Project
Illumina Body Map 2: averaged replicates
Navigation
Downloads
Results for NR1H4
Z-value: 1.06
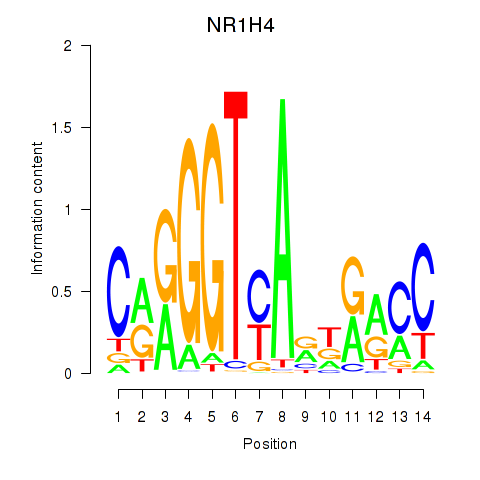
Transcription factors associated with NR1H4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NR1H4
|
ENSG00000012504.9 | nuclear receptor subfamily 1 group H member 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NR1H4 | hg19_v2_chr12_+_100867733_100867750 | 0.18 | 3.2e-01 | Click! |
Activity profile of NR1H4 motif
Sorted Z-values of NR1H4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_116663127 | 3.75 |
ENST00000433069.1
ENST00000542499.1 |
APOA5
|
apolipoprotein A-V |
| chr9_-_137809718 | 3.14 |
ENST00000371806.3
|
FCN1
|
ficolin (collagen/fibrinogen domain containing) 1 |
| chr20_+_30639991 | 2.41 |
ENST00000534862.1
ENST00000538448.1 ENST00000375862.2 |
HCK
|
hemopoietic cell kinase |
| chr14_-_25103472 | 2.41 |
ENST00000216341.4
ENST00000382542.1 ENST00000382540.1 |
GZMB
|
granzyme B (granzyme 2, cytotoxic T-lymphocyte-associated serine esterase 1) |
| chr20_+_30640004 | 2.37 |
ENST00000520553.1
ENST00000518730.1 ENST00000375852.2 |
HCK
|
hemopoietic cell kinase |
| chr19_+_7741968 | 2.33 |
ENST00000597445.1
|
C19orf59
|
chromosome 19 open reading frame 59 |
| chr17_+_43318434 | 2.20 |
ENST00000587489.1
|
FMNL1
|
formin-like 1 |
| chr22_+_22786288 | 2.17 |
ENST00000390301.2
|
IGLV1-36
|
immunoglobulin lambda variable 1-36 |
| chr14_-_25103388 | 2.09 |
ENST00000526004.1
ENST00000415355.3 |
GZMB
|
granzyme B (granzyme 2, cytotoxic T-lymphocyte-associated serine esterase 1) |
| chr20_+_62369623 | 2.01 |
ENST00000467211.1
|
RP4-583P15.14
|
RP4-583P15.14 |
| chr3_+_111260856 | 1.88 |
ENST00000352690.4
|
CD96
|
CD96 molecule |
| chr14_+_22356029 | 1.85 |
ENST00000390437.2
|
TRAV12-2
|
T cell receptor alpha variable 12-2 |
| chr3_+_111260954 | 1.85 |
ENST00000283285.5
|
CD96
|
CD96 molecule |
| chr11_-_64511575 | 1.76 |
ENST00000431822.1
ENST00000377486.3 ENST00000394432.3 |
RASGRP2
|
RAS guanyl releasing protein 2 (calcium and DAG-regulated) |
| chr18_-_67624160 | 1.75 |
ENST00000581982.1
ENST00000280200.4 |
CD226
|
CD226 molecule |
| chr14_+_22314715 | 1.74 |
ENST00000390434.3
|
TRAV8-2
|
T cell receptor alpha variable 8-2 |
| chr5_+_133450365 | 1.73 |
ENST00000342854.5
ENST00000321603.6 ENST00000321584.4 ENST00000378564.1 ENST00000395029.1 |
TCF7
|
transcription factor 7 (T-cell specific, HMG-box) |
| chr17_-_73840614 | 1.69 |
ENST00000586108.1
|
UNC13D
|
unc-13 homolog D (C. elegans) |
| chr12_-_7281469 | 1.68 |
ENST00000542370.1
ENST00000266560.3 |
RBP5
|
retinol binding protein 5, cellular |
| chr3_+_111260980 | 1.67 |
ENST00000438817.2
|
CD96
|
CD96 molecule |
| chr19_+_1071203 | 1.64 |
ENST00000543365.1
|
HMHA1
|
histocompatibility (minor) HA-1 |
| chr17_-_73840415 | 1.63 |
ENST00000592386.1
ENST00000412096.2 ENST00000586147.1 |
UNC13D
|
unc-13 homolog D (C. elegans) |
| chr12_-_68845165 | 1.61 |
ENST00000360485.3
ENST00000441255.2 |
RP11-81H14.2
|
RP11-81H14.2 |
| chr10_-_95360983 | 1.52 |
ENST00000371464.3
|
RBP4
|
retinol binding protein 4, plasma |
| chr22_+_23247030 | 1.48 |
ENST00000390324.2
|
IGLJ3
|
immunoglobulin lambda joining 3 |
| chr2_+_192542879 | 1.44 |
ENST00000409510.1
|
NABP1
|
nucleic acid binding protein 1 |
| chr17_-_73840774 | 1.43 |
ENST00000207549.4
|
UNC13D
|
unc-13 homolog D (C. elegans) |
| chr17_-_73839792 | 1.42 |
ENST00000590762.1
|
UNC13D
|
unc-13 homolog D (C. elegans) |
| chr7_+_2671663 | 1.39 |
ENST00000407643.1
|
TTYH3
|
tweety family member 3 |
| chr16_+_23847339 | 1.36 |
ENST00000303531.7
|
PRKCB
|
protein kinase C, beta |
| chr2_+_192543153 | 1.26 |
ENST00000425611.2
|
NABP1
|
nucleic acid binding protein 1 |
| chr2_+_192543694 | 1.25 |
ENST00000435931.1
|
NABP1
|
nucleic acid binding protein 1 |
| chr19_-_42636617 | 1.24 |
ENST00000529067.1
ENST00000529952.1 ENST00000533720.1 ENST00000389341.5 ENST00000342301.4 |
POU2F2
|
POU class 2 homeobox 2 |
| chr1_-_25256368 | 1.22 |
ENST00000308873.6
|
RUNX3
|
runt-related transcription factor 3 |
| chr16_+_23847267 | 1.20 |
ENST00000321728.7
|
PRKCB
|
protein kinase C, beta |
| chr19_-_11689752 | 1.20 |
ENST00000592659.1
ENST00000592828.1 ENST00000218758.5 ENST00000412435.2 |
ACP5
|
acid phosphatase 5, tartrate resistant |
| chr3_+_52828805 | 1.19 |
ENST00000416872.2
ENST00000449956.2 |
ITIH3
|
inter-alpha-trypsin inhibitor heavy chain 3 |
| chr1_+_28206150 | 1.18 |
ENST00000456990.1
|
THEMIS2
|
thymocyte selection associated family member 2 |
| chr3_+_122296443 | 1.12 |
ENST00000464300.2
|
PARP15
|
poly (ADP-ribose) polymerase family, member 15 |
| chr18_-_67623906 | 1.08 |
ENST00000583955.1
|
CD226
|
CD226 molecule |
| chr12_+_6898674 | 1.07 |
ENST00000541982.1
ENST00000539492.1 |
CD4
|
CD4 molecule |
| chr19_+_1026566 | 1.05 |
ENST00000348419.3
ENST00000565096.2 ENST00000562958.2 ENST00000562075.2 ENST00000607102.1 |
CNN2
|
calponin 2 |
| chr22_+_45072925 | 1.04 |
ENST00000006251.7
|
PRR5
|
proline rich 5 (renal) |
| chr2_-_182545603 | 1.04 |
ENST00000295108.3
|
NEUROD1
|
neuronal differentiation 1 |
| chr12_+_6898638 | 1.03 |
ENST00000011653.4
|
CD4
|
CD4 molecule |
| chr2_+_192542850 | 1.01 |
ENST00000410026.2
|
NABP1
|
nucleic acid binding protein 1 |
| chr12_+_109273806 | 1.00 |
ENST00000228476.3
ENST00000547768.1 |
DAO
|
D-amino-acid oxidase |
| chr16_+_2014941 | 0.99 |
ENST00000531523.1
|
SNHG9
|
small nucleolar RNA host gene 9 (non-protein coding) |
| chr2_-_64371546 | 0.99 |
ENST00000358912.4
|
PELI1
|
pellino E3 ubiquitin protein ligase 1 |
| chr18_-_67624412 | 0.97 |
ENST00000580335.1
|
CD226
|
CD226 molecule |
| chr16_+_23847355 | 0.97 |
ENST00000498058.1
|
PRKCB
|
protein kinase C, beta |
| chr2_+_233877324 | 0.95 |
ENST00000409905.1
|
AC106876.2
|
Uncharacterized protein |
| chr9_+_127023704 | 0.93 |
ENST00000373596.1
ENST00000425237.1 |
NEK6
|
NIMA-related kinase 6 |
| chr8_-_131028660 | 0.88 |
ENST00000401979.2
ENST00000517654.1 ENST00000522361.1 ENST00000518167.1 |
FAM49B
|
family with sequence similarity 49, member B |
| chr14_+_21492331 | 0.86 |
ENST00000533984.1
ENST00000532213.2 |
AL161668.5
|
AL161668.5 |
| chr19_+_1026298 | 0.85 |
ENST00000263097.4
|
CNN2
|
calponin 2 |
| chr7_+_2671568 | 0.85 |
ENST00000258796.7
|
TTYH3
|
tweety family member 3 |
| chr22_+_45072958 | 0.84 |
ENST00000403581.1
|
PRR5
|
proline rich 5 (renal) |
| chr11_-_65624415 | 0.83 |
ENST00000524553.1
ENST00000527344.1 |
CFL1
|
cofilin 1 (non-muscle) |
| chr11_-_65625014 | 0.81 |
ENST00000534784.1
|
CFL1
|
cofilin 1 (non-muscle) |
| chr19_+_10197463 | 0.81 |
ENST00000590378.1
ENST00000397881.3 |
C19orf66
|
chromosome 19 open reading frame 66 |
| chr1_+_156024525 | 0.81 |
ENST00000368305.4
|
LAMTOR2
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 2 |
| chr1_+_29241027 | 0.80 |
ENST00000373797.1
|
EPB41
|
erythrocyte membrane protein band 4.1 (elliptocytosis 1, RH-linked) |
| chr1_+_162351503 | 0.79 |
ENST00000458626.2
|
C1orf226
|
chromosome 1 open reading frame 226 |
| chr1_-_27240455 | 0.77 |
ENST00000254227.3
|
NR0B2
|
nuclear receptor subfamily 0, group B, member 2 |
| chr22_+_20105012 | 0.74 |
ENST00000331821.3
ENST00000411892.1 |
RANBP1
|
RAN binding protein 1 |
| chr19_-_36304201 | 0.73 |
ENST00000301175.3
|
PRODH2
|
proline dehydrogenase (oxidase) 2 |
| chr19_+_19496624 | 0.71 |
ENST00000494516.2
ENST00000360315.3 ENST00000252577.5 |
GATAD2A
|
GATA zinc finger domain containing 2A |
| chr22_+_20104947 | 0.71 |
ENST00000402752.1
|
RANBP1
|
RAN binding protein 1 |
| chr16_+_81478775 | 0.70 |
ENST00000537098.3
|
CMIP
|
c-Maf inducing protein |
| chr1_-_149908217 | 0.68 |
ENST00000369140.3
|
MTMR11
|
myotubularin related protein 11 |
| chr11_-_62380199 | 0.68 |
ENST00000419857.1
ENST00000394773.2 |
EML3
|
echinoderm microtubule associated protein like 3 |
| chr1_+_156024552 | 0.68 |
ENST00000368304.5
ENST00000368302.3 |
LAMTOR2
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 2 |
| chr11_-_118305921 | 0.66 |
ENST00000532619.1
|
RP11-770J1.4
|
RP11-770J1.4 |
| chr8_-_131028782 | 0.66 |
ENST00000519020.1
|
FAM49B
|
family with sequence similarity 49, member B |
| chr14_-_65769392 | 0.65 |
ENST00000555736.1
|
CTD-2509G16.5
|
CTD-2509G16.5 |
| chr22_+_20105259 | 0.65 |
ENST00000416427.1
ENST00000421656.1 ENST00000423859.1 ENST00000418705.2 |
RANBP1
|
RAN binding protein 1 |
| chr12_+_120875887 | 0.65 |
ENST00000229379.2
|
COX6A1
|
cytochrome c oxidase subunit VIa polypeptide 1 |
| chr19_-_11688951 | 0.63 |
ENST00000589792.1
|
ACP5
|
acid phosphatase 5, tartrate resistant |
| chr17_-_4852243 | 0.62 |
ENST00000225655.5
|
PFN1
|
profilin 1 |
| chr12_-_8088871 | 0.61 |
ENST00000075120.7
|
SLC2A3
|
solute carrier family 2 (facilitated glucose transporter), member 3 |
| chr3_+_14989186 | 0.60 |
ENST00000435454.1
ENST00000323373.6 |
NR2C2
|
nuclear receptor subfamily 2, group C, member 2 |
| chr2_-_74780176 | 0.60 |
ENST00000409549.1
|
LOXL3
|
lysyl oxidase-like 3 |
| chr3_-_126236605 | 0.59 |
ENST00000290868.2
|
UROC1
|
urocanate hydratase 1 |
| chr5_+_132208014 | 0.58 |
ENST00000296877.2
|
LEAP2
|
liver expressed antimicrobial peptide 2 |
| chr17_-_4852332 | 0.58 |
ENST00000572383.1
|
PFN1
|
profilin 1 |
| chr1_+_25599018 | 0.58 |
ENST00000417538.2
ENST00000357542.4 ENST00000568195.1 ENST00000342055.5 ENST00000423810.2 |
RHD
|
Rh blood group, D antigen |
| chr6_-_33548006 | 0.58 |
ENST00000374467.3
|
BAK1
|
BCL2-antagonist/killer 1 |
| chr19_+_11546440 | 0.57 |
ENST00000589126.1
ENST00000588269.1 ENST00000587509.1 ENST00000592741.1 ENST00000593101.1 ENST00000587327.1 |
PRKCSH
|
protein kinase C substrate 80K-H |
| chr12_+_56511943 | 0.57 |
ENST00000257940.2
ENST00000552345.1 ENST00000551880.1 ENST00000546903.1 ENST00000551790.1 |
ZC3H10
ESYT1
|
zinc finger CCCH-type containing 10 extended synaptotagmin-like protein 1 |
| chr19_+_11546153 | 0.55 |
ENST00000591946.1
ENST00000252455.2 ENST00000412601.1 |
PRKCSH
|
protein kinase C substrate 80K-H |
| chr22_+_50714103 | 0.55 |
ENST00000595015.1
|
AL022328.1
|
Uncharacterized protein |
| chr3_-_126236588 | 0.55 |
ENST00000383579.3
|
UROC1
|
urocanate hydratase 1 |
| chr17_-_26879567 | 0.53 |
ENST00000581945.1
ENST00000444148.1 ENST00000301032.4 ENST00000335765.4 |
UNC119
|
unc-119 homolog (C. elegans) |
| chr14_+_57671888 | 0.53 |
ENST00000391612.1
|
AL391152.1
|
AL391152.1 |
| chr3_-_194119083 | 0.53 |
ENST00000401815.1
|
GP5
|
glycoprotein V (platelet) |
| chr14_+_22957581 | 0.53 |
ENST00000390487.1
|
TRAJ50
|
T cell receptor alpha joining 50 |
| chr3_+_50316458 | 0.52 |
ENST00000316436.3
|
LSMEM2
|
leucine-rich single-pass membrane protein 2 |
| chr19_+_11546093 | 0.52 |
ENST00000591462.1
|
PRKCSH
|
protein kinase C substrate 80K-H |
| chr14_-_54908043 | 0.51 |
ENST00000556113.1
ENST00000553660.1 ENST00000395573.4 ENST00000557690.1 ENST00000216416.4 |
CNIH1
|
cornichon family AMPA receptor auxiliary protein 1 |
| chr12_+_120875910 | 0.51 |
ENST00000551806.1
|
AL021546.6
|
Glutamyl-tRNA(Gln) amidotransferase subunit C, mitochondrial |
| chr1_-_16539094 | 0.51 |
ENST00000270747.3
|
ARHGEF19
|
Rho guanine nucleotide exchange factor (GEF) 19 |
| chr3_-_57113314 | 0.50 |
ENST00000338458.4
ENST00000468727.1 |
ARHGEF3
|
Rho guanine nucleotide exchange factor (GEF) 3 |
| chr19_+_10196981 | 0.50 |
ENST00000591813.1
|
C19orf66
|
chromosome 19 open reading frame 66 |
| chr17_+_73975292 | 0.50 |
ENST00000397640.1
ENST00000416485.1 ENST00000588202.1 ENST00000590676.1 ENST00000586891.1 |
TEN1
|
TEN1 CST complex subunit |
| chr8_-_110346614 | 0.49 |
ENST00000239690.4
|
NUDCD1
|
NudC domain containing 1 |
| chr6_-_33547975 | 0.49 |
ENST00000442998.2
ENST00000360661.5 |
BAK1
|
BCL2-antagonist/killer 1 |
| chr19_+_19496728 | 0.48 |
ENST00000537887.1
ENST00000417582.2 |
GATAD2A
|
GATA zinc finger domain containing 2A |
| chr8_-_131028869 | 0.48 |
ENST00000518283.1
ENST00000519110.1 |
FAM49B
|
family with sequence similarity 49, member B |
| chr1_+_12123414 | 0.48 |
ENST00000263932.2
|
TNFRSF8
|
tumor necrosis factor receptor superfamily, member 8 |
| chr2_+_219433281 | 0.47 |
ENST00000273064.6
ENST00000509807.2 ENST00000542068.1 |
RQCD1
|
RCD1 required for cell differentiation1 homolog (S. pombe) |
| chr12_-_6451235 | 0.47 |
ENST00000440083.2
ENST00000162749.2 |
TNFRSF1A
|
tumor necrosis factor receptor superfamily, member 1A |
| chr12_-_6451186 | 0.47 |
ENST00000540022.1
ENST00000536194.1 |
TNFRSF1A
|
tumor necrosis factor receptor superfamily, member 1A |
| chr12_-_107168696 | 0.46 |
ENST00000551505.1
|
RP11-144F15.1
|
Uncharacterized protein |
| chr1_-_225616515 | 0.46 |
ENST00000338179.2
ENST00000425080.1 |
LBR
|
lamin B receptor |
| chr6_+_96025341 | 0.45 |
ENST00000369293.1
ENST00000358812.4 |
MANEA
|
mannosidase, endo-alpha |
| chr8_+_110346546 | 0.45 |
ENST00000521662.1
ENST00000521688.1 ENST00000520147.1 |
ENY2
|
enhancer of yellow 2 homolog (Drosophila) |
| chr11_-_65430554 | 0.44 |
ENST00000308639.9
ENST00000406246.3 |
RELA
|
v-rel avian reticuloendotheliosis viral oncogene homolog A |
| chr16_-_69166460 | 0.43 |
ENST00000523421.1
ENST00000448552.2 ENST00000306585.6 ENST00000567763.1 ENST00000522497.1 ENST00000522091.1 ENST00000519520.1 |
CHTF8
|
CTF8, chromosome transmission fidelity factor 8 homolog (S. cerevisiae) |
| chr2_+_207024306 | 0.43 |
ENST00000236957.5
ENST00000392221.1 ENST00000392222.2 ENST00000445505.1 |
EEF1B2
|
eukaryotic translation elongation factor 1 beta 2 |
| chr11_-_58345569 | 0.42 |
ENST00000528954.1
ENST00000528489.1 |
LPXN
|
leupaxin |
| chr18_+_3448455 | 0.42 |
ENST00000549780.1
|
TGIF1
|
TGFB-induced factor homeobox 1 |
| chr3_+_190105909 | 0.42 |
ENST00000456423.1
|
CLDN16
|
claudin 16 |
| chr1_+_26872324 | 0.41 |
ENST00000531382.1
|
RPS6KA1
|
ribosomal protein S6 kinase, 90kDa, polypeptide 1 |
| chr1_+_151483855 | 0.41 |
ENST00000427934.2
ENST00000271636.7 |
CGN
|
cingulin |
| chr17_-_1083078 | 0.41 |
ENST00000574266.1
ENST00000302538.5 |
ABR
|
active BCR-related |
| chrX_+_106449862 | 0.40 |
ENST00000372453.3
ENST00000535523.1 |
PIH1D3
|
PIH1 domain containing 3 |
| chr12_+_100661156 | 0.40 |
ENST00000360820.2
|
SCYL2
|
SCY1-like 2 (S. cerevisiae) |
| chr8_-_101321584 | 0.39 |
ENST00000523167.1
|
RNF19A
|
ring finger protein 19A, RBR E3 ubiquitin protein ligase |
| chr1_+_25598989 | 0.39 |
ENST00000454452.2
|
RHD
|
Rh blood group, D antigen |
| chr12_-_8088773 | 0.38 |
ENST00000544291.1
|
SLC2A3
|
solute carrier family 2 (facilitated glucose transporter), member 3 |
| chr13_+_27844464 | 0.38 |
ENST00000241463.4
|
RASL11A
|
RAS-like, family 11, member A |
| chr2_-_214014959 | 0.37 |
ENST00000442445.1
ENST00000457361.1 ENST00000342002.2 |
IKZF2
|
IKAROS family zinc finger 2 (Helios) |
| chr1_+_1334895 | 0.37 |
ENST00000448629.2
ENST00000444362.1 |
RP4-758J18.2
|
HCG20425, isoform CRA_a; Uncharacterized protein; cDNA FLJ53815 |
| chr1_+_203765437 | 0.37 |
ENST00000550078.1
|
ZBED6
|
zinc finger, BED-type containing 6 |
| chr1_+_12123463 | 0.36 |
ENST00000417814.2
|
TNFRSF8
|
tumor necrosis factor receptor superfamily, member 8 |
| chr1_-_36789755 | 0.36 |
ENST00000270824.1
|
EVA1B
|
eva-1 homolog B (C. elegans) |
| chrX_-_106449656 | 0.35 |
ENST00000372466.4
ENST00000421752.1 ENST00000372461.3 |
NUP62CL
|
nucleoporin 62kDa C-terminal like |
| chr9_+_116343192 | 0.34 |
ENST00000471324.2
|
RGS3
|
regulator of G-protein signaling 3 |
| chr3_-_38071122 | 0.33 |
ENST00000334661.4
|
PLCD1
|
phospholipase C, delta 1 |
| chr12_-_6451014 | 0.32 |
ENST00000366159.4
ENST00000539372.1 |
TNFRSF1A
|
tumor necrosis factor receptor superfamily, member 1A |
| chr17_-_3595042 | 0.30 |
ENST00000552723.1
|
P2RX5
|
purinergic receptor P2X, ligand-gated ion channel, 5 |
| chr22_-_20104700 | 0.30 |
ENST00000439169.2
ENST00000445045.1 ENST00000404751.3 ENST00000252136.7 ENST00000403707.3 |
TRMT2A
|
tRNA methyltransferase 2 homolog A (S. cerevisiae) |
| chr17_+_45286387 | 0.30 |
ENST00000572316.1
ENST00000354968.1 ENST00000576874.1 ENST00000536623.2 |
MYL4
|
myosin, light chain 4, alkali; atrial, embryonic |
| chr11_-_4414880 | 0.29 |
ENST00000254436.7
ENST00000543625.1 |
TRIM21
|
tripartite motif containing 21 |
| chr4_+_26322409 | 0.29 |
ENST00000514807.1
ENST00000348160.4 ENST00000509158.1 ENST00000355476.3 |
RBPJ
|
recombination signal binding protein for immunoglobulin kappa J region |
| chr19_+_1495362 | 0.28 |
ENST00000395479.4
|
REEP6
|
receptor accessory protein 6 |
| chr2_+_219433588 | 0.27 |
ENST00000295701.5
|
RQCD1
|
RCD1 required for cell differentiation1 homolog (S. pombe) |
| chr5_+_162930114 | 0.27 |
ENST00000280969.5
|
MAT2B
|
methionine adenosyltransferase II, beta |
| chr20_-_45981138 | 0.27 |
ENST00000446994.2
|
ZMYND8
|
zinc finger, MYND-type containing 8 |
| chr12_-_16430619 | 0.26 |
ENST00000344941.3
|
SLC15A5
|
solute carrier family 15, member 5 |
| chr7_-_22233442 | 0.26 |
ENST00000401957.2
|
RAPGEF5
|
Rap guanine nucleotide exchange factor (GEF) 5 |
| chr7_-_71802595 | 0.25 |
ENST00000446128.1
|
CALN1
|
calneuron 1 |
| chrX_-_152989798 | 0.25 |
ENST00000441714.1
ENST00000442093.1 ENST00000429550.1 ENST00000345046.6 |
BCAP31
|
B-cell receptor-associated protein 31 |
| chr16_-_73178346 | 0.25 |
ENST00000358463.2
|
C16orf47
|
chromosome 16 open reading frame 47 |
| chr17_-_59940830 | 0.25 |
ENST00000259008.2
|
BRIP1
|
BRCA1 interacting protein C-terminal helicase 1 |
| chr6_+_152011628 | 0.25 |
ENST00000404742.1
ENST00000440973.1 |
ESR1
|
estrogen receptor 1 |
| chr3_-_47484661 | 0.25 |
ENST00000495603.2
|
SCAP
|
SREBF chaperone |
| chr17_-_43568062 | 0.25 |
ENST00000421073.2
ENST00000584420.1 ENST00000589780.1 ENST00000430334.3 |
PLEKHM1
|
pleckstrin homology domain containing, family M (with RUN domain) member 1 |
| chr14_-_34931458 | 0.24 |
ENST00000298130.4
|
SPTSSA
|
serine palmitoyltransferase, small subunit A |
| chr11_-_67188642 | 0.24 |
ENST00000546202.1
ENST00000542876.1 |
PPP1CA
|
protein phosphatase 1, catalytic subunit, alpha isozyme |
| chr19_-_51220176 | 0.24 |
ENST00000359082.3
ENST00000293441.1 |
SHANK1
|
SH3 and multiple ankyrin repeat domains 1 |
| chr15_+_101256294 | 0.23 |
ENST00000559755.1
|
RP11-66B24.5
|
RP11-66B24.5 |
| chr12_-_46121554 | 0.22 |
ENST00000609803.1
|
LINC00938
|
long intergenic non-protein coding RNA 938 |
| chr17_+_36858694 | 0.22 |
ENST00000563897.1
|
CTB-58E17.1
|
CTB-58E17.1 |
| chr1_-_1334685 | 0.22 |
ENST00000400809.3
ENST00000408918.4 |
CCNL2
|
cyclin L2 |
| chr8_-_116504448 | 0.22 |
ENST00000518018.1
|
TRPS1
|
trichorhinophalangeal syndrome I |
| chr3_-_15540055 | 0.21 |
ENST00000605797.1
ENST00000435459.2 |
COLQ
|
collagen-like tail subunit (single strand of homotrimer) of asymmetric acetylcholinesterase |
| chr3_-_69249863 | 0.21 |
ENST00000478263.1
ENST00000462512.1 |
FRMD4B
|
FERM domain containing 4B |
| chr12_-_58135903 | 0.21 |
ENST00000257897.3
|
AGAP2
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 2 |
| chr10_+_64564469 | 0.20 |
ENST00000373783.1
|
ADO
|
2-aminoethanethiol (cysteamine) dioxygenase |
| chr12_-_56727676 | 0.20 |
ENST00000547572.1
ENST00000257931.5 ENST00000440411.3 |
PAN2
|
PAN2 poly(A) specific ribonuclease subunit homolog (S. cerevisiae) |
| chr19_+_40871734 | 0.20 |
ENST00000359274.5
|
PLD3
|
phospholipase D family, member 3 |
| chr2_-_214015111 | 0.20 |
ENST00000433134.1
|
IKZF2
|
IKAROS family zinc finger 2 (Helios) |
| chr11_-_65429891 | 0.20 |
ENST00000527874.1
|
RELA
|
v-rel avian reticuloendotheliosis viral oncogene homolog A |
| chr12_-_56727487 | 0.20 |
ENST00000548043.1
ENST00000425394.2 |
PAN2
|
PAN2 poly(A) specific ribonuclease subunit homolog (S. cerevisiae) |
| chr18_+_19668021 | 0.20 |
ENST00000579830.1
|
RP11-595B24.2
|
Uncharacterized protein |
| chr2_-_47143160 | 0.20 |
ENST00000409800.1
ENST00000409218.1 |
MCFD2
|
multiple coagulation factor deficiency 2 |
| chr17_-_73975444 | 0.20 |
ENST00000293217.5
ENST00000537812.1 |
ACOX1
|
acyl-CoA oxidase 1, palmitoyl |
| chr6_-_38607673 | 0.19 |
ENST00000481247.1
|
BTBD9
|
BTB (POZ) domain containing 9 |
| chr14_-_69444957 | 0.19 |
ENST00000556571.1
ENST00000553659.1 ENST00000555616.1 |
ACTN1
|
actinin, alpha 1 |
| chr15_+_83776137 | 0.19 |
ENST00000322019.9
|
TM6SF1
|
transmembrane 6 superfamily member 1 |
| chr5_+_138629628 | 0.19 |
ENST00000508689.1
ENST00000514528.1 |
MATR3
|
matrin 3 |
| chr6_-_38607628 | 0.18 |
ENST00000498633.1
|
BTBD9
|
BTB (POZ) domain containing 9 |
| chr19_+_6361841 | 0.18 |
ENST00000596605.1
|
CLPP
|
caseinolytic mitochondrial matrix peptidase proteolytic subunit |
| chr22_+_21321447 | 0.18 |
ENST00000434714.1
|
AIFM3
|
apoptosis-inducing factor, mitochondrion-associated, 3 |
| chr6_+_30594619 | 0.18 |
ENST00000318999.7
ENST00000376485.4 ENST00000376478.2 ENST00000319027.5 ENST00000376483.4 ENST00000329992.8 ENST00000330083.5 |
ATAT1
|
alpha tubulin acetyltransferase 1 |
| chr1_+_1334914 | 0.18 |
ENST00000576232.1
ENST00000570344.1 |
RP4-758J18.2
|
HCG20425, isoform CRA_a; Uncharacterized protein; cDNA FLJ53815 |
| chr5_-_33892046 | 0.17 |
ENST00000352040.3
|
ADAMTS12
|
ADAM metallopeptidase with thrombospondin type 1 motif, 12 |
| chr17_+_17685422 | 0.17 |
ENST00000395774.1
|
RAI1
|
retinoic acid induced 1 |
| chr17_-_73257667 | 0.17 |
ENST00000538886.1
ENST00000580799.1 ENST00000351904.7 ENST00000537686.1 |
GGA3
|
golgi-associated, gamma adaptin ear containing, ARF binding protein 3 |
| chrX_-_153141783 | 0.17 |
ENST00000458029.1
|
L1CAM
|
L1 cell adhesion molecule |
| chr17_+_7533439 | 0.16 |
ENST00000441599.2
ENST00000380450.4 ENST00000416273.3 ENST00000575903.1 ENST00000576830.1 ENST00000571153.1 ENST00000575618.1 ENST00000576152.1 |
SHBG
|
sex hormone-binding globulin |
| chr3_+_160117062 | 0.16 |
ENST00000497311.1
|
SMC4
|
structural maintenance of chromosomes 4 |
| chr3_+_160117418 | 0.16 |
ENST00000465903.1
ENST00000485645.1 ENST00000360111.2 ENST00000472991.1 ENST00000467468.1 ENST00000469762.1 ENST00000489573.1 ENST00000462787.1 ENST00000490207.1 ENST00000485867.1 |
SMC4
|
structural maintenance of chromosomes 4 |
| chrX_-_47930980 | 0.16 |
ENST00000442455.3
ENST00000428686.1 ENST00000276054.4 |
ZNF630
|
zinc finger protein 630 |
| chr19_-_14201776 | 0.16 |
ENST00000269724.5
|
SAMD1
|
sterile alpha motif domain containing 1 |
| chr19_-_49250054 | 0.16 |
ENST00000602105.1
ENST00000332955.2 |
IZUMO1
|
izumo sperm-egg fusion 1 |
| chr3_-_147123837 | 0.15 |
ENST00000462748.2
|
ZIC4
|
Zic family member 4 |
| chr17_-_27038765 | 0.15 |
ENST00000581289.1
ENST00000301039.2 |
PROCA1
|
protein interacting with cyclin A1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of NR1H4
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 5.4 | GO:0002728 | negative regulation of natural killer cell cytokine production(GO:0002728) |
| 1.6 | 4.8 | GO:0002522 | leukocyte migration involved in immune response(GO:0002522) |
| 1.5 | 6.2 | GO:0002432 | granuloma formation(GO:0002432) |
| 1.0 | 3.1 | GO:0002752 | cell surface pattern recognition receptor signaling pathway(GO:0002752) |
| 0.9 | 3.6 | GO:0035408 | histone H3-T6 phosphorylation(GO:0035408) |
| 0.8 | 3.8 | GO:0010902 | positive regulation of very-low-density lipoprotein particle remodeling(GO:0010902) |
| 0.6 | 3.8 | GO:0002727 | natural killer cell cytokine production(GO:0002370) regulation of natural killer cell cytokine production(GO:0002727) positive regulation of natural killer cell cytokine production(GO:0002729) |
| 0.6 | 4.5 | GO:0008626 | granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 0.5 | 2.1 | GO:0035397 | helper T cell enhancement of adaptive immune response(GO:0035397) |
| 0.4 | 1.1 | GO:0048597 | B cell negative selection(GO:0002352) post-embryonic camera-type eye morphogenesis(GO:0048597) |
| 0.3 | 1.0 | GO:0060730 | regulation of intestinal epithelial structure maintenance(GO:0060730) |
| 0.3 | 1.0 | GO:0046416 | D-amino acid metabolic process(GO:0046416) |
| 0.3 | 2.0 | GO:0035709 | memory T cell activation(GO:0035709) regulation of memory T cell activation(GO:2000567) positive regulation of memory T cell activation(GO:2000568) |
| 0.3 | 1.7 | GO:0030538 | embryonic genitalia morphogenesis(GO:0030538) |
| 0.3 | 0.8 | GO:0045553 | TRAIL biosynthetic process(GO:0045553) regulation of TRAIL biosynthetic process(GO:0045554) positive regulation of TRAIL biosynthetic process(GO:0045556) |
| 0.3 | 2.1 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.3 | 0.8 | GO:0006117 | acetaldehyde metabolic process(GO:0006117) |
| 0.3 | 1.5 | GO:0048807 | female genitalia morphogenesis(GO:0048807) |
| 0.2 | 1.6 | GO:2000784 | regulation of unidimensional cell growth(GO:0051510) negative regulation of unidimensional cell growth(GO:0051511) establishment of cell polarity regulating cell shape(GO:0071964) regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000769) positive regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000771) regulation of establishment of cell polarity regulating cell shape(GO:2000782) positive regulation of establishment of cell polarity regulating cell shape(GO:2000784) positive regulation of barbed-end actin filament capping(GO:2000814) |
| 0.2 | 1.8 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.2 | 1.0 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.2 | 1.1 | GO:0019556 | histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 0.1 | 0.3 | GO:0090086 | negative regulation of protein deubiquitination(GO:0090086) |
| 0.1 | 1.0 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.1 | 0.5 | GO:2001287 | negative regulation of caveolin-mediated endocytosis(GO:2001287) |
| 0.1 | 0.6 | GO:0018057 | peptidyl-lysine oxidation(GO:0018057) |
| 0.1 | 0.7 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.1 | 1.9 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.1 | 1.2 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.1 | 2.2 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.1 | 0.3 | GO:1904154 | positive regulation of retrograde protein transport, ER to cytosol(GO:1904154) |
| 0.1 | 1.2 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.1 | 0.2 | GO:0050894 | determination of affect(GO:0050894) |
| 0.1 | 1.3 | GO:0071550 | death-inducing signaling complex assembly(GO:0071550) |
| 0.1 | 0.3 | GO:0000706 | meiotic DNA double-strand break processing(GO:0000706) double-strand break repair involved in meiotic recombination(GO:1990918) |
| 0.1 | 0.3 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.1 | 6.2 | GO:0042795 | snRNA transcription(GO:0009301) snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.1 | 0.4 | GO:1900242 | regulation of synaptic vesicle endocytosis(GO:1900242) |
| 0.1 | 0.4 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) |
| 0.0 | 0.1 | GO:2001113 | negative regulation of hepatocyte growth factor receptor signaling pathway(GO:1902203) regulation of cellular response to hepatocyte growth factor stimulus(GO:2001112) negative regulation of cellular response to hepatocyte growth factor stimulus(GO:2001113) |
| 0.0 | 0.2 | GO:0036496 | regulation of translational initiation by eIF2 alpha dephosphorylation(GO:0036496) |
| 0.0 | 0.4 | GO:2000370 | receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) positive regulation of clathrin-mediated endocytosis(GO:2000370) |
| 0.0 | 0.1 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.0 | 0.4 | GO:0050859 | negative regulation of B cell receptor signaling pathway(GO:0050859) |
| 0.0 | 1.2 | GO:0048935 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.0 | 1.6 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.3 | GO:1901297 | positive regulation of ephrin receptor signaling pathway(GO:1901189) positive regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901297) positive regulation of canonical Wnt signaling pathway involved in heart development(GO:1905068) |
| 0.0 | 0.2 | GO:0006581 | acetylcholine catabolic process in synaptic cleft(GO:0001507) acetylcholine catabolic process(GO:0006581) |
| 0.0 | 0.5 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.0 | 0.3 | GO:0045541 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.0 | 0.1 | GO:2000619 | negative regulation of histone H4-K16 acetylation(GO:2000619) |
| 0.0 | 0.8 | GO:1904776 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.0 | 0.8 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 1.0 | GO:0015695 | organic cation transport(GO:0015695) |
| 0.0 | 1.5 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.0 | 0.3 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.0 | 0.2 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.0 | 0.3 | GO:0060523 | prostate epithelial cord elongation(GO:0060523) |
| 0.0 | 0.5 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.0 | 2.0 | GO:0071349 | interleukin-12-mediated signaling pathway(GO:0035722) cellular response to interleukin-12(GO:0071349) |
| 0.0 | 0.4 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.0 | 0.4 | GO:1903830 | magnesium ion transmembrane transport(GO:1903830) |
| 0.0 | 1.2 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.0 | 0.2 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.3 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 | 0.1 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 | 0.2 | GO:0000098 | sulfur amino acid catabolic process(GO:0000098) |
| 0.0 | 0.1 | GO:2000360 | egg activation(GO:0007343) negative regulation of binding of sperm to zona pellucida(GO:2000360) |
| 0.0 | 0.5 | GO:0032211 | negative regulation of telomere maintenance via telomerase(GO:0032211) |
| 0.0 | 1.6 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 0.9 | GO:0031572 | G2 DNA damage checkpoint(GO:0031572) |
| 0.0 | 0.0 | GO:0051935 | amino acid neurotransmitter reuptake(GO:0051933) glutamate reuptake(GO:0051935) |
| 0.0 | 0.1 | GO:2000809 | positive regulation of synaptic vesicle clustering(GO:2000809) |
| 0.0 | 0.1 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.0 | 2.2 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.0 | 0.4 | GO:0045943 | positive regulation of transcription from RNA polymerase I promoter(GO:0045943) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 5.0 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.3 | 3.1 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.3 | 6.2 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.3 | 0.8 | GO:0035525 | NF-kappaB p50/p65 complex(GO:0035525) |
| 0.1 | 3.8 | GO:0042627 | chylomicron(GO:0042627) |
| 0.1 | 2.2 | GO:0032059 | bleb(GO:0032059) |
| 0.1 | 1.5 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.1 | 0.4 | GO:0031251 | PAN complex(GO:0031251) |
| 0.1 | 0.3 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.1 | 1.1 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.1 | 2.1 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.1 | 1.2 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.1 | 1.9 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.1 | 4.5 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.2 | GO:0009368 | endopeptidase Clp complex(GO:0009368) |
| 0.0 | 0.4 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.0 | 0.3 | GO:0005784 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.0 | 0.6 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 1.7 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.0 | 1.6 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 0.4 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 1.2 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.3 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 0.8 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 0.8 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.0 | 0.5 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.3 | GO:0000796 | condensin complex(GO:0000796) |
| 0.0 | 0.3 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.0 | 0.7 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.0 | 4.2 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 2.2 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.5 | GO:0000783 | telomere cap complex(GO:0000782) nuclear telomere cap complex(GO:0000783) |
| 0.0 | 1.9 | GO:0035580 | specific granule lumen(GO:0035580) |
| 0.0 | 2.8 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 0.2 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 1.3 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.1 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.0 | 2.0 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 0.3 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.0 | 0.5 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 0.2 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.2 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.2 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.0 | 1.0 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.0 | 0.1 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 3.6 | GO:0035403 | histone kinase activity (H3-T6 specific)(GO:0035403) |
| 0.5 | 3.8 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.3 | 2.0 | GO:0023025 | MHC class Ib protein complex binding(GO:0023025) MHC class Ib protein binding, via antigen binding groove(GO:0023030) |
| 0.3 | 2.1 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.2 | 1.0 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.2 | 0.7 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.2 | 1.3 | GO:0043120 | tumor necrosis factor binding(GO:0043120) |
| 0.2 | 0.5 | GO:0050613 | delta14-sterol reductase activity(GO:0050613) |
| 0.1 | 2.2 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.1 | 3.2 | GO:0019841 | retinol binding(GO:0019841) |
| 0.1 | 2.2 | GO:0005522 | profilin binding(GO:0005522) |
| 0.1 | 1.8 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.1 | 1.0 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.1 | 1.2 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.1 | 0.8 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.1 | 2.2 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.1 | 2.1 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.1 | 0.3 | GO:0032038 | myosin II heavy chain binding(GO:0032038) |
| 0.1 | 4.7 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.1 | 1.8 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.1 | 1.4 | GO:0016641 | oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor(GO:0016641) |
| 0.0 | 1.1 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 0.8 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 1.0 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 1.2 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.0 | 0.3 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 0.9 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 0.2 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.3 | GO:0038052 | RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.0 | 4.8 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 2.3 | GO:0051219 | phosphoprotein binding(GO:0051219) |
| 0.0 | 0.3 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.0 | 0.3 | GO:0004931 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.0 | 0.8 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.2 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 1.0 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.0 | 6.1 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 0.1 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.0 | 0.5 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
| 0.0 | 0.1 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.0 | 0.3 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.2 | GO:0005497 | androgen binding(GO:0005497) |
| 0.0 | 0.4 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.2 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.4 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.3 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 0.1 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.0 | 0.5 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.0 | 0.2 | GO:0035326 | enhancer binding(GO:0035326) |
| 0.0 | 1.0 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 4.7 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.4 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.3 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.2 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 1.5 | GO:0032947 | protein complex scaffold(GO:0032947) |
| 0.0 | 2.2 | GO:0003823 | antigen binding(GO:0003823) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 8.4 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.1 | 4.7 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.1 | 2.3 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.1 | 1.1 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.1 | 1.6 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.1 | 2.0 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
| 0.0 | 0.8 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 1.8 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.8 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.6 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 2.3 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 1.2 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.0 | 0.8 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 1.2 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 2.0 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 1.2 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 0.4 | REACTOME SIGNALING BY TGF BETA RECEPTOR COMPLEX | Genes involved in Signaling by TGF-beta Receptor Complex |
| 0.3 | 2.1 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.1 | 3.8 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.1 | 4.7 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.1 | 5.6 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.1 | 3.6 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.1 | 1.6 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.1 | 9.2 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.1 | 0.8 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.1 | 1.0 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 0.5 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 1.6 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.0 | 1.3 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 1.0 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 1.8 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 1.0 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 1.2 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 1.8 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.3 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 0.7 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.5 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 2.1 | REACTOME LATE PHASE OF HIV LIFE CYCLE | Genes involved in Late Phase of HIV Life Cycle |
| 0.0 | 0.3 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |