Project
Illumina Body Map 2: averaged replicates
Navigation
Downloads
Results for MEF2D_MEF2A
Z-value: 7.99
Transcription factors associated with MEF2D_MEF2A
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
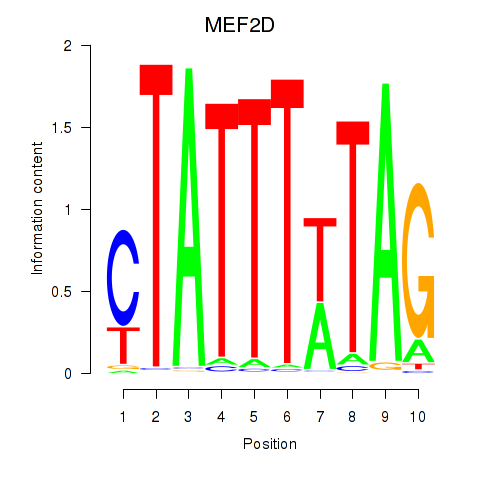
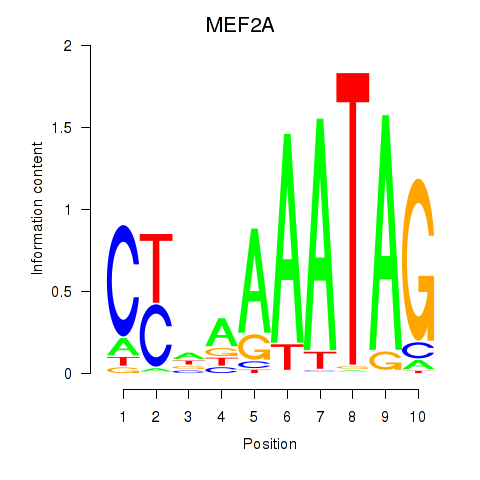
MEF2D
|
ENSG00000116604.13 | myocyte enhancer factor 2D |
|
MEF2A
|
ENSG00000068305.13 | myocyte enhancer factor 2A |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| MEF2A | hg19_v2_chr15_+_100106244_100106292 | 0.42 | 1.6e-02 | Click! |
| MEF2D | hg19_v2_chr1_-_156460391_156460417 | 0.40 | 2.5e-02 | Click! |
Activity profile of MEF2D_MEF2A motif
Sorted Z-values of MEF2D_MEF2A motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_19223523 | 72.82 |
ENST00000265968.3
|
CSRP3
|
cysteine and glycine-rich protein 3 (cardiac LIM protein) |
| chr7_-_113559104 | 68.71 |
ENST00000284601.3
|
PPP1R3A
|
protein phosphatase 1, regulatory subunit 3A |
| chr1_-_201391149 | 59.71 |
ENST00000555948.1
ENST00000556362.1 |
TNNI1
|
troponin I type 1 (skeletal, slow) |
| chr2_+_168043793 | 59.44 |
ENST00000409273.1
ENST00000409605.1 |
XIRP2
|
xin actin-binding repeat containing 2 |
| chr17_+_37821593 | 59.34 |
ENST00000578283.1
|
TCAP
|
titin-cap |
| chr6_-_123957942 | 59.30 |
ENST00000398178.3
|
TRDN
|
triadin |
| chr6_-_123958051 | 55.87 |
ENST00000546248.1
|
TRDN
|
triadin |
| chr2_+_88367299 | 55.78 |
ENST00000419482.2
ENST00000444564.2 |
SMYD1
|
SET and MYND domain containing 1 |
| chr16_-_31439735 | 54.77 |
ENST00000287490.4
|
COX6A2
|
cytochrome c oxidase subunit VIa polypeptide 2 |
| chr10_-_115423792 | 53.97 |
ENST00000369360.3
ENST00000360478.3 ENST00000359988.3 ENST00000369358.4 |
NRAP
|
nebulin-related anchoring protein |
| chr12_-_111358372 | 51.77 |
ENST00000548438.1
ENST00000228841.8 |
MYL2
|
myosin, light chain 2, regulatory, cardiac, slow |
| chr6_-_123958111 | 51.22 |
ENST00000542443.1
|
TRDN
|
triadin |
| chr2_-_211168332 | 50.56 |
ENST00000341685.4
|
MYL1
|
myosin, light chain 1, alkali; skeletal, fast |
| chrX_-_21776281 | 48.60 |
ENST00000379494.3
|
SMPX
|
small muscle protein, X-linked |
| chr3_-_39234074 | 48.36 |
ENST00000340369.3
ENST00000421646.1 ENST00000396251.1 |
XIRP1
|
xin actin-binding repeat containing 1 |
| chr2_+_88367368 | 47.88 |
ENST00000438570.1
|
SMYD1
|
SET and MYND domain containing 1 |
| chr5_+_53751445 | 41.61 |
ENST00000302005.1
|
HSPB3
|
heat shock 27kDa protein 3 |
| chr1_-_917497 | 41.31 |
ENST00000433179.2
|
C1orf170
|
chromosome 1 open reading frame 170 |
| chr10_+_50507232 | 40.45 |
ENST00000374144.3
|
C10orf71
|
chromosome 10 open reading frame 71 |
| chr5_+_191592 | 38.21 |
ENST00000328278.3
|
LRRC14B
|
leucine rich repeat containing 14B |
| chr1_-_917466 | 36.99 |
ENST00000341290.2
|
C1orf170
|
chromosome 1 open reading frame 170 |
| chr10_+_50507181 | 36.79 |
ENST00000323868.4
|
C10orf71
|
chromosome 10 open reading frame 71 |
| chr4_+_120056939 | 33.62 |
ENST00000307128.5
|
MYOZ2
|
myozenin 2 |
| chr10_-_75415825 | 33.38 |
ENST00000394810.2
|
SYNPO2L
|
synaptopodin 2-like |
| chr8_+_1993152 | 32.65 |
ENST00000262113.4
|
MYOM2
|
myomesin 2 |
| chr22_-_36013368 | 31.52 |
ENST00000442617.1
ENST00000397326.2 ENST00000397328.1 ENST00000451685.1 |
MB
|
myoglobin |
| chrX_-_15332665 | 31.26 |
ENST00000537676.1
ENST00000344384.4 |
ASB11
|
ankyrin repeat and SOCS box containing 11 |
| chr11_+_1942580 | 31.00 |
ENST00000381558.1
|
TNNT3
|
troponin T type 3 (skeletal, fast) |
| chr3_-_52486841 | 30.69 |
ENST00000496590.1
|
TNNC1
|
troponin C type 1 (slow) |
| chr8_+_1993173 | 29.87 |
ENST00000523438.1
|
MYOM2
|
myomesin 2 |
| chr22_+_26138108 | 29.85 |
ENST00000536101.1
ENST00000335473.7 ENST00000407587.2 |
MYO18B
|
myosin XVIIIB |
| chr1_-_100643765 | 29.56 |
ENST00000370137.1
ENST00000370138.1 ENST00000342895.3 |
LRRC39
|
leucine rich repeat containing 39 |
| chr10_+_88428370 | 29.51 |
ENST00000372066.3
ENST00000263066.6 ENST00000372056.4 ENST00000310944.6 ENST00000361373.4 ENST00000542786.1 |
LDB3
|
LIM domain binding 3 |
| chr17_-_9694614 | 29.24 |
ENST00000330255.5
ENST00000571134.1 |
DHRS7C
|
dehydrogenase/reductase (SDR family) member 7C |
| chr6_-_123958141 | 29.20 |
ENST00000334268.4
|
TRDN
|
triadin |
| chr14_-_23877474 | 28.82 |
ENST00000405093.3
|
MYH6
|
myosin, heavy chain 6, cardiac muscle, alpha |
| chr12_-_49393092 | 28.71 |
ENST00000421952.2
|
DDN
|
dendrin |
| chr2_-_211179883 | 28.46 |
ENST00000352451.3
|
MYL1
|
myosin, light chain 1, alkali; skeletal, fast |
| chr3_+_121311966 | 28.40 |
ENST00000338040.4
|
FBXO40
|
F-box protein 40 |
| chr1_-_26394114 | 28.36 |
ENST00000374272.3
|
TRIM63
|
tripartite motif containing 63, E3 ubiquitin protein ligase |
| chr11_+_1940925 | 28.17 |
ENST00000453458.1
ENST00000381557.2 ENST00000381589.3 ENST00000381579.3 ENST00000381563.4 ENST00000344578.4 |
TNNT3
|
troponin T type 3 (skeletal, fast) |
| chr7_-_150884332 | 27.90 |
ENST00000275838.1
ENST00000422024.1 ENST00000434669.1 |
ASB10
|
ankyrin repeat and SOCS box containing 10 |
| chr16_+_31225337 | 27.52 |
ENST00000322122.3
|
TRIM72
|
tripartite motif containing 72 |
| chr22_-_36018569 | 26.99 |
ENST00000419229.1
ENST00000406324.1 |
MB
|
myoglobin |
| chr20_+_61147651 | 26.29 |
ENST00000370527.3
ENST00000370524.2 |
C20orf166
|
chromosome 20 open reading frame 166 |
| chr11_+_1860200 | 25.31 |
ENST00000381911.1
|
TNNI2
|
troponin I type 2 (skeletal, fast) |
| chr5_+_78985673 | 25.09 |
ENST00000446378.2
|
CMYA5
|
cardiomyopathy associated 5 |
| chr12_+_101962128 | 24.74 |
ENST00000550514.1
|
MYBPC1
|
myosin binding protein C, slow type |
| chr9_-_97356075 | 24.46 |
ENST00000375337.3
|
FBP2
|
fructose-1,6-bisphosphatase 2 |
| chr10_+_88428206 | 24.44 |
ENST00000429277.2
ENST00000458213.2 ENST00000352360.5 |
LDB3
|
LIM domain binding 3 |
| chrX_-_63450480 | 24.27 |
ENST00000362002.2
|
ASB12
|
ankyrin repeat and SOCS box containing 12 |
| chr3_+_35683651 | 24.22 |
ENST00000187397.4
|
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa |
| chr4_+_119810134 | 23.96 |
ENST00000434046.2
|
SYNPO2
|
synaptopodin 2 |
| chr1_+_145413268 | 23.31 |
ENST00000421822.2
ENST00000336751.5 ENST00000497365.1 ENST00000475797.1 |
HFE2
|
hemochromatosis type 2 (juvenile) |
| chr18_-_3219847 | 23.27 |
ENST00000261606.7
|
MYOM1
|
myomesin 1 |
| chr2_+_170366203 | 23.16 |
ENST00000284669.1
|
KLHL41
|
kelch-like family member 41 |
| chr1_-_115238207 | 23.08 |
ENST00000520113.2
ENST00000369538.3 ENST00000353928.6 |
AMPD1
|
adenosine monophosphate deaminase 1 |
| chr17_-_10421853 | 22.32 |
ENST00000226207.5
|
MYH1
|
myosin, heavy chain 1, skeletal muscle, adult |
| chr18_-_3220106 | 22.22 |
ENST00000356443.4
ENST00000400569.3 |
MYOM1
|
myomesin 1 |
| chr10_-_75410771 | 21.91 |
ENST00000372873.4
|
SYNPO2L
|
synaptopodin 2-like |
| chr4_+_119809984 | 21.88 |
ENST00000307142.4
ENST00000448416.2 ENST00000429713.2 |
SYNPO2
|
synaptopodin 2 |
| chr1_+_160160283 | 21.82 |
ENST00000368079.3
|
CASQ1
|
calsequestrin 1 (fast-twitch, skeletal muscle) |
| chr1_+_160160346 | 21.75 |
ENST00000368078.3
|
CASQ1
|
calsequestrin 1 (fast-twitch, skeletal muscle) |
| chr16_+_7382745 | 21.64 |
ENST00000436368.2
ENST00000311745.5 ENST00000355637.4 ENST00000340209.4 |
RBFOX1
|
RNA binding protein, fox-1 homolog (C. elegans) 1 |
| chr17_+_7184986 | 21.26 |
ENST00000317370.8
ENST00000571308.1 |
SLC2A4
|
solute carrier family 2 (facilitated glucose transporter), member 4 |
| chr11_+_57308979 | 21.10 |
ENST00000457912.1
|
SMTNL1
|
smoothelin-like 1 |
| chr7_-_150884873 | 20.97 |
ENST00000377867.3
|
ASB10
|
ankyrin repeat and SOCS box containing 10 |
| chr17_+_4855053 | 20.63 |
ENST00000518175.1
|
ENO3
|
enolase 3 (beta, muscle) |
| chr19_+_3933085 | 19.89 |
ENST00000168977.2
ENST00000599576.1 |
NMRK2
|
nicotinamide riboside kinase 2 |
| chr15_+_42651691 | 19.65 |
ENST00000357568.3
ENST00000349748.3 ENST00000318023.7 ENST00000397163.3 |
CAPN3
|
calpain 3, (p94) |
| chr2_+_220299547 | 19.63 |
ENST00000312358.7
|
SPEG
|
SPEG complex locus |
| chr2_+_103378472 | 19.44 |
ENST00000412401.2
|
TMEM182
|
transmembrane protein 182 |
| chr19_+_3933579 | 18.56 |
ENST00000593949.1
|
NMRK2
|
nicotinamide riboside kinase 2 |
| chr3_+_42727011 | 18.12 |
ENST00000287777.4
|
KLHL40
|
kelch-like family member 40 |
| chr6_+_53948221 | 18.01 |
ENST00000460844.2
|
MLIP
|
muscular LMNA-interacting protein |
| chr12_+_101988627 | 17.48 |
ENST00000547405.1
ENST00000452455.2 ENST00000441232.1 ENST00000360610.2 ENST00000392934.3 ENST00000547509.1 ENST00000361685.2 ENST00000549145.1 ENST00000553190.1 |
MYBPC1
|
myosin binding protein C, slow type |
| chr5_-_142077569 | 17.43 |
ENST00000407758.1
ENST00000441680.2 ENST00000419524.2 |
FGF1
|
fibroblast growth factor 1 (acidic) |
| chr6_+_53948328 | 17.16 |
ENST00000370876.2
|
MLIP
|
muscular LMNA-interacting protein |
| chrX_+_153046456 | 17.09 |
ENST00000393786.3
ENST00000370104.1 ENST00000370108.3 ENST00000370101.3 ENST00000430541.1 ENST00000370100.1 |
SRPK3
|
SRSF protein kinase 3 |
| chr6_+_41021027 | 17.00 |
ENST00000244669.2
|
APOBEC2
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 2 |
| chr22_-_51017084 | 16.31 |
ENST00000360719.2
ENST00000457250.1 ENST00000440709.1 |
CPT1B
|
carnitine palmitoyltransferase 1B (muscle) |
| chr1_-_116311402 | 16.00 |
ENST00000261448.5
|
CASQ2
|
calsequestrin 2 (cardiac muscle) |
| chr22_-_51016846 | 15.89 |
ENST00000312108.7
ENST00000395650.2 |
CPT1B
|
carnitine palmitoyltransferase 1B (muscle) |
| chr11_+_1860832 | 15.50 |
ENST00000252898.7
|
TNNI2
|
troponin I type 2 (skeletal, fast) |
| chr7_-_44105158 | 15.24 |
ENST00000297283.3
|
PGAM2
|
phosphoglycerate mutase 2 (muscle) |
| chr3_+_135741576 | 15.20 |
ENST00000334546.2
|
PPP2R3A
|
protein phosphatase 2, regulatory subunit B'', alpha |
| chr1_-_116311323 | 15.13 |
ENST00000456138.2
|
CASQ2
|
calsequestrin 2 (cardiac muscle) |
| chr15_+_65369082 | 15.11 |
ENST00000432196.2
|
KBTBD13
|
kelch repeat and BTB (POZ) domain containing 13 |
| chr10_-_69455873 | 14.67 |
ENST00000433211.2
|
CTNNA3
|
catenin (cadherin-associated protein), alpha 3 |
| chr12_-_22094336 | 14.60 |
ENST00000326684.4
|
ABCC9
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 9 |
| chr5_+_155753745 | 14.32 |
ENST00000435422.3
ENST00000337851.4 ENST00000447401.1 |
SGCD
|
sarcoglycan, delta (35kDa dystrophin-associated glycoprotein) |
| chr3_-_155011483 | 14.25 |
ENST00000489090.1
|
RP11-451G4.2
|
RP11-451G4.2 |
| chr10_-_69425399 | 14.15 |
ENST00000330298.6
|
CTNNA3
|
catenin (cadherin-associated protein), alpha 3 |
| chr1_+_160085567 | 13.90 |
ENST00000392233.3
|
ATP1A2
|
ATPase, Na+/K+ transporting, alpha 2 polypeptide |
| chr1_-_201390846 | 13.88 |
ENST00000367312.1
ENST00000555340.2 ENST00000361379.4 |
TNNI1
|
troponin I type 1 (skeletal, slow) |
| chr3_-_196242233 | 13.78 |
ENST00000397537.2
|
SMCO1
|
single-pass membrane protein with coiled-coil domains 1 |
| chr1_+_160085501 | 13.65 |
ENST00000361216.3
|
ATP1A2
|
ATPase, Na+/K+ transporting, alpha 2 polypeptide |
| chr1_+_228395755 | 13.61 |
ENST00000284548.11
ENST00000570156.2 ENST00000422127.1 ENST00000366707.4 ENST00000366709.4 |
OBSCN
|
obscurin, cytoskeletal calmodulin and titin-interacting RhoGEF |
| chr2_+_239047337 | 13.56 |
ENST00000409223.1
ENST00000305959.4 |
KLHL30
|
kelch-like family member 30 |
| chr12_-_22094159 | 13.56 |
ENST00000538350.1
|
ABCC9
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 9 |
| chr17_-_79900255 | 13.49 |
ENST00000330655.3
ENST00000582198.1 |
MYADML2
PYCR1
|
myeloid-associated differentiation marker-like 2 pyrroline-5-carboxylate reductase 1 |
| chr2_-_97509749 | 13.33 |
ENST00000331001.2
ENST00000318357.4 |
ANKRD23
|
ankyrin repeat domain 23 |
| chr16_-_4292071 | 13.10 |
ENST00000399609.3
|
SRL
|
sarcalumenin |
| chr17_-_41132088 | 13.05 |
ENST00000591916.1
ENST00000451885.2 ENST00000454303.1 |
PTGES3L
PTGES3L-AARSD1
|
prostaglandin E synthase 3 (cytosolic)-like PTGES3L-AARSD1 readthrough |
| chr11_+_1860682 | 12.94 |
ENST00000381906.1
|
TNNI2
|
troponin I type 2 (skeletal, fast) |
| chr2_-_97509729 | 12.94 |
ENST00000418232.1
|
ANKRD23
|
ankyrin repeat domain 23 |
| chr15_-_83474806 | 12.88 |
ENST00000541889.1
ENST00000334574.8 ENST00000561368.1 |
FSD2
|
fibronectin type III and SPRY domain containing 2 |
| chr12_+_81101277 | 12.86 |
ENST00000228641.3
|
MYF6
|
myogenic factor 6 (herculin) |
| chr12_+_119616447 | 12.77 |
ENST00000281938.2
|
HSPB8
|
heat shock 22kDa protein 8 |
| chr11_+_1940786 | 12.76 |
ENST00000278317.6
ENST00000381561.4 ENST00000381548.3 ENST00000360603.3 ENST00000381549.3 |
TNNT3
|
troponin T type 3 (skeletal, fast) |
| chr3_+_127634069 | 12.74 |
ENST00000405109.1
|
KBTBD12
|
kelch repeat and BTB (POZ) domain containing 12 |
| chr11_-_47374246 | 12.74 |
ENST00000545968.1
ENST00000399249.2 ENST00000256993.4 |
MYBPC3
|
myosin binding protein C, cardiac |
| chr1_-_216896780 | 12.71 |
ENST00000459955.1
ENST00000366937.1 ENST00000408911.3 ENST00000391890.3 |
ESRRG
|
estrogen-related receptor gamma |
| chr7_+_143013198 | 12.48 |
ENST00000343257.2
|
CLCN1
|
chloride channel, voltage-sensitive 1 |
| chr11_+_3666335 | 12.38 |
ENST00000250693.1
|
ART1
|
ADP-ribosyltransferase 1 |
| chr8_-_70745575 | 12.08 |
ENST00000524945.1
|
SLCO5A1
|
solute carrier organic anion transporter family, member 5A1 |
| chr1_-_40137710 | 11.85 |
ENST00000235628.1
|
NT5C1A
|
5'-nucleotidase, cytosolic IA |
| chr14_-_21493884 | 11.78 |
ENST00000556974.1
ENST00000554419.1 ENST00000298687.5 ENST00000397858.1 ENST00000360463.3 ENST00000350792.3 ENST00000397847.2 |
NDRG2
|
NDRG family member 2 |
| chr17_-_42188598 | 11.72 |
ENST00000591714.1
|
HDAC5
|
histone deacetylase 5 |
| chr13_+_76378305 | 11.71 |
ENST00000526371.1
ENST00000526528.1 |
LMO7
|
LIM domain 7 |
| chr1_-_31845914 | 11.70 |
ENST00000373713.2
|
FABP3
|
fatty acid binding protein 3, muscle and heart (mammary-derived growth inhibitor) |
| chr4_-_100575781 | 11.65 |
ENST00000511828.1
|
RP11-766F14.2
|
Protein LOC285556 |
| chr5_-_16509101 | 11.48 |
ENST00000399793.2
|
FAM134B
|
family with sequence similarity 134, member B |
| chr17_-_27949911 | 11.43 |
ENST00000492276.2
ENST00000345068.5 ENST00000584602.1 |
CORO6
|
coronin 6 |
| chr3_+_127634312 | 11.33 |
ENST00000407609.3
|
KBTBD12
|
kelch repeat and BTB (POZ) domain containing 12 |
| chr7_+_28448995 | 11.11 |
ENST00000424599.1
|
CREB5
|
cAMP responsive element binding protein 5 |
| chr4_+_186064395 | 11.00 |
ENST00000281456.6
|
SLC25A4
|
solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 4 |
| chr14_-_21493649 | 10.93 |
ENST00000553442.1
ENST00000555869.1 ENST00000556457.1 ENST00000397844.2 ENST00000554415.1 |
NDRG2
|
NDRG family member 2 |
| chr2_-_179537145 | 10.84 |
ENST00000448510.2
|
TTN
|
titin |
| chr17_+_65040678 | 10.76 |
ENST00000226021.3
|
CACNG1
|
calcium channel, voltage-dependent, gamma subunit 1 |
| chr2_-_219696519 | 10.58 |
ENST00000545803.1
ENST00000430489.1 ENST00000392098.3 |
PRKAG3
|
protein kinase, AMP-activated, gamma 3 non-catalytic subunit |
| chr3_+_179370517 | 10.52 |
ENST00000263966.3
|
USP13
|
ubiquitin specific peptidase 13 (isopeptidase T-3) |
| chr10_-_72141330 | 10.38 |
ENST00000395011.1
ENST00000395010.1 |
LRRC20
|
leucine rich repeat containing 20 |
| chr2_-_219696511 | 10.29 |
ENST00000439262.2
|
PRKAG3
|
protein kinase, AMP-activated, gamma 3 non-catalytic subunit |
| chr6_-_3912207 | 10.15 |
ENST00000566733.1
|
RP1-140K8.5
|
RP1-140K8.5 |
| chr4_+_41362796 | 10.11 |
ENST00000508501.1
ENST00000512946.1 ENST00000313860.7 ENST00000512632.1 ENST00000512820.1 |
LIMCH1
|
LIM and calponin homology domains 1 |
| chr12_+_52431016 | 10.09 |
ENST00000553200.1
|
NR4A1
|
nuclear receptor subfamily 4, group A, member 1 |
| chr3_-_46904946 | 9.87 |
ENST00000292327.4
|
MYL3
|
myosin, light chain 3, alkali; ventricular, skeletal, slow |
| chr17_-_10372875 | 9.85 |
ENST00000255381.2
|
MYH4
|
myosin, heavy chain 4, skeletal muscle |
| chr3_-_46904918 | 9.72 |
ENST00000395869.1
|
MYL3
|
myosin, light chain 3, alkali; ventricular, skeletal, slow |
| chr4_+_95373037 | 9.65 |
ENST00000359265.4
ENST00000437932.1 ENST00000380180.3 ENST00000318007.5 ENST00000450793.1 ENST00000538141.1 ENST00000317968.4 ENST00000512274.1 ENST00000503974.1 ENST00000504489.1 ENST00000542407.1 |
PDLIM5
|
PDZ and LIM domain 5 |
| chr9_-_85882145 | 9.63 |
ENST00000328788.1
|
FRMD3
|
FERM domain containing 3 |
| chr3_+_35683775 | 9.62 |
ENST00000452563.1
|
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa |
| chr2_+_179149636 | 9.49 |
ENST00000409631.1
|
OSBPL6
|
oxysterol binding protein-like 6 |
| chr16_+_30075463 | 9.41 |
ENST00000562168.1
ENST00000569545.1 |
ALDOA
|
aldolase A, fructose-bisphosphate |
| chr16_+_30075595 | 9.38 |
ENST00000563060.2
|
ALDOA
|
aldolase A, fructose-bisphosphate |
| chr3_-_57326704 | 9.26 |
ENST00000487349.1
ENST00000389601.3 |
ASB14
|
ankyrin repeat and SOCS box containing 14 |
| chr9_-_130639997 | 9.17 |
ENST00000373176.1
|
AK1
|
adenylate kinase 1 |
| chr3_-_192445289 | 9.08 |
ENST00000430714.1
ENST00000418610.1 ENST00000448795.1 ENST00000445105.2 |
FGF12
|
fibroblast growth factor 12 |
| chr2_+_220283091 | 9.00 |
ENST00000373960.3
|
DES
|
desmin |
| chr8_+_67039278 | 8.94 |
ENST00000276573.7
ENST00000350034.4 |
TRIM55
|
tripartite motif containing 55 |
| chr3_+_138067521 | 8.73 |
ENST00000494949.1
|
MRAS
|
muscle RAS oncogene homolog |
| chr16_+_6069072 | 8.73 |
ENST00000547605.1
ENST00000550418.1 ENST00000553186.1 |
RBFOX1
|
RNA binding protein, fox-1 homolog (C. elegans) 1 |
| chr19_+_35630022 | 8.69 |
ENST00000589209.1
|
FXYD1
|
FXYD domain containing ion transport regulator 1 |
| chr9_+_100263912 | 8.62 |
ENST00000259365.4
|
TMOD1
|
tropomodulin 1 |
| chr12_-_57644952 | 8.61 |
ENST00000554578.1
ENST00000546246.2 ENST00000553489.1 ENST00000332782.2 |
STAC3
|
SH3 and cysteine rich domain 3 |
| chr1_-_216896674 | 8.51 |
ENST00000475275.1
ENST00000469486.1 ENST00000481543.1 |
ESRRG
|
estrogen-related receptor gamma |
| chr3_+_35683816 | 8.30 |
ENST00000438577.1
|
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa |
| chr13_+_76378407 | 8.22 |
ENST00000447038.1
|
LMO7
|
LIM domain 7 |
| chr1_+_150254936 | 8.12 |
ENST00000447007.1
ENST00000369095.1 ENST00000369094.1 |
C1orf51
|
chromosome 1 open reading frame 51 |
| chr14_-_70546897 | 8.08 |
ENST00000394330.2
ENST00000533541.1 ENST00000216568.7 |
SLC8A3
|
solute carrier family 8 (sodium/calcium exchanger), member 3 |
| chr4_-_23891658 | 8.07 |
ENST00000507380.1
|
PPARGC1A
|
peroxisome proliferator-activated receptor gamma, coactivator 1 alpha |
| chr8_+_67039131 | 8.04 |
ENST00000315962.4
ENST00000353317.5 |
TRIM55
|
tripartite motif containing 55 |
| chr16_+_30075783 | 8.04 |
ENST00000412304.2
|
ALDOA
|
aldolase A, fructose-bisphosphate |
| chr13_+_76413852 | 8.03 |
ENST00000533809.2
|
LMO7
|
LIM domain 7 |
| chr6_-_76203345 | 7.93 |
ENST00000393004.2
|
FILIP1
|
filamin A interacting protein 1 |
| chr5_+_173316341 | 7.93 |
ENST00000520867.1
ENST00000334035.5 |
CPEB4
|
cytoplasmic polyadenylation element binding protein 4 |
| chr16_+_30076052 | 7.93 |
ENST00000563987.1
|
ALDOA
|
aldolase A, fructose-bisphosphate |
| chrX_-_138304939 | 7.87 |
ENST00000448673.1
|
FGF13
|
fibroblast growth factor 13 |
| chr19_+_35630344 | 7.84 |
ENST00000455515.2
|
FXYD1
|
FXYD domain containing ion transport regulator 1 |
| chr4_+_41362615 | 7.82 |
ENST00000509638.1
|
LIMCH1
|
LIM and calponin homology domains 1 |
| chr4_-_44653636 | 7.79 |
ENST00000415895.4
ENST00000332990.5 |
YIPF7
|
Yip1 domain family, member 7 |
| chr6_-_76203454 | 7.79 |
ENST00000237172.7
|
FILIP1
|
filamin A interacting protein 1 |
| chr4_+_114066764 | 7.77 |
ENST00000511380.1
|
ANK2
|
ankyrin 2, neuronal |
| chr1_+_170633047 | 7.62 |
ENST00000239461.6
ENST00000497230.2 |
PRRX1
|
paired related homeobox 1 |
| chr11_+_112832090 | 7.62 |
ENST00000533760.1
|
NCAM1
|
neural cell adhesion molecule 1 |
| chr10_+_86184676 | 7.44 |
ENST00000543283.1
|
CCSER2
|
coiled-coil serine-rich protein 2 |
| chr17_-_27467418 | 7.24 |
ENST00000528564.1
|
MYO18A
|
myosin XVIIIA |
| chr10_-_14880002 | 7.12 |
ENST00000465530.1
|
CDNF
|
cerebral dopamine neurotrophic factor |
| chr13_+_76210448 | 7.08 |
ENST00000377499.5
|
LMO7
|
LIM domain 7 |
| chrX_+_70521584 | 7.04 |
ENST00000373829.3
ENST00000538820.1 |
ITGB1BP2
|
integrin beta 1 binding protein (melusin) 2 |
| chr6_-_45983581 | 7.02 |
ENST00000339561.6
|
CLIC5
|
chloride intracellular channel 5 |
| chr19_+_35629702 | 7.02 |
ENST00000351325.4
|
FXYD1
|
FXYD domain containing ion transport regulator 1 |
| chr1_-_203144941 | 7.00 |
ENST00000255416.4
|
MYBPH
|
myosin binding protein H |
| chr11_+_112832202 | 6.97 |
ENST00000534015.1
|
NCAM1
|
neural cell adhesion molecule 1 |
| chr22_+_17956618 | 6.95 |
ENST00000262608.8
|
CECR2
|
cat eye syndrome chromosome region, candidate 2 |
| chr5_+_133772383 | 6.86 |
ENST00000513329.1
|
AC005355.1
|
AC005355.1 |
| chr9_-_33402506 | 6.84 |
ENST00000377425.4
ENST00000537089.1 ENST00000297988.1 ENST00000539936.1 ENST00000541274.1 |
AQP7
|
aquaporin 7 |
| chr6_-_45983549 | 6.75 |
ENST00000544153.1
|
CLIC5
|
chloride intracellular channel 5 |
| chr3_+_138067314 | 6.64 |
ENST00000423968.2
|
MRAS
|
muscle RAS oncogene homolog |
| chrX_-_71933888 | 6.62 |
ENST00000373542.4
ENST00000339490.3 ENST00000541944.1 ENST00000373539.3 ENST00000373545.3 |
PHKA1
|
phosphorylase kinase, alpha 1 (muscle) |
| chr19_-_49576198 | 6.58 |
ENST00000221444.1
|
KCNA7
|
potassium voltage-gated channel, shaker-related subfamily, member 7 |
| chr7_+_95115210 | 6.55 |
ENST00000428113.1
ENST00000325885.5 |
ASB4
|
ankyrin repeat and SOCS box containing 4 |
| chr14_-_62217779 | 6.40 |
ENST00000554254.1
|
HIF1A-AS2
|
HIF1A antisense RNA 2 |
| chr8_-_17533838 | 6.35 |
ENST00000400046.1
|
MTUS1
|
microtubule associated tumor suppressor 1 |
| chr14_-_21493123 | 6.32 |
ENST00000556147.1
ENST00000554489.1 ENST00000555657.1 ENST00000557274.1 ENST00000555158.1 ENST00000554833.1 ENST00000555384.1 ENST00000556420.1 ENST00000554893.1 ENST00000553503.1 ENST00000555733.1 ENST00000553867.1 ENST00000397856.3 ENST00000397855.3 ENST00000556008.1 ENST00000557182.1 ENST00000554483.1 ENST00000556688.1 ENST00000397853.3 ENST00000556329.2 ENST00000554143.1 ENST00000397851.2 ENST00000555142.1 ENST00000557676.1 ENST00000556924.1 |
NDRG2
|
NDRG family member 2 |
| chr3_+_35721182 | 6.29 |
ENST00000413378.1
ENST00000417925.1 |
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa |
| chr7_+_15728003 | 6.20 |
ENST00000442176.1
|
AC005550.4
|
AC005550.4 |
| chr3_+_35680339 | 6.20 |
ENST00000450234.1
|
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa |
| chr1_-_23751189 | 6.13 |
ENST00000374601.3
ENST00000450454.2 |
TCEA3
|
transcription elongation factor A (SII), 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of MEF2D_MEF2A
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 48.9 | 195.6 | GO:1901846 | positive regulation of cell communication by electrical coupling involved in cardiac conduction(GO:1901846) |
| 17.9 | 143.0 | GO:0035995 | detection of muscle stretch(GO:0035995) |
| 10.8 | 108.0 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 9.2 | 36.8 | GO:0032972 | regulation of muscle filament sliding speed(GO:0032972) |
| 9.2 | 27.6 | GO:1903280 | negative regulation of calcium:sodium antiporter activity(GO:1903280) |
| 8.7 | 43.6 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 8.1 | 40.7 | GO:0014878 | response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) |
| 7.3 | 58.5 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 6.1 | 24.5 | GO:0005986 | sucrose biosynthetic process(GO:0005986) |
| 5.4 | 472.1 | GO:0030049 | muscle filament sliding(GO:0030049) actin-myosin filament sliding(GO:0033275) |
| 4.1 | 24.7 | GO:0036309 | protein localization to M-band(GO:0036309) |
| 3.8 | 15.2 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 3.5 | 10.5 | GO:0044313 | protein K6-linked deubiquitination(GO:0044313) |
| 3.1 | 115.3 | GO:0045663 | positive regulation of myoblast differentiation(GO:0045663) |
| 2.9 | 23.5 | GO:0010731 | protein glutathionylation(GO:0010731) regulation of protein glutathionylation(GO:0010732) negative regulation of protein glutathionylation(GO:0010734) |
| 2.8 | 31.1 | GO:0071313 | cellular response to caffeine(GO:0071313) |
| 2.8 | 19.6 | GO:0014718 | positive regulation of satellite cell activation involved in skeletal muscle regeneration(GO:0014718) |
| 2.6 | 18.1 | GO:0098528 | skeletal muscle fiber differentiation(GO:0098528) |
| 2.3 | 11.7 | GO:2001245 | regulation of phosphatidylcholine biosynthetic process(GO:2001245) |
| 2.3 | 34.8 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 2.2 | 60.3 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 2.2 | 28.8 | GO:0086073 | bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 2.1 | 23.1 | GO:0032264 | IMP salvage(GO:0032264) |
| 2.0 | 26.3 | GO:0060297 | regulation of sarcomere organization(GO:0060297) |
| 1.9 | 23.2 | GO:2000291 | regulation of myoblast proliferation(GO:2000291) |
| 1.9 | 13.5 | GO:0055129 | L-proline biosynthetic process(GO:0055129) |
| 1.8 | 11.0 | GO:0015853 | adenine transport(GO:0015853) |
| 1.8 | 32.9 | GO:0090361 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 1.8 | 20.1 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 1.8 | 7.2 | GO:1903028 | positive regulation of opsonization(GO:1903028) |
| 1.7 | 17.0 | GO:0006216 | cytidine catabolic process(GO:0006216) cytidine deamination(GO:0009972) cytidine metabolic process(GO:0046087) |
| 1.7 | 10.0 | GO:0045200 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 1.6 | 9.8 | GO:2001295 | malonyl-CoA biosynthetic process(GO:2001295) |
| 1.6 | 31.1 | GO:0006853 | carnitine shuttle(GO:0006853) |
| 1.4 | 4.1 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 1.4 | 60.9 | GO:0045214 | sarcomere organization(GO:0045214) |
| 1.3 | 17.4 | GO:0060681 | branch elongation involved in ureteric bud branching(GO:0060681) |
| 1.3 | 3.9 | GO:0001544 | initiation of primordial ovarian follicle growth(GO:0001544) |
| 1.3 | 3.8 | GO:0032242 | regulation of nucleoside transport(GO:0032242) |
| 1.2 | 38.4 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 1.1 | 9.1 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 1.1 | 5.6 | GO:0000117 | regulation of transcription involved in G2/M transition of mitotic cell cycle(GO:0000117) |
| 1.0 | 23.3 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.9 | 5.7 | GO:0055011 | atrial cardiac muscle cell differentiation(GO:0055011) atrial cardiac muscle cell development(GO:0055014) |
| 0.9 | 2.8 | GO:0033058 | directional locomotion(GO:0033058) |
| 0.9 | 36.1 | GO:0010880 | regulation of release of sequestered calcium ion into cytosol by sarcoplasmic reticulum(GO:0010880) |
| 0.9 | 18.3 | GO:0090051 | negative regulation of cell migration involved in sprouting angiogenesis(GO:0090051) |
| 0.9 | 11.8 | GO:0009128 | purine nucleoside monophosphate catabolic process(GO:0009128) |
| 0.9 | 8.1 | GO:0015793 | glycerol transport(GO:0015793) |
| 0.9 | 8.1 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.9 | 2.7 | GO:0006059 | hexitol metabolic process(GO:0006059) |
| 0.9 | 12.5 | GO:0098870 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.9 | 59.4 | GO:0055008 | cardiac muscle tissue morphogenesis(GO:0055008) |
| 0.9 | 101.1 | GO:0032233 | positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.8 | 26.9 | GO:0003298 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.8 | 13.8 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.7 | 6.3 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.7 | 4.9 | GO:0010961 | cellular magnesium ion homeostasis(GO:0010961) |
| 0.7 | 97.7 | GO:0005977 | glycogen metabolic process(GO:0005977) |
| 0.7 | 26.3 | GO:0044381 | glucose import in response to insulin stimulus(GO:0044381) |
| 0.6 | 2.5 | GO:0099640 | axo-dendritic protein transport(GO:0099640) |
| 0.6 | 2.4 | GO:0006480 | N-terminal protein amino acid methylation(GO:0006480) |
| 0.6 | 1.7 | GO:0090108 | positive regulation of high-density lipoprotein particle assembly(GO:0090108) positive regulation of secretion of lysosomal enzymes(GO:0090340) |
| 0.6 | 2.8 | GO:0015862 | uridine transport(GO:0015862) |
| 0.6 | 3.4 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.6 | 3.9 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.5 | 20.3 | GO:0061718 | NADH regeneration(GO:0006735) glycolytic process through fructose-6-phosphate(GO:0061615) glycolytic process through glucose-6-phosphate(GO:0061620) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.5 | 6.7 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.5 | 5.4 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.5 | 1.9 | GO:0035523 | protein K29-linked deubiquitination(GO:0035523) |
| 0.5 | 1.4 | GO:0061394 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) |
| 0.5 | 7.9 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.4 | 21.1 | GO:0045907 | positive regulation of vasoconstriction(GO:0045907) |
| 0.4 | 5.3 | GO:0060087 | relaxation of vascular smooth muscle(GO:0060087) |
| 0.4 | 1.3 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.4 | 1.3 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.4 | 60.7 | GO:0006941 | striated muscle contraction(GO:0006941) |
| 0.4 | 26.7 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.4 | 4.6 | GO:0045078 | positive regulation of interferon-gamma biosynthetic process(GO:0045078) |
| 0.4 | 16.3 | GO:0030239 | myofibril assembly(GO:0030239) |
| 0.4 | 4.1 | GO:0048664 | neuron fate determination(GO:0048664) |
| 0.4 | 0.8 | GO:1903644 | regulation of chaperone-mediated protein folding(GO:1903644) |
| 0.4 | 3.1 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.4 | 1.6 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.4 | 0.7 | GO:0003219 | cardiac right ventricle formation(GO:0003219) |
| 0.4 | 4.6 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.4 | 1.4 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.4 | 2.8 | GO:0032485 | Ral protein signal transduction(GO:0032484) regulation of Ral protein signal transduction(GO:0032485) |
| 0.3 | 5.9 | GO:2001214 | positive regulation of vasculogenesis(GO:2001214) |
| 0.3 | 1.0 | GO:0033594 | response to hydroxyisoflavone(GO:0033594) |
| 0.3 | 12.3 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.3 | 7.5 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.3 | 1.4 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.3 | 1.9 | GO:0032468 | Golgi calcium ion homeostasis(GO:0032468) |
| 0.3 | 3.5 | GO:0097106 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) postsynaptic density organization(GO:0097106) cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 0.3 | 2.2 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.3 | 1.3 | GO:0003094 | glomerular filtration(GO:0003094) renal filtration(GO:0097205) |
| 0.3 | 211.5 | GO:0043687 | post-translational protein modification(GO:0043687) |
| 0.3 | 12.4 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.3 | 18.6 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.3 | 1.5 | GO:0007512 | adult heart development(GO:0007512) |
| 0.3 | 3.5 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.3 | 56.8 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.2 | 7.0 | GO:0006309 | apoptotic DNA fragmentation(GO:0006309) |
| 0.2 | 1.5 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.2 | 4.9 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.2 | 2.6 | GO:0035860 | glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.2 | 12.4 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.2 | 3.6 | GO:0000733 | DNA strand renaturation(GO:0000733) |
| 0.2 | 16.5 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.2 | 1.7 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.2 | 2.6 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.2 | 16.5 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.2 | 3.5 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 0.2 | 1.8 | GO:2000074 | regulation of type B pancreatic cell development(GO:2000074) |
| 0.2 | 2.4 | GO:0030497 | fatty acid elongation(GO:0030497) |
| 0.2 | 4.8 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.2 | 2.5 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.1 | 4.5 | GO:0030207 | chondroitin sulfate catabolic process(GO:0030207) |
| 0.1 | 33.0 | GO:0006936 | muscle contraction(GO:0006936) |
| 0.1 | 35.9 | GO:0006986 | response to unfolded protein(GO:0006986) |
| 0.1 | 16.9 | GO:0032091 | negative regulation of protein binding(GO:0032091) |
| 0.1 | 17.7 | GO:0031032 | actomyosin structure organization(GO:0031032) |
| 0.1 | 18.6 | GO:0060333 | interferon-gamma-mediated signaling pathway(GO:0060333) |
| 0.1 | 1.7 | GO:1903546 | protein localization to photoreceptor outer segment(GO:1903546) |
| 0.1 | 9.2 | GO:0015949 | nucleobase-containing small molecule interconversion(GO:0015949) |
| 0.1 | 0.5 | GO:0036367 | adaptation of rhodopsin mediated signaling(GO:0016062) light adaption(GO:0036367) |
| 0.1 | 14.3 | GO:0048738 | cardiac muscle tissue development(GO:0048738) |
| 0.1 | 1.8 | GO:0016127 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.1 | 3.4 | GO:0019054 | modulation by virus of host process(GO:0019054) |
| 0.1 | 0.8 | GO:1903527 | positive regulation of membrane tubulation(GO:1903527) |
| 0.1 | 2.1 | GO:2000675 | negative regulation of type B pancreatic cell apoptotic process(GO:2000675) |
| 0.1 | 1.0 | GO:0060347 | heart trabecula formation(GO:0060347) |
| 0.1 | 0.1 | GO:0070252 | actin-mediated cell contraction(GO:0070252) |
| 0.1 | 0.8 | GO:0070193 | synaptonemal complex assembly(GO:0007130) synaptonemal complex organization(GO:0070193) |
| 0.1 | 15.2 | GO:1990823 | response to leukemia inhibitory factor(GO:1990823) cellular response to leukemia inhibitory factor(GO:1990830) |
| 0.1 | 0.6 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.1 | 4.0 | GO:0019228 | neuronal action potential(GO:0019228) |
| 0.1 | 2.8 | GO:0048265 | response to pain(GO:0048265) |
| 0.1 | 0.7 | GO:0030222 | eosinophil differentiation(GO:0030222) |
| 0.1 | 0.5 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.1 | 1.2 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.1 | 2.1 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.1 | 0.9 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.1 | 1.0 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.1 | 3.3 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.1 | 3.6 | GO:0032024 | positive regulation of insulin secretion(GO:0032024) |
| 0.1 | 0.4 | GO:0019566 | arabinose metabolic process(GO:0019566) L-arabinose metabolic process(GO:0046373) |
| 0.1 | 2.7 | GO:0060338 | regulation of type I interferon-mediated signaling pathway(GO:0060338) |
| 0.1 | 4.0 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.1 | 0.5 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.1 | 5.1 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.0 | 0.4 | GO:0021914 | negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021914) |
| 0.0 | 1.4 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 6.8 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
| 0.0 | 2.4 | GO:1901379 | regulation of potassium ion transmembrane transport(GO:1901379) |
| 0.0 | 5.2 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.0 | 0.9 | GO:1900745 | positive regulation of p38MAPK cascade(GO:1900745) |
| 0.0 | 2.5 | GO:1901222 | regulation of NIK/NF-kappaB signaling(GO:1901222) |
| 0.0 | 12.3 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 0.0 | 0.9 | GO:0071542 | dopaminergic neuron differentiation(GO:0071542) |
| 0.0 | 1.4 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.0 | 0.5 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.0 | 2.3 | GO:0007040 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.0 | 2.2 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.0 | 0.9 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.0 | 3.3 | GO:0015918 | sterol transport(GO:0015918) |
| 0.0 | 0.9 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 2.7 | GO:0007044 | cell-substrate junction assembly(GO:0007044) |
| 0.0 | 2.0 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.0 | 16.0 | GO:0016567 | protein ubiquitination(GO:0016567) |
| 0.0 | 0.0 | GO:1902714 | negative regulation of interferon-gamma secretion(GO:1902714) |
| 0.0 | 0.5 | GO:1900077 | negative regulation of insulin receptor signaling pathway(GO:0046627) negative regulation of cellular response to insulin stimulus(GO:1900077) |
| 0.0 | 1.0 | GO:0030521 | androgen receptor signaling pathway(GO:0030521) |
| 0.0 | 0.2 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 1.2 | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.0 | 0.5 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.0 | 0.7 | GO:0048593 | camera-type eye morphogenesis(GO:0048593) |
| 0.0 | 0.3 | GO:0007188 | adenylate cyclase-modulating G-protein coupled receptor signaling pathway(GO:0007188) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 17.7 | 283.4 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 17.1 | 102.6 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 14.6 | 58.2 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 9.8 | 236.0 | GO:0005861 | troponin complex(GO:0005861) |
| 6.8 | 291.5 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 4.7 | 60.8 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 3.8 | 162.8 | GO:0031430 | M band(GO:0031430) |
| 3.6 | 28.8 | GO:0005916 | fascia adherens(GO:0005916) |
| 3.5 | 28.2 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 3.4 | 60.3 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 2.9 | 20.6 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 2.1 | 19.3 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 2.0 | 503.5 | GO:0031674 | I band(GO:0031674) |
| 1.9 | 30.3 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 1.8 | 51.1 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 1.5 | 23.3 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 1.4 | 14.3 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 1.4 | 6.8 | GO:0034680 | integrin alpha10-beta1 complex(GO:0034680) |
| 1.1 | 28.7 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 1.0 | 49.1 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.8 | 14.0 | GO:0005865 | striated muscle thin filament(GO:0005865) myofilament(GO:0036379) |
| 0.7 | 47.8 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.7 | 18.9 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.7 | 4.0 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 0.7 | 6.6 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.7 | 6.6 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.6 | 2.5 | GO:0031592 | centrosomal corona(GO:0031592) |
| 0.6 | 10.6 | GO:0031010 | ISWI-type complex(GO:0031010) |
| 0.5 | 2.7 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.5 | 1.4 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 0.5 | 1.4 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.5 | 53.2 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.4 | 1.3 | GO:0043614 | multi-eIF complex(GO:0043614) |
| 0.4 | 1.3 | GO:0020005 | symbiont-containing vacuole(GO:0020003) symbiont-containing vacuole membrane(GO:0020005) |
| 0.4 | 19.0 | GO:0016528 | sarcoplasm(GO:0016528) |
| 0.4 | 5.6 | GO:0032039 | integrator complex(GO:0032039) |
| 0.4 | 12.3 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.3 | 1.7 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.3 | 14.9 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.3 | 114.9 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.3 | 5.3 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.3 | 0.5 | GO:0019898 | extrinsic component of membrane(GO:0019898) |
| 0.2 | 7.0 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.2 | 33.9 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.2 | 7.9 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.2 | 10.8 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.2 | 1.7 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.2 | 35.2 | GO:0016605 | PML body(GO:0016605) |
| 0.2 | 6.8 | GO:0030673 | axolemma(GO:0030673) |
| 0.1 | 3.3 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.1 | 2.4 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.1 | 11.5 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.1 | 2.8 | GO:0005922 | connexon complex(GO:0005922) |
| 0.1 | 5.3 | GO:0016459 | myosin complex(GO:0016459) |
| 0.1 | 22.5 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.1 | 3.4 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.1 | 0.8 | GO:0030893 | meiotic cohesin complex(GO:0030893) |
| 0.1 | 0.5 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.1 | 20.8 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 6.1 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.1 | 2.7 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.1 | 0.7 | GO:0098645 | network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) |
| 0.1 | 9.7 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.1 | 3.5 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.0 | 3.4 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.0 | 1.6 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 0.7 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 10.0 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 8.6 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 5.5 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 0.4 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.1 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.0 | 20.6 | GO:0015629 | actin cytoskeleton(GO:0015629) |
| 0.0 | 11.2 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.5 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 3.4 | GO:0043657 | host(GO:0018995) host cell(GO:0043657) |
| 0.0 | 1.0 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 3.9 | GO:0005930 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.0 | 0.3 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.7 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 9.2 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 2.6 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 2.2 | GO:0035580 | specific granule lumen(GO:0035580) |
| 0.0 | 4.3 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 4.3 | GO:0005938 | cell cortex(GO:0005938) |
| 0.0 | 4.9 | GO:0070062 | extracellular exosome(GO:0070062) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 18.0 | 71.9 | GO:0030899 | calcium-dependent ATPase activity(GO:0030899) |
| 15.5 | 93.0 | GO:0051373 | FATZ binding(GO:0051373) |
| 12.1 | 72.8 | GO:0031433 | telethonin binding(GO:0031433) |
| 9.6 | 38.4 | GO:0061769 | ribosylnicotinamide kinase activity(GO:0050262) ribosylnicotinate kinase activity(GO:0061769) |
| 9.2 | 342.1 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 7.0 | 28.2 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 6.1 | 24.5 | GO:0042132 | fructose 1,6-bisphosphate 1-phosphatase activity(GO:0042132) |
| 6.0 | 84.4 | GO:0031014 | troponin T binding(GO:0031014) |
| 5.0 | 94.4 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 4.6 | 91.2 | GO:0031432 | titin binding(GO:0031432) |
| 4.0 | 32.2 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) |
| 3.9 | 11.7 | GO:0070538 | oleic acid binding(GO:0070538) |
| 3.8 | 34.6 | GO:0017018 | myosin phosphatase activity(GO:0017018) |
| 3.8 | 15.2 | GO:0004082 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 3.0 | 6.1 | GO:0030172 | troponin C binding(GO:0030172) |
| 2.9 | 23.3 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 2.9 | 34.8 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 2.7 | 11.0 | GO:0015207 | ATP:ADP antiporter activity(GO:0005471) adenine transmembrane transporter activity(GO:0015207) |
| 2.7 | 21.2 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 2.3 | 50.8 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 2.3 | 20.6 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 2.2 | 13.5 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 2.2 | 28.8 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 1.9 | 17.0 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 1.7 | 59.4 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 1.7 | 60.3 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 1.6 | 9.8 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 1.6 | 14.2 | GO:0001025 | RNA polymerase III transcription factor binding(GO:0001025) |
| 1.4 | 4.1 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 1.4 | 27.6 | GO:1990239 | steroid hormone binding(GO:1990239) |
| 1.4 | 6.8 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 1.3 | 10.5 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 1.2 | 12.4 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 1.2 | 103.7 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 1.2 | 20.9 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 1.2 | 8.1 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 1.1 | 103.2 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 1.1 | 5.4 | GO:0004572 | mannosyl-oligosaccharide 1,3-1,6-alpha-mannosidase activity(GO:0004572) |
| 1.1 | 5.3 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.9 | 27.4 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.9 | 18.7 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.8 | 6.8 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.8 | 14.4 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.8 | 3.8 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 0.7 | 187.8 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.7 | 6.6 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.6 | 2.5 | GO:0008426 | protein kinase C inhibitor activity(GO:0008426) |
| 0.6 | 17.4 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.6 | 2.4 | GO:0071885 | N-terminal protein N-methyltransferase activity(GO:0071885) |
| 0.6 | 2.9 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.6 | 1.7 | GO:0001133 | RNA polymerase II transcription factor activity, sequence-specific transcription regulatory region DNA binding(GO:0001133) |
| 0.6 | 9.7 | GO:0042805 | actinin binding(GO:0042805) |
| 0.5 | 3.1 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.5 | 27.5 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.5 | 2.4 | GO:0017099 | very-long-chain-acyl-CoA dehydrogenase activity(GO:0017099) |
| 0.5 | 11.8 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.5 | 9.2 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.5 | 8.6 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.4 | 12.5 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.4 | 1.3 | GO:0035379 | carbon dioxide transmembrane transporter activity(GO:0035379) |
| 0.4 | 27.1 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.3 | 267.5 | GO:0003779 | actin binding(GO:0003779) |
| 0.3 | 16.0 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.3 | 242.2 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.3 | 78.2 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.3 | 1.0 | GO:0046848 | hydroxyapatite binding(GO:0046848) |
| 0.3 | 3.6 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.3 | 5.1 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.3 | 3.0 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.3 | 2.7 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.3 | 1.5 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.3 | 4.1 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.3 | 2.6 | GO:0016167 | glial cell-derived neurotrophic factor receptor activity(GO:0016167) |
| 0.3 | 1.7 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.3 | 0.8 | GO:0010698 | acetyltransferase activator activity(GO:0010698) |
| 0.3 | 4.9 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.2 | 2.5 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.2 | 12.4 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.2 | 19.2 | GO:0070888 | E-box binding(GO:0070888) |
| 0.2 | 13.8 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.2 | 1.6 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.2 | 3.6 | GO:0016279 | protein-lysine N-methyltransferase activity(GO:0016279) |
| 0.2 | 5.3 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.2 | 0.6 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.2 | 4.9 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.2 | 22.8 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.2 | 10.8 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.2 | 2.7 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.2 | 2.1 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.2 | 0.5 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 0.2 | 5.6 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.2 | 1.9 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.2 | 4.0 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.1 | 18.4 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 4.0 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.1 | 14.4 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.1 | 7.9 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.1 | 1.9 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.1 | 8.9 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.1 | 2.8 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.1 | 5.7 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.1 | 3.4 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.1 | 0.9 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.1 | 1.9 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.1 | 1.0 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.1 | 21.6 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.1 | 3.3 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.1 | 2.4 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.1 | 5.9 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.1 | 0.6 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.1 | 0.4 | GO:0046556 | alpha-L-arabinofuranosidase activity(GO:0046556) |
| 0.1 | 2.8 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.1 | 0.3 | GO:0015056 | corticotrophin-releasing factor receptor activity(GO:0015056) |
| 0.1 | 1.4 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 0.9 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.1 | 1.3 | GO:0042923 | neuropeptide binding(GO:0042923) |
| 0.1 | 1.8 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.1 | 1.0 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 5.1 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.0 | 0.2 | GO:0004782 | sulfinoalanine decarboxylase activity(GO:0004782) |
| 0.0 | 5.4 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 1.4 | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups(GO:0016765) |
| 0.0 | 4.0 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 0.5 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 1.3 | GO:0017022 | myosin binding(GO:0017022) |
| 0.0 | 1.2 | GO:0022832 | voltage-gated ion channel activity(GO:0005244) voltage-gated channel activity(GO:0022832) |
| 0.0 | 3.7 | GO:0042626 | ATPase activity, coupled to transmembrane movement of substances(GO:0042626) |
| 0.0 | 1.3 | GO:0004177 | aminopeptidase activity(GO:0004177) |
| 0.0 | 0.2 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 2.4 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 2.4 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 0.6 | GO:0016788 | hydrolase activity, acting on ester bonds(GO:0016788) |
| 0.0 | 1.3 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 2.6 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 1.2 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 2.2 | GO:0002020 | protease binding(GO:0002020) |
| 0.0 | 5.0 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 2.5 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 1.6 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 51.8 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 1.0 | 58.6 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.4 | 21.2 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.3 | 26.6 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.3 | 11.1 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.2 | 20.4 | PID BMP PATHWAY | BMP receptor signaling |
| 0.2 | 24.6 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.2 | 26.9 | PID FGF PATHWAY | FGF signaling pathway |
| 0.2 | 3.7 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.2 | 13.9 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.1 | 6.8 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.1 | 3.4 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.1 | 4.8 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.1 | 9.4 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.1 | 15.4 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.1 | 1.3 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.1 | 5.1 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.1 | 4.2 | ST P38 MAPK PATHWAY | p38 MAPK Pathway |
| 0.1 | 13.5 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.1 | 3.3 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.1 | 3.1 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.1 | 2.4 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 3.4 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 4.6 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 2.2 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 2.9 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 1.1 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 1.4 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 0.6 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.0 | 1.0 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 1.8 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 0.7 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.5 | PID INSULIN PATHWAY | Insulin Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.2 | 473.7 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 1.4 | 62.9 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 1.1 | 83.6 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 1.0 | 17.4 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.8 | 23.1 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.7 | 11.8 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.6 | 18.7 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.5 | 31.9 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.5 | 8.1 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.4 | 29.5 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.4 | 13.5 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.3 | 23.3 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.3 | 96.6 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.3 | 1.8 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.2 | 18.6 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |
| 0.2 | 6.6 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.2 | 9.2 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.2 | 3.4 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.2 | 8.9 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.2 | 10.4 | REACTOME INTERACTIONS OF VPR WITH HOST CELLULAR PROTEINS | Genes involved in Interactions of Vpr with host cellular proteins |
| 0.2 | 7.9 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.1 | 7.0 | REACTOME DEFENSINS | Genes involved in Defensins |
| 0.1 | 13.7 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.1 | 1.8 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.1 | 3.1 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.1 | 2.8 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.1 | 5.3 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.1 | 2.5 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.1 | 3.9 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 2.7 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.1 | 1.8 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.1 | 6.6 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 3.5 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.1 | 3.7 | REACTOME DOWNSTREAM SIGNALING EVENTS OF B CELL RECEPTOR BCR | Genes involved in Downstream Signaling Events Of B Cell Receptor (BCR) |
| 0.1 | 1.7 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.1 | 3.3 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.1 | 1.0 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.1 | 4.6 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 3.4 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.1 | 9.7 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 1.5 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 1.4 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.0 | 1.4 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 1.2 | REACTOME TRANSPORT OF MATURE TRANSCRIPT TO CYTOPLASM | Genes involved in Transport of Mature Transcript to Cytoplasm |
| 0.0 | 0.5 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 2.2 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.7 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 1.6 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 0.4 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.8 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |