Project
Illumina Body Map 2: averaged replicates
Navigation
Downloads
Results for MSX2
Z-value: 1.42
Transcription factors associated with MSX2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
MSX2
|
ENSG00000120149.7 | msh homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| MSX2 | hg19_v2_chr5_+_174151536_174151610 | 0.10 | 5.9e-01 | Click! |
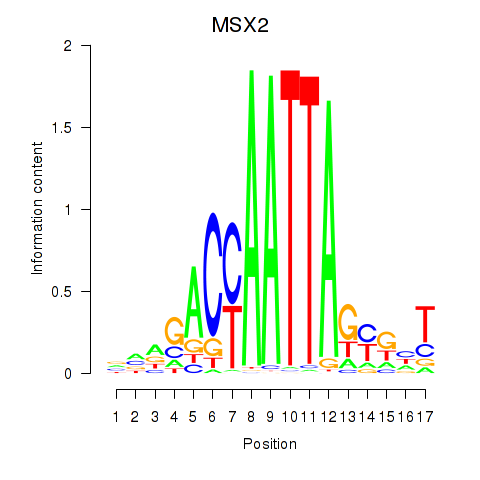
Activity profile of MSX2 motif
Sorted Z-values of MSX2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chrX_-_15332665 | 4.34 |
ENST00000537676.1
ENST00000344384.4 |
ASB11
|
ankyrin repeat and SOCS box containing 11 |
| chrX_-_21676442 | 3.60 |
ENST00000379499.2
|
KLHL34
|
kelch-like family member 34 |
| chr8_+_1993152 | 3.49 |
ENST00000262113.4
|
MYOM2
|
myomesin 2 |
| chr6_-_139613269 | 3.24 |
ENST00000358430.3
|
TXLNB
|
taxilin beta |
| chr14_+_32798462 | 3.21 |
ENST00000280979.4
|
AKAP6
|
A kinase (PRKA) anchor protein 6 |
| chr8_+_1993173 | 3.16 |
ENST00000523438.1
|
MYOM2
|
myomesin 2 |
| chr14_+_32798547 | 3.15 |
ENST00000557354.1
ENST00000557102.1 ENST00000557272.1 |
AKAP6
|
A kinase (PRKA) anchor protein 6 |
| chr6_+_53948221 | 2.91 |
ENST00000460844.2
|
MLIP
|
muscular LMNA-interacting protein |
| chrX_+_130192318 | 2.73 |
ENST00000370922.1
|
ARHGAP36
|
Rho GTPase activating protein 36 |
| chr6_+_53948328 | 2.66 |
ENST00000370876.2
|
MLIP
|
muscular LMNA-interacting protein |
| chr4_-_109684120 | 2.53 |
ENST00000512646.1
ENST00000411864.2 ENST00000296486.3 ENST00000510706.1 |
ETNPPL
|
ethanolamine-phosphate phospho-lyase |
| chr22_+_17956618 | 2.45 |
ENST00000262608.8
|
CECR2
|
cat eye syndrome chromosome region, candidate 2 |
| chr10_-_72142345 | 1.79 |
ENST00000373224.1
ENST00000446961.1 ENST00000358141.2 ENST00000357631.2 |
LRRC20
|
leucine rich repeat containing 20 |
| chr11_-_13517565 | 1.58 |
ENST00000282091.1
ENST00000529816.1 |
PTH
|
parathyroid hormone |
| chr1_+_152943122 | 1.40 |
ENST00000328051.2
|
SPRR4
|
small proline-rich protein 4 |
| chr15_+_41245160 | 1.28 |
ENST00000444189.2
ENST00000446533.3 |
CHAC1
|
ChaC, cation transport regulator homolog 1 (E. coli) |
| chr1_+_202317855 | 1.26 |
ENST00000356764.2
|
PPP1R12B
|
protein phosphatase 1, regulatory subunit 12B |
| chr17_-_10372875 | 1.20 |
ENST00000255381.2
|
MYH4
|
myosin, heavy chain 4, skeletal muscle |
| chr9_+_124329336 | 1.16 |
ENST00000394340.3
ENST00000436835.1 ENST00000259371.2 |
DAB2IP
|
DAB2 interacting protein |
| chr7_-_88425025 | 1.16 |
ENST00000297203.2
|
C7orf62
|
chromosome 7 open reading frame 62 |
| chr5_+_95998070 | 1.14 |
ENST00000421689.2
ENST00000510756.1 ENST00000512620.1 |
CAST
|
calpastatin |
| chr7_-_100183742 | 1.09 |
ENST00000310300.6
|
LRCH4
|
leucine-rich repeats and calponin homology (CH) domain containing 4 |
| chr20_+_56964169 | 1.09 |
ENST00000475243.1
|
VAPB
|
VAMP (vesicle-associated membrane protein)-associated protein B and C |
| chr1_-_151762943 | 1.06 |
ENST00000368825.3
ENST00000368823.1 ENST00000458431.2 ENST00000368827.6 ENST00000368824.3 |
TDRKH
|
tudor and KH domain containing |
| chr19_+_46236509 | 1.01 |
ENST00000457052.2
|
AC074212.3
|
Uncharacterized protein |
| chr6_+_78400375 | 1.00 |
ENST00000602452.2
|
MEI4
|
meiosis-specific 4 homolog (S. cerevisiae) |
| chr13_-_103389159 | 0.97 |
ENST00000322527.2
|
CCDC168
|
coiled-coil domain containing 168 |
| chr5_-_132200477 | 0.97 |
ENST00000296875.2
|
GDF9
|
growth differentiation factor 9 |
| chr5_+_140557371 | 0.96 |
ENST00000239444.2
|
PCDHB8
|
protocadherin beta 8 |
| chr8_+_9413410 | 0.95 |
ENST00000520408.1
ENST00000310430.6 ENST00000522110.1 |
TNKS
|
tankyrase, TRF1-interacting ankyrin-related ADP-ribose polymerase |
| chr6_+_34204642 | 0.95 |
ENST00000347617.6
ENST00000401473.3 ENST00000311487.5 ENST00000447654.1 ENST00000395004.3 |
HMGA1
|
high mobility group AT-hook 1 |
| chr1_-_151762900 | 0.94 |
ENST00000440583.2
|
TDRKH
|
tudor and KH domain containing |
| chr3_-_114866084 | 0.92 |
ENST00000357258.3
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr4_-_46996424 | 0.92 |
ENST00000264318.3
|
GABRA4
|
gamma-aminobutyric acid (GABA) A receptor, alpha 4 |
| chr4_-_138453606 | 0.90 |
ENST00000412923.2
ENST00000344876.4 ENST00000507846.1 ENST00000510305.1 |
PCDH18
|
protocadherin 18 |
| chr18_+_5748793 | 0.85 |
ENST00000566533.1
ENST00000562452.2 |
RP11-945C19.1
|
RP11-945C19.1 |
| chr6_-_167040731 | 0.85 |
ENST00000265678.4
|
RPS6KA2
|
ribosomal protein S6 kinase, 90kDa, polypeptide 2 |
| chr4_-_88449771 | 0.84 |
ENST00000535835.1
|
SPARCL1
|
SPARC-like 1 (hevin) |
| chr17_+_12569306 | 0.80 |
ENST00000425538.1
|
MYOCD
|
myocardin |
| chr4_-_69536346 | 0.80 |
ENST00000338206.5
|
UGT2B15
|
UDP glucuronosyltransferase 2 family, polypeptide B15 |
| chr1_-_21377383 | 0.79 |
ENST00000374935.3
|
EIF4G3
|
eukaryotic translation initiation factor 4 gamma, 3 |
| chr5_+_95997918 | 0.79 |
ENST00000395812.2
ENST00000395813.1 ENST00000359176.4 ENST00000325674.7 |
CAST
|
calpastatin |
| chr16_-_52061283 | 0.79 |
ENST00000566314.1
|
C16orf97
|
chromosome 16 open reading frame 97 |
| chr7_+_100183927 | 0.78 |
ENST00000241071.6
ENST00000360609.2 |
FBXO24
|
F-box protein 24 |
| chr1_-_232651312 | 0.77 |
ENST00000262861.4
|
SIPA1L2
|
signal-induced proliferation-associated 1 like 2 |
| chr1_-_36916066 | 0.77 |
ENST00000315643.9
|
OSCP1
|
organic solute carrier partner 1 |
| chr5_+_95997885 | 0.75 |
ENST00000511097.2
|
CAST
|
calpastatin |
| chr18_-_48346298 | 0.75 |
ENST00000398439.3
|
MRO
|
maestro |
| chr1_-_94050668 | 0.75 |
ENST00000539242.1
|
BCAR3
|
breast cancer anti-estrogen resistance 3 |
| chr20_+_31805131 | 0.73 |
ENST00000375454.3
ENST00000375452.3 |
BPIFA3
|
BPI fold containing family A, member 3 |
| chr1_-_36916011 | 0.72 |
ENST00000356637.5
ENST00000354267.3 ENST00000235532.5 |
OSCP1
|
organic solute carrier partner 1 |
| chr6_-_167040693 | 0.72 |
ENST00000366863.2
|
RPS6KA2
|
ribosomal protein S6 kinase, 90kDa, polypeptide 2 |
| chr1_-_36915880 | 0.69 |
ENST00000445843.3
|
OSCP1
|
organic solute carrier partner 1 |
| chr3_+_140396881 | 0.69 |
ENST00000286349.3
|
TRIM42
|
tripartite motif containing 42 |
| chr1_-_8877692 | 0.68 |
ENST00000400908.2
|
RERE
|
arginine-glutamic acid dipeptide (RE) repeats |
| chr18_+_63273310 | 0.68 |
ENST00000579862.1
|
RP11-775G23.1
|
RP11-775G23.1 |
| chrX_+_89176881 | 0.67 |
ENST00000283891.5
ENST00000561129.2 |
TGIF2LX
|
TGFB-induced factor homeobox 2-like, X-linked |
| chr1_+_202317815 | 0.66 |
ENST00000608999.1
ENST00000336894.4 ENST00000480184.1 |
PPP1R12B
|
protein phosphatase 1, regulatory subunit 12B |
| chr20_+_56964253 | 0.66 |
ENST00000395802.3
|
VAPB
|
VAMP (vesicle-associated membrane protein)-associated protein B and C |
| chr12_-_10955226 | 0.66 |
ENST00000240687.2
|
TAS2R7
|
taste receptor, type 2, member 7 |
| chr19_+_42817450 | 0.65 |
ENST00000301204.3
|
TMEM145
|
transmembrane protein 145 |
| chr2_-_200335983 | 0.64 |
ENST00000457245.1
|
SATB2
|
SATB homeobox 2 |
| chr14_-_23504337 | 0.61 |
ENST00000361611.6
|
PSMB5
|
proteasome (prosome, macropain) subunit, beta type, 5 |
| chr3_-_164796269 | 0.61 |
ENST00000264382.3
|
SI
|
sucrase-isomaltase (alpha-glucosidase) |
| chr6_+_25652432 | 0.60 |
ENST00000377961.2
|
SCGN
|
secretagogin, EF-hand calcium binding protein |
| chr7_+_130794846 | 0.60 |
ENST00000421797.2
|
MKLN1
|
muskelin 1, intracellular mediator containing kelch motifs |
| chr8_+_92261516 | 0.59 |
ENST00000276609.3
ENST00000309536.2 |
SLC26A7
|
solute carrier family 26 (anion exchanger), member 7 |
| chr20_-_50418947 | 0.59 |
ENST00000371539.3
|
SALL4
|
spalt-like transcription factor 4 |
| chr8_-_117886563 | 0.59 |
ENST00000519837.1
ENST00000522699.1 |
RAD21
|
RAD21 homolog (S. pombe) |
| chr4_-_138453559 | 0.58 |
ENST00000511115.1
|
PCDH18
|
protocadherin 18 |
| chr10_+_99205959 | 0.58 |
ENST00000352634.4
ENST00000353979.3 ENST00000370842.2 ENST00000345745.5 |
ZDHHC16
|
zinc finger, DHHC-type containing 16 |
| chr5_+_31532373 | 0.56 |
ENST00000325366.9
ENST00000355907.3 ENST00000507818.2 |
C5orf22
|
chromosome 5 open reading frame 22 |
| chr20_-_50419055 | 0.56 |
ENST00000217086.4
|
SALL4
|
spalt-like transcription factor 4 |
| chr14_-_23504432 | 0.55 |
ENST00000425762.2
|
PSMB5
|
proteasome (prosome, macropain) subunit, beta type, 5 |
| chrX_+_77154935 | 0.55 |
ENST00000481445.1
|
COX7B
|
cytochrome c oxidase subunit VIIb |
| chr11_-_123814580 | 0.55 |
ENST00000321252.2
|
OR6T1
|
olfactory receptor, family 6, subfamily T, member 1 |
| chr16_+_69345243 | 0.54 |
ENST00000254950.11
|
VPS4A
|
vacuolar protein sorting 4 homolog A (S. cerevisiae) |
| chr19_-_47137942 | 0.51 |
ENST00000300873.4
|
GNG8
|
guanine nucleotide binding protein (G protein), gamma 8 |
| chr7_+_130794878 | 0.50 |
ENST00000416992.2
|
MKLN1
|
muskelin 1, intracellular mediator containing kelch motifs |
| chr9_-_95056010 | 0.50 |
ENST00000443024.2
|
IARS
|
isoleucyl-tRNA synthetase |
| chr4_-_46126093 | 0.49 |
ENST00000295452.4
|
GABRG1
|
gamma-aminobutyric acid (GABA) A receptor, gamma 1 |
| chr17_-_12920907 | 0.49 |
ENST00000609757.1
ENST00000581499.2 ENST00000580504.1 |
ELAC2
|
elaC ribonuclease Z 2 |
| chr1_+_244515930 | 0.49 |
ENST00000366537.1
ENST00000308105.4 |
C1orf100
|
chromosome 1 open reading frame 100 |
| chr6_+_25652501 | 0.49 |
ENST00000334979.6
|
SCGN
|
secretagogin, EF-hand calcium binding protein |
| chrX_-_139587225 | 0.49 |
ENST00000370536.2
|
SOX3
|
SRY (sex determining region Y)-box 3 |
| chr19_-_19249255 | 0.48 |
ENST00000587583.2
ENST00000450333.2 ENST00000587096.1 ENST00000162044.9 ENST00000592369.1 ENST00000587915.1 |
TMEM161A
|
transmembrane protein 161A |
| chr5_-_31532160 | 0.48 |
ENST00000511367.2
ENST00000513349.1 |
DROSHA
|
drosha, ribonuclease type III |
| chr8_-_117886612 | 0.48 |
ENST00000520992.1
|
RAD21
|
RAD21 homolog (S. pombe) |
| chr6_-_111927449 | 0.48 |
ENST00000368761.5
ENST00000392556.4 ENST00000340026.6 |
TRAF3IP2
|
TRAF3 interacting protein 2 |
| chr17_-_43045439 | 0.47 |
ENST00000253407.3
|
C1QL1
|
complement component 1, q subcomponent-like 1 |
| chr12_+_64798826 | 0.47 |
ENST00000540203.1
|
XPOT
|
exportin, tRNA |
| chr3_+_147795932 | 0.46 |
ENST00000490465.1
|
RP11-639B1.1
|
RP11-639B1.1 |
| chr12_-_95397442 | 0.45 |
ENST00000547157.1
ENST00000547986.1 ENST00000327772.2 |
NDUFA12
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 12 |
| chr1_+_151735431 | 0.44 |
ENST00000321531.5
ENST00000315067.8 |
OAZ3
|
ornithine decarboxylase antizyme 3 |
| chr20_-_50418972 | 0.44 |
ENST00000395997.3
|
SALL4
|
spalt-like transcription factor 4 |
| chr1_-_190446759 | 0.43 |
ENST00000367462.3
|
BRINP3
|
bone morphogenetic protein/retinoic acid inducible neural-specific 3 |
| chr11_-_40315640 | 0.42 |
ENST00000278198.2
|
LRRC4C
|
leucine rich repeat containing 4C |
| chr19_-_46088068 | 0.40 |
ENST00000263275.4
ENST00000323060.3 |
OPA3
|
optic atrophy 3 (autosomal recessive, with chorea and spastic paraplegia) |
| chr6_-_111927062 | 0.40 |
ENST00000359831.4
|
TRAF3IP2
|
TRAF3 interacting protein 2 |
| chr5_+_140797296 | 0.39 |
ENST00000398594.2
|
PCDHGB7
|
protocadherin gamma subfamily B, 7 |
| chr2_-_73460334 | 0.39 |
ENST00000258083.2
|
PRADC1
|
protease-associated domain containing 1 |
| chr18_+_39535239 | 0.39 |
ENST00000585528.1
|
PIK3C3
|
phosphatidylinositol 3-kinase, catalytic subunit type 3 |
| chr14_-_36990061 | 0.38 |
ENST00000546983.1
|
NKX2-1
|
NK2 homeobox 1 |
| chr7_+_91570165 | 0.36 |
ENST00000356239.3
ENST00000359028.2 ENST00000358100.2 |
AKAP9
|
A kinase (PRKA) anchor protein 9 |
| chr8_-_12612962 | 0.36 |
ENST00000398246.3
|
LONRF1
|
LON peptidase N-terminal domain and ring finger 1 |
| chr11_-_124190184 | 0.35 |
ENST00000357438.2
|
OR8D2
|
olfactory receptor, family 8, subfamily D, member 2 |
| chr9_-_107299137 | 0.35 |
ENST00000374781.2
|
OR13C3
|
olfactory receptor, family 13, subfamily C, member 3 |
| chr2_+_172864490 | 0.35 |
ENST00000315796.4
|
METAP1D
|
methionyl aminopeptidase type 1D (mitochondrial) |
| chr20_-_54967187 | 0.35 |
ENST00000422322.1
ENST00000371356.2 ENST00000451915.1 ENST00000347343.2 ENST00000395911.1 ENST00000395907.1 ENST00000441357.1 ENST00000456249.1 ENST00000420474.1 ENST00000395909.4 ENST00000395914.1 ENST00000312783.6 ENST00000395915.3 ENST00000395913.3 |
AURKA
|
aurora kinase A |
| chr12_-_57039739 | 0.32 |
ENST00000552959.1
ENST00000551020.1 ENST00000553007.2 ENST00000552919.1 ENST00000552104.1 ENST00000262030.3 |
ATP5B
|
ATP synthase, H+ transporting, mitochondrial F1 complex, beta polypeptide |
| chr1_-_197115818 | 0.32 |
ENST00000367409.4
ENST00000294732.7 |
ASPM
|
asp (abnormal spindle) homolog, microcephaly associated (Drosophila) |
| chr2_+_187371440 | 0.32 |
ENST00000445547.1
|
ZC3H15
|
zinc finger CCCH-type containing 15 |
| chr11_-_71823796 | 0.31 |
ENST00000545680.1
ENST00000543587.1 ENST00000538393.1 ENST00000535234.1 ENST00000227618.4 ENST00000535503.1 |
ANAPC15
|
anaphase promoting complex subunit 15 |
| chr12_+_28410128 | 0.31 |
ENST00000381259.1
ENST00000381256.1 |
CCDC91
|
coiled-coil domain containing 91 |
| chr1_-_228613026 | 0.30 |
ENST00000366696.1
|
HIST3H3
|
histone cluster 3, H3 |
| chr17_+_44679808 | 0.30 |
ENST00000571172.1
|
NSF
|
N-ethylmaleimide-sensitive factor |
| chr5_-_31532238 | 0.30 |
ENST00000507438.1
|
DROSHA
|
drosha, ribonuclease type III |
| chr11_-_71823715 | 0.29 |
ENST00000545944.1
ENST00000502597.2 |
ANAPC15
|
anaphase promoting complex subunit 15 |
| chr7_-_99716940 | 0.29 |
ENST00000440225.1
|
TAF6
|
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa |
| chr19_+_17378278 | 0.28 |
ENST00000596335.1
ENST00000601436.1 ENST00000595632.1 |
BABAM1
|
BRISC and BRCA1 A complex member 1 |
| chr10_+_99205894 | 0.28 |
ENST00000370854.3
ENST00000393760.1 ENST00000414567.1 ENST00000370846.4 |
ZDHHC16
|
zinc finger, DHHC-type containing 16 |
| chr10_-_99205607 | 0.27 |
ENST00000477692.2
ENST00000485122.2 ENST00000370886.5 ENST00000370885.4 ENST00000370902.3 ENST00000370884.5 |
EXOSC1
|
exosome component 1 |
| chr19_+_17378159 | 0.27 |
ENST00000598188.1
ENST00000359435.4 ENST00000599474.1 ENST00000599057.1 ENST00000601043.1 ENST00000447614.2 |
BABAM1
|
BRISC and BRCA1 A complex member 1 |
| chr11_+_17298342 | 0.27 |
ENST00000530964.1
|
NUCB2
|
nucleobindin 2 |
| chr10_-_15762124 | 0.25 |
ENST00000378076.3
|
ITGA8
|
integrin, alpha 8 |
| chr1_-_23694794 | 0.25 |
ENST00000374608.3
|
ZNF436
|
zinc finger protein 436 |
| chr3_-_147997869 | 0.25 |
ENST00000460324.1
|
RP11-501O2.4
|
RP11-501O2.4 |
| chr10_-_105110831 | 0.25 |
ENST00000337211.4
|
PCGF6
|
polycomb group ring finger 6 |
| chr7_-_99716952 | 0.24 |
ENST00000523306.1
ENST00000344095.4 ENST00000417349.1 ENST00000493322.1 ENST00000520135.1 ENST00000418432.2 ENST00000460673.2 ENST00000452041.1 ENST00000452438.2 ENST00000451699.1 ENST00000453269.2 |
TAF6
|
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa |
| chr4_+_71587669 | 0.23 |
ENST00000381006.3
ENST00000226328.4 |
RUFY3
|
RUN and FYVE domain containing 3 |
| chr3_-_141747950 | 0.22 |
ENST00000497579.1
|
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr10_-_105110890 | 0.21 |
ENST00000369847.3
|
PCGF6
|
polycomb group ring finger 6 |
| chr17_-_80250663 | 0.20 |
ENST00000581489.1
|
RP13-516M14.4
|
RP13-516M14.4 |
| chr7_-_99717463 | 0.19 |
ENST00000437822.2
|
TAF6
|
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa |
| chr17_-_46716647 | 0.18 |
ENST00000608940.1
|
RP11-357H14.17
|
RP11-357H14.17 |
| chrX_-_69269788 | 0.18 |
ENST00000276101.3
|
AWAT2
|
acyl-CoA wax alcohol acyltransferase 2 |
| chr15_+_80351910 | 0.18 |
ENST00000261749.6
ENST00000561060.1 |
ZFAND6
|
zinc finger, AN1-type domain 6 |
| chr19_+_42817527 | 0.17 |
ENST00000598766.1
|
TMEM145
|
transmembrane protein 145 |
| chr1_+_198985262 | 0.17 |
ENST00000432488.1
|
RP11-16L9.4
|
RP11-16L9.4 |
| chr17_+_67759813 | 0.16 |
ENST00000587241.1
|
AC003051.1
|
AC003051.1 |
| chr18_+_39535152 | 0.16 |
ENST00000262039.4
ENST00000398870.3 ENST00000586545.1 |
PIK3C3
|
phosphatidylinositol 3-kinase, catalytic subunit type 3 |
| chr11_+_17298543 | 0.16 |
ENST00000533926.1
|
NUCB2
|
nucleobindin 2 |
| chr5_+_162864575 | 0.15 |
ENST00000512163.1
ENST00000393929.1 ENST00000340828.2 ENST00000511683.2 ENST00000510097.1 ENST00000511490.2 ENST00000510664.1 |
CCNG1
|
cyclin G1 |
| chr12_+_9102632 | 0.14 |
ENST00000539240.1
|
KLRG1
|
killer cell lectin-like receptor subfamily G, member 1 |
| chr3_+_186353756 | 0.14 |
ENST00000431018.1
ENST00000450521.1 ENST00000539949.1 |
FETUB
|
fetuin B |
| chr19_-_10121144 | 0.13 |
ENST00000264828.3
|
COL5A3
|
collagen, type V, alpha 3 |
| chr12_+_52695617 | 0.12 |
ENST00000293525.5
|
KRT86
|
keratin 86 |
| chr1_-_151431909 | 0.10 |
ENST00000361398.3
ENST00000271715.2 |
POGZ
|
pogo transposable element with ZNF domain |
| chr11_-_121986923 | 0.09 |
ENST00000560104.1
|
BLID
|
BH3-like motif containing, cell death inducer |
| chr1_-_151431674 | 0.09 |
ENST00000531094.1
|
POGZ
|
pogo transposable element with ZNF domain |
| chr9_-_5339873 | 0.08 |
ENST00000223862.1
ENST00000223858.4 |
RLN1
|
relaxin 1 |
| chrX_+_113818545 | 0.08 |
ENST00000371951.1
ENST00000276198.1 ENST00000371950.3 |
HTR2C
|
5-hydroxytryptamine (serotonin) receptor 2C, G protein-coupled |
| chr17_-_38956205 | 0.07 |
ENST00000306658.7
|
KRT28
|
keratin 28 |
| chr1_+_211813019 | 0.07 |
ENST00000430123.1
|
RP11-354K1.1
|
RP11-354K1.1 |
| chr1_-_151431647 | 0.07 |
ENST00000368863.2
ENST00000409503.1 ENST00000491586.1 ENST00000533351.1 ENST00000540984.1 |
POGZ
|
pogo transposable element with ZNF domain |
| chr12_-_120315074 | 0.07 |
ENST00000261833.7
ENST00000392521.2 |
CIT
|
citron (rho-interacting, serine/threonine kinase 21) |
| chr11_-_58207646 | 0.07 |
ENST00000302572.2
|
OR5B12
|
olfactory receptor, family 5, subfamily B, member 12 |
| chr11_-_71823266 | 0.05 |
ENST00000538919.1
ENST00000539395.1 ENST00000542531.1 |
ANAPC15
|
anaphase promoting complex subunit 15 |
| chr12_+_117099342 | 0.04 |
ENST00000546835.1
|
RP11-497G19.1
|
RP11-497G19.1 |
| chr7_-_56174161 | 0.04 |
ENST00000395422.3
|
CHCHD2
|
coiled-coil-helix-coiled-coil-helix domain containing 2 |
| chr11_+_17298522 | 0.03 |
ENST00000529313.1
|
NUCB2
|
nucleobindin 2 |
| chr15_-_98417780 | 0.02 |
ENST00000503874.3
|
LINC00923
|
long intergenic non-protein coding RNA 923 |
| chr3_-_11685345 | 0.01 |
ENST00000430365.2
|
VGLL4
|
vestigial like 4 (Drosophila) |
| chr21_-_32253874 | 0.01 |
ENST00000332378.4
|
KRTAP11-1
|
keratin associated protein 11-1 |
| chr20_+_54967409 | 0.00 |
ENST00000415828.1
ENST00000217109.4 ENST00000428552.1 |
CSTF1
|
cleavage stimulation factor, 3' pre-RNA, subunit 1, 50kDa |
| chr4_-_19458597 | 0.00 |
ENST00000505347.1
|
RP11-3J1.1
|
RP11-3J1.1 |
| chr3_-_112127981 | 0.00 |
ENST00000486726.2
|
RP11-231E6.1
|
RP11-231E6.1 |
| chr20_+_54967663 | 0.00 |
ENST00000452950.1
|
CSTF1
|
cleavage stimulation factor, 3' pre-RNA, subunit 1, 50kDa |
Network of associatons between targets according to the STRING database.
First level regulatory network of MSX2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 6.4 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.7 | 6.7 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.5 | 1.6 | GO:1900158 | negative regulation of bone mineralization involved in bone maturation(GO:1900158) |
| 0.3 | 1.0 | GO:2000870 | regulation of progesterone secretion(GO:2000870) |
| 0.2 | 1.2 | GO:0043553 | vascular endothelial growth factor receptor-2 signaling pathway(GO:0036324) negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.2 | 1.0 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 0.2 | 1.0 | GO:0090402 | oncogene-induced cell senescence(GO:0090402) |
| 0.2 | 0.9 | GO:2001023 | regulation of response to drug(GO:2001023) |
| 0.2 | 0.5 | GO:1903774 | positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 0.2 | 1.0 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.2 | 1.3 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.2 | 0.5 | GO:0072684 | mitochondrial tRNA 3'-trailer cleavage, endonucleolytic(GO:0072684) |
| 0.2 | 0.5 | GO:0099558 | maintenance of synapse structure(GO:0099558) |
| 0.2 | 1.4 | GO:0006987 | activation of signaling protein activity involved in unfolded protein response(GO:0006987) |
| 0.1 | 0.7 | GO:2000721 | positive regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000721) |
| 0.1 | 2.7 | GO:2000675 | negative regulation of type B pancreatic cell apoptotic process(GO:2000675) |
| 0.1 | 0.5 | GO:0006428 | isoleucyl-tRNA aminoacylation(GO:0006428) |
| 0.1 | 0.3 | GO:0007057 | spindle assembly involved in female meiosis I(GO:0007057) |
| 0.1 | 0.7 | GO:0021691 | cerebellar Purkinje cell layer maturation(GO:0021691) |
| 0.1 | 0.3 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.1 | 1.6 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.1 | 0.6 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.1 | 0.4 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.1 | 1.1 | GO:0045876 | positive regulation of sister chromatid cohesion(GO:0045876) |
| 0.1 | 2.5 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.1 | 2.5 | GO:0006309 | apoptotic DNA fragmentation(GO:0006309) |
| 0.1 | 2.0 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.1 | 1.3 | GO:0006751 | glutathione catabolic process(GO:0006751) |
| 0.1 | 0.5 | GO:0030242 | pexophagy(GO:0030242) |
| 0.1 | 0.8 | GO:0030422 | production of siRNA involved in RNA interference(GO:0030422) |
| 0.1 | 0.4 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.0 | 0.6 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.0 | 1.1 | GO:0045199 | maintenance of apical/basal cell polarity(GO:0035090) maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.0 | 0.3 | GO:0035494 | SNARE complex disassembly(GO:0035494) |
| 0.0 | 0.4 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.0 | 0.3 | GO:0006933 | negative regulation of cell adhesion involved in substrate-bound cell migration(GO:0006933) |
| 0.0 | 0.7 | GO:0090266 | regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.0 | 0.9 | GO:0002230 | positive regulation of defense response to virus by host(GO:0002230) |
| 0.0 | 0.5 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.0 | 0.2 | GO:0010025 | wax biosynthetic process(GO:0010025) wax metabolic process(GO:0010166) |
| 0.0 | 0.4 | GO:0018206 | peptidyl-methionine modification(GO:0018206) |
| 0.0 | 0.5 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.1 | GO:0043397 | corticotropin-releasing hormone secretion(GO:0043396) regulation of corticotropin-releasing hormone secretion(GO:0043397) |
| 0.0 | 0.9 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 0.7 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.0 | 0.3 | GO:0090160 | Golgi to lysosome transport(GO:0090160) |
| 0.0 | 1.4 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.7 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 2.2 | GO:1903955 | positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.0 | 1.0 | GO:0045739 | positive regulation of DNA repair(GO:0045739) |
| 0.0 | 0.5 | GO:0007530 | sex determination(GO:0007530) |
| 0.0 | 1.2 | GO:0030049 | muscle filament sliding(GO:0030049) actin-myosin filament sliding(GO:0033275) |
| 0.0 | 0.3 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.0 | 0.3 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.0 | 1.2 | GO:0006521 | regulation of cellular amino acid metabolic process(GO:0006521) |
| 0.0 | 0.4 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.6 | GO:0044301 | climbing fiber(GO:0044301) |
| 0.4 | 6.4 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.3 | 0.8 | GO:1903095 | microprocessor complex(GO:0070877) ribonuclease III complex(GO:1903095) |
| 0.2 | 7.9 | GO:0032982 | myosin filament(GO:0032982) |
| 0.2 | 2.0 | GO:0071547 | piP-body(GO:0071547) |
| 0.1 | 2.5 | GO:0031010 | ISWI-type complex(GO:0031010) |
| 0.1 | 0.5 | GO:0034271 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.1 | 1.1 | GO:0000798 | nuclear cohesin complex(GO:0000798) |
| 0.1 | 0.3 | GO:0042585 | germinal vesicle(GO:0042585) |
| 0.1 | 1.0 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.1 | 1.2 | GO:0019774 | proteasome core complex, beta-subunit complex(GO:0019774) |
| 0.1 | 0.6 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.1 | 1.6 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 1.0 | GO:0000800 | lateral element(GO:0000800) |
| 0.0 | 0.3 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) |
| 0.0 | 0.3 | GO:0034678 | integrin alpha8-beta1 complex(GO:0034678) |
| 0.0 | 0.1 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 2.2 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 1.3 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 6.3 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.5 | GO:0090543 | ESCRT III complex(GO:0000815) Flemming body(GO:0090543) |
| 0.0 | 0.8 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 3.3 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.5 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 0.0 | 0.9 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.0 | 1.9 | GO:0031672 | A band(GO:0031672) |
| 0.0 | 0.5 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.0 | 0.5 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.3 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 0.3 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 1.4 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.7 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.5 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 0.6 | 1.7 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.3 | 1.3 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.3 | 6.4 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.2 | 2.7 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.2 | 6.7 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.2 | 1.2 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.2 | 0.6 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 0.1 | 0.7 | GO:0008267 | poly-glutamine tract binding(GO:0008267) |
| 0.1 | 0.8 | GO:0032296 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) |
| 0.1 | 0.5 | GO:0004822 | isoleucine-tRNA ligase activity(GO:0004822) |
| 0.1 | 0.5 | GO:0042781 | 3'-tRNA processing endoribonuclease activity(GO:0042781) |
| 0.1 | 0.4 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.1 | 0.9 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.1 | 1.0 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.1 | 1.6 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.0 | 0.3 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.0 | 0.3 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.0 | 1.2 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.2 | GO:0050252 | retinol O-fatty-acyltransferase activity(GO:0050252) |
| 0.0 | 0.7 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.6 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.0 | 1.2 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.0 | 0.5 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.8 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.0 | 3.5 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 1.0 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.4 | GO:0044213 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) intronic transcription regulatory region DNA binding(GO:0044213) |
| 0.0 | 1.6 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.5 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 0.5 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.4 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 1.0 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.3 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.0 | 1.2 | PID RAS PATHWAY | Regulation of Ras family activation |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.1 | 0.3 | REACTOME APC C CDH1 MEDIATED DEGRADATION OF CDC20 AND OTHER APC C CDH1 TARGETED PROTEINS IN LATE MITOSIS EARLY G1 | Genes involved in APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 |
| 0.0 | 0.6 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 1.7 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 1.9 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF RAS | Genes involved in CREB phosphorylation through the activation of Ras |
| 0.0 | 0.9 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.8 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.0 | 0.3 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 1.6 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.0 | 0.5 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.0 | 0.3 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 1.6 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.5 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 3.8 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.5 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 1.4 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 1.0 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |