Project
Illumina Body Map 2: averaged replicates
Navigation
Downloads
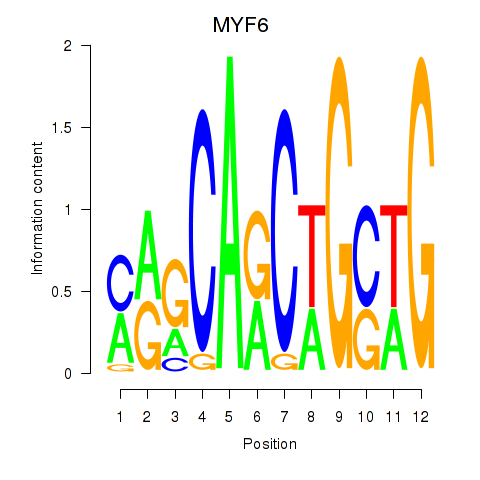
Results for MYF6
Z-value: 2.73
Transcription factors associated with MYF6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
MYF6
|
ENSG00000111046.3 | myogenic factor 6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| MYF6 | hg19_v2_chr12_+_81101277_81101321 | -0.24 | 1.8e-01 | Click! |
Activity profile of MYF6 motif
Sorted Z-values of MYF6 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_-_108518986 | 8.17 |
ENST00000375915.2
|
FAM155A
|
family with sequence similarity 155, member A |
| chr2_+_79740118 | 7.15 |
ENST00000496558.1
ENST00000451966.1 |
CTNNA2
|
catenin (cadherin-associated protein), alpha 2 |
| chr22_-_21905120 | 5.57 |
ENST00000331505.5
|
RIMBP3C
|
RIMS binding protein 3C |
| chr2_+_115199876 | 5.46 |
ENST00000436732.1
ENST00000410059.1 |
DPP10
|
dipeptidyl-peptidase 10 (non-functional) |
| chr2_+_105471969 | 5.14 |
ENST00000361360.2
|
POU3F3
|
POU class 3 homeobox 3 |
| chr4_-_168155700 | 4.51 |
ENST00000357545.4
ENST00000512648.1 |
SPOCK3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr4_-_168155730 | 4.36 |
ENST00000502330.1
ENST00000357154.3 |
SPOCK3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr7_+_96635277 | 3.86 |
ENST00000007660.5
|
DLX6
|
distal-less homeobox 6 |
| chrX_-_92928557 | 3.77 |
ENST00000373079.3
ENST00000475430.2 |
NAP1L3
|
nucleosome assembly protein 1-like 3 |
| chr10_+_25463951 | 3.56 |
ENST00000376351.3
|
GPR158
|
G protein-coupled receptor 158 |
| chrX_+_100333709 | 3.53 |
ENST00000372930.4
|
TMEM35
|
transmembrane protein 35 |
| chr6_-_31514516 | 3.50 |
ENST00000303892.5
ENST00000483251.1 |
ATP6V1G2
|
ATPase, H+ transporting, lysosomal 13kDa, V1 subunit G2 |
| chr1_-_72748140 | 3.44 |
ENST00000434200.1
|
NEGR1
|
neuronal growth regulator 1 |
| chr5_-_146258205 | 3.42 |
ENST00000394413.3
|
PPP2R2B
|
protein phosphatase 2, regulatory subunit B, beta |
| chr6_-_31514333 | 3.36 |
ENST00000376151.4
|
ATP6V1G2
|
ATPase, H+ transporting, lysosomal 13kDa, V1 subunit G2 |
| chr5_-_146258291 | 3.34 |
ENST00000394411.4
ENST00000453001.1 |
PPP2R2B
|
protein phosphatase 2, regulatory subunit B, beta |
| chr17_+_37784749 | 3.32 |
ENST00000394265.1
ENST00000394267.2 |
PPP1R1B
|
protein phosphatase 1, regulatory (inhibitor) subunit 1B |
| chr2_-_220174166 | 3.31 |
ENST00000409251.3
ENST00000451506.1 ENST00000295718.2 ENST00000446182.1 |
PTPRN
|
protein tyrosine phosphatase, receptor type, N |
| chr16_+_23847339 | 3.20 |
ENST00000303531.7
|
PRKCB
|
protein kinase C, beta |
| chr15_-_94614049 | 3.19 |
ENST00000556447.1
ENST00000555772.1 |
CTD-3049M7.1
|
CTD-3049M7.1 |
| chr3_-_116163830 | 3.18 |
ENST00000333617.4
|
LSAMP
|
limbic system-associated membrane protein |
| chr4_-_168155577 | 3.18 |
ENST00000541354.1
ENST00000509854.1 ENST00000512681.1 ENST00000421836.2 ENST00000510741.1 ENST00000510403.1 |
SPOCK3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr1_+_10509971 | 3.14 |
ENST00000320498.4
|
CORT
|
cortistatin |
| chr14_+_29236269 | 3.12 |
ENST00000313071.4
|
FOXG1
|
forkhead box G1 |
| chr8_+_24771265 | 3.06 |
ENST00000518131.1
ENST00000437366.2 |
NEFM
|
neurofilament, medium polypeptide |
| chr2_-_220173685 | 3.06 |
ENST00000423636.2
ENST00000442029.1 ENST00000412847.1 |
PTPRN
|
protein tyrosine phosphatase, receptor type, N |
| chr2_-_230579185 | 3.05 |
ENST00000341772.4
|
DNER
|
delta/notch-like EGF repeat containing |
| chr8_-_68658578 | 2.97 |
ENST00000518549.1
ENST00000297770.4 ENST00000297769.4 |
CPA6
|
carboxypeptidase A6 |
| chr4_-_168155417 | 2.92 |
ENST00000511269.1
ENST00000506697.1 ENST00000512042.1 |
SPOCK3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr2_+_187558698 | 2.91 |
ENST00000304698.5
|
FAM171B
|
family with sequence similarity 171, member B |
| chr16_+_56623433 | 2.91 |
ENST00000570176.1
|
MT3
|
metallothionein 3 |
| chr5_+_173472607 | 2.80 |
ENST00000303177.3
ENST00000519867.1 |
NSG2
|
Neuron-specific protein family member 2 |
| chr11_-_9113137 | 2.67 |
ENST00000520467.1
ENST00000309263.3 ENST00000457346.2 |
SCUBE2
|
signal peptide, CUB domain, EGF-like 2 |
| chr16_-_74808710 | 2.67 |
ENST00000219368.3
ENST00000544337.1 |
FA2H
|
fatty acid 2-hydroxylase |
| chr4_-_168155300 | 2.65 |
ENST00000541637.1
|
SPOCK3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr11_-_133402410 | 2.61 |
ENST00000524381.1
|
OPCML
|
opioid binding protein/cell adhesion molecule-like |
| chr6_-_127837739 | 2.61 |
ENST00000368268.2
|
SOGA3
|
SOGA family member 3 |
| chr12_-_99548270 | 2.60 |
ENST00000546568.1
ENST00000332712.7 ENST00000546960.1 |
ANKS1B
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr3_+_85008089 | 2.59 |
ENST00000383699.3
|
CADM2
|
cell adhesion molecule 2 |
| chr6_+_69942298 | 2.56 |
ENST00000238918.8
|
BAI3
|
brain-specific angiogenesis inhibitor 3 |
| chr7_+_119913688 | 2.53 |
ENST00000331113.4
|
KCND2
|
potassium voltage-gated channel, Shal-related subfamily, member 2 |
| chrX_-_20074895 | 2.51 |
ENST00000543767.1
|
MAP7D2
|
MAP7 domain containing 2 |
| chr12_-_65515334 | 2.51 |
ENST00000286574.4
|
WIF1
|
WNT inhibitory factor 1 |
| chrX_+_12156582 | 2.46 |
ENST00000380682.1
|
FRMPD4
|
FERM and PDZ domain containing 4 |
| chr11_-_9113093 | 2.44 |
ENST00000450649.2
|
SCUBE2
|
signal peptide, CUB domain, EGF-like 2 |
| chr3_-_187388173 | 2.44 |
ENST00000287641.3
|
SST
|
somatostatin |
| chr3_-_116164306 | 2.43 |
ENST00000490035.2
|
LSAMP
|
limbic system-associated membrane protein |
| chr5_-_139422654 | 2.43 |
ENST00000289409.4
ENST00000358522.3 ENST00000378238.4 ENST00000289422.7 ENST00000361474.1 ENST00000545385.1 ENST00000394770.1 ENST00000541337.1 |
NRG2
|
neuregulin 2 |
| chr11_-_117748138 | 2.39 |
ENST00000527717.1
|
FXYD6
|
FXYD domain containing ion transport regulator 6 |
| chr16_-_68406161 | 2.39 |
ENST00000568373.1
ENST00000563226.1 |
SMPD3
|
sphingomyelin phosphodiesterase 3, neutral membrane (neutral sphingomyelinase II) |
| chr19_+_3224700 | 2.38 |
ENST00000292672.2
ENST00000541430.2 |
CELF5
|
CUGBP, Elav-like family member 5 |
| chr11_-_117747607 | 2.35 |
ENST00000540359.1
ENST00000539526.1 |
FXYD6
|
FXYD domain containing ion transport regulator 6 |
| chr22_+_29876197 | 2.34 |
ENST00000310624.6
|
NEFH
|
neurofilament, heavy polypeptide |
| chr9_+_140033862 | 2.30 |
ENST00000350902.5
ENST00000371550.4 ENST00000371546.4 ENST00000371555.4 ENST00000371553.3 ENST00000371559.4 ENST00000371560.3 |
GRIN1
|
glutamate receptor, ionotropic, N-methyl D-aspartate 1 |
| chr2_+_148778570 | 2.30 |
ENST00000407073.1
|
MBD5
|
methyl-CpG binding domain protein 5 |
| chr4_-_46391567 | 2.27 |
ENST00000507460.1
|
GABRA2
|
gamma-aminobutyric acid (GABA) A receptor, alpha 2 |
| chr11_-_117747434 | 2.25 |
ENST00000529335.2
ENST00000530956.1 ENST00000260282.4 |
FXYD6
|
FXYD domain containing ion transport regulator 6 |
| chr10_-_90967063 | 2.22 |
ENST00000371852.2
|
CH25H
|
cholesterol 25-hydroxylase |
| chr2_-_219925189 | 2.20 |
ENST00000295731.6
|
IHH
|
indian hedgehog |
| chr2_-_75426826 | 2.20 |
ENST00000305249.5
|
TACR1
|
tachykinin receptor 1 |
| chr4_-_87278857 | 2.17 |
ENST00000509464.1
ENST00000511167.1 |
MAPK10
|
mitogen-activated protein kinase 10 |
| chr5_+_137774706 | 2.15 |
ENST00000378339.2
ENST00000254901.5 ENST00000506158.1 |
REEP2
|
receptor accessory protein 2 |
| chr1_-_177134024 | 2.15 |
ENST00000367654.3
|
ASTN1
|
astrotactin 1 |
| chr3_+_167453493 | 2.15 |
ENST00000295777.5
ENST00000472747.2 |
SERPINI1
|
serpin peptidase inhibitor, clade I (neuroserpin), member 1 |
| chr6_+_69942915 | 2.15 |
ENST00000604969.1
ENST00000603207.1 |
BAI3
|
brain-specific angiogenesis inhibitor 3 |
| chr11_-_130298888 | 2.14 |
ENST00000257359.6
|
ADAMTS8
|
ADAM metallopeptidase with thrombospondin type 1 motif, 8 |
| chr5_+_140201183 | 2.13 |
ENST00000529619.1
ENST00000529859.1 ENST00000378126.3 |
PCDHA5
|
protocadherin alpha 5 |
| chr21_+_34442439 | 2.13 |
ENST00000382348.1
ENST00000333063.5 |
OLIG1
|
oligodendrocyte transcription factor 1 |
| chr16_+_24266874 | 2.13 |
ENST00000005284.3
|
CACNG3
|
calcium channel, voltage-dependent, gamma subunit 3 |
| chr3_+_167453026 | 2.13 |
ENST00000472941.1
|
SERPINI1
|
serpin peptidase inhibitor, clade I (neuroserpin), member 1 |
| chr12_-_99548645 | 2.11 |
ENST00000549025.2
|
ANKS1B
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr2_-_158184211 | 2.11 |
ENST00000397283.2
|
ERMN
|
ermin, ERM-like protein |
| chr1_+_50575292 | 2.09 |
ENST00000371821.1
ENST00000371819.1 |
ELAVL4
|
ELAV like neuron-specific RNA binding protein 4 |
| chr5_+_126626498 | 2.09 |
ENST00000503335.2
ENST00000508365.1 ENST00000418761.2 ENST00000274473.6 |
MEGF10
|
multiple EGF-like-domains 10 |
| chrX_+_21392553 | 2.08 |
ENST00000279451.4
|
CNKSR2
|
connector enhancer of kinase suppressor of Ras 2 |
| chr8_-_89339705 | 2.06 |
ENST00000286614.6
|
MMP16
|
matrix metallopeptidase 16 (membrane-inserted) |
| chr6_-_127780510 | 2.03 |
ENST00000487331.2
ENST00000483725.3 |
KIAA0408
|
KIAA0408 |
| chr11_-_46408107 | 2.01 |
ENST00000433765.2
|
CHRM4
|
cholinergic receptor, muscarinic 4 |
| chr19_+_54385439 | 2.01 |
ENST00000536044.1
ENST00000540413.1 ENST00000263431.3 ENST00000419486.1 |
PRKCG
|
protein kinase C, gamma |
| chr17_-_42991062 | 1.99 |
ENST00000587997.1
|
GFAP
|
glial fibrillary acidic protein |
| chr19_-_47137942 | 1.98 |
ENST00000300873.4
|
GNG8
|
guanine nucleotide binding protein (G protein), gamma 8 |
| chr14_+_62584197 | 1.97 |
ENST00000334389.4
|
LINC00643
|
long intergenic non-protein coding RNA 643 |
| chr4_-_46391367 | 1.97 |
ENST00000503806.1
ENST00000356504.1 ENST00000514090.1 ENST00000506961.1 |
GABRA2
|
gamma-aminobutyric acid (GABA) A receptor, alpha 2 |
| chr2_+_185463093 | 1.97 |
ENST00000302277.6
|
ZNF804A
|
zinc finger protein 804A |
| chr1_-_145076068 | 1.96 |
ENST00000369345.4
|
PDE4DIP
|
phosphodiesterase 4D interacting protein |
| chr1_+_1361494 | 1.96 |
ENST00000378821.3
|
TMEM88B
|
transmembrane protein 88B |
| chr10_-_48438974 | 1.95 |
ENST00000224605.2
|
GDF10
|
growth differentiation factor 10 |
| chr9_-_120177342 | 1.94 |
ENST00000361209.2
|
ASTN2
|
astrotactin 2 |
| chr12_-_16759440 | 1.93 |
ENST00000537304.1
|
LMO3
|
LIM domain only 3 (rhombotin-like 2) |
| chr8_-_23712312 | 1.93 |
ENST00000290271.2
|
STC1
|
stanniocalcin 1 |
| chr19_+_54926621 | 1.90 |
ENST00000376530.3
ENST00000445095.1 ENST00000391739.3 ENST00000376531.3 |
TTYH1
|
tweety family member 1 |
| chr11_+_369804 | 1.87 |
ENST00000329962.6
|
B4GALNT4
|
beta-1,4-N-acetyl-galactosaminyl transferase 4 |
| chr12_+_79258444 | 1.87 |
ENST00000261205.4
|
SYT1
|
synaptotagmin I |
| chr16_+_335680 | 1.86 |
ENST00000435833.1
|
PDIA2
|
protein disulfide isomerase family A, member 2 |
| chrX_-_151619746 | 1.86 |
ENST00000370314.4
|
GABRA3
|
gamma-aminobutyric acid (GABA) A receptor, alpha 3 |
| chr19_-_19006920 | 1.85 |
ENST00000429504.2
ENST00000427170.2 |
CERS1
|
ceramide synthase 1 |
| chr7_-_124405681 | 1.83 |
ENST00000303921.2
|
GPR37
|
G protein-coupled receptor 37 (endothelin receptor type B-like) |
| chr19_+_12949251 | 1.81 |
ENST00000251472.4
|
MAST1
|
microtubule associated serine/threonine kinase 1 |
| chr2_-_11810284 | 1.79 |
ENST00000306928.5
|
NTSR2
|
neurotensin receptor 2 |
| chr15_+_32933866 | 1.79 |
ENST00000300175.4
ENST00000413748.2 ENST00000494364.1 ENST00000497208.1 |
SCG5
|
secretogranin V (7B2 protein) |
| chr11_-_117747327 | 1.79 |
ENST00000584230.1
ENST00000527429.1 ENST00000584394.1 ENST00000532984.1 |
FXYD6
FXYD6-FXYD2
|
FXYD domain containing ion transport regulator 6 FXYD6-FXYD2 readthrough |
| chr10_-_99771079 | 1.77 |
ENST00000309155.3
|
CRTAC1
|
cartilage acidic protein 1 |
| chr5_-_44388899 | 1.77 |
ENST00000264664.4
|
FGF10
|
fibroblast growth factor 10 |
| chr19_-_38747172 | 1.76 |
ENST00000347262.4
ENST00000591585.1 ENST00000301242.4 |
PPP1R14A
|
protein phosphatase 1, regulatory (inhibitor) subunit 14A |
| chr7_-_107880508 | 1.76 |
ENST00000425651.2
|
NRCAM
|
neuronal cell adhesion molecule |
| chr4_-_46391805 | 1.76 |
ENST00000540012.1
|
GABRA2
|
gamma-aminobutyric acid (GABA) A receptor, alpha 2 |
| chr13_-_36705425 | 1.75 |
ENST00000255448.4
ENST00000360631.3 ENST00000379892.4 |
DCLK1
|
doublecortin-like kinase 1 |
| chr4_-_46391931 | 1.74 |
ENST00000381620.4
|
GABRA2
|
gamma-aminobutyric acid (GABA) A receptor, alpha 2 |
| chr16_-_29910365 | 1.74 |
ENST00000346932.5
ENST00000350527.3 ENST00000537485.1 ENST00000568380.1 |
SEZ6L2
|
seizure related 6 homolog (mouse)-like 2 |
| chr16_+_86229728 | 1.72 |
ENST00000601250.1
|
LINC01082
|
long intergenic non-protein coding RNA 1082 |
| chr2_-_70129841 | 1.72 |
ENST00000599032.1
|
AC019206.1
|
Uncharacterized protein |
| chr12_-_6579833 | 1.72 |
ENST00000396308.3
|
VAMP1
|
vesicle-associated membrane protein 1 (synaptobrevin 1) |
| chr1_+_43613566 | 1.72 |
ENST00000409396.1
|
FAM183A
|
family with sequence similarity 183, member A |
| chr21_+_22370717 | 1.71 |
ENST00000284894.7
|
NCAM2
|
neural cell adhesion molecule 2 |
| chr6_-_150185156 | 1.71 |
ENST00000239367.2
ENST00000367368.2 |
LRP11
|
low density lipoprotein receptor-related protein 11 |
| chr19_+_54926601 | 1.70 |
ENST00000301194.4
|
TTYH1
|
tweety family member 1 |
| chr10_-_73848531 | 1.68 |
ENST00000373109.2
|
SPOCK2
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 2 |
| chrX_-_102531717 | 1.68 |
ENST00000372680.1
|
TCEAL5
|
transcription elongation factor A (SII)-like 5 |
| chr1_+_43613612 | 1.67 |
ENST00000335282.4
|
FAM183A
|
family with sequence similarity 183, member A |
| chr21_-_42219065 | 1.66 |
ENST00000400454.1
|
DSCAM
|
Down syndrome cell adhesion molecule |
| chrX_-_152160740 | 1.66 |
ENST00000361887.5
ENST00000439251.1 ENST00000452693.1 |
PNMA5
|
paraneoplastic Ma antigen family member 5 |
| chr12_-_16758873 | 1.66 |
ENST00000535535.1
|
LMO3
|
LIM domain only 3 (rhombotin-like 2) |
| chr21_+_22370608 | 1.66 |
ENST00000400546.1
|
NCAM2
|
neural cell adhesion molecule 2 |
| chr13_-_51417812 | 1.64 |
ENST00000504404.1
|
DLEU7
|
deleted in lymphocytic leukemia, 7 |
| chr7_-_137028498 | 1.64 |
ENST00000393083.2
|
PTN
|
pleiotrophin |
| chrX_-_132095419 | 1.64 |
ENST00000370836.2
ENST00000521489.1 |
HS6ST2
|
heparan sulfate 6-O-sulfotransferase 2 |
| chr4_-_46392290 | 1.63 |
ENST00000515082.1
|
GABRA2
|
gamma-aminobutyric acid (GABA) A receptor, alpha 2 |
| chr7_-_137028534 | 1.63 |
ENST00000348225.2
|
PTN
|
pleiotrophin |
| chr1_-_226076843 | 1.62 |
ENST00000272134.5
|
LEFTY1
|
left-right determination factor 1 |
| chr1_+_77748756 | 1.62 |
ENST00000478407.1
|
AK5
|
adenylate kinase 5 |
| chr9_-_140064496 | 1.61 |
ENST00000371542.3
|
LRRC26
|
leucine rich repeat containing 26 |
| chr4_-_87281196 | 1.60 |
ENST00000359221.3
|
MAPK10
|
mitogen-activated protein kinase 10 |
| chr22_+_42372970 | 1.60 |
ENST00000291236.11
|
SEPT3
|
septin 3 |
| chr16_+_23847267 | 1.60 |
ENST00000321728.7
|
PRKCB
|
protein kinase C, beta |
| chr12_-_6580094 | 1.59 |
ENST00000361716.3
|
VAMP1
|
vesicle-associated membrane protein 1 (synaptobrevin 1) |
| chr16_+_333152 | 1.58 |
ENST00000219406.6
ENST00000404312.1 ENST00000456379.1 |
PDIA2
|
protein disulfide isomerase family A, member 2 |
| chr4_-_5021164 | 1.58 |
ENST00000506508.1
ENST00000509419.1 ENST00000307746.4 |
CYTL1
|
cytokine-like 1 |
| chr8_+_99439214 | 1.57 |
ENST00000287042.4
|
KCNS2
|
potassium voltage-gated channel, delayed-rectifier, subfamily S, member 2 |
| chr6_-_110500905 | 1.57 |
ENST00000392587.2
|
WASF1
|
WAS protein family, member 1 |
| chr6_-_110500826 | 1.57 |
ENST00000265601.3
ENST00000447287.1 ENST00000444391.1 |
WASF1
|
WAS protein family, member 1 |
| chr3_+_159943362 | 1.56 |
ENST00000326474.3
|
C3orf80
|
chromosome 3 open reading frame 80 |
| chr9_-_139948468 | 1.56 |
ENST00000312665.5
|
ENTPD2
|
ectonucleoside triphosphate diphosphohydrolase 2 |
| chrX_+_100474711 | 1.54 |
ENST00000402866.1
|
DRP2
|
dystrophin related protein 2 |
| chrX_-_153151586 | 1.53 |
ENST00000370060.1
ENST00000370055.1 ENST00000420165.1 |
L1CAM
|
L1 cell adhesion molecule |
| chr8_-_10336885 | 1.53 |
ENST00000520494.1
|
RP11-981G7.3
|
RP11-981G7.3 |
| chr19_-_51220176 | 1.52 |
ENST00000359082.3
ENST00000293441.1 |
SHANK1
|
SH3 and multiple ankyrin repeat domains 1 |
| chr19_-_6501778 | 1.52 |
ENST00000596291.1
|
TUBB4A
|
tubulin, beta 4A class IVa |
| chr18_+_56892724 | 1.51 |
ENST00000456142.3
ENST00000530323.1 |
GRP
|
gastrin-releasing peptide |
| chr5_+_140864649 | 1.51 |
ENST00000306593.1
|
PCDHGC4
|
protocadherin gamma subfamily C, 4 |
| chr12_-_16758835 | 1.50 |
ENST00000541295.1
|
LMO3
|
LIM domain only 3 (rhombotin-like 2) |
| chr4_+_7194247 | 1.50 |
ENST00000507866.2
|
SORCS2
|
sortilin-related VPS10 domain containing receptor 2 |
| chr12_+_106994905 | 1.50 |
ENST00000357881.4
|
RFX4
|
regulatory factor X, 4 (influences HLA class II expression) |
| chr2_+_14772810 | 1.50 |
ENST00000295092.2
ENST00000331243.4 |
FAM84A
|
family with sequence similarity 84, member A |
| chr5_-_147162263 | 1.49 |
ENST00000333010.6
ENST00000265272.5 |
JAKMIP2
|
janus kinase and microtubule interacting protein 2 |
| chr16_+_230435 | 1.49 |
ENST00000199708.2
|
HBQ1
|
hemoglobin, theta 1 |
| chr4_-_87281224 | 1.48 |
ENST00000395169.3
ENST00000395161.2 |
MAPK10
|
mitogen-activated protein kinase 10 |
| chr14_+_100150622 | 1.48 |
ENST00000261835.3
|
CYP46A1
|
cytochrome P450, family 46, subfamily A, polypeptide 1 |
| chr12_+_1929644 | 1.48 |
ENST00000299194.1
|
LRTM2
|
leucine-rich repeats and transmembrane domains 2 |
| chr16_+_56225248 | 1.47 |
ENST00000262493.6
|
GNAO1
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide O |
| chr11_+_1074870 | 1.47 |
ENST00000441003.2
ENST00000359061.5 |
MUC2
|
mucin 2, oligomeric mucus/gel-forming |
| chr8_-_133493200 | 1.47 |
ENST00000388996.4
|
KCNQ3
|
potassium voltage-gated channel, KQT-like subfamily, member 3 |
| chr19_-_19006890 | 1.46 |
ENST00000247005.6
|
GDF1
|
growth differentiation factor 1 |
| chr15_-_63674034 | 1.46 |
ENST00000344366.3
ENST00000422263.2 |
CA12
|
carbonic anhydrase XII |
| chr3_-_9595480 | 1.46 |
ENST00000287585.6
|
LHFPL4
|
lipoma HMGIC fusion partner-like 4 |
| chr9_-_117880477 | 1.45 |
ENST00000534839.1
ENST00000340094.3 ENST00000535648.1 ENST00000346706.3 ENST00000345230.3 ENST00000350763.4 |
TNC
|
tenascin C |
| chr19_-_55686399 | 1.45 |
ENST00000587067.1
|
SYT5
|
synaptotagmin V |
| chr10_-_79398250 | 1.45 |
ENST00000286627.5
|
KCNMA1
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr11_-_75379612 | 1.44 |
ENST00000526740.1
|
MAP6
|
microtubule-associated protein 6 |
| chr12_+_79258547 | 1.44 |
ENST00000457153.2
|
SYT1
|
synaptotagmin I |
| chr5_-_132113063 | 1.44 |
ENST00000378719.2
|
SEPT8
|
septin 8 |
| chr1_-_177133998 | 1.44 |
ENST00000367657.3
|
ASTN1
|
astrotactin 1 |
| chr20_-_23402028 | 1.42 |
ENST00000398425.3
ENST00000432543.2 ENST00000377026.4 |
NAPB
|
N-ethylmaleimide-sensitive factor attachment protein, beta |
| chr11_-_1619524 | 1.42 |
ENST00000412090.1
|
KRTAP5-2
|
keratin associated protein 5-2 |
| chr15_-_63674218 | 1.42 |
ENST00000178638.3
|
CA12
|
carbonic anhydrase XII |
| chr8_-_140715294 | 1.41 |
ENST00000303015.1
ENST00000520439.1 |
KCNK9
|
potassium channel, subfamily K, member 9 |
| chr12_-_99548524 | 1.41 |
ENST00000549558.2
ENST00000550693.2 ENST00000549493.2 |
ANKS1B
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr12_-_6579808 | 1.39 |
ENST00000535180.1
ENST00000400911.3 |
VAMP1
|
vesicle-associated membrane protein 1 (synaptobrevin 1) |
| chr4_+_95972822 | 1.39 |
ENST00000509540.1
ENST00000440890.2 |
BMPR1B
|
bone morphogenetic protein receptor, type IB |
| chrX_+_100474906 | 1.38 |
ENST00000541709.1
|
DRP2
|
dystrophin related protein 2 |
| chr19_-_35625765 | 1.38 |
ENST00000591633.1
|
LGI4
|
leucine-rich repeat LGI family, member 4 |
| chr1_-_156390128 | 1.38 |
ENST00000368242.3
|
C1orf61
|
chromosome 1 open reading frame 61 |
| chr5_+_173472744 | 1.38 |
ENST00000521585.1
|
NSG2
|
Neuron-specific protein family member 2 |
| chr14_+_79745682 | 1.38 |
ENST00000557594.1
|
NRXN3
|
neurexin 3 |
| chr5_-_169816638 | 1.38 |
ENST00000521859.1
ENST00000274629.4 |
KCNMB1
|
potassium large conductance calcium-activated channel, subfamily M, beta member 1 |
| chr12_+_72667203 | 1.37 |
ENST00000547300.1
|
TRHDE
|
thyrotropin-releasing hormone degrading enzyme |
| chrX_+_100474763 | 1.37 |
ENST00000395209.3
|
DRP2
|
dystrophin related protein 2 |
| chr7_+_153749732 | 1.37 |
ENST00000377770.3
|
DPP6
|
dipeptidyl-peptidase 6 |
| chr8_+_15397732 | 1.37 |
ENST00000382020.4
ENST00000506802.1 ENST00000509380.1 ENST00000503731.1 |
TUSC3
|
tumor suppressor candidate 3 |
| chr16_+_2867228 | 1.36 |
ENST00000005995.3
ENST00000574813.1 |
PRSS21
|
protease, serine, 21 (testisin) |
| chr8_+_91803695 | 1.35 |
ENST00000417640.2
|
NECAB1
|
N-terminal EF-hand calcium binding protein 1 |
| chr3_-_58652523 | 1.35 |
ENST00000489857.1
ENST00000358781.2 |
FAM3D
|
family with sequence similarity 3, member D |
| chr1_+_233749739 | 1.34 |
ENST00000366621.3
|
KCNK1
|
potassium channel, subfamily K, member 1 |
| chr17_+_11144580 | 1.34 |
ENST00000441885.3
ENST00000432116.3 ENST00000409168.3 |
SHISA6
|
shisa family member 6 |
| chr12_+_119772502 | 1.33 |
ENST00000536742.1
ENST00000327554.2 |
CCDC60
|
coiled-coil domain containing 60 |
| chr15_-_43910998 | 1.33 |
ENST00000450892.2
|
STRC
|
stereocilin |
| chr7_-_158380371 | 1.31 |
ENST00000389418.4
ENST00000389416.4 |
PTPRN2
|
protein tyrosine phosphatase, receptor type, N polypeptide 2 |
| chr4_-_44450814 | 1.31 |
ENST00000360029.3
|
KCTD8
|
potassium channel tetramerization domain containing 8 |
| chr12_+_79371565 | 1.31 |
ENST00000551304.1
|
SYT1
|
synaptotagmin I |
| chr12_+_72666407 | 1.30 |
ENST00000261180.4
|
TRHDE
|
thyrotropin-releasing hormone degrading enzyme |
| chr17_-_7123021 | 1.30 |
ENST00000399510.2
|
DLG4
|
discs, large homolog 4 (Drosophila) |
Network of associatons between targets according to the STRING database.
First level regulatory network of MYF6
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 5.4 | GO:0033693 | neurofilament bundle assembly(GO:0033693) |
| 1.6 | 6.5 | GO:0035408 | histone H3-T6 phosphorylation(GO:0035408) |
| 1.6 | 6.4 | GO:1990502 | dense core granule maturation(GO:1990502) |
| 1.2 | 3.5 | GO:1904397 | negative regulation of neuromuscular junction development(GO:1904397) |
| 1.0 | 5.1 | GO:0072023 | thick ascending limb development(GO:0072023) metanephric thick ascending limb development(GO:0072233) |
| 1.0 | 2.9 | GO:0097212 | cadmium ion homeostasis(GO:0055073) lysosomal membrane organization(GO:0097212) negative regulation of hydrogen peroxide catabolic process(GO:2000296) regulation of oxygen metabolic process(GO:2000374) |
| 0.9 | 2.7 | GO:0006173 | dADP biosynthetic process(GO:0006173) |
| 0.9 | 6.1 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.8 | 20.2 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.8 | 2.4 | GO:0060435 | bronchiole development(GO:0060435) |
| 0.8 | 2.3 | GO:0016539 | intein-mediated protein splicing(GO:0016539) protein splicing(GO:0030908) |
| 0.8 | 4.6 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 0.8 | 2.3 | GO:0043449 | cellular alkene metabolic process(GO:0043449) |
| 0.7 | 1.5 | GO:0060447 | bud outgrowth involved in lung branching(GO:0060447) |
| 0.7 | 1.4 | GO:1903413 | cellular response to bile acid(GO:1903413) |
| 0.6 | 1.8 | GO:0072720 | cellular response to mycotoxin(GO:0036146) response to dithiothreitol(GO:0072720) |
| 0.6 | 1.8 | GO:0017186 | peptidyl-pyroglutamic acid biosynthetic process, using glutaminyl-peptide cyclotransferase(GO:0017186) |
| 0.5 | 7.4 | GO:0060083 | smooth muscle contraction involved in micturition(GO:0060083) |
| 0.5 | 1.5 | GO:0050894 | determination of affect(GO:0050894) |
| 0.5 | 2.4 | GO:0008628 | hormone-mediated apoptotic signaling pathway(GO:0008628) |
| 0.5 | 7.4 | GO:0021942 | radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.5 | 6.4 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.4 | 2.2 | GO:0032425 | positive regulation of mismatch repair(GO:0032425) |
| 0.4 | 6.8 | GO:0098703 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.4 | 2.5 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.4 | 2.5 | GO:0051835 | positive regulation of synapse structural plasticity(GO:0051835) |
| 0.4 | 2.0 | GO:1903613 | regulation of protein tyrosine phosphatase activity(GO:1903613) positive regulation of protein tyrosine phosphatase activity(GO:1903615) |
| 0.4 | 5.3 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.4 | 1.5 | GO:0021849 | neuroblast division in subventricular zone(GO:0021849) |
| 0.4 | 2.2 | GO:0050916 | sensory perception of sweet taste(GO:0050916) |
| 0.3 | 1.0 | GO:0051466 | positive regulation of corticotropin-releasing hormone secretion(GO:0051466) |
| 0.3 | 1.0 | GO:0090274 | regulation of somatostatin secretion(GO:0090273) positive regulation of somatostatin secretion(GO:0090274) |
| 0.3 | 2.0 | GO:0060399 | positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.3 | 0.7 | GO:0051464 | positive regulation of cortisol secretion(GO:0051464) |
| 0.3 | 2.6 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 0.3 | 2.9 | GO:0055064 | chloride ion homeostasis(GO:0055064) |
| 0.3 | 1.3 | GO:0000711 | meiotic DNA repair synthesis(GO:0000711) |
| 0.3 | 1.9 | GO:0090258 | negative regulation of mitochondrial fission(GO:0090258) |
| 0.3 | 1.2 | GO:0003358 | noradrenergic neuron development(GO:0003358) |
| 0.3 | 1.5 | GO:1900738 | positive regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900738) |
| 0.3 | 3.3 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 0.3 | 0.9 | GO:0050720 | interleukin-1 beta biosynthetic process(GO:0050720) |
| 0.3 | 15.3 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.3 | 6.9 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 0.3 | 3.7 | GO:0070257 | positive regulation of mucus secretion(GO:0070257) |
| 0.3 | 2.3 | GO:0043932 | ossification involved in bone remodeling(GO:0043932) |
| 0.3 | 1.1 | GO:0038170 | somatostatin receptor signaling pathway(GO:0038169) somatostatin signaling pathway(GO:0038170) |
| 0.3 | 3.4 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.3 | 1.6 | GO:0009181 | purine nucleoside diphosphate catabolic process(GO:0009137) purine ribonucleoside diphosphate catabolic process(GO:0009181) |
| 0.3 | 0.8 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 0.3 | 1.0 | GO:0046946 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.3 | 1.0 | GO:0048686 | modulation by virus of host transcription(GO:0019056) regulation of sprouting of injured axon(GO:0048686) positive regulation of sprouting of injured axon(GO:0048687) regulation of axon extension involved in regeneration(GO:0048690) positive regulation of axon extension involved in regeneration(GO:0048691) modulation by symbiont of host transcription(GO:0052026) |
| 0.3 | 3.5 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.2 | 2.0 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.2 | 2.2 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 0.2 | 1.4 | GO:1901843 | positive regulation of high voltage-gated calcium channel activity(GO:1901843) |
| 0.2 | 1.0 | GO:0072268 | pattern specification involved in metanephros development(GO:0072268) |
| 0.2 | 6.9 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.2 | 2.8 | GO:0032286 | central nervous system myelin maintenance(GO:0032286) |
| 0.2 | 1.6 | GO:0046208 | spermine catabolic process(GO:0046208) |
| 0.2 | 3.4 | GO:0050910 | detection of mechanical stimulus involved in sensory perception of sound(GO:0050910) |
| 0.2 | 8.3 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.2 | 5.9 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.2 | 0.2 | GO:0045843 | negative regulation of striated muscle tissue development(GO:0045843) negative regulation of muscle organ development(GO:0048635) negative regulation of muscle tissue development(GO:1901862) |
| 0.2 | 1.3 | GO:0050925 | negative regulation of negative chemotaxis(GO:0050925) apoptotic process involved in luteolysis(GO:0061364) |
| 0.2 | 2.1 | GO:0048022 | negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) |
| 0.2 | 1.7 | GO:0048104 | establishment of body hair or bristle planar orientation(GO:0048104) establishment of body hair planar orientation(GO:0048105) |
| 0.2 | 0.6 | GO:0051389 | inactivation of MAPKK activity(GO:0051389) |
| 0.2 | 0.8 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.2 | 1.4 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.2 | 1.1 | GO:0097490 | sympathetic neuron projection extension(GO:0097490) sympathetic neuron projection guidance(GO:0097491) |
| 0.2 | 0.9 | GO:0072137 | condensed mesenchymal cell proliferation(GO:0072137) |
| 0.2 | 0.9 | GO:0045065 | cytotoxic T cell differentiation(GO:0045065) |
| 0.2 | 0.2 | GO:2001038 | regulation of cellular response to drug(GO:2001038) |
| 0.2 | 0.7 | GO:1903998 | regulation of eating behavior(GO:1903998) |
| 0.2 | 0.7 | GO:0033685 | negative regulation of luteinizing hormone secretion(GO:0033685) |
| 0.2 | 0.7 | GO:0002426 | immunoglobulin production in mucosal tissue(GO:0002426) |
| 0.2 | 3.9 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.2 | 1.8 | GO:0045964 | positive regulation of catecholamine metabolic process(GO:0045915) positive regulation of dopamine metabolic process(GO:0045964) |
| 0.2 | 1.8 | GO:1902961 | positive regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902961) positive regulation of aspartic-type peptidase activity(GO:1905247) |
| 0.2 | 2.0 | GO:0035754 | B cell chemotaxis(GO:0035754) |
| 0.2 | 6.7 | GO:0090383 | phagosome acidification(GO:0090383) |
| 0.2 | 0.6 | GO:0031443 | fast-twitch skeletal muscle fiber contraction(GO:0031443) |
| 0.2 | 1.1 | GO:0015798 | myo-inositol transport(GO:0015798) |
| 0.2 | 0.8 | GO:0055020 | apoptotic process involved in endocardial cushion morphogenesis(GO:0003277) intermediate mesoderm development(GO:0048389) intermediate mesoderm morphogenesis(GO:0048390) intermediate mesoderm formation(GO:0048391) intermediate mesodermal cell differentiation(GO:0048392) regulation of cardiac muscle fiber development(GO:0055018) positive regulation of cardiac muscle fiber development(GO:0055020) bud dilation involved in lung branching(GO:0060503) BMP signaling pathway involved in ureter morphogenesis(GO:0061149) renal system segmentation(GO:0061150) BMP signaling pathway involved in renal system segmentation(GO:0061151) pulmonary artery endothelial tube morphogenesis(GO:0061155) regulation of transcription from RNA polymerase II promoter involved in mesonephros development(GO:0061216) pattern specification involved in mesonephros development(GO:0061227) BMP signaling pathway involved in nephric duct formation(GO:0071893) negative regulation of branch elongation involved in ureteric bud branching(GO:0072096) negative regulation of branch elongation involved in ureteric bud branching by BMP signaling pathway(GO:0072097) anterior/posterior pattern specification involved in kidney development(GO:0072098) anterior/posterior pattern specification involved in ureteric bud development(GO:0072099) specification of ureteric bud anterior/posterior symmetry(GO:0072100) specification of ureteric bud anterior/posterior symmetry by BMP signaling pathway(GO:0072101) ureter epithelial cell differentiation(GO:0072192) negative regulation of mesenchymal cell proliferation involved in ureter development(GO:0072200) positive regulation of cell proliferation involved in outflow tract morphogenesis(GO:1901964) cardiac jelly development(GO:1905072) regulation of metanephric S-shaped body morphogenesis(GO:2000004) negative regulation of metanephric S-shaped body morphogenesis(GO:2000005) regulation of metanephric comma-shaped body morphogenesis(GO:2000006) negative regulation of metanephric comma-shaped body morphogenesis(GO:2000007) |
| 0.2 | 0.8 | GO:2000768 | glomerular parietal epithelial cell differentiation(GO:0072139) positive regulation of nephron tubule epithelial cell differentiation(GO:2000768) |
| 0.2 | 0.8 | GO:2000821 | regulation of grooming behavior(GO:2000821) |
| 0.1 | 0.4 | GO:1904235 | regulation of substrate-dependent cell migration, cell attachment to substrate(GO:1904235) positive regulation of substrate-dependent cell migration, cell attachment to substrate(GO:1904237) |
| 0.1 | 9.0 | GO:0008038 | neuron recognition(GO:0008038) |
| 0.1 | 3.3 | GO:0035988 | chondrocyte proliferation(GO:0035988) |
| 0.1 | 1.7 | GO:0009414 | response to water deprivation(GO:0009414) |
| 0.1 | 0.3 | GO:1902474 | positive regulation of protein localization to synapse(GO:1902474) |
| 0.1 | 1.0 | GO:0098704 | fructose import(GO:0032445) carbohydrate import into cell(GO:0097319) carbohydrate import across plasma membrane(GO:0098704) fructose import across plasma membrane(GO:1990539) |
| 0.1 | 0.4 | GO:0072061 | inner medullary collecting duct development(GO:0072061) |
| 0.1 | 0.7 | GO:0097021 | lymphocyte migration into lymphoid organs(GO:0097021) |
| 0.1 | 0.6 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.1 | 1.5 | GO:0021914 | negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021914) |
| 0.1 | 0.1 | GO:0032416 | negative regulation of sodium:proton antiporter activity(GO:0032416) |
| 0.1 | 0.4 | GO:2000870 | regulation of progesterone secretion(GO:2000870) |
| 0.1 | 0.4 | GO:0050923 | chemorepulsion of branchiomotor axon(GO:0021793) regulation of negative chemotaxis(GO:0050923) |
| 0.1 | 2.7 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
| 0.1 | 0.1 | GO:0060364 | frontal suture morphogenesis(GO:0060364) |
| 0.1 | 2.0 | GO:0007035 | vacuolar acidification(GO:0007035) |
| 0.1 | 1.0 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) |
| 0.1 | 1.2 | GO:0035494 | SNARE complex disassembly(GO:0035494) |
| 0.1 | 2.9 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.1 | 0.9 | GO:0071442 | positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.1 | 0.4 | GO:0097535 | lymphoid lineage cell migration(GO:0097534) lymphoid lineage cell migration into thymus(GO:0097535) |
| 0.1 | 0.4 | GO:1900369 | transcription, RNA-templated(GO:0001172) negative regulation of RNA interference(GO:1900369) |
| 0.1 | 1.2 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.1 | 2.2 | GO:0050774 | negative regulation of dendrite morphogenesis(GO:0050774) |
| 0.1 | 0.5 | GO:0003335 | corneocyte development(GO:0003335) |
| 0.1 | 2.6 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.1 | 2.2 | GO:0090154 | positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 0.1 | 0.9 | GO:2001137 | positive regulation of endocytic recycling(GO:2001137) |
| 0.1 | 4.2 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.1 | 0.4 | GO:0009956 | radial pattern formation(GO:0009956) |
| 0.1 | 1.7 | GO:0097151 | positive regulation of inhibitory postsynaptic potential(GO:0097151) modulation of inhibitory postsynaptic potential(GO:0098828) |
| 0.1 | 1.0 | GO:1900454 | positive regulation of long term synaptic depression(GO:1900454) |
| 0.1 | 0.7 | GO:0032377 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.1 | 0.1 | GO:0002582 | positive regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002582) positive regulation of antigen processing and presentation of peptide antigen(GO:0002585) positive regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002588) |
| 0.1 | 0.3 | GO:1903966 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.1 | 0.3 | GO:0007228 | signal transduction downstream of smoothened(GO:0007227) positive regulation of hh target transcription factor activity(GO:0007228) |
| 0.1 | 2.9 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.1 | 1.4 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.1 | 0.4 | GO:0046833 | snRNA export from nucleus(GO:0006408) positive regulation of RNA export from nucleus(GO:0046833) |
| 0.1 | 0.6 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 0.1 | 1.1 | GO:0006707 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.1 | 0.6 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.1 | 0.2 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.1 | 0.4 | GO:1901662 | phylloquinone metabolic process(GO:0042374) phylloquinone catabolic process(GO:0042376) quinone catabolic process(GO:1901662) |
| 0.1 | 3.3 | GO:0034755 | iron ion transmembrane transport(GO:0034755) |
| 0.1 | 1.7 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.1 | 1.7 | GO:0021514 | ventral spinal cord interneuron differentiation(GO:0021514) |
| 0.1 | 0.4 | GO:2000671 | regulation of motor neuron apoptotic process(GO:2000671) negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.1 | 0.3 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.1 | 0.6 | GO:0021800 | cerebral cortex tangential migration(GO:0021800) |
| 0.1 | 0.2 | GO:0010841 | positive regulation of circadian sleep/wake cycle, wakefulness(GO:0010841) |
| 0.1 | 1.8 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.1 | 0.3 | GO:0000412 | histone peptidyl-prolyl isomerization(GO:0000412) |
| 0.1 | 0.2 | GO:0032827 | negative regulation of natural killer cell differentiation(GO:0032824) negative regulation of natural killer cell differentiation involved in immune response(GO:0032827) positive regulation of cytolysis in other organism(GO:0051714) |
| 0.1 | 2.1 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.1 | 1.0 | GO:0002028 | regulation of sodium ion transport(GO:0002028) |
| 0.1 | 0.2 | GO:0006667 | sphinganine metabolic process(GO:0006667) |
| 0.1 | 5.8 | GO:0090004 | positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.1 | 0.3 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.1 | 5.0 | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
| 0.1 | 1.0 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.1 | 0.6 | GO:0016080 | synaptic vesicle targeting(GO:0016080) |
| 0.1 | 1.6 | GO:1903818 | positive regulation of voltage-gated potassium channel activity(GO:1903818) |
| 0.1 | 0.6 | GO:0046545 | development of primary female sexual characteristics(GO:0046545) |
| 0.1 | 1.1 | GO:0042355 | fucose catabolic process(GO:0019317) L-fucose metabolic process(GO:0042354) L-fucose catabolic process(GO:0042355) |
| 0.1 | 1.0 | GO:0016081 | synaptic vesicle docking(GO:0016081) |
| 0.1 | 0.4 | GO:0035751 | regulation of lysosomal lumen pH(GO:0035751) |
| 0.1 | 0.5 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.1 | 1.8 | GO:1902259 | regulation of delayed rectifier potassium channel activity(GO:1902259) |
| 0.1 | 0.5 | GO:0046676 | negative regulation of insulin secretion(GO:0046676) |
| 0.1 | 0.9 | GO:0061469 | response to corticotropin-releasing hormone(GO:0043435) regulation of type B pancreatic cell proliferation(GO:0061469) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.1 | 1.5 | GO:0060081 | membrane hyperpolarization(GO:0060081) |
| 0.1 | 1.3 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.1 | 0.6 | GO:0017157 | regulation of exocytosis(GO:0017157) |
| 0.1 | 0.6 | GO:0003084 | positive regulation of systemic arterial blood pressure(GO:0003084) |
| 0.1 | 0.4 | GO:0006344 | maintenance of chromatin silencing(GO:0006344) |
| 0.1 | 0.6 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.1 | 0.3 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.1 | 1.8 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.1 | 0.6 | GO:1900452 | regulation of long term synaptic depression(GO:1900452) |
| 0.1 | 0.6 | GO:0003383 | apical constriction(GO:0003383) |
| 0.1 | 1.2 | GO:0035690 | cellular response to drug(GO:0035690) |
| 0.1 | 0.6 | GO:0002759 | regulation of antimicrobial humoral response(GO:0002759) |
| 0.1 | 0.2 | GO:1902805 | positive regulation of synaptic vesicle endocytosis(GO:1900244) positive regulation of synaptic vesicle transport(GO:1902805) positive regulation of synaptic vesicle recycling(GO:1903423) |
| 0.1 | 0.8 | GO:1900120 | regulation of receptor binding(GO:1900120) |
| 0.1 | 0.4 | GO:0060075 | regulation of resting membrane potential(GO:0060075) |
| 0.1 | 0.2 | GO:0097105 | presynaptic membrane assembly(GO:0097105) |
| 0.1 | 0.2 | GO:0019074 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.1 | 0.6 | GO:2000576 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.1 | 0.2 | GO:1990051 | activation of protein kinase C activity(GO:1990051) |
| 0.1 | 0.8 | GO:0032494 | response to peptidoglycan(GO:0032494) |
| 0.1 | 1.1 | GO:1904706 | negative regulation of vascular smooth muscle cell proliferation(GO:1904706) |
| 0.1 | 0.8 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 0.1 | 0.3 | GO:0035407 | histone H3-T11 phosphorylation(GO:0035407) |
| 0.1 | 0.9 | GO:0009607 | response to biotic stimulus(GO:0009607) |
| 0.1 | 0.2 | GO:0044313 | protein K6-linked deubiquitination(GO:0044313) |
| 0.1 | 0.4 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.1 | 0.2 | GO:0046521 | sphingoid catabolic process(GO:0046521) |
| 0.1 | 0.9 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.1 | 0.3 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.1 | 0.2 | GO:1904425 | negative regulation of GTP binding(GO:1904425) |
| 0.0 | 0.1 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.2 | GO:0060363 | cranial suture morphogenesis(GO:0060363) |
| 0.0 | 0.9 | GO:0039692 | single stranded viral RNA replication via double stranded DNA intermediate(GO:0039692) |
| 0.0 | 0.2 | GO:0042369 | vitamin D catabolic process(GO:0042369) |
| 0.0 | 1.5 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.0 | 0.8 | GO:0050974 | detection of mechanical stimulus involved in sensory perception(GO:0050974) |
| 0.0 | 0.1 | GO:0070309 | lens fiber cell morphogenesis(GO:0070309) |
| 0.0 | 0.8 | GO:1904714 | regulation of chaperone-mediated autophagy(GO:1904714) |
| 0.0 | 0.5 | GO:0098915 | membrane repolarization during atrial cardiac muscle cell action potential(GO:0098914) membrane repolarization during ventricular cardiac muscle cell action potential(GO:0098915) |
| 0.0 | 1.4 | GO:0007618 | mating(GO:0007618) |
| 0.0 | 9.7 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 1.0 | GO:0045651 | positive regulation of macrophage differentiation(GO:0045651) |
| 0.0 | 0.1 | GO:1905123 | regulation of glucosylceramidase activity(GO:1905123) |
| 0.0 | 0.9 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.0 | 6.4 | GO:1903779 | regulation of cardiac conduction(GO:1903779) |
| 0.0 | 3.3 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 4.7 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 2.7 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.0 | 0.3 | GO:1900619 | acetylcholine metabolic process(GO:0008291) acetate ester metabolic process(GO:1900619) |
| 0.0 | 1.2 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.0 | 0.4 | GO:0097084 | vascular smooth muscle cell development(GO:0097084) |
| 0.0 | 0.9 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.0 | 1.4 | GO:0000226 | microtubule cytoskeleton organization(GO:0000226) |
| 0.0 | 4.8 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.0 | 0.2 | GO:0061762 | CAMKK-AMPK signaling cascade(GO:0061762) |
| 0.0 | 1.8 | GO:0060674 | placenta blood vessel development(GO:0060674) |
| 0.0 | 1.5 | GO:0014072 | response to isoquinoline alkaloid(GO:0014072) response to morphine(GO:0043278) |
| 0.0 | 1.4 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.0 | 0.7 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.0 | 0.1 | GO:1903697 | negative regulation of microvillus assembly(GO:1903697) |
| 0.0 | 0.7 | GO:2000786 | positive regulation of autophagosome assembly(GO:2000786) |
| 0.0 | 2.0 | GO:0045124 | regulation of bone resorption(GO:0045124) |
| 0.0 | 0.3 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.0 | 0.3 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.0 | 0.2 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.0 | 0.5 | GO:0034315 | regulation of Arp2/3 complex-mediated actin nucleation(GO:0034315) |
| 0.0 | 0.8 | GO:0048485 | sympathetic nervous system development(GO:0048485) |
| 0.0 | 1.1 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.0 | 0.1 | GO:0002525 | acute inflammatory response to non-antigenic stimulus(GO:0002525) regulation of acute inflammatory response to non-antigenic stimulus(GO:0002877) positive regulation of acute inflammatory response to non-antigenic stimulus(GO:0002879) |
| 0.0 | 0.7 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.0 | 2.1 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 1.1 | GO:1900153 | regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900151) positive regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900153) |
| 0.0 | 0.8 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.0 | 0.5 | GO:2000615 | regulation of histone H3-K9 acetylation(GO:2000615) |
| 0.0 | 0.3 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.0 | 0.1 | GO:2001274 | negative regulation of glucose import in response to insulin stimulus(GO:2001274) |
| 0.0 | 0.2 | GO:0090341 | negative regulation of secretion of lysosomal enzymes(GO:0090341) |
| 0.0 | 0.3 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.2 | GO:2000676 | positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.0 | 0.9 | GO:0071380 | cellular response to prostaglandin E stimulus(GO:0071380) |
| 0.0 | 0.1 | GO:0010002 | cardioblast differentiation(GO:0010002) |
| 0.0 | 1.0 | GO:0050901 | leukocyte tethering or rolling(GO:0050901) |
| 0.0 | 0.4 | GO:0042048 | olfactory behavior(GO:0042048) |
| 0.0 | 1.7 | GO:1901379 | regulation of potassium ion transmembrane transport(GO:1901379) |
| 0.0 | 0.7 | GO:2000171 | negative regulation of dendrite development(GO:2000171) |
| 0.0 | 0.5 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.0 | 0.5 | GO:0030050 | vesicle transport along actin filament(GO:0030050) |
| 0.0 | 0.6 | GO:0003417 | growth plate cartilage development(GO:0003417) |
| 0.0 | 0.2 | GO:0032808 | lacrimal gland development(GO:0032808) |
| 0.0 | 0.2 | GO:1902019 | regulation of cilium movement involved in cell motility(GO:0060295) regulation of cilium beat frequency involved in ciliary motility(GO:0060296) regulation of cilium-dependent cell motility(GO:1902019) |
| 0.0 | 1.0 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.0 | 0.5 | GO:0070571 | negative regulation of axon regeneration(GO:0048681) negative regulation of neuron projection regeneration(GO:0070571) |
| 0.0 | 0.6 | GO:0030238 | male sex determination(GO:0030238) |
| 0.0 | 0.6 | GO:2000169 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) regulation of peptidyl-cysteine S-nitrosylation(GO:2000169) |
| 0.0 | 0.8 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.0 | 1.4 | GO:0046928 | regulation of neurotransmitter secretion(GO:0046928) |
| 0.0 | 0.2 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.0 | 0.2 | GO:1904823 | pyrimidine nucleobase transport(GO:0015855) purine nucleobase transmembrane transport(GO:1904823) |
| 0.0 | 0.1 | GO:0021648 | vestibulocochlear nerve morphogenesis(GO:0021648) vestibulocochlear nerve formation(GO:0021650) |
| 0.0 | 0.2 | GO:0048619 | hindgut morphogenesis(GO:0007442) embryonic hindgut morphogenesis(GO:0048619) |
| 0.0 | 0.2 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.0 | 0.7 | GO:0097205 | renal filtration(GO:0097205) |
| 0.0 | 3.0 | GO:0042472 | inner ear morphogenesis(GO:0042472) |
| 0.0 | 0.9 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.0 | 0.1 | GO:0021537 | telencephalon development(GO:0021537) |
| 0.0 | 0.4 | GO:0035860 | glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.0 | 0.5 | GO:0001514 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.0 | 0.3 | GO:0031290 | retinal ganglion cell axon guidance(GO:0031290) |
| 0.0 | 0.2 | GO:0035630 | bone mineralization involved in bone maturation(GO:0035630) |
| 0.0 | 0.5 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.0 | 0.2 | GO:0045950 | negative regulation of mitotic recombination(GO:0045950) |
| 0.0 | 0.2 | GO:0097411 | hypoxia-inducible factor-1alpha signaling pathway(GO:0097411) |
| 0.0 | 0.6 | GO:0031069 | hair follicle morphogenesis(GO:0031069) |
| 0.0 | 0.2 | GO:0030856 | regulation of epithelial cell differentiation(GO:0030856) |
| 0.0 | 0.4 | GO:0010921 | regulation of phosphatase activity(GO:0010921) |
| 0.0 | 0.2 | GO:0040038 | polar body extrusion after meiotic divisions(GO:0040038) formin-nucleated actin cable assembly(GO:0070649) |
| 0.0 | 0.2 | GO:0051415 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.0 | 3.7 | GO:0006813 | potassium ion transport(GO:0006813) |
| 0.0 | 0.1 | GO:2001206 | positive regulation of osteoclast development(GO:2001206) |
| 0.0 | 0.2 | GO:0061299 | retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.0 | 0.1 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 0.0 | 0.1 | GO:0072344 | rescue of stalled ribosome(GO:0072344) |
| 0.0 | 0.2 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.0 | 3.3 | GO:0006836 | neurotransmitter transport(GO:0006836) |
| 0.0 | 0.8 | GO:0014009 | glial cell proliferation(GO:0014009) |
| 0.0 | 0.2 | GO:0002911 | lymphocyte anergy(GO:0002249) regulation of T cell anergy(GO:0002667) T cell anergy(GO:0002870) regulation of lymphocyte anergy(GO:0002911) |
| 0.0 | 0.0 | GO:0006537 | glutamate biosynthetic process(GO:0006537) |
| 0.0 | 0.3 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.0 | 3.5 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 0.4 | GO:0061436 | establishment of skin barrier(GO:0061436) |
| 0.0 | 0.3 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.0 | 3.1 | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.0 | 1.3 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 1.0 | GO:0031640 | killing of cells of other organism(GO:0031640) disruption of cells of other organism(GO:0044364) |
| 0.0 | 0.1 | GO:0097368 | establishment of Sertoli cell barrier(GO:0097368) |
| 0.0 | 0.2 | GO:0042426 | choline catabolic process(GO:0042426) |
| 0.0 | 0.9 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.0 | 0.3 | GO:0090026 | positive regulation of monocyte chemotaxis(GO:0090026) |
| 0.0 | 0.1 | GO:0000117 | regulation of transcription involved in G2/M transition of mitotic cell cycle(GO:0000117) |
| 0.0 | 0.8 | GO:0034219 | carbohydrate transmembrane transport(GO:0034219) |
| 0.0 | 0.9 | GO:0008542 | visual learning(GO:0008542) |
| 0.0 | 0.7 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.0 | 0.3 | GO:0042501 | serine phosphorylation of STAT protein(GO:0042501) |
| 0.0 | 0.4 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.0 | 0.6 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.0 | 0.1 | GO:1904383 | response to sodium phosphate(GO:1904383) response to insulin-like growth factor stimulus(GO:1990418) |
| 0.0 | 0.1 | GO:0060684 | epithelial-mesenchymal cell signaling(GO:0060684) |
| 0.0 | 0.0 | GO:0030853 | negative regulation of granulocyte differentiation(GO:0030853) |
| 0.0 | 0.4 | GO:0050885 | neuromuscular process controlling balance(GO:0050885) |
| 0.0 | 0.4 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 1.0 | GO:0045540 | regulation of cholesterol biosynthetic process(GO:0045540) |
| 0.0 | 0.9 | GO:0030317 | sperm motility(GO:0030317) |
| 0.0 | 0.4 | GO:0043068 | positive regulation of apoptotic process(GO:0043065) positive regulation of programmed cell death(GO:0043068) |
| 0.0 | 0.1 | GO:2000121 | regulation of removal of superoxide radicals(GO:2000121) |
| 0.0 | 0.4 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.6 | GO:0007340 | acrosome reaction(GO:0007340) |
| 0.0 | 0.1 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.0 | 0.3 | GO:0070734 | histone H3-K27 methylation(GO:0070734) |
| 0.0 | 0.8 | GO:0001764 | neuron migration(GO:0001764) |
| 0.0 | 0.2 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.0 | 0.3 | GO:0035767 | endothelial cell chemotaxis(GO:0035767) |
| 0.0 | 0.4 | GO:0051642 | centrosome localization(GO:0051642) |
| 0.0 | 0.3 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 0.5 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 0.5 | GO:0002244 | hematopoietic progenitor cell differentiation(GO:0002244) |
| 0.0 | 0.0 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.0 | 0.4 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.2 | GO:0008589 | regulation of smoothened signaling pathway(GO:0008589) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 3.1 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 0.8 | 2.5 | GO:0070703 | inner mucus layer(GO:0070702) outer mucus layer(GO:0070703) |
| 0.8 | 3.1 | GO:0060187 | cell pole(GO:0060187) |
| 0.7 | 2.9 | GO:0097450 | astrocyte end-foot(GO:0097450) |
| 0.5 | 4.6 | GO:0060200 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.5 | 8.2 | GO:0097418 | neurofibrillary tangle(GO:0097418) |
| 0.4 | 14.8 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.4 | 3.6 | GO:0030868 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.3 | 1.9 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.3 | 1.0 | GO:0034686 | integrin alphav-beta8 complex(GO:0034686) |
| 0.3 | 1.3 | GO:0005694 | chromosome(GO:0005694) |
| 0.3 | 1.3 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.3 | 3.6 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.3 | 3.6 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.3 | 7.4 | GO:0016471 | vacuolar proton-transporting V-type ATPase complex(GO:0016471) |
| 0.2 | 3.9 | GO:0060091 | kinocilium(GO:0060091) |
| 0.2 | 1.4 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.2 | 0.7 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.2 | 1.3 | GO:0030673 | axolemma(GO:0030673) |
| 0.2 | 3.1 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.2 | 0.5 | GO:0005940 | septin ring(GO:0005940) septin collar(GO:0032173) |
| 0.2 | 2.4 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.2 | 0.9 | GO:0032044 | DSIF complex(GO:0032044) |
| 0.1 | 2.2 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 6.6 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.1 | 6.5 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 2.1 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.1 | 0.6 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.1 | 0.8 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.1 | 0.5 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.1 | 0.6 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.1 | 0.4 | GO:0031379 | RNA-directed RNA polymerase complex(GO:0031379) |
| 0.1 | 0.5 | GO:0097224 | sperm connecting piece(GO:0097224) |
| 0.1 | 19.6 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.1 | 1.0 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.1 | 1.2 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.1 | 0.7 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.1 | 0.4 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.1 | 1.4 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.1 | 0.6 | GO:0098536 | deuterosome(GO:0098536) |
| 0.1 | 0.2 | GO:0065010 | extracellular membrane-bounded organelle(GO:0065010) |
| 0.1 | 2.3 | GO:0010369 | chromocenter(GO:0010369) |
| 0.1 | 14.4 | GO:0043679 | axon terminus(GO:0043679) |
| 0.1 | 1.1 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.1 | 28.2 | GO:0014069 | postsynaptic density(GO:0014069) postsynaptic specialization(GO:0099572) |
| 0.1 | 1.1 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.1 | 1.8 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.1 | 0.4 | GO:0005846 | nuclear cap binding complex(GO:0005846) |
| 0.1 | 0.3 | GO:0070931 | Golgi-associated vesicle lumen(GO:0070931) |
| 0.1 | 0.2 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.1 | 15.2 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 3.7 | GO:0043005 | neuron projection(GO:0043005) |
| 0.1 | 32.9 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.1 | 4.0 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.1 | 0.9 | GO:0097227 | sperm annulus(GO:0097227) |
| 0.0 | 1.5 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.9 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 1.6 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.3 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.0 | 0.6 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 0.4 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 0.9 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.2 | GO:0034667 | integrin alpha3-beta1 complex(GO:0034667) |
| 0.0 | 0.2 | GO:0000923 | equatorial microtubule organizing center(GO:0000923) |
| 0.0 | 4.5 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 5.1 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.5 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 0.2 | GO:0097013 | phagocytic vesicle lumen(GO:0097013) |
| 0.0 | 0.4 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 0.6 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.3 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.0 | 0.6 | GO:0043218 | compact myelin(GO:0043218) |
| 0.0 | 0.5 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 0.4 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 2.1 | GO:0098793 | presynapse(GO:0098793) |
| 0.0 | 0.6 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 1.0 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 1.1 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.7 | GO:0031985 | Golgi cisterna(GO:0031985) |
| 0.0 | 0.6 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 0.3 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 1.1 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 0.2 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.0 | 1.0 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 2.4 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.5 | GO:0045178 | basal part of cell(GO:0045178) |
| 0.0 | 0.2 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.0 | 3.0 | GO:0044309 | dendritic spine(GO:0043197) neuron spine(GO:0044309) |
| 0.0 | 6.0 | GO:0070382 | exocytic vesicle(GO:0070382) |
| 0.0 | 0.2 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.2 | GO:0097208 | alveolar lamellar body(GO:0097208) |
| 0.0 | 1.2 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.0 | 0.1 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 0.7 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 1.9 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 0.0 | GO:0034680 | integrin alpha10-beta1 complex(GO:0034680) |
| 0.0 | 0.2 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.0 | 2.0 | GO:0005815 | microtubule organizing center(GO:0005815) |
| 0.0 | 0.7 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.0 | 0.1 | GO:0031213 | RSF complex(GO:0031213) |
| 0.0 | 0.4 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.1 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.3 | GO:0005858 | axonemal dynein complex(GO:0005858) |
| 0.0 | 0.7 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.6 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 1.9 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 0.1 | GO:0072558 | NLRP1 inflammasome complex(GO:0072558) |
| 0.0 | 0.5 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 1.1 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 0.3 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 5.7 | GO:0044297 | cell body(GO:0044297) |
| 0.0 | 2.2 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 1.1 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 1.3 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 0.2 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.0 | 1.1 | GO:0036126 | sperm flagellum(GO:0036126) |
| 0.0 | 0.7 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.5 | GO:0030529 | intracellular ribonucleoprotein complex(GO:0030529) ribonucleoprotein complex(GO:1990904) |
| 0.0 | 0.9 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 0.3 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.0 | 0.1 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 6.8 | GO:0015275 | stretch-activated, cation-selective, calcium channel activity(GO:0015275) |
| 1.6 | 6.5 | GO:0035403 | histone kinase activity (H3-T6 specific)(GO:0035403) |
| 1.0 | 2.0 | GO:0004698 | calcium-dependent protein kinase C activity(GO:0004698) |
| 0.8 | 3.3 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.8 | 12.9 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 0.7 | 3.3 | GO:0031751 | D4 dopamine receptor binding(GO:0031751) |
| 0.7 | 20.4 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.6 | 3.6 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.6 | 1.8 | GO:0016603 | glutaminyl-peptide cyclotransferase activity(GO:0016603) |
| 0.6 | 1.8 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.6 | 5.3 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.5 | 1.6 | GO:0052895 | norspermine:oxygen oxidoreductase activity(GO:0052894) N1-acetylspermine:oxygen oxidoreductase (N1-acetylspermidine-forming) activity(GO:0052895) |
| 0.5 | 1.5 | GO:0031877 | somatostatin receptor binding(GO:0031877) |
| 0.5 | 1.9 | GO:0099609 | microtubule lateral binding(GO:0099609) |
| 0.4 | 2.2 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.4 | 5.3 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.4 | 1.6 | GO:0017095 | heparan sulfate 6-O-sulfotransferase activity(GO:0017095) |
| 0.4 | 4.3 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.4 | 2.3 | GO:0099529 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.4 | 1.1 | GO:0005365 | myo-inositol transmembrane transporter activity(GO:0005365) |
| 0.4 | 1.8 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.4 | 1.1 | GO:0050429 | calcium-dependent phospholipase C activity(GO:0050429) |
| 0.3 | 2.0 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.3 | 2.6 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.3 | 1.3 | GO:0031811 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 0.3 | 4.2 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.3 | 0.3 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.3 | 0.9 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 0.3 | 3.1 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.3 | 2.8 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.3 | 2.1 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.3 | 0.8 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.3 | 1.0 | GO:0033823 | procollagen-lysine 5-dioxygenase activity(GO:0008475) procollagen glucosyltransferase activity(GO:0033823) |
| 0.3 | 1.0 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.2 | 2.2 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.2 | 1.2 | GO:0004991 | parathyroid hormone receptor activity(GO:0004991) |
| 0.2 | 0.7 | GO:0047225 | acetylgalactosaminyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0047225) |
| 0.2 | 1.1 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.2 | 0.8 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.2 | 2.9 | GO:0046870 | cadmium ion binding(GO:0046870) |
| 0.2 | 1.0 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 0.2 | 1.9 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.2 | 1.9 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.2 | 0.7 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.2 | 3.1 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.2 | 0.8 | GO:0004052 | arachidonate 12-lipoxygenase activity(GO:0004052) |
| 0.2 | 5.5 | GO:0005112 | Notch binding(GO:0005112) |
| 0.2 | 2.6 | GO:0005113 | patched binding(GO:0005113) |
| 0.2 | 3.5 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.2 | 3.9 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.2 | 1.0 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.2 | 1.4 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.2 | 1.6 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.2 | 5.7 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.1 | 0.4 | GO:0000253 | 3-keto sterol reductase activity(GO:0000253) |
| 0.1 | 1.0 | GO:0005353 | fructose transmembrane transporter activity(GO:0005353) |
| 0.1 | 0.8 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 0.1 | 2.7 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.1 | 1.5 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.1 | 4.0 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.1 | 4.7 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.1 | 0.4 | GO:0003968 | RNA-directed RNA polymerase activity(GO:0003968) |
| 0.1 | 0.5 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.1 | 1.0 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.1 | 0.7 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.1 | 1.4 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.1 | 0.7 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.1 | 0.8 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.1 | 1.8 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.1 | 0.4 | GO:0022842 | leak channel activity(GO:0022840) narrow pore channel activity(GO:0022842) |
| 0.1 | 1.4 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.1 | 0.3 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.1 | 2.9 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 3.3 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.1 | 1.1 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.1 | 7.0 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.1 | 0.5 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.1 | 0.7 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.1 | 0.9 | GO:0050656 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) 3'-phosphoadenosine 5'-phosphosulfate binding(GO:0050656) |
| 0.1 | 1.2 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.1 | 2.1 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.1 | 0.8 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.1 | 5.3 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.1 | 3.1 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 0.4 | GO:0004140 | dephospho-CoA kinase activity(GO:0004140) |
| 0.1 | 1.4 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.1 | 0.7 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.1 | 0.8 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.1 | 1.3 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.1 | 1.0 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.1 | 0.6 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.1 | 4.7 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.1 | 0.6 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.1 | 3.4 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.1 | 1.2 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.1 | 0.3 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 0.1 | 0.7 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.1 | 3.1 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.1 | 3.1 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.1 | 2.5 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.1 | 0.5 | GO:0086089 | voltage-gated potassium channel activity involved in atrial cardiac muscle cell action potential repolarization(GO:0086089) |
| 0.1 | 0.5 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.1 | 0.8 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.1 | 0.8 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.1 | 0.3 | GO:0033265 | choline binding(GO:0033265) |
| 0.1 | 1.1 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.1 | 2.0 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.1 | 8.1 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.1 | 1.8 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.1 | 1.0 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.1 | 0.2 | GO:0070576 | vitamin D 24-hydroxylase activity(GO:0070576) |
| 0.1 | 0.3 | GO:0004082 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.1 | 0.2 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.1 | 3.2 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.1 | 0.9 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.1 | 0.2 | GO:0008745 | N-acetylmuramoyl-L-alanine amidase activity(GO:0008745) |
| 0.1 | 6.2 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.1 | 3.4 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.1 | 0.9 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.1 | 8.9 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.1 | 1.9 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.1 | 1.1 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.1 | 0.5 | GO:0001025 | RNA polymerase III transcription factor binding(GO:0001025) |
| 0.1 | 0.3 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 0.1 | 11.6 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 0.4 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.1 | 1.9 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.1 | 0.3 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 0.1 | 1.0 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.1 | 0.2 | GO:0016768 | spermine synthase activity(GO:0016768) |
| 0.0 | 1.1 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.0 | 0.4 | GO:0016167 | glial cell-derived neurotrophic factor receptor activity(GO:0016167) |
| 0.0 | 0.3 | GO:0047493 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.0 | 0.7 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.4 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.0 | 1.2 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.9 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) |
| 0.0 | 0.6 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.0 | 1.4 | GO:0015114 | phosphate ion transmembrane transporter activity(GO:0015114) |
| 0.0 | 0.2 | GO:0015389 | pyrimidine- and adenine-specific:sodium symporter activity(GO:0015389) |
| 0.0 | 1.0 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.3 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.0 | 0.7 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 0.5 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.0 | 1.9 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.4 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 0.7 | GO:0070513 | death domain binding(GO:0070513) |
| 0.0 | 1.3 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.1 | GO:0070506 | high-density lipoprotein particle receptor activity(GO:0070506) |
| 0.0 | 0.8 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.3 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 0.3 | GO:0016215 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) |
| 0.0 | 2.7 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 1.2 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 1.2 | GO:0005326 | neurotransmitter transporter activity(GO:0005326) |
| 0.0 | 0.2 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.6 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.3 | GO:0042923 | neuropeptide binding(GO:0042923) |
| 0.0 | 4.5 | GO:0016820 | hydrolase activity, acting on acid anhydrides, catalyzing transmembrane movement of substances(GO:0016820) |
| 0.0 | 0.3 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 1.7 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 2.8 | GO:0016247 | channel regulator activity(GO:0016247) |
| 0.0 | 0.2 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.9 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.0 | 0.1 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.0 | 0.2 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.2 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.9 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.9 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 2.5 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.5 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 0.2 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.0 | 0.7 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.0 | 0.5 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.2 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 1.2 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.1 | GO:0010858 | calcium-dependent protein kinase regulator activity(GO:0010858) |
| 0.0 | 0.2 | GO:0004075 | biotin carboxylase activity(GO:0004075) biotin binding(GO:0009374) |
| 0.0 | 0.2 | GO:0032810 | sterol response element binding(GO:0032810) |
| 0.0 | 0.1 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.0 | 0.5 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.3 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.0 | 0.3 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 0.1 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 0.6 | GO:0000217 | DNA secondary structure binding(GO:0000217) |
| 0.0 | 0.8 | GO:0046983 | protein dimerization activity(GO:0046983) |
| 0.0 | 0.1 | GO:0008481 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.0 | 0.0 | GO:0004090 | carbonyl reductase (NADPH) activity(GO:0004090) |
| 0.0 | 0.2 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 0.0 | 0.1 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.0 | 1.3 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 0.5 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 1.8 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 0.2 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.1 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.0 | 0.2 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.0 | GO:0035650 | AP-1 adaptor complex binding(GO:0035650) |
| 0.0 | 0.8 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.4 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 0.8 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 2.5 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 0.0 | GO:0016639 | oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 0.0 | 1.1 | GO:0008186 | ATP-dependent RNA helicase activity(GO:0004004) RNA-dependent ATPase activity(GO:0008186) |
| 0.0 | 0.3 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 0.6 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.0 | 5.8 | GO:0005525 | GTP binding(GO:0005525) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 20.4 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.2 | 6.5 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.1 | 9.7 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.1 | 3.9 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.1 | 1.9 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.1 | 6.8 | PID ATR PATHWAY | ATR signaling pathway |
| 0.1 | 3.1 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.1 | 3.2 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.1 | 1.6 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.1 | 1.5 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.1 | 2.6 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.1 | 0.3 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.1 | 4.9 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.2 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 1.6 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 1.7 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 3.3 | SIG CHEMOTAXIS | Genes related to chemotaxis |
| 0.0 | 3.2 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 2.2 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 16.8 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 1.4 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 2.2 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 4.3 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.3 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 2.1 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 0.5 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.0 | 0.2 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 0.3 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 2.9 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.5 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 0.6 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 0.4 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.7 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 0.9 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.0 | 0.1 | PID AVB3 INTEGRIN PATHWAY | Integrins in angiogenesis |
| 0.0 | 0.5 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.6 | PID AURORA A PATHWAY | Aurora A signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 14.8 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.3 | 6.3 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.2 | 2.3 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.2 | 3.9 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.2 | 3.1 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.2 | 8.9 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.2 | 4.7 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.2 | 6.7 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.2 | 5.3 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.1 | 9.0 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 2.7 | REACTOME FGFR1 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR1 ligand binding and activation |
| 0.1 | 1.4 | REACTOME REGULATION OF INSULIN SECRETION | Genes involved in Regulation of Insulin Secretion |
| 0.1 | 3.3 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.1 | 5.6 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.1 | 4.2 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 3.3 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS | Genes involved in Synthesis of bile acids and bile salts |
| 0.1 | 1.7 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.1 | 3.1 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.1 | 1.7 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.1 | 2.0 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.1 | 3.1 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.1 | 3.2 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.1 | 0.3 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.1 | 2.5 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.1 | 1.9 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 2.5 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.0 | 2.9 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.0 | 11.2 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.6 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 0.6 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 1.8 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 1.0 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.5 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.0 | 2.2 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.8 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.4 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 1.1 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.8 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 0.2 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.4 | REACTOME FRS2 MEDIATED CASCADE | Genes involved in FRS2-mediated cascade |
| 0.0 | 0.3 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 1.3 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.0 | 0.8 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 0.5 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.0 | 0.5 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.0 | 1.1 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.5 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.7 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.7 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 0.2 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.6 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 1.6 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |