
| chr22_+_26138108 | 36.19 |
ENST00000536101.1
ENST00000335473.7 ENST00000407587.2 |
MYO18B
|
myosin XVIIIB |
| chr10_-_75410771 | 35.77 |
ENST00000372873.4
|
SYNPO2L
|
synaptopodin 2-like |
| chr6_-_123958141 | 34.79 |
ENST00000334268.4
|
TRDN
|
triadin |
| chr10_+_50507232 | 34.18 |
ENST00000374144.3
|
C10orf71
|
chromosome 10 open reading frame 71 |
| chr16_-_31439735 | 32.75 |
ENST00000287490.4
|
COX6A2
|
cytochrome c oxidase subunit VIa polypeptide 2 |
| chr10_+_50507181 | 31.93 |
ENST00000323868.4
|
C10orf71
|
chromosome 10 open reading frame 71 |
| chr1_-_26394114 | 30.40 |
ENST00000374272.3
|
TRIM63
|
tripartite motif containing 63, E3 ubiquitin protein ligase |
| chr11_+_1942580 | 30.20 |
ENST00000381558.1
|
TNNT3
|
troponin T type 3 (skeletal, fast) |
| chr2_+_170366203 | 29.01 |
ENST00000284669.1
|
KLHL41
|
kelch-like family member 41 |
| chr2_+_88367299 | 28.96 |
ENST00000419482.2
ENST00000444564.2 |
SMYD1
|
SET and MYND domain containing 1 |
| chr3_-_52486841 | 28.90 |
ENST00000496590.1
|
TNNC1
|
troponin C type 1 (slow) |
| chr1_-_917497 | 28.64 |
ENST00000433179.2
|
C1orf170
|
chromosome 1 open reading frame 170 |
| chr4_+_120056939 | 27.87 |
ENST00000307128.5
|
MYOZ2
|
myozenin 2 |
| chr10_-_115423792 | 27.08 |
ENST00000369360.3
ENST00000360478.3 ENST00000359988.3 ENST00000369358.4 |
NRAP
|
nebulin-related anchoring protein |
| chr10_-_75415825 | 26.83 |
ENST00000394810.2
|
SYNPO2L
|
synaptopodin 2-like |
| chr1_+_145413268 | 26.46 |
ENST00000421822.2
ENST00000336751.5 ENST00000497365.1 ENST00000475797.1 |
HFE2
|
hemochromatosis type 2 (juvenile) |
| chr2_-_211168332 | 25.78 |
ENST00000341685.4
|
MYL1
|
myosin, light chain 1, alkali; skeletal, fast |
| chr1_+_228395755 | 25.69 |
ENST00000284548.11
ENST00000570156.2 ENST00000422127.1 ENST00000366707.4 ENST00000366709.4 |
OBSCN
|
obscurin, cytoskeletal calmodulin and titin-interacting RhoGEF |
| chr17_+_4855053 | 25.43 |
ENST00000518175.1
|
ENO3
|
enolase 3 (beta, muscle) |
| chr2_+_88367368 | 25.35 |
ENST00000438570.1
|
SMYD1
|
SET and MYND domain containing 1 |
| chrX_-_15332665 | 25.03 |
ENST00000537676.1
ENST00000344384.4 |
ASB11
|
ankyrin repeat and SOCS box containing 11 |
| chr1_-_917466 | 24.74 |
ENST00000341290.2
|
C1orf170
|
chromosome 1 open reading frame 170 |
| chr14_-_23877474 | 24.47 |
ENST00000405093.3
|
MYH6
|
myosin, heavy chain 6, cardiac muscle, alpha |
| chr5_+_53751445 | 24.32 |
ENST00000302005.1
|
HSPB3
|
heat shock 27kDa protein 3 |
| chr17_-_9694614 | 22.46 |
ENST00000330255.5
ENST00000571134.1 |
DHRS7C
|
dehydrogenase/reductase (SDR family) member 7C |
| chr11_+_1940925 | 21.96 |
ENST00000453458.1
ENST00000381557.2 ENST00000381589.3 ENST00000381579.3 ENST00000381563.4 ENST00000344578.4 |
TNNT3
|
troponin T type 3 (skeletal, fast) |
| chr11_+_1860200 | 21.37 |
ENST00000381911.1
|
TNNI2
|
troponin I type 2 (skeletal, fast) |
| chr11_+_1940786 | 20.99 |
ENST00000278317.6
ENST00000381561.4 ENST00000381548.3 ENST00000360603.3 ENST00000381549.3 |
TNNT3
|
troponin T type 3 (skeletal, fast) |
| chr12_+_101962128 | 20.61 |
ENST00000550514.1
|
MYBPC1
|
myosin binding protein C, slow type |
| chr3_-_46904946 | 20.37 |
ENST00000292327.4
|
MYL3
|
myosin, light chain 3, alkali; ventricular, skeletal, slow |
| chr10_+_88428370 | 20.34 |
ENST00000372066.3
ENST00000263066.6 ENST00000372056.4 ENST00000310944.6 ENST00000361373.4 ENST00000542786.1 |
LDB3
|
LIM domain binding 3 |
| chr3_-_46904918 | 20.26 |
ENST00000395869.1
|
MYL3
|
myosin, light chain 3, alkali; ventricular, skeletal, slow |
| chr8_+_1993152 | 19.97 |
ENST00000262113.4
|
MYOM2
|
myomesin 2 |
| chr2_+_239047337 | 19.00 |
ENST00000409223.1
ENST00000305959.4 |
KLHL30
|
kelch-like family member 30 |
| chr1_+_160160283 | 18.89 |
ENST00000368079.3
|
CASQ1
|
calsequestrin 1 (fast-twitch, skeletal muscle) |
| chr7_-_44105158 | 18.84 |
ENST00000297283.3
|
PGAM2
|
phosphoglycerate mutase 2 (muscle) |
| chr1_+_160160346 | 18.81 |
ENST00000368078.3
|
CASQ1
|
calsequestrin 1 (fast-twitch, skeletal muscle) |
| chr8_-_33457453 | 18.28 |
ENST00000523956.1
ENST00000256261.4 |
DUSP26
|
dual specificity phosphatase 26 (putative) |
| chr8_+_1993173 | 18.19 |
ENST00000523438.1
|
MYOM2
|
myomesin 2 |
| chr7_-_150884332 | 18.00 |
ENST00000275838.1
ENST00000422024.1 ENST00000434669.1 |
ASB10
|
ankyrin repeat and SOCS box containing 10 |
| chr1_-_201390846 | 17.77 |
ENST00000367312.1
ENST00000555340.2 ENST00000361379.4 |
TNNI1
|
troponin I type 1 (skeletal, slow) |
| chr1_-_116311402 | 17.41 |
ENST00000261448.5
|
CASQ2
|
calsequestrin 2 (cardiac muscle) |
| chr1_-_116311323 | 17.26 |
ENST00000456138.2
|
CASQ2
|
calsequestrin 2 (cardiac muscle) |
| chr11_+_1860832 | 17.00 |
ENST00000252898.7
|
TNNI2
|
troponin I type 2 (skeletal, fast) |
| chr10_+_88428206 | 16.08 |
ENST00000429277.2
ENST00000458213.2 ENST00000352360.5 |
LDB3
|
LIM domain binding 3 |
| chr1_-_31845914 | 16.00 |
ENST00000373713.2
|
FABP3
|
fatty acid binding protein 3, muscle and heart (mammary-derived growth inhibitor) |
| chr16_+_31225337 | 15.98 |
ENST00000322122.3
|
TRIM72
|
tripartite motif containing 72 |
| chr20_+_61147651 | 15.76 |
ENST00000370527.3
ENST00000370524.2 |
C20orf166
|
chromosome 20 open reading frame 166 |
| chr17_-_27949911 | 15.68 |
ENST00000492276.2
ENST00000345068.5 ENST00000584602.1 |
CORO6
|
coronin 6 |
| chr18_-_3219847 | 15.55 |
ENST00000261606.7
|
MYOM1
|
myomesin 1 |
| chr2_-_179537145 | 15.50 |
ENST00000448510.2
|
TTN
|
titin |
| chr3_+_135741576 | 15.37 |
ENST00000334546.2
|
PPP2R3A
|
protein phosphatase 2, regulatory subunit B'', alpha |
| chrX_-_63450480 | 15.16 |
ENST00000362002.2
|
ASB12
|
ankyrin repeat and SOCS box containing 12 |
| chr7_-_150884873 | 15.13 |
ENST00000377867.3
|
ASB10
|
ankyrin repeat and SOCS box containing 10 |
| chr11_+_1860682 | 14.90 |
ENST00000381906.1
|
TNNI2
|
troponin I type 2 (skeletal, fast) |
| chr18_-_3220106 | 14.54 |
ENST00000356443.4
ENST00000400569.3 |
MYOM1
|
myomesin 1 |
| chr1_-_24438664 | 14.48 |
ENST00000374434.3
ENST00000330966.7 ENST00000329601.7 |
MYOM3
|
myomesin 3 |
| chrX_+_135279179 | 14.35 |
ENST00000370676.3
|
FHL1
|
four and a half LIM domains 1 |
| chr16_+_7382745 | 14.31 |
ENST00000436368.2
ENST00000311745.5 ENST00000355637.4 ENST00000340209.4 |
RBFOX1
|
RNA binding protein, fox-1 homolog (C. elegans) 1 |
| chr3_+_42727011 | 14.07 |
ENST00000287777.4
|
KLHL40
|
kelch-like family member 40 |
| chr12_-_49393092 | 13.91 |
ENST00000421952.2
|
DDN
|
dendrin |
| chr6_-_45983581 | 13.91 |
ENST00000339561.6
|
CLIC5
|
chloride intracellular channel 5 |
| chrX_+_153046456 | 13.82 |
ENST00000393786.3
ENST00000370104.1 ENST00000370108.3 ENST00000370101.3 ENST00000430541.1 ENST00000370100.1 |
SRPK3
|
SRSF protein kinase 3 |
| chr15_+_65369082 | 13.42 |
ENST00000432196.2
|
KBTBD13
|
kelch repeat and BTB (POZ) domain containing 13 |
| chr14_-_94421923 | 13.38 |
ENST00000555507.1
|
ASB2
|
ankyrin repeat and SOCS box containing 2 |
| chr1_-_100643765 | 13.24 |
ENST00000370137.1
ENST00000370138.1 ENST00000342895.3 |
LRRC39
|
leucine rich repeat containing 39 |
| chr11_+_3666335 | 13.21 |
ENST00000250693.1
|
ART1
|
ADP-ribosyltransferase 1 |
| chr6_-_45983549 | 13.20 |
ENST00000544153.1
|
CLIC5
|
chloride intracellular channel 5 |
| chr8_-_70745575 | 12.43 |
ENST00000524945.1
|
SLCO5A1
|
solute carrier organic anion transporter family, member 5A1 |
| chr3_+_179370517 | 12.35 |
ENST00000263966.3
|
USP13
|
ubiquitin specific peptidase 13 (isopeptidase T-3) |
| chr11_-_47374246 | 12.34 |
ENST00000545968.1
ENST00000399249.2 ENST00000256993.4 |
MYBPC3
|
myosin binding protein C, cardiac |
| chr4_+_114066764 | 12.32 |
ENST00000511380.1
|
ANK2
|
ankyrin 2, neuronal |
| chr14_-_94421605 | 11.94 |
ENST00000556062.1
|
ASB2
|
ankyrin repeat and SOCS box containing 2 |
| chr12_+_109569155 | 11.93 |
ENST00000539864.1
|
ACACB
|
acetyl-CoA carboxylase beta |
| chr10_-_69455873 | 11.83 |
ENST00000433211.2
|
CTNNA3
|
catenin (cadherin-associated protein), alpha 3 |
| chr5_-_16509101 | 11.53 |
ENST00000399793.2
|
FAM134B
|
family with sequence similarity 134, member B |
| chr6_-_76203345 | 11.53 |
ENST00000393004.2
|
FILIP1
|
filamin A interacting protein 1 |
| chr22_-_51017084 | 11.47 |
ENST00000360719.2
ENST00000457250.1 ENST00000440709.1 |
CPT1B
|
carnitine palmitoyltransferase 1B (muscle) |
| chrX_+_135278908 | 11.47 |
ENST00000539015.1
ENST00000370683.1 |
FHL1
|
four and a half LIM domains 1 |
| chr17_+_7184986 | 11.45 |
ENST00000317370.8
ENST00000571308.1 |
SLC2A4
|
solute carrier family 2 (facilitated glucose transporter), member 4 |
| chr6_-_76203454 | 11.25 |
ENST00000237172.7
|
FILIP1
|
filamin A interacting protein 1 |
| chr7_+_80267949 | 11.22 |
ENST00000482059.2
|
CD36
|
CD36 molecule (thrombospondin receptor) |
| chr22_-_51016846 | 11.19 |
ENST00000312108.7
ENST00000395650.2 |
CPT1B
|
carnitine palmitoyltransferase 1B (muscle) |
| chr2_+_220299547 | 11.00 |
ENST00000312358.7
|
SPEG
|
SPEG complex locus |
| chr12_-_57644952 | 10.82 |
ENST00000554578.1
ENST00000546246.2 ENST00000553489.1 ENST00000332782.2 |
STAC3
|
SH3 and cysteine rich domain 3 |
| chr17_+_65040678 | 10.62 |
ENST00000226021.3
|
CACNG1
|
calcium channel, voltage-dependent, gamma subunit 1 |
| chr11_+_57308979 | 10.59 |
ENST00000457912.1
|
SMTNL1
|
smoothelin-like 1 |
| chr7_+_80231466 | 10.55 |
ENST00000309881.7
ENST00000534394.1 |
CD36
|
CD36 molecule (thrombospondin receptor) |
| chr7_-_56160625 | 10.52 |
ENST00000446428.1
ENST00000432123.1 ENST00000452681.2 ENST00000537360.1 |
PHKG1
|
phosphorylase kinase, gamma 1 (muscle) |
| chr4_-_186697044 | 10.43 |
ENST00000437304.2
|
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr7_+_80267973 | 10.20 |
ENST00000394788.3
ENST00000447544.2 |
CD36
|
CD36 molecule (thrombospondin receptor) |
| chr9_-_33402506 | 10.02 |
ENST00000377425.4
ENST00000537089.1 ENST00000297988.1 ENST00000539936.1 ENST00000541274.1 |
AQP7
|
aquaporin 7 |
| chr1_-_115238207 | 9.87 |
ENST00000520113.2
ENST00000369538.3 ENST00000353928.6 |
AMPD1
|
adenosine monophosphate deaminase 1 |
| chr15_-_83474806 | 9.86 |
ENST00000541889.1
ENST00000334574.8 ENST00000561368.1 |
FSD2
|
fibronectin type III and SPRY domain containing 2 |
| chr16_+_6069072 | 9.78 |
ENST00000547605.1
ENST00000550418.1 ENST00000553186.1 |
RBFOX1
|
RNA binding protein, fox-1 homolog (C. elegans) 1 |
| chr3_+_8543533 | 9.76 |
ENST00000454244.1
|
LMCD1
|
LIM and cysteine-rich domains 1 |
| chr13_+_76378305 | 9.74 |
ENST00000526371.1
ENST00000526528.1 |
LMO7
|
LIM domain 7 |
| chr3_+_8543393 | 9.52 |
ENST00000157600.3
ENST00000415597.1 ENST00000535732.1 |
LMCD1
|
LIM and cysteine-rich domains 1 |
| chr2_+_103378472 | 9.47 |
ENST00000412401.2
|
TMEM182
|
transmembrane protein 182 |
| chr7_-_56160666 | 9.45 |
ENST00000297373.2
|
PHKG1
|
phosphorylase kinase, gamma 1 (muscle) |
| chr4_+_110834033 | 9.19 |
ENST00000509793.1
ENST00000265171.5 |
EGF
|
epidermal growth factor |
| chr3_-_192445289 | 9.16 |
ENST00000430714.1
ENST00000418610.1 ENST00000448795.1 ENST00000445105.2 |
FGF12
|
fibroblast growth factor 12 |
| chr1_-_201346761 | 8.94 |
ENST00000455702.1
ENST00000422165.1 ENST00000367318.5 ENST00000367320.2 ENST00000438742.1 ENST00000412633.1 ENST00000458432.2 ENST00000421663.2 ENST00000367322.1 ENST00000509001.1 |
TNNT2
|
troponin T type 2 (cardiac) |
| chr17_-_10276319 | 8.73 |
ENST00000252172.4
ENST00000418404.3 |
MYH13
|
myosin, heavy chain 13, skeletal muscle |
| chr19_-_49576198 | 8.59 |
ENST00000221444.1
|
KCNA7
|
potassium voltage-gated channel, shaker-related subfamily, member 7 |
| chr10_-_29923893 | 8.57 |
ENST00000355867.4
|
SVIL
|
supervillin |
| chr4_+_169418255 | 8.53 |
ENST00000505667.1
ENST00000511948.1 |
PALLD
|
palladin, cytoskeletal associated protein |
| chr21_+_35552978 | 8.35 |
ENST00000428914.2
ENST00000609062.1 ENST00000609947.1 |
LINC00310
|
long intergenic non-protein coding RNA 310 |
| chr4_+_3465027 | 8.30 |
ENST00000389653.2
ENST00000507039.1 ENST00000340083.5 |
DOK7
|
docking protein 7 |
| chr3_+_8543561 | 8.00 |
ENST00000397386.3
|
LMCD1
|
LIM and cysteine-rich domains 1 |
| chr7_-_82073031 | 8.00 |
ENST00000356253.5
ENST00000423588.1 |
CACNA2D1
|
calcium channel, voltage-dependent, alpha 2/delta subunit 1 |
| chr7_-_82073109 | 7.56 |
ENST00000356860.3
|
CACNA2D1
|
calcium channel, voltage-dependent, alpha 2/delta subunit 1 |
| chr7_+_95115210 | 7.41 |
ENST00000428113.1
ENST00000325885.5 |
ASB4
|
ankyrin repeat and SOCS box containing 4 |
| chr15_+_42651691 | 7.31 |
ENST00000357568.3
ENST00000349748.3 ENST00000318023.7 ENST00000397163.3 |
CAPN3
|
calpain 3, (p94) |
| chr2_-_237172988 | 7.14 |
ENST00000409749.3
ENST00000430053.1 |
ASB18
|
ankyrin repeat and SOCS box containing 18 |
| chr19_+_35630344 | 7.01 |
ENST00000455515.2
|
FXYD1
|
FXYD domain containing ion transport regulator 1 |
| chr3_+_127634069 | 6.82 |
ENST00000405109.1
|
KBTBD12
|
kelch repeat and BTB (POZ) domain containing 12 |
| chr10_-_61900762 | 6.80 |
ENST00000355288.2
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G) |
| chr7_+_143013198 | 6.63 |
ENST00000343257.2
|
CLCN1
|
chloride channel, voltage-sensitive 1 |
| chr13_+_76378407 | 6.60 |
ENST00000447038.1
|
LMO7
|
LIM domain 7 |
| chr3_+_127634312 | 6.52 |
ENST00000407609.3
|
KBTBD12
|
kelch repeat and BTB (POZ) domain containing 12 |
| chr5_+_96077888 | 6.51 |
ENST00000509259.1
ENST00000503828.1 |
CAST
|
calpastatin |
| chr2_-_179914760 | 6.47 |
ENST00000420890.2
ENST00000409284.1 ENST00000443758.1 ENST00000446116.1 |
CCDC141
|
coiled-coil domain containing 141 |
| chr12_-_70004942 | 6.29 |
ENST00000361484.3
|
LRRC10
|
leucine rich repeat containing 10 |
| chr12_+_52431016 | 6.29 |
ENST00000553200.1
|
NR4A1
|
nuclear receptor subfamily 4, group A, member 1 |
| chr17_-_10372875 | 6.07 |
ENST00000255381.2
|
MYH4
|
myosin, heavy chain 4, skeletal muscle |
| chr4_+_95373037 | 5.98 |
ENST00000359265.4
ENST00000437932.1 ENST00000380180.3 ENST00000318007.5 ENST00000450793.1 ENST00000538141.1 ENST00000317968.4 ENST00000512274.1 ENST00000503974.1 ENST00000504489.1 ENST00000542407.1 |
PDLIM5
|
PDZ and LIM domain 5 |
| chr4_-_186696561 | 5.90 |
ENST00000445115.1
ENST00000451701.1 ENST00000457247.1 ENST00000435480.1 ENST00000425679.1 ENST00000457934.1 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr3_+_159570722 | 5.89 |
ENST00000482804.1
|
SCHIP1
|
schwannomin interacting protein 1 |
| chr13_+_76378357 | 5.68 |
ENST00000489941.2
ENST00000525373.1 |
LMO7
|
LIM domain 7 |
| chr1_-_205782304 | 5.63 |
ENST00000367137.3
|
SLC41A1
|
solute carrier family 41 (magnesium transporter), member 1 |
| chr3_+_69812877 | 5.50 |
ENST00000457080.1
ENST00000328528.6 |
MITF
|
microphthalmia-associated transcription factor |
| chr4_-_186696636 | 5.42 |
ENST00000444771.1
|
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr5_-_142065223 | 5.42 |
ENST00000378046.1
|
FGF1
|
fibroblast growth factor 1 (acidic) |
| chr5_-_142065612 | 5.42 |
ENST00000360966.5
ENST00000411960.1 |
FGF1
|
fibroblast growth factor 1 (acidic) |
| chr13_+_76334567 | 5.38 |
ENST00000321797.8
|
LMO7
|
LIM domain 7 |
| chr4_-_186696425 | 5.37 |
ENST00000430503.1
ENST00000319454.6 ENST00000450341.1 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr5_-_145252531 | 5.37 |
ENST00000377976.1
|
GRXCR2
|
glutaredoxin, cysteine rich 2 |
| chr1_-_154164534 | 5.32 |
ENST00000271850.7
ENST00000368530.2 |
TPM3
|
tropomyosin 3 |
| chr8_-_72268721 | 5.30 |
ENST00000419131.1
ENST00000388743.2 |
EYA1
|
eyes absent homolog 1 (Drosophila) |
| chr16_+_30076052 | 5.30 |
ENST00000563987.1
|
ALDOA
|
aldolase A, fructose-bisphosphate |
| chr16_+_30075463 | 5.23 |
ENST00000562168.1
ENST00000569545.1 |
ALDOA
|
aldolase A, fructose-bisphosphate |
| chr2_+_30569506 | 5.15 |
ENST00000421976.2
|
AC109642.1
|
AC109642.1 |
| chr1_-_178838404 | 5.14 |
ENST00000444255.1
|
ANGPTL1
|
angiopoietin-like 1 |
| chr14_-_104408645 | 5.12 |
ENST00000557640.1
|
RD3L
|
retinal degeneration 3-like |
| chr4_+_41614720 | 5.09 |
ENST00000509277.1
|
LIMCH1
|
LIM and calponin homology domains 1 |
| chr16_+_30075595 | 5.07 |
ENST00000563060.2
|
ALDOA
|
aldolase A, fructose-bisphosphate |
| chr12_-_22094336 | 5.05 |
ENST00000326684.4
|
ABCC9
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 9 |
| chr8_-_72268889 | 5.01 |
ENST00000388742.4
|
EYA1
|
eyes absent homolog 1 (Drosophila) |
| chr4_+_41362796 | 4.97 |
ENST00000508501.1
ENST00000512946.1 ENST00000313860.7 ENST00000512632.1 ENST00000512820.1 |
LIMCH1
|
LIM and calponin homology domains 1 |
| chr19_+_35630022 | 4.95 |
ENST00000589209.1
|
FXYD1
|
FXYD domain containing ion transport regulator 1 |
| chr3_+_35683651 | 4.94 |
ENST00000187397.4
|
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa |
| chr9_+_70971815 | 4.85 |
ENST00000396392.1
ENST00000396396.1 |
PGM5
|
phosphoglucomutase 5 |
| chr4_+_41614909 | 4.81 |
ENST00000509454.1
ENST00000396595.3 ENST00000381753.4 |
LIMCH1
|
LIM and calponin homology domains 1 |
| chr3_+_138067521 | 4.79 |
ENST00000494949.1
|
MRAS
|
muscle RAS oncogene homolog |
| chr1_+_170633047 | 4.78 |
ENST00000239461.6
ENST00000497230.2 |
PRRX1
|
paired related homeobox 1 |
| chr5_+_113391696 | 4.78 |
ENST00000512927.1
|
RP11-371M22.1
|
RP11-371M22.1 |
| chr4_+_41362615 | 4.74 |
ENST00000509638.1
|
LIMCH1
|
LIM and calponin homology domains 1 |
| chr2_-_100925967 | 4.73 |
ENST00000409647.1
|
LONRF2
|
LON peptidase N-terminal domain and ring finger 2 |
| chr3_+_69928256 | 4.71 |
ENST00000394355.2
|
MITF
|
microphthalmia-associated transcription factor |
| chr4_-_23891658 | 4.70 |
ENST00000507380.1
|
PPARGC1A
|
peroxisome proliferator-activated receptor gamma, coactivator 1 alpha |
| chr2_+_182850743 | 4.70 |
ENST00000409702.1
|
PPP1R1C
|
protein phosphatase 1, regulatory (inhibitor) subunit 1C |
| chr12_-_22094159 | 4.60 |
ENST00000538350.1
|
ABCC9
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 9 |
| chr13_+_76334795 | 4.56 |
ENST00000526202.1
ENST00000465261.2 |
LMO7
|
LIM domain 7 |
| chr16_+_30075783 | 4.53 |
ENST00000412304.2
|
ALDOA
|
aldolase A, fructose-bisphosphate |
| chr7_+_75931861 | 4.48 |
ENST00000248553.6
|
HSPB1
|
heat shock 27kDa protein 1 |
| chr11_+_12766583 | 4.38 |
ENST00000361985.2
|
TEAD1
|
TEA domain family member 1 (SV40 transcriptional enhancer factor) |
| chr18_+_3252206 | 4.36 |
ENST00000578562.2
|
MYL12A
|
myosin, light chain 12A, regulatory, non-sarcomeric |
| chr17_-_79900255 | 4.35 |
ENST00000330655.3
ENST00000582198.1 |
MYADML2
PYCR1
|
myeloid-associated differentiation marker-like 2 pyrroline-5-carboxylate reductase 1 |
| chr17_-_42188598 | 4.33 |
ENST00000591714.1
|
HDAC5
|
histone deacetylase 5 |
| chr14_+_76776957 | 4.32 |
ENST00000512784.1
|
ESRRB
|
estrogen-related receptor beta |
| chr9_+_100263912 | 4.28 |
ENST00000259365.4
|
TMOD1
|
tropomodulin 1 |
| chr3_+_138067314 | 4.21 |
ENST00000423968.2
|
MRAS
|
muscle RAS oncogene homolog |
| chr10_+_52751010 | 4.14 |
ENST00000373985.1
|
PRKG1
|
protein kinase, cGMP-dependent, type I |
| chr4_-_186696515 | 4.13 |
ENST00000456596.1
ENST00000414724.1 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr14_-_23652849 | 4.05 |
ENST00000316902.7
ENST00000469263.1 ENST00000525062.1 ENST00000524758.1 |
SLC7A8
|
solute carrier family 7 (amino acid transporter light chain, L system), member 8 |
| chr15_-_70994612 | 4.04 |
ENST00000558758.1
ENST00000379983.2 ENST00000560441.1 |
UACA
|
uveal autoantigen with coiled-coil domains and ankyrin repeats |
| chr8_-_72268968 | 3.99 |
ENST00000388740.3
|
EYA1
|
eyes absent homolog 1 (Drosophila) |
| chrM_+_5824 | 3.94 |
ENST00000361624.2
|
MT-CO1
|
mitochondrially encoded cytochrome c oxidase I |
| chr15_+_78558523 | 3.90 |
ENST00000446172.2
|
DNAJA4
|
DnaJ (Hsp40) homolog, subfamily A, member 4 |
| chr3_+_138067666 | 3.78 |
ENST00000475711.1
ENST00000464896.1 |
MRAS
|
muscle RAS oncogene homolog |
| chr19_+_35629702 | 3.76 |
ENST00000351325.4
|
FXYD1
|
FXYD domain containing ion transport regulator 1 |
| chr18_+_3252265 | 3.75 |
ENST00000580887.1
ENST00000536605.1 |
MYL12A
|
myosin, light chain 12A, regulatory, non-sarcomeric |
| chr10_-_14880002 | 3.75 |
ENST00000465530.1
|
CDNF
|
cerebral dopamine neurotrophic factor |
| chr1_+_74701062 | 3.68 |
ENST00000326637.3
|
TNNI3K
|
TNNI3 interacting kinase |
| chr10_+_63661053 | 3.67 |
ENST00000279873.7
|
ARID5B
|
AT rich interactive domain 5B (MRF1-like) |
| chr15_+_63889577 | 3.62 |
ENST00000534939.1
ENST00000539570.3 |
FBXL22
|
F-box and leucine-rich repeat protein 22 |
| chrX_-_138304939 | 3.58 |
ENST00000448673.1
|
FGF13
|
fibroblast growth factor 13 |
Network of associatons between targets according to the STRING database.
First level regulatory network of MEF2C
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 40.7 | 162.7 | GO:1901846 | positive regulation of cell communication by electrical coupling involved in cardiac conduction(GO:1901846) |
| 14.6 | 116.8 | GO:0035995 | detection of muscle stretch(GO:0035995) |
| 9.5 | 37.8 | GO:0032972 | regulation of muscle filament sliding speed(GO:0032972) |
| 9.4 | 37.7 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 7.3 | 36.5 | GO:0014878 | response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) |
| 7.2 | 115.7 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 6.9 | 41.4 | GO:0036309 | protein localization to M-band(GO:0036309) |
| 5.8 | 34.7 | GO:0071313 | cellular response to caffeine(GO:0071313) |
| 5.1 | 41.1 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 4.9 | 53.5 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 4.7 | 18.8 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 4.6 | 18.3 | GO:1902310 | positive regulation of peptidyl-serine dephosphorylation(GO:1902310) |
| 4.1 | 12.4 | GO:0044313 | protein K6-linked deubiquitination(GO:0044313) |
| 3.8 | 65.1 | GO:0014883 | transition between fast and slow fiber(GO:0014883) |
| 3.3 | 233.0 | GO:0030049 | muscle filament sliding(GO:0030049) actin-myosin filament sliding(GO:0033275) |
| 3.2 | 16.0 | GO:2001245 | regulation of phosphatidylcholine biosynthetic process(GO:2001245) |
| 2.7 | 32.0 | GO:2000332 | blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 2.0 | 14.1 | GO:0098528 | skeletal muscle fiber differentiation(GO:0098528) |
| 2.0 | 11.9 | GO:2001295 | malonyl-CoA biosynthetic process(GO:2001295) |
| 2.0 | 15.7 | GO:0010734 | protein glutathionylation(GO:0010731) regulation of protein glutathionylation(GO:0010732) negative regulation of protein glutathionylation(GO:0010734) |
| 1.9 | 15.6 | GO:0086048 | membrane depolarization during bundle of His cell action potential(GO:0086048) |
| 1.8 | 9.2 | GO:0090370 | negative regulation of cholesterol efflux(GO:0090370) |
| 1.6 | 29.0 | GO:0031272 | regulation of pseudopodium assembly(GO:0031272) |
| 1.5 | 27.1 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 1.5 | 54.3 | GO:0045663 | positive regulation of myoblast differentiation(GO:0045663) |
| 1.4 | 36.7 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 1.3 | 14.3 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 1.3 | 8.9 | GO:0010961 | cellular magnesium ion homeostasis(GO:0010961) |
| 1.3 | 13.8 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 1.2 | 10.5 | GO:0015793 | glycerol transport(GO:0015793) |
| 1.1 | 9.2 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 1.1 | 21.3 | GO:0006853 | carnitine shuttle(GO:0006853) |
| 1.1 | 4.5 | GO:0099640 | axo-dendritic protein transport(GO:0099640) |
| 1.1 | 26.5 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 1.1 | 20.1 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 1.0 | 7.3 | GO:0014718 | positive regulation of satellite cell activation involved in skeletal muscle regeneration(GO:0014718) |
| 1.0 | 6.2 | GO:0010650 | positive regulation of cell communication by electrical coupling(GO:0010650) |
| 1.0 | 39.5 | GO:0061049 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 1.0 | 10.8 | GO:0060681 | branch elongation involved in ureteric bud branching(GO:0060681) |
| 0.9 | 25.9 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.9 | 11.8 | GO:0086073 | bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 0.9 | 9.9 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.9 | 4.3 | GO:0000117 | regulation of transcription involved in G2/M transition of mitotic cell cycle(GO:0000117) |
| 0.8 | 36.2 | GO:0070296 | sarcoplasmic reticulum calcium ion transport(GO:0070296) |
| 0.7 | 36.4 | GO:0045214 | sarcomere organization(GO:0045214) |
| 0.7 | 4.1 | GO:0045200 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.6 | 3.2 | GO:0015862 | uridine transport(GO:0015862) |
| 0.6 | 3.7 | GO:0071542 | dopaminergic neuron differentiation(GO:0071542) |
| 0.6 | 4.4 | GO:0055129 | L-proline biosynthetic process(GO:0055129) |
| 0.6 | 64.2 | GO:0032233 | positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.6 | 37.2 | GO:0055008 | cardiac muscle tissue morphogenesis(GO:0055008) |
| 0.6 | 1.7 | GO:0007500 | mesodermal cell fate determination(GO:0007500) |
| 0.6 | 2.9 | GO:0090402 | oncogene-induced cell senescence(GO:0090402) |
| 0.6 | 1.7 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.5 | 25.4 | GO:0061718 | NADH regeneration(GO:0006735) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.5 | 66.5 | GO:0005977 | glycogen metabolic process(GO:0005977) |
| 0.5 | 2.9 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.5 | 4.8 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.5 | 6.6 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.5 | 11.6 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.5 | 3.7 | GO:0086069 | bundle of His cell to Purkinje myocyte communication(GO:0086069) |
| 0.5 | 1.8 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.4 | 7.2 | GO:2001214 | positive regulation of vasculogenesis(GO:2001214) |
| 0.4 | 2.4 | GO:0015840 | urea transport(GO:0015840) |
| 0.4 | 1.6 | GO:1903028 | positive regulation of opsonization(GO:1903028) |
| 0.4 | 15.4 | GO:0030239 | myofibril assembly(GO:0030239) |
| 0.4 | 26.1 | GO:0043268 | positive regulation of potassium ion transport(GO:0043268) |
| 0.3 | 2.1 | GO:1903527 | positive regulation of membrane tubulation(GO:1903527) |
| 0.3 | 4.1 | GO:0044557 | relaxation of smooth muscle(GO:0044557) relaxation of vascular smooth muscle(GO:0060087) |
| 0.3 | 6.5 | GO:2000675 | negative regulation of type B pancreatic cell apoptotic process(GO:2000675) |
| 0.3 | 25.8 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.3 | 10.8 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.3 | 2.4 | GO:0060235 | lens induction in camera-type eye(GO:0060235) |
| 0.3 | 11.6 | GO:0044381 | glucose import in response to insulin stimulus(GO:0044381) |
| 0.3 | 5.2 | GO:0090051 | negative regulation of cell migration involved in sprouting angiogenesis(GO:0090051) |
| 0.3 | 0.9 | GO:0003219 | cardiac right ventricle formation(GO:0003219) |
| 0.3 | 10.6 | GO:0045907 | positive regulation of vasoconstriction(GO:0045907) |
| 0.3 | 13.2 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.2 | 1.4 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.2 | 2.6 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.2 | 12.4 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.2 | 4.8 | GO:0042474 | middle ear morphogenesis(GO:0042474) |
| 0.2 | 3.9 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.2 | 31.4 | GO:0032091 | negative regulation of protein binding(GO:0032091) |
| 0.2 | 2.5 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.2 | 137.5 | GO:0043687 | post-translational protein modification(GO:0043687) |
| 0.2 | 1.4 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.2 | 11.8 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.2 | 2.2 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.2 | 13.8 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.1 | 8.3 | GO:0061098 | positive regulation of protein tyrosine kinase activity(GO:0061098) |
| 0.1 | 0.5 | GO:1904425 | negative regulation of GTP binding(GO:1904425) |
| 0.1 | 2.0 | GO:0039536 | negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.1 | 4.0 | GO:0055013 | cardiac muscle cell development(GO:0055013) |
| 0.1 | 2.7 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.1 | 4.1 | GO:0015695 | organic cation transport(GO:0015695) |
| 0.1 | 5.4 | GO:0007605 | sensory perception of sound(GO:0007605) |
| 0.1 | 16.3 | GO:0031032 | actomyosin structure organization(GO:0031032) |
| 0.1 | 4.7 | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.1 | 3.3 | GO:0060644 | mammary gland epithelial cell differentiation(GO:0060644) |
| 0.1 | 1.1 | GO:0045040 | protein import into mitochondrial outer membrane(GO:0045040) |
| 0.1 | 0.7 | GO:0015838 | quaternary ammonium group transport(GO:0015697) amino-acid betaine transport(GO:0015838) |
| 0.1 | 8.1 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.1 | 0.2 | GO:0033058 | directional locomotion(GO:0033058) |
| 0.1 | 21.2 | GO:0006986 | response to unfolded protein(GO:0006986) |
| 0.1 | 3.4 | GO:0031529 | ruffle organization(GO:0031529) |
| 0.1 | 3.7 | GO:0016577 | histone demethylation(GO:0016577) |
| 0.1 | 0.9 | GO:1904424 | regulation of GTP binding(GO:1904424) |
| 0.1 | 3.1 | GO:0007405 | neuroblast proliferation(GO:0007405) |
| 0.1 | 1.8 | GO:0051894 | positive regulation of focal adhesion assembly(GO:0051894) |
| 0.1 | 4.4 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.1 | 10.2 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.1 | 1.7 | GO:2000178 | negative regulation of neural precursor cell proliferation(GO:2000178) |
| 0.1 | 7.5 | GO:1990830 | response to leukemia inhibitory factor(GO:1990823) cellular response to leukemia inhibitory factor(GO:1990830) |
| 0.0 | 1.3 | GO:0036149 | phosphatidylinositol acyl-chain remodeling(GO:0036149) |
| 0.0 | 0.4 | GO:2000074 | regulation of type B pancreatic cell development(GO:2000074) |
| 0.0 | 4.7 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) |
| 0.0 | 3.2 | GO:0046426 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 0.7 | GO:0090360 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 0.0 | 0.1 | GO:0071376 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.0 | 2.3 | GO:0006941 | striated muscle contraction(GO:0006941) |
| 0.0 | 0.9 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.2 | GO:0015692 | lead ion transport(GO:0015692) |
| 0.0 | 3.4 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
| 0.0 | 20.9 | GO:0016567 | protein ubiquitination(GO:0016567) |
| 0.0 | 1.8 | GO:1901222 | regulation of NIK/NF-kappaB signaling(GO:1901222) |
| 0.0 | 1.2 | GO:0000038 | very long-chain fatty acid metabolic process(GO:0000038) |
| 0.0 | 0.5 | GO:1900745 | positive regulation of p38MAPK cascade(GO:1900745) |
| 0.0 | 1.7 | GO:0007044 | cell-substrate junction assembly(GO:0007044) |
| 0.0 | 0.4 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.0 | 0.7 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 2.1 | GO:0050821 | protein stabilization(GO:0050821) |
| 0.0 | 1.5 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 | 0.8 | GO:0090002 | establishment of protein localization to plasma membrane(GO:0090002) |
| 0.0 | 0.8 | GO:0045540 | regulation of cholesterol biosynthetic process(GO:0045540) |
| 0.0 | 1.3 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 0.1 | GO:0015684 | ferrous iron transport(GO:0015684) ferrous iron transmembrane transport(GO:1903874) |
| 0.0 | 1.2 | GO:0031100 | organ regeneration(GO:0031100) |
| 0.0 | 2.1 | GO:0007586 | digestion(GO:0007586) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 15.2 | 197.4 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 10.6 | 42.4 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 9.6 | 229.4 | GO:0005861 | troponin complex(GO:0005861) |
| 5.4 | 37.7 | GO:0014802 | terminal cisterna(GO:0014802) |
| 4.5 | 194.4 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 4.5 | 27.1 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 4.1 | 53.5 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 3.6 | 25.4 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 3.3 | 32.7 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 3.1 | 131.5 | GO:0031430 | M band(GO:0031430) |
| 2.0 | 36.7 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 2.0 | 20.0 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 2.0 | 37.6 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 1.9 | 11.4 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 1.7 | 26.5 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 1.5 | 11.8 | GO:0005916 | fascia adherens(GO:0005916) |
| 1.4 | 338.6 | GO:0030018 | Z disc(GO:0030018) |
| 1.4 | 14.1 | GO:0031672 | A band(GO:0031672) |
| 1.2 | 9.6 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 1.2 | 15.6 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 1.1 | 5.7 | GO:0034680 | integrin alpha10-beta1 complex(GO:0034680) |
| 0.8 | 37.4 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.7 | 32.0 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.7 | 4.4 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 0.7 | 48.2 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.7 | 5.3 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.6 | 15.7 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.5 | 13.9 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.5 | 8.6 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.5 | 4.3 | GO:0036379 | myofilament(GO:0036379) |
| 0.4 | 45.8 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.3 | 15.7 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.3 | 33.7 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.3 | 3.2 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.3 | 4.3 | GO:0032039 | integrator complex(GO:0032039) |
| 0.2 | 13.2 | GO:0016528 | sarcoplasm(GO:0016528) |
| 0.2 | 10.6 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.2 | 2.6 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.2 | 75.5 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.2 | 6.1 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.2 | 6.3 | GO:0030017 | sarcomere(GO:0030017) |
| 0.2 | 3.1 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.1 | 0.4 | GO:0020003 | symbiont-containing vacuole(GO:0020003) symbiont-containing vacuole membrane(GO:0020005) |
| 0.1 | 6.6 | GO:0016459 | myosin complex(GO:0016459) |
| 0.1 | 1.2 | GO:0098642 | network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) |
| 0.1 | 2.0 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.1 | 1.1 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.1 | 5.4 | GO:0032420 | stereocilium(GO:0032420) |
| 0.1 | 2.7 | GO:0098827 | endoplasmic reticulum subcompartment(GO:0098827) |
| 0.1 | 2.9 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.1 | 2.5 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.1 | 0.8 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.1 | 0.9 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.1 | 0.2 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.1 | 1.6 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.1 | 5.1 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.1 | 35.4 | GO:0015629 | actin cytoskeleton(GO:0015629) |
| 0.1 | 1.7 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 5.6 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.0 | 2.9 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.0 | 7.2 | GO:0030665 | clathrin-coated vesicle membrane(GO:0030665) |
| 0.0 | 10.0 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 3.0 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 9.3 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 27.3 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 0.9 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 14.3 | GO:0005924 | cell-substrate adherens junction(GO:0005924) focal adhesion(GO:0005925) |
| 0.0 | 2.4 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 0.2 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.0 | 0.5 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 2.4 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 2.4 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.3 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 2.1 | GO:0010008 | endosome membrane(GO:0010008) |
| 0.0 | 3.5 | GO:0005730 | nucleolus(GO:0005730) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 18.3 | 73.2 | GO:0030899 | calcium-dependent ATPase activity(GO:0030899) |
| 12.3 | 73.7 | GO:0051373 | FATZ binding(GO:0051373) |
| 8.1 | 40.6 | GO:0032038 | myosin II heavy chain binding(GO:0032038) |
| 6.5 | 71.0 | GO:0031433 | telethonin binding(GO:0031433) |
| 6.0 | 190.7 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 5.9 | 41.5 | GO:0031013 | troponin I binding(GO:0031013) |
| 5.3 | 16.0 | GO:0070538 | oleic acid binding(GO:0070538) |
| 5.2 | 15.6 | GO:0086057 | voltage-gated calcium channel activity involved in bundle of His cell action potential(GO:0086057) |
| 4.7 | 18.8 | GO:0046538 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 4.5 | 63.4 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 4.4 | 53.3 | GO:0031014 | troponin T binding(GO:0031014) |
| 3.3 | 26.5 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 3.2 | 63.4 | GO:0031432 | titin binding(GO:0031432) |
| 2.9 | 32.0 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 2.8 | 22.7 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) |
| 2.8 | 25.4 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 2.7 | 24.5 | GO:0017018 | myosin phosphatase activity(GO:0017018) |
| 2.6 | 18.3 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 2.4 | 9.6 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 2.0 | 10.0 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 2.0 | 20.0 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 2.0 | 11.9 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 1.7 | 20.1 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 1.5 | 12.4 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 1.4 | 46.6 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 1.3 | 29.7 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 1.3 | 13.2 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 1.1 | 4.5 | GO:0008426 | protein kinase C inhibitor activity(GO:0008426) |
| 1.0 | 36.7 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 1.0 | 3.0 | GO:0017130 | poly(C) RNA binding(GO:0017130) |
| 0.9 | 11.8 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 0.8 | 4.1 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.7 | 31.3 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.7 | 4.4 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.7 | 5.7 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.7 | 4.8 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.7 | 184.5 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.7 | 21.0 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.6 | 54.2 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.6 | 10.7 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.6 | 6.5 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.6 | 5.2 | GO:0001025 | RNA polymerase III transcription factor binding(GO:0001025) |
| 0.6 | 1.7 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.5 | 11.4 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.5 | 7.9 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.5 | 27.1 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.5 | 3.2 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.4 | 4.8 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.4 | 31.1 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.4 | 2.0 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.4 | 10.8 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.4 | 16.2 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.4 | 2.7 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.4 | 20.7 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.4 | 6.0 | GO:0042805 | actinin binding(GO:0042805) |
| 0.3 | 33.2 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.3 | 250.7 | GO:0003779 | actin binding(GO:0003779) |
| 0.3 | 4.1 | GO:0019534 | toxin transporter activity(GO:0019534) |
| 0.3 | 1.7 | GO:0001010 | transcription factor activity, sequence-specific DNA binding transcription factor recruiting(GO:0001010) |
| 0.3 | 4.7 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.3 | 201.6 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.2 | 12.4 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.2 | 5.6 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.2 | 2.7 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.2 | 6.6 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.2 | 2.2 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.2 | 10.6 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.1 | 0.4 | GO:0015168 | glycerol transmembrane transporter activity(GO:0015168) |
| 0.1 | 6.4 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.1 | 3.4 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.1 | 2.0 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.1 | 4.4 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.1 | 10.2 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 15.7 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.1 | 4.3 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.1 | 1.5 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.1 | 6.4 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.1 | 2.2 | GO:0031702 | angiotensin receptor binding(GO:0031701) type 1 angiotensin receptor binding(GO:0031702) |
| 0.1 | 3.2 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.1 | 1.1 | GO:0015266 | protein channel activity(GO:0015266) |
| 0.1 | 0.3 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 0.1 | 5.6 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 14.2 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.0 | 0.9 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 0.5 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.0 | 0.9 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 5.8 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 2.5 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 0.1 | GO:0015056 | corticotrophin-releasing factor receptor activity(GO:0015056) |
| 0.0 | 1.3 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.0 | 0.2 | GO:0015094 | cadmium ion transmembrane transporter activity(GO:0015086) cobalt ion transmembrane transporter activity(GO:0015087) lead ion transmembrane transporter activity(GO:0015094) ferrous iron uptake transmembrane transporter activity(GO:0015639) |
| 0.0 | 2.5 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.9 | GO:0035257 | nuclear hormone receptor binding(GO:0035257) |
| 0.0 | 0.2 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.0 | 1.4 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 0.3 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 2.6 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.1 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.0 | 1.7 | GO:0043130 | ubiquitin binding(GO:0043130) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 53.5 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.6 | 32.9 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.3 | 26.5 | PID BMP PATHWAY | BMP receptor signaling |
| 0.2 | 21.4 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.2 | 27.6 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.2 | 6.3 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.1 | 6.6 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 16.5 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.1 | 5.7 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.1 | 2.2 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.1 | 10.4 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.1 | 4.9 | ST P38 MAPK PATHWAY | p38 MAPK Pathway |
| 0.1 | 8.2 | PID FGF PATHWAY | FGF signaling pathway |
| 0.1 | 2.5 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.1 | 1.7 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.1 | 8.3 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.1 | 1.7 | PID EPO PATHWAY | EPO signaling pathway |
| 0.0 | 4.5 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 1.9 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 2.9 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 3.2 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.0 | 0.9 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 0.7 | PID AP1 PATHWAY | AP-1 transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.0 | 376.6 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.9 | 56.8 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.8 | 34.6 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.7 | 10.5 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.6 | 20.0 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.6 | 10.8 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.4 | 11.4 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.4 | 9.9 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.3 | 2.9 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.3 | 138.4 | REACTOME CLASS I MHC MEDIATED ANTIGEN PROCESSING PRESENTATION | Genes involved in Class I MHC mediated antigen processing & presentation |
| 0.3 | 9.2 | REACTOME SHC1 EVENTS IN EGFR SIGNALING | Genes involved in SHC1 events in EGFR signaling |
| 0.3 | 26.5 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.3 | 26.7 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.2 | 10.2 | REACTOME DEFENSINS | Genes involved in Defensins |
| 0.2 | 4.5 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.2 | 1.7 | REACTOME ARMS MEDIATED ACTIVATION | Genes involved in ARMS-mediated activation |
| 0.1 | 4.4 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.1 | 1.6 | REACTOME DOWNSTREAM SIGNALING EVENTS OF B CELL RECEPTOR BCR | Genes involved in Downstream Signaling Events Of B Cell Receptor (BCR) |
| 0.1 | 7.2 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.1 | 8.6 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 2.0 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.1 | 5.6 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.1 | 3.2 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 4.1 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.1 | 1.7 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.1 | 4.7 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.1 | 0.4 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 3.2 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 1.8 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 2.0 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 1.8 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 2.9 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 1.3 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 3.9 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.9 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 1.1 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.2 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.0 | 0.4 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |