Project
Illumina Body Map 2: averaged replicates
Navigation
Downloads
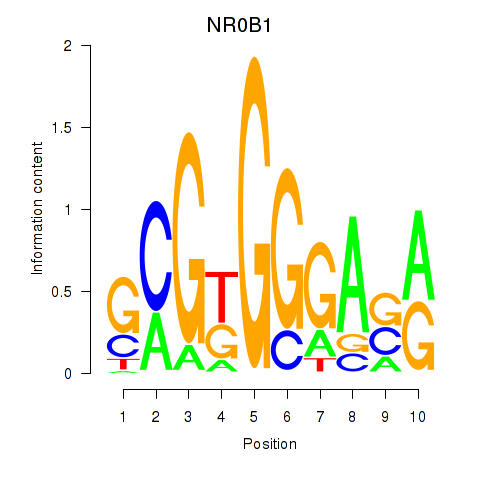
Results for NR0B1
Z-value: 1.53
Transcription factors associated with NR0B1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NR0B1
|
ENSG00000169297.6 | nuclear receptor subfamily 0 group B member 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NR0B1 | hg19_v2_chrX_-_30326445_30326605 | 0.14 | 4.3e-01 | Click! |
Activity profile of NR0B1 motif
Sorted Z-values of NR0B1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_+_4845379 | 3.52 |
ENST00000588606.1
ENST00000586005.1 |
SMIM22
|
small integral membrane protein 22 |
| chr2_+_105471969 | 3.09 |
ENST00000361360.2
|
POU3F3
|
POU class 3 homeobox 3 |
| chr2_+_220492373 | 2.71 |
ENST00000317151.3
|
SLC4A3
|
solute carrier family 4 (anion exchanger), member 3 |
| chr3_-_123168551 | 2.65 |
ENST00000462833.1
|
ADCY5
|
adenylate cyclase 5 |
| chr6_-_94129244 | 2.15 |
ENST00000369303.4
ENST00000369297.1 |
EPHA7
|
EPH receptor A7 |
| chr2_+_220492116 | 2.04 |
ENST00000373760.2
|
SLC4A3
|
solute carrier family 4 (anion exchanger), member 3 |
| chr15_-_65067773 | 2.01 |
ENST00000300069.4
|
RBPMS2
|
RNA binding protein with multiple splicing 2 |
| chr11_-_12030746 | 1.94 |
ENST00000533813.1
|
DKK3
|
dickkopf WNT signaling pathway inhibitor 3 |
| chr4_+_150999418 | 1.93 |
ENST00000296550.7
|
DCLK2
|
doublecortin-like kinase 2 |
| chr20_-_18038521 | 1.92 |
ENST00000278780.6
|
OVOL2
|
ovo-like zinc finger 2 |
| chrX_-_128788914 | 1.86 |
ENST00000429967.1
ENST00000307484.6 |
APLN
|
apelin |
| chr2_+_234545092 | 1.86 |
ENST00000344644.5
|
UGT1A10
|
UDP glucuronosyltransferase 1 family, polypeptide A10 |
| chr2_-_219925189 | 1.79 |
ENST00000295731.6
|
IHH
|
indian hedgehog |
| chr11_-_12030629 | 1.77 |
ENST00000396505.2
|
DKK3
|
dickkopf WNT signaling pathway inhibitor 3 |
| chr11_-_12030681 | 1.75 |
ENST00000529338.1
|
DKK3
|
dickkopf WNT signaling pathway inhibitor 3 |
| chr1_-_147245484 | 1.70 |
ENST00000271348.2
|
GJA5
|
gap junction protein, alpha 5, 40kDa |
| chr10_+_112404132 | 1.64 |
ENST00000369519.3
|
RBM20
|
RNA binding motif protein 20 |
| chr9_+_128509624 | 1.62 |
ENST00000342287.5
ENST00000373487.4 |
PBX3
|
pre-B-cell leukemia homeobox 3 |
| chrX_-_114468605 | 1.58 |
ENST00000538422.1
ENST00000317135.8 |
LRCH2
|
leucine-rich repeats and calponin homology (CH) domain containing 2 |
| chr1_-_147245445 | 1.39 |
ENST00000430508.1
|
GJA5
|
gap junction protein, alpha 5, 40kDa |
| chr10_+_60936347 | 1.36 |
ENST00000373880.4
|
PHYHIPL
|
phytanoyl-CoA 2-hydroxylase interacting protein-like |
| chr9_+_71320596 | 1.35 |
ENST00000265382.3
|
PIP5K1B
|
phosphatidylinositol-4-phosphate 5-kinase, type I, beta |
| chr9_+_71320557 | 1.35 |
ENST00000541509.1
|
PIP5K1B
|
phosphatidylinositol-4-phosphate 5-kinase, type I, beta |
| chr10_-_21463116 | 1.28 |
ENST00000417816.2
|
NEBL
|
nebulette |
| chr15_-_73661605 | 1.26 |
ENST00000261917.3
|
HCN4
|
hyperpolarization activated cyclic nucleotide-gated potassium channel 4 |
| chr19_-_45909585 | 1.23 |
ENST00000593226.1
ENST00000418234.2 |
PPP1R13L
|
protein phosphatase 1, regulatory subunit 13 like |
| chr1_-_40098672 | 1.23 |
ENST00000535435.1
|
HEYL
|
hes-related family bHLH transcription factor with YRPW motif-like |
| chr2_+_155555201 | 1.21 |
ENST00000544049.1
|
KCNJ3
|
potassium inwardly-rectifying channel, subfamily J, member 3 |
| chr3_+_181429704 | 1.19 |
ENST00000431565.2
ENST00000325404.1 |
SOX2
|
SRY (sex determining region Y)-box 2 |
| chr12_+_41086297 | 1.18 |
ENST00000551295.2
|
CNTN1
|
contactin 1 |
| chr2_+_220306745 | 1.18 |
ENST00000431523.1
ENST00000396698.1 ENST00000396695.2 |
SPEG
|
SPEG complex locus |
| chr13_-_38443860 | 1.16 |
ENST00000426868.2
ENST00000379681.3 ENST00000338947.5 ENST00000355779.2 ENST00000358477.2 ENST00000379673.2 |
TRPC4
|
transient receptor potential cation channel, subfamily C, member 4 |
| chr1_-_196577489 | 1.11 |
ENST00000609185.1
ENST00000451324.2 ENST00000367433.5 ENST00000367431.4 |
KCNT2
|
potassium channel, subfamily T, member 2 |
| chr6_+_17393839 | 1.08 |
ENST00000489374.1
ENST00000378990.2 |
CAP2
|
CAP, adenylate cyclase-associated protein, 2 (yeast) |
| chr11_-_119993979 | 1.07 |
ENST00000524816.3
ENST00000525327.1 |
TRIM29
|
tripartite motif containing 29 |
| chr6_+_17393888 | 1.05 |
ENST00000493172.1
ENST00000465994.1 |
CAP2
|
CAP, adenylate cyclase-associated protein, 2 (yeast) |
| chr10_+_18689637 | 1.04 |
ENST00000377315.4
|
CACNB2
|
calcium channel, voltage-dependent, beta 2 subunit |
| chr2_+_234580525 | 1.04 |
ENST00000609637.1
|
UGT1A1
|
UDP glucuronosyltransferase 1 family, polypeptide A8 |
| chr9_-_35691017 | 1.02 |
ENST00000378292.3
|
TPM2
|
tropomyosin 2 (beta) |
| chr10_-_17659234 | 1.02 |
ENST00000466335.1
|
PTPLA
|
protein tyrosine phosphatase-like (proline instead of catalytic arginine), member A |
| chr8_-_28347737 | 1.01 |
ENST00000517673.1
ENST00000518734.1 ENST00000346498.2 ENST00000380254.2 |
FBXO16
|
F-box protein 16 |
| chrX_-_153151586 | 0.98 |
ENST00000370060.1
ENST00000370055.1 ENST00000420165.1 |
L1CAM
|
L1 cell adhesion molecule |
| chr1_-_23810664 | 0.98 |
ENST00000336689.3
ENST00000437606.2 |
ASAP3
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 3 |
| chr14_+_96968707 | 0.97 |
ENST00000216277.8
ENST00000557320.1 ENST00000557471.1 |
PAPOLA
|
poly(A) polymerase alpha |
| chr8_+_95653302 | 0.96 |
ENST00000423620.2
ENST00000433389.2 |
ESRP1
|
epithelial splicing regulatory protein 1 |
| chr2_+_14775215 | 0.95 |
ENST00000581929.1
|
AC011897.1
|
Uncharacterized protein |
| chr3_+_111578640 | 0.95 |
ENST00000393925.3
|
PHLDB2
|
pleckstrin homology-like domain, family B, member 2 |
| chr6_+_44238203 | 0.95 |
ENST00000451188.2
|
TMEM151B
|
transmembrane protein 151B |
| chr3_-_183979251 | 0.94 |
ENST00000296238.3
|
CAMK2N2
|
calcium/calmodulin-dependent protein kinase II inhibitor 2 |
| chr6_+_126070726 | 0.93 |
ENST00000368364.3
|
HEY2
|
hes-related family bHLH transcription factor with YRPW motif 2 |
| chr2_+_234545148 | 0.92 |
ENST00000373445.1
|
UGT1A10
|
UDP glucuronosyltransferase 1 family, polypeptide A10 |
| chr2_+_234580499 | 0.91 |
ENST00000354728.4
|
UGT1A9
|
UDP glucuronosyltransferase 1 family, polypeptide A9 |
| chr9_+_129986734 | 0.91 |
ENST00000444677.1
|
GARNL3
|
GTPase activating Rap/RanGAP domain-like 3 |
| chr7_+_89841024 | 0.89 |
ENST00000394626.1
|
STEAP2
|
STEAP family member 2, metalloreductase |
| chr12_+_58005204 | 0.89 |
ENST00000286494.4
|
ARHGEF25
|
Rho guanine nucleotide exchange factor (GEF) 25 |
| chr2_+_14772810 | 0.88 |
ENST00000295092.2
ENST00000331243.4 |
FAM84A
|
family with sequence similarity 84, member A |
| chr6_-_112575687 | 0.88 |
ENST00000521398.1
ENST00000424408.2 ENST00000243219.3 |
LAMA4
|
laminin, alpha 4 |
| chr1_+_117452669 | 0.87 |
ENST00000393203.2
|
PTGFRN
|
prostaglandin F2 receptor inhibitor |
| chr19_-_36523709 | 0.87 |
ENST00000592017.1
ENST00000360535.4 |
CLIP3
|
CAP-GLY domain containing linker protein 3 |
| chr14_-_89021077 | 0.85 |
ENST00000556564.1
|
PTPN21
|
protein tyrosine phosphatase, non-receptor type 21 |
| chr7_+_89841144 | 0.85 |
ENST00000394622.2
ENST00000394632.1 ENST00000426158.1 ENST00000394621.2 ENST00000402625.2 |
STEAP2
|
STEAP family member 2, metalloreductase |
| chr11_-_2160180 | 0.84 |
ENST00000381406.4
|
IGF2
|
insulin-like growth factor 2 (somatomedin A) |
| chr3_+_111578131 | 0.82 |
ENST00000498699.1
|
PHLDB2
|
pleckstrin homology-like domain, family B, member 2 |
| chr7_-_140340576 | 0.81 |
ENST00000275884.6
ENST00000475837.1 |
DENND2A
|
DENN/MADD domain containing 2A |
| chr9_+_128509663 | 0.81 |
ENST00000373489.5
ENST00000373483.2 |
PBX3
|
pre-B-cell leukemia homeobox 3 |
| chr13_-_33859819 | 0.81 |
ENST00000336934.5
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13 |
| chr6_-_112575758 | 0.80 |
ENST00000431543.2
ENST00000453937.2 ENST00000368638.4 ENST00000389463.4 |
LAMA4
|
laminin, alpha 4 |
| chr16_+_2014941 | 0.80 |
ENST00000531523.1
|
SNHG9
|
small nucleolar RNA host gene 9 (non-protein coding) |
| chr5_+_38258511 | 0.79 |
ENST00000354891.3
ENST00000322350.5 |
EGFLAM
|
EGF-like, fibronectin type III and laminin G domains |
| chr1_-_223537401 | 0.78 |
ENST00000343846.3
ENST00000454695.2 ENST00000484758.2 |
SUSD4
|
sushi domain containing 4 |
| chr12_+_56473939 | 0.78 |
ENST00000450146.2
|
ERBB3
|
v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 3 |
| chr14_-_23822061 | 0.76 |
ENST00000397260.3
|
SLC22A17
|
solute carrier family 22, member 17 |
| chr3_+_111578027 | 0.76 |
ENST00000431670.2
ENST00000412622.1 |
PHLDB2
|
pleckstrin homology-like domain, family B, member 2 |
| chr12_+_56473628 | 0.75 |
ENST00000549282.1
ENST00000549061.1 ENST00000267101.3 |
ERBB3
|
v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 3 |
| chr4_+_88928777 | 0.75 |
ENST00000237596.2
|
PKD2
|
polycystic kidney disease 2 (autosomal dominant) |
| chr19_-_44143939 | 0.75 |
ENST00000222374.2
|
CADM4
|
cell adhesion molecule 4 |
| chr19_+_49661037 | 0.75 |
ENST00000427978.2
|
TRPM4
|
transient receptor potential cation channel, subfamily M, member 4 |
| chr3_-_169381183 | 0.72 |
ENST00000494292.1
|
MECOM
|
MDS1 and EVI1 complex locus |
| chr1_-_223536679 | 0.72 |
ENST00000608996.1
|
SUSD4
|
sushi domain containing 4 |
| chr3_+_111578583 | 0.72 |
ENST00000478922.1
ENST00000477695.1 |
PHLDB2
|
pleckstrin homology-like domain, family B, member 2 |
| chr19_+_8429031 | 0.71 |
ENST00000301455.2
ENST00000541807.1 ENST00000393962.2 |
ANGPTL4
|
angiopoietin-like 4 |
| chr6_+_7541808 | 0.68 |
ENST00000379802.3
|
DSP
|
desmoplakin |
| chr10_-_17659357 | 0.68 |
ENST00000326961.6
ENST00000361271.3 |
PTPLA
|
protein tyrosine phosphatase-like (proline instead of catalytic arginine), member A |
| chr3_-_122712079 | 0.67 |
ENST00000393583.2
|
SEMA5B
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5B |
| chr6_+_7541845 | 0.67 |
ENST00000418664.2
|
DSP
|
desmoplakin |
| chr20_+_55204351 | 0.66 |
ENST00000201031.2
|
TFAP2C
|
transcription factor AP-2 gamma (activating enhancer binding protein 2 gamma) |
| chr12_-_6484715 | 0.65 |
ENST00000228916.2
|
SCNN1A
|
sodium channel, non-voltage-gated 1 alpha subunit |
| chr19_-_36523529 | 0.64 |
ENST00000593074.1
|
CLIP3
|
CAP-GLY domain containing linker protein 3 |
| chr12_+_65672702 | 0.64 |
ENST00000538045.1
ENST00000535239.1 |
MSRB3
|
methionine sulfoxide reductase B3 |
| chr5_+_79331164 | 0.64 |
ENST00000350881.2
|
THBS4
|
thrombospondin 4 |
| chr14_-_23822080 | 0.63 |
ENST00000397267.1
ENST00000354772.3 |
SLC22A17
|
solute carrier family 22, member 17 |
| chr17_+_77751931 | 0.62 |
ENST00000310942.4
ENST00000269399.5 |
CBX2
|
chromobox homolog 2 |
| chr15_-_72490114 | 0.61 |
ENST00000309731.7
|
GRAMD2
|
GRAM domain containing 2 |
| chr10_-_13570533 | 0.61 |
ENST00000396900.2
ENST00000396898.2 |
BEND7
|
BEN domain containing 7 |
| chr22_+_51112800 | 0.61 |
ENST00000414786.2
|
SHANK3
|
SH3 and multiple ankyrin repeat domains 3 |
| chr6_-_112575838 | 0.60 |
ENST00000455073.1
|
LAMA4
|
laminin, alpha 4 |
| chr12_+_49212514 | 0.59 |
ENST00000301050.2
ENST00000548279.1 ENST00000547230.1 |
CACNB3
|
calcium channel, voltage-dependent, beta 3 subunit |
| chr14_+_96968802 | 0.58 |
ENST00000556619.1
ENST00000392990.2 |
PAPOLA
|
poly(A) polymerase alpha |
| chr4_-_135122903 | 0.57 |
ENST00000421491.3
ENST00000529122.2 |
PABPC4L
|
poly(A) binding protein, cytoplasmic 4-like |
| chr2_+_5832799 | 0.57 |
ENST00000322002.3
|
SOX11
|
SRY (sex determining region Y)-box 11 |
| chr2_+_201170999 | 0.56 |
ENST00000439395.1
ENST00000444012.1 |
SPATS2L
|
spermatogenesis associated, serine-rich 2-like |
| chr22_+_29279552 | 0.56 |
ENST00000544604.2
|
ZNRF3
|
zinc and ring finger 3 |
| chrX_+_9754461 | 0.56 |
ENST00000380913.3
|
SHROOM2
|
shroom family member 2 |
| chr19_-_46272106 | 0.56 |
ENST00000560168.1
|
SIX5
|
SIX homeobox 5 |
| chr20_+_306221 | 0.56 |
ENST00000342665.2
|
SOX12
|
SRY (sex determining region Y)-box 12 |
| chr4_-_168155730 | 0.56 |
ENST00000502330.1
ENST00000357154.3 |
SPOCK3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr2_+_201171577 | 0.56 |
ENST00000409397.2
|
SPATS2L
|
spermatogenesis associated, serine-rich 2-like |
| chr12_+_50355647 | 0.55 |
ENST00000293599.6
|
AQP5
|
aquaporin 5 |
| chr20_+_306177 | 0.55 |
ENST00000544632.1
|
SOX12
|
SRY (sex determining region Y)-box 12 |
| chrX_-_130423240 | 0.54 |
ENST00000370910.1
ENST00000370901.4 |
IGSF1
|
immunoglobulin superfamily, member 1 |
| chr2_-_183903133 | 0.54 |
ENST00000361354.4
|
NCKAP1
|
NCK-associated protein 1 |
| chr15_-_82338460 | 0.54 |
ENST00000558133.1
ENST00000329713.4 |
MEX3B
|
mex-3 RNA binding family member B |
| chr2_+_27301435 | 0.53 |
ENST00000380320.4
|
EMILIN1
|
elastin microfibril interfacer 1 |
| chr1_+_26146674 | 0.53 |
ENST00000525713.1
ENST00000374301.3 |
MTFR1L
|
mitochondrial fission regulator 1-like |
| chr6_+_143381979 | 0.53 |
ENST00000367598.5
ENST00000447498.1 ENST00000357847.4 ENST00000344492.5 ENST00000367596.1 ENST00000494282.2 ENST00000275235.4 |
AIG1
|
androgen-induced 1 |
| chr2_+_66662249 | 0.53 |
ENST00000560281.2
|
MEIS1
|
Meis homeobox 1 |
| chr7_-_8302298 | 0.53 |
ENST00000446305.1
|
ICA1
|
islet cell autoantigen 1, 69kDa |
| chr13_+_98794810 | 0.53 |
ENST00000595437.1
|
FARP1
|
FERM, RhoGEF (ARHGEF) and pleckstrin domain protein 1 (chondrocyte-derived) |
| chr16_-_10674528 | 0.52 |
ENST00000359543.3
|
EMP2
|
epithelial membrane protein 2 |
| chr12_-_39837192 | 0.51 |
ENST00000361961.3
ENST00000395670.3 |
KIF21A
|
kinesin family member 21A |
| chr16_+_15528332 | 0.51 |
ENST00000566490.1
|
C16orf45
|
chromosome 16 open reading frame 45 |
| chr1_+_26146397 | 0.50 |
ENST00000374303.2
ENST00000533762.1 ENST00000529116.1 ENST00000474295.1 ENST00000488327.2 ENST00000472643.1 ENST00000526894.1 ENST00000524618.1 ENST00000374307.5 |
MTFR1L
|
mitochondrial fission regulator 1-like |
| chrX_-_119445306 | 0.50 |
ENST00000371369.4
ENST00000440464.1 ENST00000519908.1 |
TMEM255A
|
transmembrane protein 255A |
| chr4_-_168155417 | 0.50 |
ENST00000511269.1
ENST00000506697.1 ENST00000512042.1 |
SPOCK3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr4_+_159593418 | 0.50 |
ENST00000507475.1
ENST00000307738.5 |
ETFDH
|
electron-transferring-flavoprotein dehydrogenase |
| chr8_+_95653427 | 0.49 |
ENST00000454170.2
|
ESRP1
|
epithelial splicing regulatory protein 1 |
| chr12_+_41086215 | 0.49 |
ENST00000547702.1
ENST00000551424.1 |
CNTN1
|
contactin 1 |
| chr16_+_89894875 | 0.49 |
ENST00000393062.2
|
SPIRE2
|
spire-type actin nucleation factor 2 |
| chr19_+_55795493 | 0.48 |
ENST00000309383.1
|
BRSK1
|
BR serine/threonine kinase 1 |
| chr3_-_64431058 | 0.48 |
ENST00000564377.1
|
PRICKLE2
|
prickle homolog 2 (Drosophila) |
| chr16_+_55357672 | 0.48 |
ENST00000290552.7
|
IRX6
|
iroquois homeobox 6 |
| chrX_+_106163626 | 0.47 |
ENST00000336803.1
|
CLDN2
|
claudin 2 |
| chr12_+_27677085 | 0.47 |
ENST00000545334.1
ENST00000540114.1 ENST00000537927.1 ENST00000318304.8 ENST00000535047.1 ENST00000542629.1 ENST00000228425.6 |
PPFIBP1
|
PTPRF interacting protein, binding protein 1 (liprin beta 1) |
| chr2_+_201171064 | 0.46 |
ENST00000451764.2
|
SPATS2L
|
spermatogenesis associated, serine-rich 2-like |
| chrX_-_119445263 | 0.46 |
ENST00000309720.5
|
TMEM255A
|
transmembrane protein 255A |
| chr3_-_52567792 | 0.46 |
ENST00000307092.4
ENST00000422318.2 ENST00000459839.1 |
NT5DC2
|
5'-nucleotidase domain containing 2 |
| chrX_+_19362011 | 0.46 |
ENST00000379806.5
ENST00000545074.1 ENST00000540249.1 ENST00000423505.1 ENST00000417819.1 ENST00000422285.2 ENST00000355808.5 ENST00000379805.3 |
PDHA1
|
pyruvate dehydrogenase (lipoamide) alpha 1 |
| chr9_+_129987488 | 0.45 |
ENST00000446764.1
|
GARNL3
|
GTPase activating Rap/RanGAP domain-like 3 |
| chr3_-_169381166 | 0.45 |
ENST00000486748.1
|
MECOM
|
MDS1 and EVI1 complex locus |
| chr2_-_96812751 | 0.45 |
ENST00000449242.1
|
AC012307.2
|
AC012307.2 |
| chr19_+_41036371 | 0.44 |
ENST00000392023.1
|
SPTBN4
|
spectrin, beta, non-erythrocytic 4 |
| chr6_-_53530474 | 0.44 |
ENST00000370905.3
|
KLHL31
|
kelch-like family member 31 |
| chr17_+_40811283 | 0.44 |
ENST00000251412.7
|
TUBG2
|
tubulin, gamma 2 |
| chr1_+_155247207 | 0.43 |
ENST00000368358.3
|
HCN3
|
hyperpolarization activated cyclic nucleotide-gated potassium channel 3 |
| chr19_+_12305824 | 0.43 |
ENST00000415793.1
ENST00000440004.1 ENST00000426044.1 |
CTD-2666L21.1
|
CTD-2666L21.1 |
| chr3_+_140770183 | 0.43 |
ENST00000310546.2
|
SPSB4
|
splA/ryanodine receptor domain and SOCS box containing 4 |
| chr1_+_110754094 | 0.43 |
ENST00000369787.3
ENST00000413138.3 ENST00000438661.2 |
KCNC4
|
potassium voltage-gated channel, Shaw-related subfamily, member 4 |
| chr11_-_117667806 | 0.42 |
ENST00000527706.1
ENST00000321322.6 |
DSCAML1
|
Down syndrome cell adhesion molecule like 1 |
| chr2_+_204193149 | 0.42 |
ENST00000422511.2
|
ABI2
|
abl-interactor 2 |
| chr5_-_42811986 | 0.41 |
ENST00000511224.1
ENST00000507920.1 ENST00000510965.1 |
SEPP1
|
selenoprotein P, plasma, 1 |
| chr2_+_220306238 | 0.41 |
ENST00000435853.1
|
SPEG
|
SPEG complex locus |
| chr19_+_49660997 | 0.41 |
ENST00000598691.1
ENST00000252826.5 |
TRPM4
|
transient receptor potential cation channel, subfamily M, member 4 |
| chr1_-_46598371 | 0.40 |
ENST00000372006.1
ENST00000425892.1 ENST00000420542.1 ENST00000354242.4 ENST00000340332.6 |
PIK3R3
|
phosphoinositide-3-kinase, regulatory subunit 3 (gamma) |
| chr7_-_8302164 | 0.40 |
ENST00000447326.1
ENST00000406470.2 |
ICA1
|
islet cell autoantigen 1, 69kDa |
| chr4_-_87278857 | 0.40 |
ENST00000509464.1
ENST00000511167.1 |
MAPK10
|
mitogen-activated protein kinase 10 |
| chr2_+_159313452 | 0.40 |
ENST00000389757.3
ENST00000389759.3 |
PKP4
|
plakophilin 4 |
| chr17_+_7308172 | 0.39 |
ENST00000575301.1
|
NLGN2
|
neuroligin 2 |
| chr12_+_56661033 | 0.39 |
ENST00000433805.2
|
COQ10A
|
coenzyme Q10 homolog A (S. cerevisiae) |
| chr6_-_110500826 | 0.38 |
ENST00000265601.3
ENST00000447287.1 ENST00000444391.1 |
WASF1
|
WAS protein family, member 1 |
| chr19_-_51466681 | 0.38 |
ENST00000456750.2
|
KLK6
|
kallikrein-related peptidase 6 |
| chrX_-_130423386 | 0.38 |
ENST00000370903.3
|
IGSF1
|
immunoglobulin superfamily, member 1 |
| chr12_+_56661461 | 0.38 |
ENST00000546544.1
ENST00000553234.1 |
COQ10A
|
coenzyme Q10 homolog A (S. cerevisiae) |
| chr3_+_128720424 | 0.38 |
ENST00000480450.1
ENST00000436022.2 |
EFCC1
|
EF-hand and coiled-coil domain containing 1 |
| chr11_-_63536113 | 0.38 |
ENST00000433688.1
ENST00000546282.2 |
C11orf95
RP11-466C23.4
|
chromosome 11 open reading frame 95 RP11-466C23.4 |
| chr12_+_107168342 | 0.38 |
ENST00000392837.4
|
RIC8B
|
RIC8 guanine nucleotide exchange factor B |
| chr7_+_103969104 | 0.37 |
ENST00000424859.1
ENST00000535008.1 ENST00000401970.2 ENST00000543266.1 |
LHFPL3
|
lipoma HMGIC fusion partner-like 3 |
| chr11_-_118023490 | 0.37 |
ENST00000324727.4
|
SCN4B
|
sodium channel, voltage-gated, type IV, beta subunit |
| chr13_+_33590553 | 0.36 |
ENST00000380099.3
|
KL
|
klotho |
| chr1_+_180601139 | 0.35 |
ENST00000367590.4
ENST00000367589.3 |
XPR1
|
xenotropic and polytropic retrovirus receptor 1 |
| chr12_+_107168418 | 0.35 |
ENST00000392839.2
ENST00000548914.1 ENST00000355478.2 ENST00000552619.1 ENST00000549643.1 |
RIC8B
|
RIC8 guanine nucleotide exchange factor B |
| chr14_-_27066960 | 0.35 |
ENST00000539517.2
|
NOVA1
|
neuro-oncological ventral antigen 1 |
| chr1_-_177133998 | 0.35 |
ENST00000367657.3
|
ASTN1
|
astrotactin 1 |
| chr20_+_49348109 | 0.35 |
ENST00000396039.1
|
PARD6B
|
par-6 family cell polarity regulator beta |
| chr5_+_121647764 | 0.35 |
ENST00000261368.8
ENST00000379533.2 ENST00000379536.2 ENST00000379538.3 |
SNCAIP
|
synuclein, alpha interacting protein |
| chr7_+_149411860 | 0.34 |
ENST00000486744.1
|
KRBA1
|
KRAB-A domain containing 1 |
| chr17_+_68101117 | 0.33 |
ENST00000587698.1
ENST00000587892.1 |
KCNJ16
|
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr7_-_8301682 | 0.33 |
ENST00000396675.3
ENST00000430867.1 |
ICA1
|
islet cell autoantigen 1, 69kDa |
| chr5_+_169780485 | 0.33 |
ENST00000377360.4
|
KCNIP1
|
Kv channel interacting protein 1 |
| chr16_+_2014993 | 0.33 |
ENST00000564014.1
|
SNHG9
|
small nucleolar RNA host gene 9 (non-protein coding) |
| chr6_-_114664180 | 0.32 |
ENST00000312719.5
|
HS3ST5
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 5 |
| chr1_+_52608046 | 0.32 |
ENST00000357206.2
ENST00000287727.3 |
ZFYVE9
|
zinc finger, FYVE domain containing 9 |
| chr2_+_204193129 | 0.32 |
ENST00000417864.1
|
ABI2
|
abl-interactor 2 |
| chr16_-_28937027 | 0.32 |
ENST00000358201.4
|
RABEP2
|
rabaptin, RAB GTPase binding effector protein 2 |
| chr1_+_244816237 | 0.31 |
ENST00000302550.11
|
DESI2
|
desumoylating isopeptidase 2 |
| chr1_+_44401479 | 0.31 |
ENST00000438616.3
|
ARTN
|
artemin |
| chr3_-_33260707 | 0.30 |
ENST00000309558.3
|
SUSD5
|
sushi domain containing 5 |
| chr4_-_177713788 | 0.30 |
ENST00000280193.2
|
VEGFC
|
vascular endothelial growth factor C |
| chr10_-_79397316 | 0.30 |
ENST00000372421.5
ENST00000457953.1 |
KCNMA1
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr7_-_8302207 | 0.29 |
ENST00000407906.1
|
ICA1
|
islet cell autoantigen 1, 69kDa |
| chr15_-_83953466 | 0.29 |
ENST00000345382.2
|
BNC1
|
basonuclin 1 |
| chr1_-_153958805 | 0.28 |
ENST00000368575.3
|
RAB13
|
RAB13, member RAS oncogene family |
| chr12_+_56473910 | 0.28 |
ENST00000411731.2
|
ERBB3
|
v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 3 |
| chr1_-_46598284 | 0.28 |
ENST00000423209.1
ENST00000262741.5 |
PIK3R3
|
phosphoinositide-3-kinase, regulatory subunit 3 (gamma) |
| chr13_+_47127293 | 0.28 |
ENST00000311191.6
|
LRCH1
|
leucine-rich repeats and calponin homology (CH) domain containing 1 |
| chr2_+_204192942 | 0.28 |
ENST00000295851.5
ENST00000261017.5 |
ABI2
|
abl-interactor 2 |
| chr15_+_74422585 | 0.27 |
ENST00000561740.1
ENST00000435464.1 ENST00000453268.2 |
ISLR2
|
immunoglobulin superfamily containing leucine-rich repeat 2 |
| chr2_+_166152283 | 0.27 |
ENST00000375427.2
|
SCN2A
|
sodium channel, voltage-gated, type II, alpha subunit |
| chr7_-_559853 | 0.26 |
ENST00000405692.2
|
PDGFA
|
platelet-derived growth factor alpha polypeptide |
| chr8_-_42396721 | 0.26 |
ENST00000518384.1
|
SLC20A2
|
solute carrier family 20 (phosphate transporter), member 2 |
| chr8_-_142011291 | 0.26 |
ENST00000521059.1
|
PTK2
|
protein tyrosine kinase 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of NR0B1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 3.1 | GO:1990029 | atrial ventricular junction remodeling(GO:0003294) atrial cardiac muscle cell to AV node cell communication by electrical coupling(GO:0086044) bundle of His cell to Purkinje myocyte communication by electrical coupling(GO:0086054) Purkinje myocyte to ventricular cardiac muscle cell communication by electrical coupling(GO:0086055) regulation of Purkinje myocyte action potential(GO:0098906) vasomotion(GO:1990029) |
| 0.8 | 2.4 | GO:0007388 | anterior compartment pattern formation(GO:0007387) posterior compartment specification(GO:0007388) |
| 0.7 | 5.5 | GO:1990262 | regulation of anti-Mullerian hormone signaling pathway(GO:1902612) negative regulation of anti-Mullerian hormone signaling pathway(GO:1902613) anti-Mullerian hormone signaling pathway(GO:1990262) |
| 0.7 | 2.0 | GO:0016539 | intein-mediated protein splicing(GO:0016539) protein splicing(GO:0030908) |
| 0.6 | 1.9 | GO:0051466 | positive regulation of corticotropin-releasing hormone secretion(GO:0051466) |
| 0.6 | 3.1 | GO:0072233 | thick ascending limb development(GO:0072023) metanephric thick ascending limb development(GO:0072233) |
| 0.4 | 4.7 | GO:0051552 | flavone metabolic process(GO:0051552) |
| 0.4 | 1.2 | GO:2000824 | negative regulation of androgen receptor activity(GO:2000824) |
| 0.4 | 0.7 | GO:0072194 | kidney smooth muscle tissue development(GO:0072194) |
| 0.3 | 1.9 | GO:0060214 | endocardium formation(GO:0060214) |
| 0.3 | 0.9 | GO:0010621 | negative regulation of transcription by transcription factor localization(GO:0010621) |
| 0.3 | 1.2 | GO:1904199 | positive regulation of regulation of vascular smooth muscle cell membrane depolarization(GO:1904199) regulation of vascular smooth muscle cell membrane depolarization(GO:1990736) |
| 0.3 | 1.4 | GO:0015688 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 0.2 | 1.7 | GO:0097461 | ferric iron import into cell(GO:0097461) ferric iron import across plasma membrane(GO:0098706) |
| 0.2 | 0.4 | GO:1904862 | inhibitory synapse assembly(GO:1904862) |
| 0.2 | 3.2 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.2 | 0.6 | GO:0061386 | closure of optic fissure(GO:0061386) |
| 0.2 | 0.6 | GO:0008057 | eye pigment granule organization(GO:0008057) |
| 0.2 | 0.5 | GO:1903515 | calcium ion transport from cytosol to endoplasmic reticulum(GO:1903515) |
| 0.2 | 2.6 | GO:1904321 | response to forskolin(GO:1904321) cellular response to forskolin(GO:1904322) |
| 0.2 | 0.6 | GO:0048170 | positive regulation of long-term neuronal synaptic plasticity(GO:0048170) |
| 0.1 | 3.1 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 0.1 | 0.6 | GO:0014859 | negative regulation of skeletal muscle cell proliferation(GO:0014859) negative regulation of skeletal muscle satellite cell proliferation(GO:1902723) |
| 0.1 | 1.3 | GO:0086046 | membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.1 | 1.9 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.1 | 0.8 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.1 | 0.3 | GO:0010512 | negative regulation of phosphatidylinositol biosynthetic process(GO:0010512) |
| 0.1 | 1.0 | GO:0098912 | membrane depolarization during AV node cell action potential(GO:0086045) membrane depolarization during atrial cardiac muscle cell action potential(GO:0098912) positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.1 | 1.3 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.1 | 1.9 | GO:0021860 | pyramidal neuron development(GO:0021860) |
| 0.1 | 1.2 | GO:0001714 | endodermal cell fate specification(GO:0001714) |
| 0.1 | 1.5 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.1 | 0.6 | GO:2000051 | Wnt receptor catabolic process(GO:0038018) negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.1 | 0.6 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.1 | 1.6 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.1 | 1.2 | GO:0098914 | membrane repolarization during atrial cardiac muscle cell action potential(GO:0098914) |
| 0.1 | 0.3 | GO:1902463 | protein localization to cell leading edge(GO:1902463) |
| 0.1 | 0.6 | GO:0086027 | AV node cell action potential(GO:0086016) AV node cell to bundle of His cell signaling(GO:0086027) |
| 0.1 | 0.6 | GO:0071603 | endothelial cell-cell adhesion(GO:0071603) |
| 0.1 | 0.3 | GO:0097359 | UDP-glucosylation(GO:0097359) |
| 0.1 | 0.4 | GO:0051620 | negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of glutathione peroxidase activity(GO:1903282) positive regulation of glutathione peroxidase activity(GO:1903284) positive regulation of hydrogen peroxide catabolic process(GO:1903285) positive regulation of peroxidase activity(GO:2000470) |
| 0.1 | 0.3 | GO:0070631 | spindle pole body duplication(GO:0030474) spindle pole body organization(GO:0051300) spindle pole body localization(GO:0070631) establishment of spindle pole body localization(GO:0070632) spindle pole body localization to nuclear envelope(GO:0071789) establishment of spindle pole body localization to nuclear envelope(GO:0071790) |
| 0.1 | 1.9 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.1 | 0.8 | GO:2000467 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) positive regulation of glycogen (starch) synthase activity(GO:2000467) |
| 0.1 | 0.7 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.1 | 0.3 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.1 | 0.3 | GO:0016598 | protein arginylation(GO:0016598) |
| 0.1 | 0.5 | GO:1903976 | negative regulation of glial cell migration(GO:1903976) |
| 0.1 | 4.8 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.1 | 0.5 | GO:0040038 | polar body extrusion after meiotic divisions(GO:0040038) formin-nucleated actin cable assembly(GO:0070649) |
| 0.1 | 1.8 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.1 | 0.3 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.1 | 0.3 | GO:0097021 | lymphocyte migration into lymphoid organs(GO:0097021) |
| 0.0 | 0.3 | GO:1901490 | regulation of lymphangiogenesis(GO:1901490) |
| 0.0 | 0.6 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 1.2 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.0 | 0.2 | GO:0044828 | negative regulation by host of viral genome replication(GO:0044828) |
| 0.0 | 0.5 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.0 | 0.4 | GO:0090080 | positive regulation of MAPKKK cascade by fibroblast growth factor receptor signaling pathway(GO:0090080) |
| 0.0 | 0.5 | GO:0070836 | caveola assembly(GO:0070836) |
| 0.0 | 0.8 | GO:0061577 | calcium ion transmembrane transport via high voltage-gated calcium channel(GO:0061577) |
| 0.0 | 2.3 | GO:0007190 | activation of adenylate cyclase activity(GO:0007190) |
| 0.0 | 0.2 | GO:0046726 | positive regulation by virus of viral protein levels in host cell(GO:0046726) |
| 0.0 | 1.3 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.0 | 0.4 | GO:0072502 | cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.0 | 0.7 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.2 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.0 | 1.0 | GO:0003215 | cardiac right ventricle morphogenesis(GO:0003215) |
| 0.0 | 0.1 | GO:0051808 | translocation of peptides or proteins into host(GO:0042000) translocation of peptides or proteins into host cell cytoplasm(GO:0044053) translocation of molecules into host(GO:0044417) translocation of peptides or proteins into other organism involved in symbiotic interaction(GO:0051808) translocation of molecules into other organism involved in symbiotic interaction(GO:0051836) |
| 0.0 | 0.4 | GO:0072718 | response to cisplatin(GO:0072718) |
| 0.0 | 0.2 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.0 | 0.1 | GO:0003365 | establishment of cell polarity involved in ameboidal cell migration(GO:0003365) |
| 0.0 | 0.2 | GO:0045602 | negative regulation of endothelial cell differentiation(GO:0045602) |
| 0.0 | 0.7 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.0 | 0.4 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.8 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.0 | 0.7 | GO:0050962 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 | 1.6 | GO:0033120 | positive regulation of RNA splicing(GO:0033120) |
| 0.0 | 0.9 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.0 | 0.1 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.0 | 0.2 | GO:0019470 | 4-hydroxyproline catabolic process(GO:0019470) |
| 0.0 | 0.3 | GO:0034465 | response to carbon monoxide(GO:0034465) |
| 0.0 | 0.2 | GO:0016080 | synaptic vesicle targeting(GO:0016080) |
| 0.0 | 0.5 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 | 0.3 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.0 | 1.4 | GO:0010765 | positive regulation of sodium ion transport(GO:0010765) |
| 0.0 | 0.7 | GO:2001275 | positive regulation of glucose import in response to insulin stimulus(GO:2001275) |
| 0.0 | 0.2 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.0 | 0.2 | GO:0032088 | negative regulation of NF-kappaB transcription factor activity(GO:0032088) |
| 0.0 | 0.1 | GO:0016267 | O-glycan processing, core 1(GO:0016267) |
| 0.0 | 0.1 | GO:0097116 | gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 0.0 | 1.3 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.0 | 0.4 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.0 | 0.3 | GO:0008627 | intrinsic apoptotic signaling pathway in response to osmotic stress(GO:0008627) |
| 0.0 | 1.0 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.0 | 1.3 | GO:0006378 | mRNA polyadenylation(GO:0006378) |
| 0.0 | 0.8 | GO:0046329 | negative regulation of JNK cascade(GO:0046329) |
| 0.0 | 0.5 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.0 | 0.5 | GO:0007020 | microtubule nucleation(GO:0007020) |
| 0.0 | 0.4 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.0 | 0.5 | GO:0060441 | epithelial tube branching involved in lung morphogenesis(GO:0060441) |
| 0.0 | 0.5 | GO:0090083 | regulation of inclusion body assembly(GO:0090083) |
| 0.0 | 0.5 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 1.0 | GO:0030049 | muscle filament sliding(GO:0030049) actin-myosin filament sliding(GO:0033275) |
| 0.0 | 0.1 | GO:0032099 | negative regulation of response to food(GO:0032096) negative regulation of appetite(GO:0032099) |
| 0.0 | 0.4 | GO:0046928 | regulation of neurotransmitter secretion(GO:0046928) |
| 0.0 | 0.0 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.0 | 1.8 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 | 0.1 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.0 | 0.2 | GO:0006108 | malate metabolic process(GO:0006108) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:0098855 | HCN channel complex(GO:0098855) |
| 0.2 | 4.0 | GO:0045180 | basal cortex(GO:0045180) |
| 0.1 | 3.1 | GO:0005922 | connexon complex(GO:0005922) |
| 0.1 | 1.9 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 1.8 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.1 | 2.7 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.1 | 0.3 | GO:0070762 | nuclear pore transmembrane ring(GO:0070762) |
| 0.1 | 2.9 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.1 | 0.3 | GO:0097224 | sperm connecting piece(GO:0097224) |
| 0.1 | 2.5 | GO:0034706 | sodium channel complex(GO:0034706) |
| 0.1 | 1.4 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 1.0 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 0.2 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.0 | 0.2 | GO:0045257 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.0 | 2.2 | GO:0005605 | basal lamina(GO:0005605) |
| 0.0 | 0.5 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.0 | 0.4 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 1.7 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 0.7 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.2 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.0 | 0.3 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.5 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.0 | 0.6 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.8 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.9 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.3 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 2.1 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 2.3 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 0.6 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.0 | 3.4 | GO:0030863 | cortical cytoskeleton(GO:0030863) |
| 0.0 | 1.2 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 0.6 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 2.2 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.6 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.1 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.0 | 0.3 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.0 | 0.6 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.2 | GO:0042641 | actomyosin(GO:0042641) |
| 0.0 | 0.2 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 0.5 | GO:0031304 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.5 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.2 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.0 | 0.9 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.0 | GO:1990923 | PET complex(GO:1990923) |
| 0.0 | 0.4 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.4 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 1.4 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 0.2 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 1.4 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 3.1 | GO:0086079 | gap junction channel activity involved in atrial cardiac muscle cell-AV node cell electrical coupling(GO:0086076) gap junction channel activity involved in bundle of His cell-Purkinje myocyte electrical coupling(GO:0086078) gap junction channel activity involved in Purkinje myocyte-ventricular cardiac muscle cell electrical coupling(GO:0086079) |
| 0.7 | 2.7 | GO:0052810 | 1-phosphatidylinositol-5-kinase activity(GO:0052810) |
| 0.5 | 2.2 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.3 | 0.9 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) |
| 0.3 | 1.5 | GO:0097001 | ceramide binding(GO:0097001) |
| 0.3 | 1.3 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.2 | 1.7 | GO:0008823 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.2 | 4.8 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.2 | 2.2 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.2 | 0.7 | GO:0048763 | HLH domain binding(GO:0043398) calcium-induced calcium release activity(GO:0048763) |
| 0.2 | 0.5 | GO:0098640 | integrin binding involved in cell-matrix adhesion(GO:0098640) |
| 0.2 | 1.2 | GO:0086089 | voltage-gated potassium channel activity involved in atrial cardiac muscle cell action potential repolarization(GO:0086089) |
| 0.2 | 0.5 | GO:0016649 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.2 | 4.8 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.2 | 0.5 | GO:0086039 | lutropin-choriogonadotropic hormone receptor binding(GO:0031775) calcium-transporting ATPase activity involved in regulation of cardiac muscle cell membrane potential(GO:0086039) |
| 0.1 | 1.8 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.1 | 0.5 | GO:0097108 | hedgehog family protein binding(GO:0097108) |
| 0.1 | 2.0 | GO:0005113 | patched binding(GO:0005113) |
| 0.1 | 1.5 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.1 | 0.9 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.1 | 1.0 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.1 | 0.9 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.1 | 0.5 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.1 | 1.4 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 0.1 | 0.6 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.1 | 3.1 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.1 | 4.7 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 0.4 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.1 | 0.4 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.1 | 0.3 | GO:0003980 | UDP-glucose:glycoprotein glucosyltransferase activity(GO:0003980) |
| 0.1 | 0.4 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.1 | 0.2 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.1 | 0.6 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.1 | 1.2 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.1 | 0.3 | GO:0004057 | arginyltransferase activity(GO:0004057) |
| 0.1 | 0.6 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.1 | 1.2 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.1 | 0.2 | GO:0003842 | 1-pyrroline-5-carboxylate dehydrogenase activity(GO:0003842) |
| 0.0 | 1.8 | GO:0005272 | sodium channel activity(GO:0005272) |
| 0.0 | 0.8 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 1.3 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 1.4 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.2 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 0.2 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.0 | 0.3 | GO:0050656 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) 3'-phosphoadenosine 5'-phosphosulfate binding(GO:0050656) |
| 0.0 | 0.6 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.4 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 2.1 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 0.6 | GO:0015250 | water channel activity(GO:0015250) |
| 0.0 | 1.0 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.0 | 0.3 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.0 | 0.3 | GO:0034597 | phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.0 | 0.8 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.8 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 0.1 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.0 | 0.7 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 1.0 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.0 | 0.3 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.0 | 0.3 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.0 | 0.9 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 0.1 | GO:0016263 | glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase activity(GO:0016263) |
| 0.0 | 0.3 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.2 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.0 | 0.2 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.5 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.5 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.4 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 1.4 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 0.3 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.0 | 2.3 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.2 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.3 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.7 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.1 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.4 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.1 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.0 | 0.4 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.3 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 0.3 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.1 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.0 | 0.3 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.0 | 0.1 | GO:0003696 | satellite DNA binding(GO:0003696) |
| 0.0 | 0.9 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.0 | 0.1 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.6 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.1 | 2.9 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.1 | 5.5 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 2.4 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 1.8 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 3.3 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 2.2 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 1.6 | ST ADRENERGIC | Adrenergic Pathway |
| 0.0 | 1.3 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 1.6 | PID ERBB1 RECEPTOR PROXIMAL PATHWAY | EGF receptor (ErbB1) signaling pathway |
| 0.0 | 1.7 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 1.3 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.3 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 1.1 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 1.1 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 1.3 | PID P53 REGULATION PATHWAY | p53 pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.9 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.1 | 3.1 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.1 | 1.2 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.1 | 2.0 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.1 | 1.7 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.1 | 1.4 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.1 | 1.5 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 1.8 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 2.8 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 1.8 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.0 | 0.6 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 1.9 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 1.7 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 0.9 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 2.1 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.8 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.4 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 0.5 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 1.0 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.5 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.4 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.5 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 1.0 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.3 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 2.3 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.3 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.3 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.3 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |